Vibrio parahaemolyticus is the most common cause of seafood-borne illness reported in the United States and is frequently associated with shellfish consumption. This study contributes to our knowledge of the biogeography and functional genomics of this species around North America.
KEYWORDS: Vibrio parahaemolyticus, MLST, phylogenetics, genomics, kSNP, cluster analysis
ABSTRACT
Vibrio parahaemolyticus is the most common cause of seafood-borne illness reported in the United States. The draft genomes of 132 North American clinical and oyster V. parahaemolyticus isolates were sequenced to investigate their phylogenetic and biogeographic relationships. The majority of oyster isolate sequence types (STs) were from a single harvest location; however, four were identified from multiple locations. There was population structure along the Gulf and Atlantic Coasts of North America, with what seemed to be a hub of genetic variability along the Gulf Coast, with some of the same STs occurring along the Atlantic Coast and one shared between the coastal waters of the Gulf and those of Washington State. Phylogenetic analyses found nine well-supported clades. Two clades were composed of isolates from both clinical and oyster sources. Four were composed of isolates entirely from clinical sources, and three were entirely from oyster sources. Each single-source clade consisted of one ST. Some human isolates lack tdh, trh, and some type III secretion system (T3SS) genes, which are established virulence genes of V. parahaemolyticus. Thus, these genes are not essential for pathogenicity. However, isolates in the monophyletic groups from clinical sources were enriched in several categories of genes compared to those from monophyletic groups of oyster isolates. These functional categories include cell signaling, transport, and metabolism. The identification of genes in these functional categories provides a basis for future in-depth pathogenicity investigations of V. parahaemolyticus.
IMPORTANCE Vibrio parahaemolyticus is the most common cause of seafood-borne illness reported in the United States and is frequently associated with shellfish consumption. This study contributes to our knowledge of the biogeography and functional genomics of this species around North America. STs shared between the Gulf Coast and the Atlantic seaboard as well as Pacific waters suggest possible transport via oceanic currents or large shipping vessels. STs frequently isolated from humans but rarely, if ever, isolated from the environment are likely more competitive in the human gut than other STs. This could be due to additional functional capabilities in areas such as cell signaling, transport, and metabolism, which may give these isolates an advantage in novel nutrient-replete environments such as the human gut.
INTRODUCTION
Vibrio parahaemolyticus is a halophilic, Gram-negative bacterium that inhabits coastal estuarine environments (1). In the United States, V. parahaemolyticus is the leading cause of seafood-borne infections (2). The incidence of V. parahaemolyticus infection continues to increase in the United States and globally (3, 4). Consumption of raw or undercooked seafood harboring V. parahaemolyticus can result in mild to acute gastroenteritis, while contact with seawater containing this bacterium occasionally results in wound infections and, rarely, sepsis (5). The pathogenicity of this bacterium has historically been linked to the presence of two hemolysin genes: the thermostable direct hemolysin (tdh) and the tdh-related hemolysin (trh) (1). However, recent studies have shown that some strains produce cytotoxic and enterotoxic effects independent of their hemolysin production (6, 7). Also, some clinical isolates have been reported to lack one or both of the virulence markers tdh and trh, and phylogeny is independent of the genes (8–10). In light of this, type III secretion systems (T3SSs) have been investigated as potential pathogenicity factors for V. parahaemolyticus (11, 12). Among isolates that carry the hemolysin genes, certain serotypes (O3:K6 and O4:K12) and/or sequence types (STs) (ST3 and -36) are thought to be more virulent, as inferred by their illness incidences (13–15). In addition to the apparent divergence in the virulence potential of V. parahaemolyticus based on serotype or ST, there is a strong correlation with isolate/source location, suggesting that geography may play a role (16). For example, the majority of shellfish-associated V. parahaemolyticus illnesses in the United States have been from the Pacific Northwest and the Northeast Atlantic coasts (15, 17) rather than the Gulf Coast, which is also a major producer of commercial shellfish and where V. parahaemolyticus thrives (1, 18–20). Together, these data indicate that there is still much to be learned about the population dynamics and virulence potential of this species.
Previous investigations of V. parahaemolyticus utilizing multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and multiple-locus variable-number tandem-repeat analysis (MLVA) have indicated high genetic diversity (9, 21–24). However, the availability of whole-genome sequencing (WGS), more powerful computing, and better algorithms has made phylogenetic analysis based on the data collected from the entire genome possible. Also, genome sequencing has made in silico MLST analysis possible (25, 26). Previous sequencing of V. parahaemolyticus demonstrated substantial genetic similarity between disease-associated clinical and certain environmental isolates, even with relatively limited strain sets (15, 27). This suggests that sequencing additional genomes from diverse clinical and environmental V. parahaemolyticus isolates will provide information for further insight into the ability to transition from an environmental niche to a human pathogen (27).
In this study, we sequenced 132 V. parahaemolyticus genomes that are geographically representative of North America and diverse regarding serotype and ST. The draft genome data were used for phylogenetic analyses, including maximum likelihood analysis and whole-genome distance analysis (WGDA), which have been successfully applied to bacterial genomes, as well as in silico MLST (26, 28, 29). These analyses were used to infer biogeographical and phylogenetic relationships of V. parahaemolyticus as a means to understand the genomic variation within V. parahaemolyticus. In addition, the data were used to further investigate the differential virulence potentials of isolates.
RESULTS
Genome description.
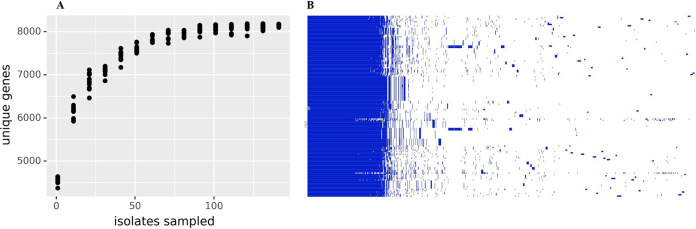
Draft genomes were collected for 132 V. parahaemolyticus isolates from oysters collected from the coasts of North America and patients in the United States. Rarefaction analysis indicated that this population contains all the genes in V. parahaemolyticus as the asymptote occurs at a pangenome size of 8,191 genes, prior to sampling all 132 genomes (Fig. 1). The core genome consisted of 3,726 genes (see Fig. S1 in the supplemental material), with much of the pangenome being composed of genes from fewer than 24 isolates (18%).
FIG 1.
Genome gene discovery and pangenome. (A) To evaluate the completeness of the gene-finding effort, a rarefaction plot was constructed by listing the COG to which each V. parahaemolyticus isolate belonged, resampling the isolates with replacement using random.randrange in Python 3.6, and then listing and counting the unique COGs. This was done for increasing numbers of isolates. For each number of isolates, 10 replicates were done. The plot was constructed with Plotnine, geom_point. (B) The pangenome was constructed to demonstrate a core genome with a highly variable auxiliary genome.
Population genetics and biogeography.
There were 61 STs identified from the 132 isolates (Table 1), of which 35 were represented by only one isolate and 21 were associated with multiple serotypes. Five isolates could not be assigned sequence types by MLST due to recA or pntA genes that could not be typed. The three most frequent STs were ST36, ST1151, and ST3. Only two sequence types, ST775 and ST34, were collected from both oyster and clinical sources.
TABLE 1.
Metadata of the 132 V. parahaemolyticus shotgun sequences
| CFSAN ID | Strain ID | Source | State or provincea | Date of isolation (day-mo-yr)b | Serotype | Sequence type | SRA accession no. | GenBank assembly ID | GenBank accession no. | BioSample accession no. | Coverage depth (%) | Total length (bp) | No. of contigs | No. of genes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CFSAN011163 | CDC_K4556-2 | Clinical | LA | 23-Oct-06 | O1:Kuk | ST744 | SRR1118644 | GCA_001727365.1 | MISQ00000000 | SAMN02368278 | 74 | 5,149,094 | 135 | 4,735 |
| CFSAN011164 | CDC_K4557 | Clinical | LA | 19-Sep-06 | O1:K33 | ST799 | SRR1840765 | GCA_001727725.1 | MISR00000000 | SAMN02368279 | 35 | 5,074,019 | 94 | 4,619 |
| CFSAN011165 | CDC_K4558-1 | Clinical | LA | 28-Aug-06 | O3:K39 | ST1143 | SRR1118643 | GCA_001727735.1 | MISS00000000 | SAMN02368280 | 34 | 5,032,007 | 120 | 4,611 |
| CFSAN011166 | CDC_K4558-2 | Clinical | LA | 28-Aug-06 | O3:Kuk | ST636 | SRR1118642 | GCA_001727745.1 | MIST00000000 | SAMN02368281 | 88 | 5,335,587 | 155 | 4,897 |
| CFSAN011167 | CDC_K4588 | Clinical | ME | 26-Jul-06 | O5:Kuk | ST746 | SRR1118641 | GCA_001727775.1 | MISU00000000 | SAMN02368282 | 57 | 5,014,336 | 118 | 4,610 |
| CFSAN011168 | CDC_K4636 | Clinical | NY | 25-Sep-06 | O10:Kuk | ST636 | SRR1815538 | GCA_001727805.1 | MISV00000000 | SAMN03358827 | 35 | 5,340,949 | 138 | 4,889 |
| CFSAN011169 | CDC_K4637-1 | Clinical | NY | 1-Oct-06 | O3:K6 | ST3 | SRR1815539 | GCA_001727815.1 | MISW00000000 | SAMN03358828 | 59 | 5,163,379 | 150 | 4,755 |
| CFSAN011170 | CDC_K4637-2 | Clinical | NY | 1-Oct-06 | O3:K6 | ST3 | SRR1815540 | GCA_001727825.1 | MISX00000000 | SAMN03358829 | 59 | 5,156,414 | 133 | 4,740 |
| CFSAN011171 | CDC_K4638 | Clinical | NY | 25-Sep-06 | O10:Kuk | ST809 | SRR1118640 | GCA_001727845.1 | MISY00000000 | SAMN02368283 | 47 | 5,077,332 | 96 | 4,619 |
| CFSAN011172 | CDC_K4639-1 | Clinical | NY | 16-Oct-06 | O4:K12 | ST36 | SRR1815541 | GCA_001727895.1 | MISZ00000000 | SAMN03358830 | 61 | 5,092,291 | 122 | 4,654 |
| CFSAN011173 | CDC_K4639-2 | Clinical | NY | 16-Oct-06 | O4:Kuk | ST36 | SRR1118639 | GCA_001727885.1 | MITA00000000 | SAMN02368284 | 57 | 5,078,734 | 114 | 4,658 |
| CFSAN011175 | CDC_K4762 | Clinical | VA | 15-Aug-06 | O5:K17 | ST674 | SRR1118637 | GCA_001727925.1 | MITB00000000 | SAMN02368286 | 78 | 5,103,196 | 108 | 4,676 |
| CFSAN011176 | CDC_K4763 | Clinical | VA | 25-Aug-06 | O5:Kuk | Untyped | SRR1840764 | GCA_001727915.1 | MITC00000000 | SAMN02368287 | 43 | 5,287,071 | 129 | 4,843 |
| CFSAN011177 | CDC_K4764-1 | Clinical | VA | 13-Oct-06 | O8:K41 | Untyped | SRR1815542 | GCA_001727965.1 | MITE00000000 | SAMN03358831 | 43 | 5,276,586 | 114 | 4,813 |
| CFSAN011178 | CDC_K4764-2 | Clinical | VA | 13-Oct-06 | O8:K41 | ST1156 | SRR1118636 | GCA_001727975.1 | MITD00000000 | SAMN02368288 | 52 | 5,033,430 | 111 | 4,616 |
| CFSAN011179 | CDC_K4775 | Clinical | GA | 24-Feb-07 | O3:K6 | ST3 | SRR1118635 | GCA_001728005.1 | MITF00000000 | SAMN02368289 | 71 | 5,158,365 | 121 | 4,699 |
| CFSAN011180 | CDC_K4842 | Clinical | MD | 16-Oct-06 | O5:K47 | ST1144 | SRR1118634 | GCA_001727995.1 | MITG00000000 | SAMN02368290 | 47 | 5,194,684 | 115 | 4,791 |
| CFSAN011181 | CDC_K4857-1 | Clinical | HI | 28-Jan-07 | O5:K17 | ST79 | SRR1118633 | GCA_001728045.1 | MITH00000000 | SAMN02368291 | 37 | 5,038,683 | 103 | 4,569 |
| CFSAN011182 | CDC_K4857-2 | Clinical | HI | 28-Jan-07 | O5:Kuk | ST79 | SRR1118632 | GCA_001728065.1 | MITI00000000 | SAMN02368292 | 47 | 5,039,775 | 108 | 4,592 |
| CFSAN011183 | CDC_K4858 | Clinical | HI | 15-Sep-06 | O4:K4 | ST283 | SRR1840763 | GCA_001728085.1 | MITJ00000000 | SAMN02368293 | 56 | 4,961,237 | 89 | 4,534 |
| CFSAN011184 | CDC_K4859 | Clinical | HI | 15-Feb-07 | O6:K18 | Untyped | SRR1118631 | GCA_001728095.1 | MITK00000000 | SAMN02368294 | 192 | 5,180,859 | 107 | 4,732 |
| CFSAN011185 | CDC_K4981 | Clinical | OK | 12-Mar-07 | O1:Kuk | ST748 | SRR1118630 | GCA_001728125.1 | MITL00000000 | SAMN02368295 | 118 | 4,928,549 | 135 | 4,500 |
| CFSAN011187 | CDC_K5009-2 | Clinical | MA | 7-Aug-06 | O4:K53 | ST749 | SRR1118629 | GCA_001728135.1 | MITM00000000 | SAMN02368297 | 94 | 5,125,537 | 130 | 4,681 |
| CFSAN011188 | CDC_K5010-1 | Clinical | MA | 16-Sep-06 | O1:Kuk | ST3 | SRR1118628 | GCA_001728155.1 | MITN00000000 | SAMN02368298 | 59 | 5,122,961 | 146 | 4,694 |
| CFSAN011189 | CDC_K5010-2 | Clinical | MA | 16-Sep-06 | O1:Kuk | ST3 | SRR1118627 | GCA_001728175.1 | MITO00000000 | SAMN02368299 | 94 | 5,118,895 | 134 | 4,676 |
| CFSAN011190 | CDC_K5058 | Clinical | TX | 15-May-07 | O3:K6 | ST3 | SRR1057385 | GCA_001728205.1 | MITP00000000 | SAMN02368300 | 60 | 5,114,568 | 84 | 4,620 |
| CFSAN011191 | CDC_K5059-1 | Clinical | TX | 10-May-07 | O5:Kuk | ST1147 | SRR1840761 | GCA_001728215.1 | MITQ00000000 | SAMN02368301 | 83 | 4,959,751 | 80 | 4,543 |
| CFSAN011192 | CDC_K5059-2 | Clinical | TX | 10-May-07 | O5:Kuk | ST1147 | SRR1057386 | GCA_001728235.1 | MITR00000000 | SAMN02368302 | 458 | 4,953,954 | 123 | 4,513 |
| CFSAN011193 | CDC_K5067 | Clinical | SD | 28-Apr-07 | O1:K56 | ST775 | SRR1118626 | GCA_001728255.1 | MITS00000000 | SAMN02368303 | 70 | 5,028,824 | 144 | 4,585 |
| CFSAN011194 | CDC_K5073 | Clinical | MD | 10-Mar-07 | O3:K56 | ST750 | SRR1118625 | GCA_001728275.1 | MITT00000000 | SAMN02368304 | 85 | 5,082,152 | 117 | 4,604 |
| CFSAN011195 | CDC_K5125 | Clinical | MS | 11-Jun-07 | O3:Kuk | ST772 | SRR1840760 | GCA_001728295.1 | MITU00000000 | SAMN02368305 | 71 | 5,134,866 | 134 | 4,699 |
| CFSAN011196 | CDC_K5126 | Clinical | MS | 21-May-07 | O3:Kuk | ST1131 | SRR1118624 | GCA_001728325.1 | MITV00000000 | SAMN02368306 | 112 | 5,143,760 | 152 | 4,680 |
| CFSAN011197 | CDC_K5276 | Clinical | NY | 20-Apr-07 | O11:Kuk | ST631 | SRR1118623 | GCA_001728335.1 | MITW00000000 | SAMN02368307 | 86 | 5,168,475 | 136 | 4,774 |
| CFSAN011198 | CDC_K5277 | Clinical | WA | No date reported | O1:Kuk | ST65 | SRR1118622 | GCA_001728345.1 | MITX00000000 | SAMN02368308 | 71 | 5,165,700 | 109 | 4,699 |
| CFSAN011199 | CDC_K5278 | Clinical | WA | 25-Jun-07 | O4:K12 | ST36 | SRR1118621 | GCA_001728405.1 | MITY00000000 | SAMN02368309 | 69 | 5,067,491 | 112 | 4,639 |
| CFSAN011200 | CDC_K5279 | Clinical | WA | No date reported | O1:Kuk | ST65 | SRR1118620 | GCA_001727655.1 | MITZ00000000 | SAMN02368310 | 76 | 5,172,883 | 93 | 4,683 |
| CFSAN011201 | CDC_K5280 | Clinical | WA | 11-Jul-07 | O4:K12 | ST36 | SRR1118619 | GCA_001727625.1 | MIUA00000000 | SAMN02368311 | 87 | 5,083,288 | 110 | 4,662 |
| CFSAN011202 | CDC_K5281 | Clinical | WA | 13-Jul-07 | O4:K12 | ST36 | SRR1118618 | GCA_001727575.1 | MIUB00000000 | SAMN02368312 | 102 | 5,077,351 | 115 | 4,661 |
| CFSAN011203 | CDC_K5282 | Clinical | HI | 24-May-07 | O5:Kuk | Untyped | SRR1118617 | GCA_001727545.1 | MIUC00000000 | SAMN02368313 | 50 | 4,963,427 | 79 | 4,506 |
| CFSAN011204 | CDC_K5306 | Clinical | GA | 23-Jul-07 | O4:K9 | ST34 | SRR1118616 | GCA_001727505.1 | MIUD00000000 | SAMN02368314 | 122 | 5,045,861 | 117 | 4,636 |
| CFSAN011205 | CDC_K5308 | Clinical | AK | 14-May-07 | O4:K63 | ST36 | SRR1118615 | GCA_001727485.1 | MIUE00000000 | SAMN02368315 | 138 | 5,095,192 | 142 | 4,669 |
| CFSAN011206 | CDC_K5323-1 | Clinical | VA | No date reported | O5:K17 | ST674 | SRR1118614 | GCA_001727475.1 | MIUF00000000 | SAMN02368316 | 30 | 5,220,531 | 98 | 4,795 |
| CFSAN011207 | CDC_K5323-2 | Clinical | VA | No date reported | O5:Kuk | ST674 | SRR1118613 | GCA_001727415.1 | MIUG00000000 | SAMN02368317 | 57 | 5,207,450 | 84 | 4,779 |
| CFSAN011208 | CDC_K5324-1 | Clinical | VA | 17-Jun-07 | O1:K20 | ST1132 | SRR1118612 | GCA_001727395.1 | MIUH00000000 | SAMN02368318 | 56 | 5,272,305 | 294 | 4,941 |
| CFSAN011209 | CDC_K5324-2 | Clinical | VA | 17-Jun-07 | O1:K20 | ST1132 | SRR1118611 | GCA_001727405.1 | MIUI00000000 | SAMN02368319 | 70 | 5,060,178 | 174 | 4,654 |
| CFSAN011210 | CDC_K5328 | Clinical | IN | No date reported | O4:K12 | ST36 | SRR1118610 | GCA_001727665.1 | MIUJ00000000 | SAMN02368320 | 85 | 5,090,169 | 118 | 4,661 |
| CFSAN011211 | CDC_K5330 | Clinical | TX | 23-Apr-07 | O5:Kuk | ST1713 | SRR1815543 | GCA_001728375.1 | MIUK00000000 | SAMN03358832 | 43 | 5,168,109 | 142 | 4,757 |
| CFSAN011212 | CDC_K5331 | Clinical | GA | 8-Aug-07 | O4:K8 | ST265 | SRR1118609 | GCA_001728425.1 | MIUL00000000 | SAMN02368321 | 90 | 5,067,258 | 95 | 4,631 |
| CFSAN011213 | CDC_K5345-1 | Clinical | IA | 7-Aug-07 | O4:K12 | ST36 | SRR1840759 | GCA_001728465.1 | MIUM00000000 | SAMN02368322 | 49 | 5,148,490 | 112 | 4,719 |
| CFSAN011214 | CDC_K5345-2 | Clinical | IA | 7-Aug-07 | O4:K12 | ST36 | SRR1118608 | GCA_001728485.1 | MIUN00000000 | SAMN02368323 | 70 | 5,125,535 | 105 | 4,687 |
| CFSAN011215 | CDC_K5346 | Clinical | PA | 21-Aug-07 | O4:K12 | ST36 | SRR1815544 | GCA_001728415.1 | MIUO00000000 | SAMN03358833 | 53 | 5,087,978 | 126 | 4,661 |
| CFSAN011216 | CDC_K5428 | Clinical | NV | 6-Jul-07 | O1:Kuk | ST199 | SRR1815545 | GCA_001727645.1 | MIUP00000000 | SAMN03358834 | 20 | 5,092,714 | 139 | 4,639 |
| CFSAN011217 | CDC_K5429 | Clinical | NV | 9-Aug-07 | O4:K12 | ST36 | SRR1118607 | GCA_001727585.1 | MIUQ00000000 | SAMN02368324 | 111 | 5,079,150 | 118 | 4,653 |
| CFSAN011218 | CDC_K5433 | Clinical | WA | 24-Jul-07 | O4:Kuk | ST36 | SRR1118606 | GCA_001727385.1 | MIUR00000000 | SAMN02368325 | 47 | 5,073,057 | 118 | 4,635 |
| CFSAN011219 | CDC_K5435 | Clinical | WA | 11-Aug-07 | O1:Kuk | ST65 | SRR1118605 | GCA_001727465.1 | MIUS00000000 | SAMN02368326 | 73 | 5,168,466 | 105 | 4,718 |
| CFSAN011221 | CDC_K5437 | Clinical | WA | 2-Sep-07 | O4:Kuk | ST36 | SRR1118604 | GCA_001727565.1 | MIUT00000000 | SAMN02368328 | 122 | 5,068,632 | 126 | 4,636 |
| CFSAN011222 | CDC_K5438 | Clinical | WA | 9-Sep-07 | O1:Kuk | ST65 | SRR1118603 | GCA_001727705.1 | MIUU00000000 | SAMN02368329 | 114 | 5,154,409 | 123 | 4,705 |
| CFSAN011223 | CDC_K5439 | Clinical | WA | 19-Sep-07 | O4:K8 | ST189 | SRR1118602 | GCA_001728495.1 | MIUV00000000 | SAMN02368330 | 36 | 5,031,538 | 106 | 4,616 |
| CFSAN011224 | CDC_K5456 | Clinical | WA | No date reported | O4:Kuk | ST36 | SRR1118601 | GCA_001728545.1 | MIUW00000000 | SAMN02368331 | 119 | 5,080,809 | 114 | 4,656 |
| CFSAN011225 | CDC_K5457 | Clinical | WA | 7-Aug-07 | O4:Kuk | ST36 | SRR1118600 | GCA_001728505.1 | MIUX00000000 | SAMN02368332 | 72 | 5,075,290 | 98 | 4,625 |
| CFSAN011226 | CDC_K5485 | Clinical | NC | 8-Jul-07 | O6:K18 | ST50 | SRR1815546 | GCA_001728565.1 | MIUY00000000 | SAMN03358835 | 23 | 5,151,277 | 128 | 4,719 |
| CFSAN011227 | CDC_K5512 | Clinical | OK | 14-Jun-07 | O4:K12 | ST36 | SRR1118599 | GCA_001728585.1 | MIUZ00000000 | SAMN02368333 | 92 | 5,082,991 | 122 | 4,670 |
| CFSAN011228 | CDC_K5528 | Clinical | GA | 6-Oct-07 | O4:K68 | ST3 | SRR1118598 | GCA_001728625.1 | MIVA00000000 | SAMN02368334 | 128 | 5,101,848 | 108 | 4,645 |
| CFSAN011229 | CDC_K5579 | Clinical | IN | No date reported | O4:K63 | ST43 | SRR1118597 | GCA_001728575.1 | MIVB00000000 | SAMN02368335 | 107 | 5,072,634 | 169 | 4,659 |
| CFSAN011230 | CDC_K5582 | Clinical | GA | 10-Oct-07 | O11:Kuk | ST631 | SRR1840757 | GCA_001728645.1 | MIVC00000000 | SAMN02368336 | 54 | 5,174,332 | 104 | 4,798 |
| CFSAN011232 | CDC_K5618 | Clinical | NY | 16-Aug-07 | O10:Kuk | ST636 | SRR1815547 | GCA_001728655.1 | MIVD00000000 | SAMN03358836 | 25 | 5,107,768 | 147 | 4,668 |
| CFSAN011233 | CDC_K5620 | Clinical | NY | 23-Aug-07 | O10:Kuk | ST636 | SRR1815548 | GCA_001728665.1 | MIVE00000000 | SAMN03358837 | 24 | 5,099,609 | 131 | 4,658 |
| CFSAN011235 | CDC_K5629 | Clinical | GA | 18-Nov-07 | O4:K13 | ST36 | SRR1118596 | GCA_001728695.1 | MIVF00000000 | SAMN02368339 | 106 | 5,077,945 | 119 | 4,659 |
| CFSAN011236 | CDC_K5635 | Clinical | MD | 3-Sep-07 | O5:K30 | ST753 | SRR1118595 | GCA_001728725.1 | MIVG00000000 | SAMN02368340 | 118 | 5,070,117 | 110 | 4,636 |
| CFSAN011237 | CDC_K5638 | Clinical | MD | No date reported | O4:K12 | ST36 | SRR1815549 | GCA_001728745.1 | MIVH00000000 | SAMN03358838 | 29 | 5,144,324 | 125 | 4,733 |
| CFSAN011238 | CDC_K5701 | Clinical | OR | 9-Sep-07 | O1:Kuk | ST65 | SRR1815550 | GCA_001728755.1 | MIVI00000000 | SAMN03358839 | 24 | 5,157,591 | 93 | 4,707 |
| CFSAN011096 | GCSL_R5 | Oyster | TX | 14-Mar-07 | O10:Kuk | ST1133 | SRR1815527 | GCA_001726165.1 | MIQI00000000 | SAMN03358816 | 43 | 5,185,921 | 95 | 4,792 |
| CFSAN011097 | GCSL_R6 | Oyster | TX | 14-Mar-07 | O10:Kuk | ST1133 | SRR1118691 | GCA_001726265.1 | MIQJ00000000 | SAMN02368223 | 80 | 5,087,599 | 107 | 4,659 |
| CFSAN011098 | GCSL_R7 | Oyster | TX | 14-Mar-07 | O10:Kuk | ST1134 | SRR1840772 | GCA_001726195.1 | MIQK00000000 | SAMN02368224 | 44 | 5,052,796 | 138 | 4,640 |
| CFSAN011099 | GCSL_R8 | Oyster | TX | 14-Mar-07 | O10:Kuk | ST1134 | SRR1118690 | GCA_001726185.1 | MIQL00000000 | SAMN02368225 | 52 | 5,200,929 | 119 | 4,807 |
| CFSAN011100 | GCSL_R10 | Oyster | FL | 19-Mar-07 | O1:Kuk | ST313 | SRR1815528 | GCA_001726175.1 | MIQM00000000 | SAMN03358817 | 38 | 5,072,012 | 158 | 4,665 |
| CFSAN011101 | GCSL_R12 | Oyster | LA | 27-Mar-07 | O4:K8 | ST32 | SRR1118689 | GCA_001726245.1 | MIQN00000000 | SAMN02368226 | 43 | 5,051,895 | 116 | 4,618 |
| CFSAN011102 | GCSL_R13 | Oyster | LA | 27-Mar-07 | O4:K10 | ST732 | SRR1118688 | GCA_001726255.1 | MIQO00000000 | SAMN02368227 | 135 | 5,122,134 | 161 | 4,694 |
| CFSAN011103 | GCSL_R16 | Oyster | FL | 30-Apr-07 | O4:K9 | ST34 | SRR1118687 | GCA_001726345.1 | MIQP00000000 | SAMN02368228 | 56 | 5,051,817 | 114 | 4,638 |
| CFSAN011104 | GCSL_R17 | Oyster | FL | 30-Apr-07 | O4:Kuk | ST536 | SRR1118686 | GCA_001726275.1 | MIQQ00000000 | SAMN02368229 | 60 | 5,010,966 | 100 | 4,576 |
| CFSAN011105 | GCSL_R21 | Oyster | TX | 4-May-07 | O5:Kuk | ST12 | SRR1118685 | GCA_001726355.1 | MIQR00000000 | SAMN02368230 | 56 | 5,124,206 | 80 | 4,685 |
| CFSAN011106 | GCSL_R26 | Oyster | NJ | 16-Jun-07 | O4:K8 | ST32 | SRR1118684 | GCA_001726335.1 | MIQS00000000 | SAMN02368231 | 119 | 5,067,308 | 134 | 4,629 |
| CFSAN011107 | GCSL_R29 | Oyster | FL | 27-May-07 | O11:Kuk | ST734 | SRR1118683 | GCA_001726325.1 | MIQT00000000 | SAMN02368232 | 40 | 4,946,371 | 104 | 4,525 |
| CFSAN011108 | GCSL_R30 | Oyster | FL | 27-May-07 | O1:Kuk | ST23 | SRR1118682 | GCA_001726405.1 | MIQU00000000 | SAMN02368233 | 45 | 5,132,857 | 131 | 4,724 |
| CFSAN011109 | GCSL_R31 | Oyster | LA | 27-May-07 | O1:Kuk | ST23 | SRR1118681 | GCA_001726415.1 | MIQV00000000 | SAMN02368234 | 102 | 5,126,812 | 132 | 4,710 |
| CFSAN011110 | GCSL_R32 | Oyster | LA | 30-May-07 | O10:Kuk | ST1142 | SRR1057384 | GCA_001726445.1 | MIQW00000000 | SAMN02368235 | 27 | 5,059,594 | 105 | 4,607 |
| CFSAN011111 | GCSL_R33 | Oyster | LA | 30-May-07 | O3:Kuk | ST28 | SRR1118680 | GCA_001726435.1 | MIQX00000000 | SAMN02368236 | 109 | 5,043,795 | 94 | 4,586 |
| CFSAN011112 | GCSL_R42 | Oyster | WA | 26-Jul-07 | O10:Kuk | ST1155 | SRR1815529 | GCA_001726485.1 | MIQY00000000 | SAMN03358818 | 39 | 4,981,892 | 105 | 4,522 |
| CFSAN011113 | GCSL_R45 | Oyster | WA | Jun/Jul-07 (exact date unknown) | O5:Kuk | ST61 | SRR1118679 | GCA_001726495.1 | MIQZ00000000 | SAMN02368237 | 130 | 4,973,945 | 101 | 4,533 |
| CFSAN011114 | GCSL_R47 | Oyster | AL | 20-Apr-07 | O4:K8 | ST32 | SRR1118678 | GCA_001726515.1 | MIRA00000000 | SAMN02368238 | 66 | 5,064,518 | 151 | 4,653 |
| CFSAN011115 | GCSL_R51 | Oyster | AL | 6-Jul-07 | O8:Kuk | ST676 | SRR1815530 | GCA_001726535.1 | MIRB00000000 | SAMN03358819 | 34 | 5,049,334 | 105 | 4,632 |
| CFSAN011116 | GCSL_R52 | Oyster | WA | 13-Jul-07 | O3:Kuk | ST735 | SRR1118677 | GCA_001726565.1 | MIRC00000000 | SAMN02368239 | 84 | 5,189,100 | 86 | 4,693 |
| CFSAN011117 | GCSL_R53 | Oyster | WA | 13-Jul-07 | O3:Kuk | ST735 | SRR1118676 | GCA_001726575.1 | MIRD00000000 | SAMN02368240 | 68 | 5,191,609 | 101 | 4,715 |
| CFSAN011118 | GCSL_R54 | Oyster | WA | 13-Jul-07 | O3:Kuk | ST735 | SRR1118675 | GCA_001726595.1 | MIRE00000000 | SAMN02368241 | 48 | 5,189,492 | 96 | 4,715 |
| CFSAN011119 | GCSL_R55 | Oyster | WA | 14-Jul-07 | O3:Kuk | ST735 | SRR1118674 | GCA_001726615.1 | MIRF00000000 | SAMN02368242 | 62 | 5,191,687 | 100 | 4,718 |
| CFSAN011120 | GCSL_R56 | Oyster | WA | 14-Jul-07 | O3:Kuk | ST735 | SRR1118673 | GCA_001726655.1 | MIRG00000000 | SAMN02368243 | 111 | 5,191,187 | 101 | 4,715 |
| CFSAN011121 | GCSL_R57 | Oyster | WA | 14-Jul-07 | O3:Kuk | ST1148 | SRR1118672 | GCA_001726645.1 | MIRH00000000 | SAMN02368244 | 72 | 5,100,768 | 101 | 4,652 |
| CFSAN011122 | GCSL_R59 | Oyster | ME | 23-Jul-07 | O5:Kuk | ST113 | SRR1815531 | GCA_001726685.1 | MIRI00000000 | SAMN03358820 | 38 | 4,953,641 | 128 | 4,512 |
| CFSAN011123 | GCSL_R60 | Oyster | ME | 23-Jul-07 | O10:Kuk | ST1135 | SRR1118671 | GCA_001726695.1 | MIRJ00000000 | SAMN02368245 | 77 | 5,264,840 | 126 | 4,802 |
| CFSAN011125 | GCSL_R62 | Oyster | ME | 23-Jul-07 | O1:Kuk | ST1136 | SRR1840771 | GCA_001726725.1 | MIRK00000000 | SAMN02368247 | 34 | 5,176,254 | 126 | 4,777 |
| CFSAN011126 | GCSL_R63 | Oyster | ME | 23-Jul-07 | O4:Kuk | Untyped | SRR1118669 | GCA_001726735.1 | MIRL00000000 | SAMN02368248 | 44 | 5,125,149 | 102 | 4,684 |
| CFSAN011128 | GCSL_R65 | Oyster | ME | 23-Jul-07 | O5:Kuk | ST1150 | SRR1118668 | GCA_001726755.1 | MIRM00000000 | SAMN02368250 | 46 | 5,065,957 | 111 | 4,652 |
| CFSAN011129 | GCSL_R74 | Oyster | VA | 23-Aug-07 | O4:K34 | ST108 | SRR1118667 | GCA_001726775.1 | MIRN00000000 | SAMN02368251 | 90 | 5,041,370 | 100 | 4,605 |
| CFSAN011130 | GCSL_R75 | Oyster | VA | 23-Aug-07 | O8:Kuk | ST676 | SRR1118666 | GCA_001726805.1 | MIRO00000000 | SAMN02368252 | 84 | 5,014,250 | 87 | 4,578 |
| CFSAN011131 | GCSL_R76 | Oyster | VA | 23-Aug-07 | O8:Kuk | ST676 | SRR1118665 | GCA_001726815.1 | MIRP00000000 | SAMN02368253 | 108 | 5,017,829 | 82 | 4,588 |
| CFSAN011132 | GCSL_R77 | Oyster | VA | 23-Aug-07 | O8:Kuk | ST676 | SRR1118664 | GCA_001726845.1 | MIRQ00000000 | SAMN02368254 | 74 | 5,014,463 | 76 | 4,581 |
| CFSAN011133 | GCSL_R86 | Oyster | FL | 13-Aug-07 | O6:Kuk | ST737 | SRR1118663 | GCA_001726885.1 | MIRR00000000 | SAMN02368255 | 70 | 4,894,547 | 116 | 4,454 |
| CFSAN011134 | GCSL_R87 | Oyster | FL | 13-Aug-07 | O8:K70 | ST320 | SRR1118662 | GCA_001726855.1 | MIRS00000000 | SAMN02368256 | 57 | 5,180,717 | 111 | 4,794 |
| CFSAN011135 | GCSL_R88 | Oyster | FL | 13-Aug-07 | O8:K70 | ST320 | SRR1840769 | GCA_001726895.1 | MIRT00000000 | SAMN02368257 | 34 | 5,184,181 | 90 | 4,781 |
| CFSAN011137 | GCSL_R95 | Oyster | PEI | 31-Jul-07 | O3:K5 | ST1151 | SRR1118660 | GCA_001726915.1 | MIRU00000000 | SAMN02368259 | 44 | 5,123,173 | 103 | 4,657 |
| CFSAN011138 | GCSL_R96 | Oyster | PEI | 31-Jul-07 | O11:Kuk | ST1152 | SRR1815532 | GCA_001726935.1 | MIRV00000000 | SAMN03358821 | 41 | 4,931,576 | 111 | 4,466 |
| CFSAN011140 | GCSL_R98 | Oyster | PEI | 31-Jul-07 | O3:K5 | ST1151 | SRR1815533 | GCA_001726965.1 | MIRW00000000 | SAMN03358822 | 41 | 5,125,449 | 102 | 4,676 |
| CFSAN011141 | GCSL_R99 | Oyster | PEI | 31-Jul-07 | O3:K5 | ST1151 | SRR1118658 | GCA_001726985.1 | MIRX00000000 | SAMN02368261 | 74 | 5,120,561 | 96 | 4,647 |
| CFSAN011143 | GCSL_R108 | Oyster | PEI | 31-Jul-07 | O3:K5 | ST1151 | SRR1815534 | GCA_001726975.1 | MIRY00000000 | SAMN03358823 | 68 | 5,156,159 | 93 | 4,700 |
| CFSAN011144 | GCSL_R109 | Oyster | PEI | 31-Jul-07 | O3:K5 | ST1151 | SRR1840768 | GCA_001727025.1 | MIRZ00000000 | SAMN02368263 | 34 | 5,124,297 | 93 | 4,665 |
| CFSAN011145 | GCSL_R110 | Oyster | PEI | 31-Jul-07 | O3:K5 | ST1151 | SRR1815535 | GCA_001727055.1 | MISA00000000 | SAMN03358824 | 54 | 5,123,801 | 96 | 4,673 |
| CFSAN011146 | GCSL_R111 | Oyster | PEI | 31-Jul-07 | O11:Kuk | ST1152 | SRR1118656 | GCA_001727065.1 | MISB00000000 | SAMN02368264 | 102 | 4,925,276 | 95 | 4,459 |
| CFSAN011147 | GCSL_R125 | Oyster | FL | 14-Oct-07 | O11:Kuk | ST739 | SRR1118655 | GCA_001727045.1 | MISC00000000 | SAMN02368265 | 138 | 5,078,459 | 150 | 4,628 |
| CFSAN011148 | GCSL_R126 | Oyster | FL | 14-Oct-07 | O4:K42 | ST1146 | SRR1118654 | GCA_001727105.1 | MISD00000000 | SAMN02368266 | 103 | 5,138,446 | 115 | 4,714 |
| CFSAN011149 | GCSL_R129 | Oyster | FL | 1-Oct-07 | O11:Kuk | ST1153 | SRR1118653 | GCA_001727115.1 | MISE00000000 | SAMN02368267 | 71 | 4,897,642 | 115 | 4,452 |
| CFSAN011150 | GCSL_R130 | Oyster | FL | 1-Oct-07 | O4:K37 | ST1140 | SRR1118652 | GCA_001975475.1 | MUCG00000000 | SAMN02368268 | 76 | 5,057,199 | 108 | 4,594 |
| CFSAN011151 | GCSL_R131 | Oyster | FL | 1-Oct-07 | O10:Kuk | ST1141 | SRR1840767 | GCA_001727125.1 | MISF00000000 | SAMN02368269 | 41 | 5,140,538 | 132 | 4,688 |
| CFSAN011152 | GCSL_R135 | Oyster | SC | 21-Nov-07 | O3:Kuk | ST741 | SRR1840766 | GCA_001727155.1 | MISG00000000 | SAMN02368270 | 34 | 5,066,023 | 141 | 4,634 |
| CFSAN011153 | GCSL_R136 | Oyster | SC | 21-Nov-07 | O1:K20 | ST775 | SRR1118651 | GCA_001727185.1 | MISH00000000 | SAMN02368271 | 78 | 5,098,402 | 139 | 4,651 |
| CFSAN011154 | GCSL_R137 | Oyster | SC | 21-Nov-07 | O1:K20 | ST775 | SRR1118650 | GCA_001727195.1 | MISI00000000 | SAMN02368272 | 71 | 5,107,378 | 127 | 4,638 |
| CFSAN011155 | GCSL_R138 | Oyster | SC | 21-Nov-07 | O1:K20 | ST775 | SRR1118649 | GCA_001727205.1 | MISJ00000000 | SAMN02368273 | 65 | 5,108,840 | 148 | 4,657 |
| CFSAN011156 | GCSL_R143 | Oyster | FL | Nov-07 (exact date unknown) | O5:Kuk | ST743 | SRR1118648 | GCA_001727265.1 | MISK00000000 | SAMN02368274 | 280 | 4,947,784 | 148 | 4,510 |
| CFSAN011157 | GCSL_R144 | Oyster | FL | Nov-07 (exact date unknown) | O5:Kuk | ST1149 | SRR1815536 | GCA_001727275.1 | MISL00000000 | SAMN03358825 | 48 | 5,085,869 | 181 | 4,641 |
| CFSAN011158 | GCSL_R145 | Oyster | FL | Nov-07 (exact date unknown) | O5:Kuk | ST1149 | SRR1118647 | GCA_001727295.1 | MISM00000000 | SAMN02368275 | 64 | 5,074,938 | 156 | 4,624 |
| CFSAN011159 | GCSL_R146 | Oyster | FL | Nov-07 (exact date unknown) | O5:Kuk | ST1149 | SRR1815537 | GCA_001727245.1 | MISN00000000 | SAMN03358826 | 50 | 5,084,933 | 172 | 4,649 |
| CFSAN011160 | GCSL_R149 | Oyster | FL | 19-Mar-07 | O1:Kuk | ST313 | SRR1118646 | GCA_001727305.1 | MISO00000000 | SAMN02368276 | 61 | 5,067,880 | 152 | 4,641 |
| CFSAN011161 | GCSL_R150 | Oyster | FL | 19-Mar-07 | O1:Kuk | ST313 | SRR1118645 | GCA_001727345.1 | MISP00000000 | SAMN02368277 | 25 | 5,066,369 | 149 | 4,663 |
The state for oyster isolates is the state where the product was harvested; for clinical isolates, this is the state that reported the isolate/illness. PEI, Prince Edward Island.
The date for oyster isolates is the date of harvest of the product; for clinical isolates, this is the date of isolation from the patient.
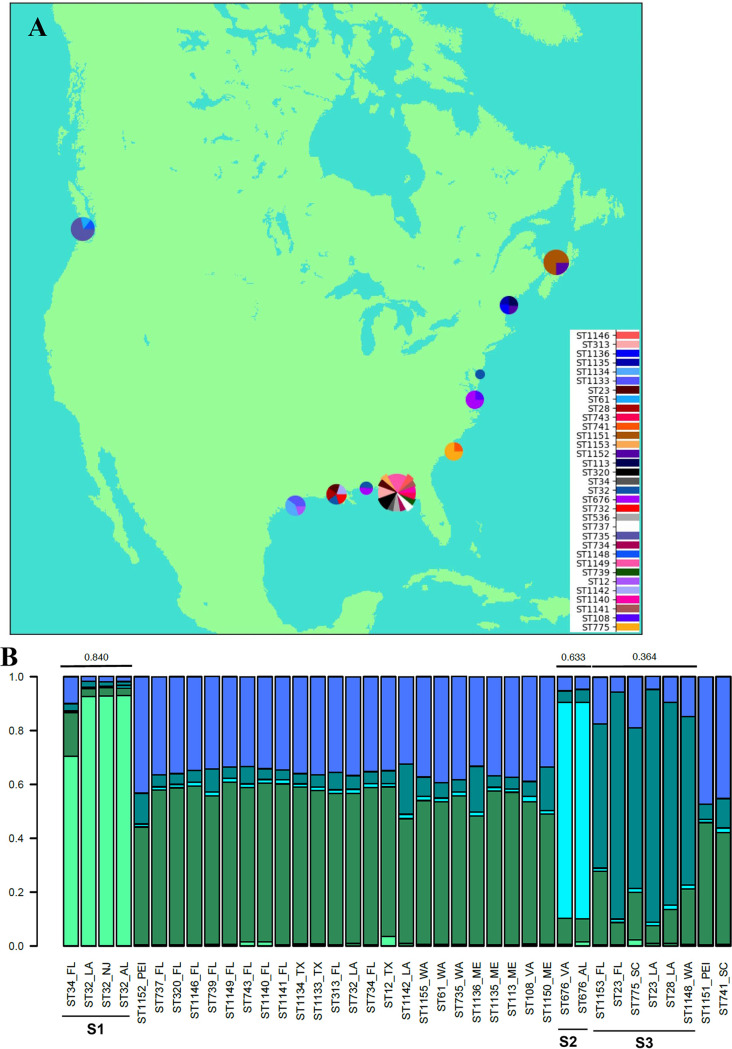
However, some geographic structure among the STs of oyster isolates is apparent. Most STs were isolated from only one site (Fig. 2A). Exceptions to this are ST676, which was isolated from Alabama and Virginia; ST23, which was isolated from Florida and Louisiana; ST32, which was isolated from Alabama, Louisiana, and New Jersey; and ST1152, which was isolated from Maine and Prince Edward Island (Canada). With the exception of Maine, all sites lacking a dominant ST were on the Gulf Coast.
FIG 2.
Geographic distribution of V. parahaemolyticus isolates from oysters. (A) Geographic distribution of the identified STs. The map was constructed using Python 2.7 with the help of Basemap from mpl_toolkits.basemap and inset_axes from mpl_toolkits.axes_grid1.inset_locator. Pie charts were added at approximate locations of collection sites as demonstrated at http://www.geophysique.be/2010/11/15/matplotlib-basemap-tutorial-05-adding-some-pie-charts/. The size of each pie chart is proportional to the total number of isolates from that location. Pie slices are proportional to the number of times that an ST was isolated at each site. (B) Genetic/geographic structure among the oyster isolates. STRUCTURE was used with MLST genes and geographic locations as priors. The maximum value of ln P(D) corresponded to a k value of 5, which was the lowest k value for which the members within each cluster with an Fst value of >0.2 shared genes. Graphs were created in R version 3.2.2 using the default plotting functions.
To further test the hypothesis that there was geographically meaningful population structure among the oyster isolates, STRUCTURE was used and identified three supported clusters (Fig. 2B). One of these (cluster S1) included ST32 and ST34 isolates primarily from the Gulf Coast (Alabama, Louisiana, and Florida) but also had a representative isolate from New Jersey. Another cluster (S2) consisted of only ST676 isolates from both Alabama and Virginia. The third cluster (S3) included ST23, ST28, ST775, ST1153, and ST1148 isolates, which had representatives from several locations in the Southeast (Florida, Louisiana, and South Carolina) plus one from Washington.
Phylogenetics and clustering.
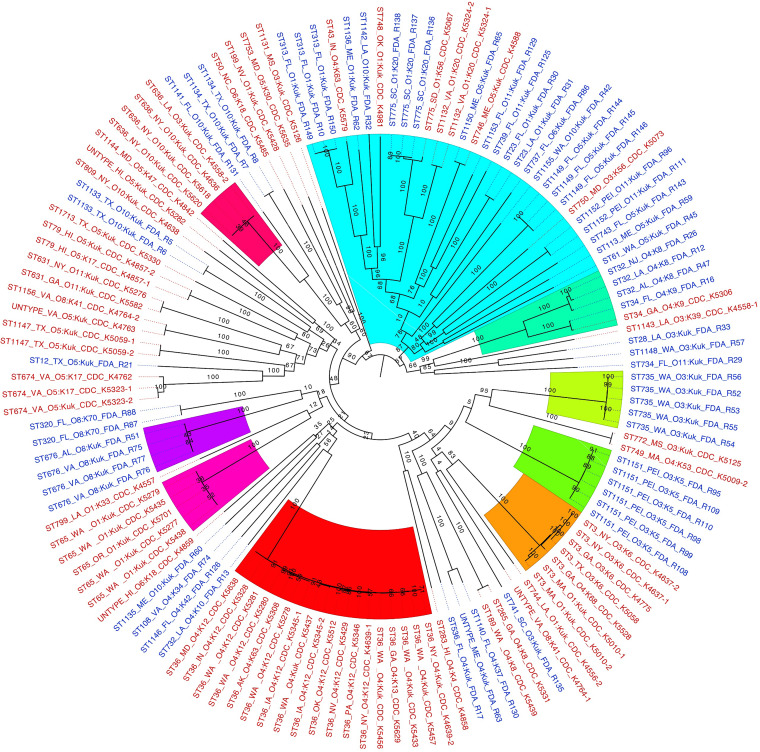
When the early-diverging bipartitions of the likelihood tree are considered, 9 partitions are identified (Fig. 3). Of these, four (partitions 3, 4, 5, and 7) are composed of only clinical isolates, three (partitions 1, 2, and 6) are composed of only oyster isolates, and two (partitions 8 and 9) contain both clinical and oyster isolates. Seven of the identified bipartitions (1 to 7) consist of only one ST. STs that include multiple serotypes and are monophyletic in the likelihood tree are ST3, ST36, and ST636. Whole-genome distance analysis was also conducted and largely supported the likelihood analysis, as eight of the well-supported partitions in the likelihood tree were also found in the distance tree (Fig. S2 and Table S1), lending more support to these clades.
FIG 3.
Likelihood tree demonstrating the phylogenetic relatedness of the 132 V. parahaemolyticus isolates. The phylogenetic relationships between isolates from clinical and oyster sources were based on a core genome of 3,726 genes. The likelihood tree was created with RAxML with the GTRCAT model with 100 bootstrap replicates based on the core genome. The 12 largest supported (bootstrap values of >70) clades are colored. Clinical isolates are labeled in red, and oyster isolates are labeled in blue. Clusters identified by STRUCTURE are identified by “SX,” where “X” is the cluster number provided in the STRUCTURE analysis.
Two of the three clusters found by STRUCTURE analysis of the oyster isolates (Fig. 2B) were monophyletic based on the phylogenetic analysis. ST32 and ST34 isolates were clustered by STRUCTURE (Fig. 2, S1) and form a well-supported monophyletic clade in the likelihood tree (Fig. 3, partition 9). ST676 isolates (Fig. 2, S2) from two locations, Virginia and Alabama, form a monophyletic group (Fig. 3, partition 6). From the third STRUCTURE cluster (Fig. 2, S3), ST23, ST775, and ST1153 isolates are all part of a large partition (Fig. 3, partition 8) but do not form a monophyletic group. ST28 and ST1148 isolates, which were also part of cluster S3 by STRUCTURE, were not part of a well-supported clade with those STs in either the likelihood or whole-genome distance analysis trees (Fig. 3; Table S1).
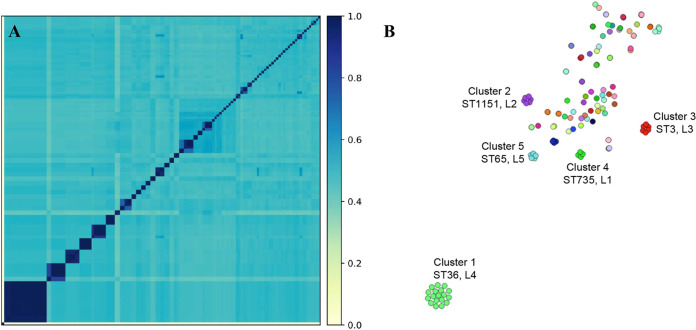
Comparison of the genomes using k-mer-based pairwise distance emphasized the higher diversity among the V. parahaemolyticus isolates (Fig. 4A) than what is commonly seen in other bacterial species. This analysis also enabled the differentiation of related groups that were distributed spatiotemporally, suggesting that they are phylogenetically linked. Further analysis with population partitioning using nucleotide k-mers (PopPUNK) identified five primary clusters, each containing isolates of a single ST (Fig. 4B): ST36 (17 isolates), ST1151 (6 isolates), ST3 (6 isolates), ST735 (5 isolates), and ST65 (5 isolates). Some smaller genomic clusters of four or fewer isolates also containing a single ST (ST676 and -775) were identified. Interestingly, PopPUNK clusters 1, 3, and 5 were composed of the same STs (ST36, -3, and -65) as the monophyletic clades of clinical isolates by the likelihood analysis (Fig. 3, partitions 4, 3, and 5, respectively) but were spatiotemporally separate in this analysis. PopPUNK clusters 2 and 4 and one of the smaller clusters (<5 isolates) were composed of ST1151, -735, and -676 found in the monophyletic clades of oyster isolates in the likelihood analysis (Fig. 3, partitions 2, 4, and 6; Table S1).
FIG 4.
Vibrio parahaemolyticus genome relatedness using whole-genome distance comparisons. (A) Genome distance using k-mers with an all-against-all comparison. (B) Genomically related isolate network using PopPUNK to demonstrate specific genomic clusters that are epidemiologically related within a cluster but genetically distinct between the groups. Colors represent the same genomic group. The distance between groups indicates relatedness as determined using whole-genome distance. Clusters are labeled with the unique cluster identification from this analysis (clusters 1 to 5), the ST that comprises the cluster, and the likelihood tree partition to which the isolates belong (“LX,” where “X” is the partition number in Fig. 3).
Potential pathogenicity genes.
To visualize a potential pattern between T3SS and TDH family genes and pathogenicity, a heat map was constructed with the isolates sorted by the total number of T3SS and TDH family genes that each possesses (Fig. 5). In most cases, when PCR detected both tdh and trh, InterPro annotated the two genes as tdh. However, there were two instances (ST636_CDC_K4558-2 and ST772_CDC_K5125) where PCR did not detect either tdh or trh but two tdh-related genes were found in the genomic sequence. In addition, there were some instances where PCR detected trh and multiple instances of tdh-related genes were found in the genomic sequence: two in some isolates (ST775_FDA_R137, ST1134_FDA_R7, and ST1134_FDA_R8) and, surprisingly, even three for some ST3 isolates (CDC_K5528, CDC_5010-1, CDC_5010-2, and CDC_4637-2). Isolates from both humans and oysters were among those predicted to have two tdh-related genes.
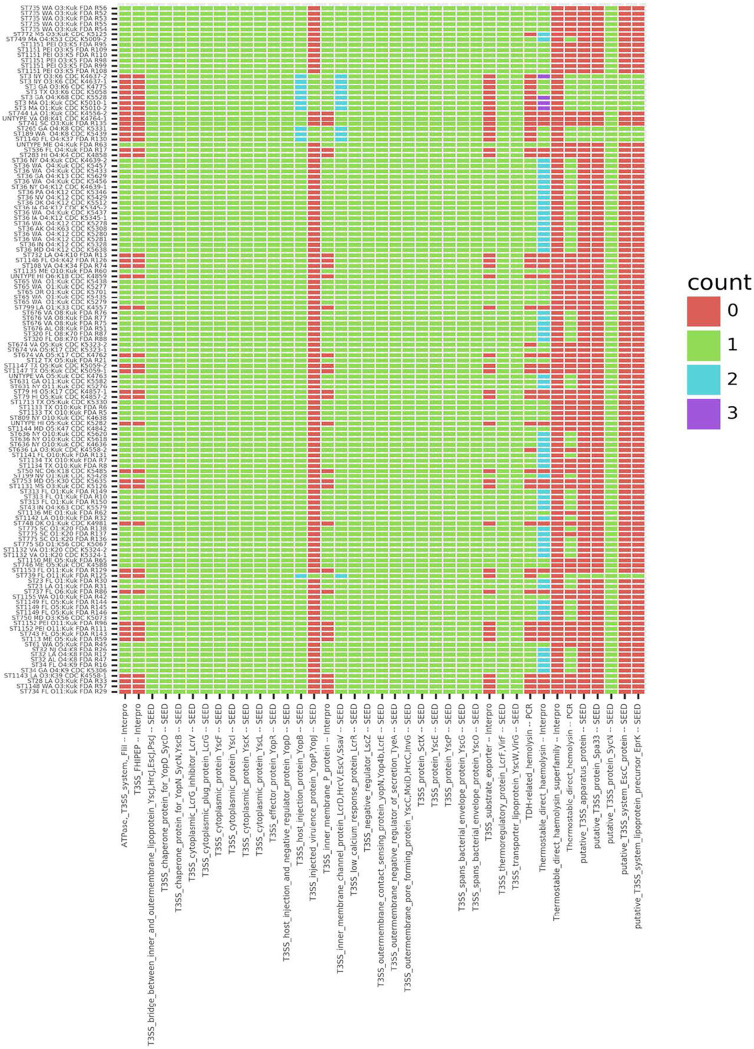
FIG 5.
Heat map of counts for SEED and InterProScan annotations. Counts of annotations for SEED T3SS genes and InterProScan annotations for TDH genes and PCR results for tdh and trh are shown. Per-isolate counts for T3SS and TDH annotated clusters along withprevious PCR results for TDH and TRH, included for comparison, were visualized within a heat map created with Plotnine, geom_tile. For ease of visualization, isolates are listed in the order of phylogeny based on the likelihood tree (i.e., isolates at the top are in partition 1 of Fig. 3).
Twenty-six T3SS genes were present in all 132 draft genomes (Fig. 5). There were also several genes found in only some genomes: the T3SS inner membrane P protein was found in 102 isolates, and the T3SS ATPase (FliL), the T3SS FHIPEP sorting domain, and the T3SS substrate exporter were found in 91 isolates. There were 11 isolates that had either additional T3SS genes or extra copies of some T3SS genes: the 7 ST3 isolates, which comprise a monophyletic clade; CDC_K5439, CDC_K5331, and FDA_R130, which cluster near the ST3 isolate clade; and the relatively unrelated isolate FDA_R125 (Fig. 3). There were 5 T3SS genes that were possessed by only these 11 isolates: the injected virulence protein YopP/YopJ, a putative T3SS apparatus protein, the putative T3SS protein Spa33, the putative T3SS system EscC protein, and the putative T3SS system lipoprotein precursor EprK. The same 11 isolates had second copies of two genes: the T3SS host injection protein YopB and the inner membrane channel protein LcrD/HrcV/EscV/SsaV.
Due to the variation in copy number within the T3SS and tdh gene groups, annotations that were prevalent in isolates within monophyletic clades of clinical isolates (ST3, ST36, and ST65) (Fig. 3, partitions 3, 4, and 5) relative to monophyletic clades of oyster isolates (ST676, ST735, and ST1151) (Fig. 3, partitions 6, 1, and 2) were examined, omitting isolates from mixed clades (Fig. 6). While some analyses strictly control for phylogeny, we wanted to be able to determine if gene abundances were consistent within the clades and not only across clades (or isolation source). In some cases, genes with an annotation were abundant, with all isolates having more than 15 genes with the annotation: major facilitator superfamily (MFS) transporters, EAL domains, and acyl-CoA N-acyltransferases. In other cases, the overall numbers were low, with all isolates having at least 1 but fewer than 10 genes: thiamine diphosphate-binding, RmlC-like cupin, integrase/recombinase N-terminal, flavin mononucleotide (FMN)-binding split barrel, and 3-keto-5-aminohexanoate cleavage enzymes. With other annotations, some isolates had none, and others had one or two: pyridoxamine 5-phosphate oxidase, FMN dependent; DUF2787; and glycosyltransferase family 11 (GT11). It was also variable as to how differentiated the clinical isolates were from the oyster isolates for all annotations (Fig. 6), the difference in numbers was statistically significant, and it was generally apparent that clinical isolates had more than oyster isolates.
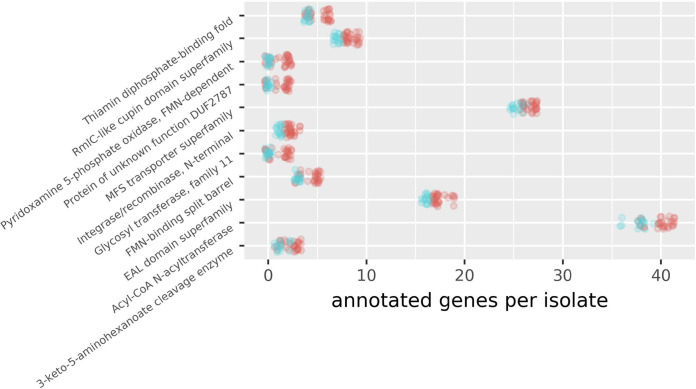
FIG 6.
Jitter plot with numbers of genes in monophyletic groups. Annotation groups with statistically significant differences between clinical and oyster isolates (P values of <0.01 after Benjamini-Hochberg adjustment [75]) were included. Genes enriched in isolates from monophyletic groups of human isolates relative to monophyletic groups of environmental isolates were found by counting the number of gene clusters that each isolate was in for each annotation. Isolate counts for each annotation for monophyletic human versus environmental isolates were then compared using Kruskal-Wallis tests. The jitter plot was illustrated with Plotnine, geom_jitter. Red indicates clinical isolates, and blue indicates oyster isolates.
DISCUSSION
Population and genetic continuity in the Gulf and East Coasts.
As reported in a previous study using a subset of these isolates, ST36 and ST3 are the most frequent STs in this isolate collection. Both ST36 and ST3 were comprised only of clinical isolates collected on various dates and from multiple regions with O4:K12/O4:Kut and pandemic-related serotypes, respectively, as previously described for these STs (9, 24). However, these data show high diversity regarding the residual STs of the isolates tested regardless of the method of analysis used (likelihood or genome distance).
STRUCTURE identified three clusters with Fst values indicative of population structure along the Gulf Coast and Atlantic Coast of North America and even including one isolate from the Pacific Coast, consistent with the results of others (30, 31). Phylogenetic support was lacking for one of these clusters detected with STRUCTURE. This is not surprising as MLST alleles represent a small portion of each genome and probably should not be used for phylogenetic purposes when larger data sets are available. However, the incongruence between the phylogenetic results and the MLST results could be indicative of horizontal gene transfer between phylogenetically distant strains (32). Indeed, genomic recombination and horizontal gene transfer were inferred using genomic distance analysis by the presence of multiple clusters (i.e., colored dots adjacent to one another and not forming a distinctive monochromatic group). The precise mechanism, as well as the selection pressure, that results in limiting the genotype diversity has not been identified thus far, although seroconversion has been investigated (33), and is a question that requires additional examination.
Oceanic currents, as well as shipping vessels, could play a role in the distribution, diversity, and genetics of V. parahaemolyticus (34, 35). The highest-diversity site was on the Gulf Coast side of Florida, probably influenced by the Yucatan and Loop currents and their small offshoots, which could be carrying organisms picked up by the Caribbean current from Caribbean islands and the northern coast of South America. Alabama seems to have much lower diversity, but there were only two isolates represented by two STs, so the detected diversity was as high as it could have been. The other Gulf Coast sites also have high diversity relative to most non-Gulf sites and could be influenced by the complex offshoots of the Loop and Mexican currents. STs found on both the Gulf and East Coasts could be an indication of connectivity due to the Florida current and the Gulf Stream. The only other site without a clear majority sequence type is Maine, which is near the convergence of the Gulf Stream and the Labrador current, presenting two possible sources of V. parahaemolyticus and a more variable environment that prevents one ST from dominating. The isolate with ST1148 could have been introduced to the Pacific Coast via ballast water, as has been previously suggested for V. parahaemolyticus (35).
Gene content and pathogenicity.
There was good general agreement between previous PCR detections of tdh-related genes and the genomic results. In most cases where there was PCR detection, tdh-related genes were annotated within the genomes, and when both tdh and trh were detected via PCR, multiple copies of tdh-related genes were found in the proteins predicted by InterPro. The few cases where more genes were annotated from the genomic data than were detected via PCR could be due to a single set of primers amplifying in two places or gene sequence diversity. The use of multiple techniques largely validated but also complemented one another. The two approaches together provide multiple lines of evidence for the existence of clinical isolates lacking any tdh or trh gene and showing that while these bacteria may use these genes during the infection process, they are not required or predictive of pathogenicity.
In addition to the hemolysins, T3SS genes are potentially associated with virulence, perhaps by targeting the cytoskeleton, as do many T3SS effectors. Eleven of our isolates, including all ST3 isolates, contained YopP/YopJ, which is involved in inducing apoptosis (36). Thus, these isolates have an additional weapon that they can use to acquire resources from eukaryotic cells. This, however, does not explain why ST36 and ST65 are so frequently isolated from humans, as these two genotypes lacked the YopP/YopJ gene. While the T3SS may be an important tool in the arsenal of V. parahaemolyticus, it does not explain the pattern of pathogenicity observed in this set of isolates.
The monophyletic clusters of clinical isolates (ST3-ST36-ST65) tend to have additional genes with specific annotations compared to isolates within monophyletic groups of environmental isolates. These groups of genes can be broadly grouped functionally into DNA handling, metabolic, and signaling. One of these annotations is for integrase/recombinase, N terminal. The ST3-ST36-ST65 isolates could use additional integrase capacity to acquire genes that could be used in the infection process (37). These isolates are also enriched in genes involved in sensing the environment, signal transduction, and gene expression. Bacteria that tend to thrive in nutrient-enriched environments typically have more genes related to gene expression and signal transduction. EAL domain proteins are involved in the synthesis of cyclic diguanylate used in intracellular signaling, which could be useful in transmitting intracellular signals related to changes in the extracellular milieu (38). Proteins of this type could help these genotypes quickly take advantage of new resources not only in the ocean but also when they find themselves in an unexpected environment, like a human. The ST3-ST36-ST65 isolates also tend to have more MFS transporters. These proteins act as antiporters and symporters that link the transport of ions with the transport of other ions or small organic solutes (39) and thus could help in nutrient absorption. Better nutrient absorption would make more-rapid growth possible.
After detecting a change in their surroundings, the ST3-ST36-ST65 isolates could use genes of metabolic function, in which they tend to be enriched, to support enhanced growth. Some genes involved in carbohydrate metabolism may have direct effects on their pathogenicity. As one example, RmlC-like cupin domains play a role in the synthesis of l-rhamnose that may be involved in bacterial pathogenesis (40). Glycosyltransferase family 11 (GT11) is a galactoside 2-l-fucosyltransferase, which transfers fucose from guanosine-diphosphate fucose to a substrate (41), homologs of which are involved in antigen synthesis in other bacterial species (42, 43). Thus, GT11 could be important in the interaction of V. parahaemolyticus with a human host.
The ST3-ST36-ST65 isolates also appear to be enriched in genes related to the processing of metabolites. Acyl-CoA N-acyltransferases function in the synthesis of fatty acids using acetyl-CoA and another lipid as the substrates and in the production of ATP from amino acids (44, 45). Both functions could be beneficial to V. parahaemolyticus during periods of low-nutrient stress, such as in coastal estuarine habitats. Other marine organisms use acyltransferases and similar proteins to store fatty acids and wax esters for use during periods of stress or dormancy (46, 47). Such a gene could be used by V. parahaemolyticus to rapidly incorporate environmental lipids for more rapid growth or to store them for use in times of stress (48). While there is much to be studied about how these genes enriched in the ST3-ST36-ST65 clade could contribute to environmental survival, transmissibility, and pathogenicity, it is clear that there are many potential benefits.
Rare in the marine environment but common in outbreaks.
It is surprising that ST3, ST36, and ST65, each of which forms a monophyletic clade, were prevalent among clinical isolates but were not recovered from oysters in this study. While seafood-associated outbreaks could be expected to be caused by genetically homogeneous organisms (49), even with the bias in favor of sampling from clinical sources, enough samples were taken from the environment that if ST3, ST36, and ST65 were common in the environment, these STs would be represented by at least a few oyster isolates. However, previous work has found ST3 and ST36 isolates in the environment (15), but that study was focused on an area where these STs were strongly predominant in patients. They may become abundant in marine environments only under specific conditions, remaining in unidentified oceanic reservoirs otherwise. The enhanced lipid storage capability, inferred from the possession of extra copies of acyl-CoA N-acyltransferase genes, of ST3-ST36-ST65 isolates may enable them to spend more time as dormant cells (50, 51) in marine habitats and be numerically inferior to other genotypes of this bacterium, making them more difficult to detect. In contrast, they seem to have some relative advantages in associating with humans. For example, they may be able to take advantage of the more-nutrient-replete conditions of the human intestine more quickly due to their enhanced transport and signaling systems as well as additional metabolic genes that allow them to outcompete other genotypes under these conditions.
Conclusions.
This study used whole-genome sequencing of 132 V. parahaemolyticus isolates to understand the distribution and genetics of this pathogen along coasts of North America. Using in silico MLST from the draft genomes, a hub of genetic diversity along the Gulf and southern Atlantic Coasts as well as lower diversity along the northern portions of the Atlantic and Pacific Coasts were identified. In addition, the data show that the results of MLST methods, which use only a small portion of the genome, are generally consistent with the results of a core genome maximum likelihood phylogeny and that the results of distance phylogenetic methods, when applied to the same genomic data, are broadly consistent with the results of the maximum likelihood analysis. These results indicate that when time, sequencing capacity, or computational capacity is limited, biologists can use faster and more affordable methods and still obtain informative, but not comprehensive, results.
Additionally, this study looked for pathogenicity genes. The tdh, trh, and T3SS genes, while present in many of our isolates, do not explain the apparent differences in pathogenicity. Instead, the isolates from monophyletic clades consisting of only human isolates were enriched in genes having to do with signal transduction, nutrient absorption, energy transduction, and energy storage. This is consistent with a model in which these strains can store energy efficiently for periods of dormancy and then quickly respond to periods of high levels of nutrients by growing quickly and outcompeting other strains of the same bacterial species. Thus, it is possible that the strains most frequently isolated from humans are difficult to detect in marine habitats because they are more likely than other strains to be in a dormant state, but when levels of nutrients become especially high, as in a human intestine, they are able to outcompete their competitors. Further experimental work is needed to elucidate the ability of strains to persist and proliferate under various conditions in relation to the genes possessed and the ecological factors that vary with geography. This experimental work should include gene expression profiling, as pathogenicity could be influenced by gene expression as well as gene content.
MATERIALS AND METHODS
Bacterial strains.
The 61 oyster and 71 clinical well-characterized isolates used in this study (Table 1) were collected from 2006 to 2007 in the United States and Canada (10, 23). These isolates were selected to be representative of the geographic diversity of North America for both environmental (oyster) and patient isolates over a given period to avoid the introduction of broad temporal variation. The isolates were stored at −80°C until analysis.
Genomic DNA extraction.
All isolates were grown in Trypticase soy agar (TSA; Thermo Fisher Scientific, Waltham, MA) overnight at 37°C. High-molecular-weight DNA was extracted using the QIAamp DNA minikit (Qiagen, Valencia, CA). The integrity of high-molecular-weight DNA was determined using a 2200 TapeStation with genomic DNA ScreenTape (Agilent Technologies, Santa Clara, CA) as previously described (52).
Whole-genome sequencing, assembly, and annotation.
Sequencing was conducted by the Weimer laboratory at the University of California—Davis through the 100K Pathogen Genome Project (http://www.genomes4health.org/) (53) as previously described (54). DNA was fragmented using the Covaris (Woburn, MA) E220 ultrasonicator. Fragmented DNA (1 μg) was used to construct sequencing libraries using the HTP library preparation kit (Kapa Biosystems, Wilmington, MA), on a Bravo NGS workstation (Agilent Technologies). Fragmented double-stranded DNA (dsDNA) molecules were end repaired (5′), adenylated (3′), and then ligated with dsDNA adapters using the NEXTflex-96 DNA barcode (Bioo Scientific, Austin, TX). The size distributions of amplified libraries were confirmed using the 2100 bioanalyzer with a high-sensitivity DNA kit (Agilent Technologies). Finally, the indexed libraries were quantified with a quantitative PCR (qPCR)-based library quantification kit (Kapa Biosystems) prior to pooling for sequencing on the Illumina HiSeq 2000 platform with PE100 plus index reads (Illumina, San Diego, CA). The genomic sequences were de novo assembled using SPAdes version 3.1.1 software (55) with a k-mer length of 32. The draft genomes were annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) (www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html).
Pangenome analysis.
The pangenome was determined as described previously by Bandoy and Weimer (56). Briefly, the genome sequences were assembled using Shovill (https://github.com/tseemann/shovill), annotated using Prokka (57), and used as the data input for pangenome analysis using Roary (58). Assembled genomes were variant called using Snippy (https://github.com/tseemann/snippy). Gene presence/absence was visualized using Phandango (59) with the associated metadata. Gene associations, metadata, and phenotypes were associated using Scoary (60).
Population genetics and biogeography.
Genomes were analyzed for MLST sequence types (STs) based on the allele types of the housekeeping genes dnaE, gyrB, dtdS, recA, pyrC, pntA, and tnaA through the MLST database for V. parahaemolyticus at http://pubmlst.org/vparahaemolyticus/ (21). STRUCTURE, which is a method for inferring population structure without incorporating any a priori information as to membership (61, 62), was run using the MLST alleles from oyster isolates as the input and the geographic collection sites as priors. The burn-in consisted of 30,000 Markov chain Monte Carlo (MCMC) iterations followed by 60,000 additional repetitions that were utilized. Values for k from 2 to 16 were tested. The highest ln P(D) value corresponded to a k value of 5. This k value resulted in all well-supported clusters (Fst > 0.2) sharing alleles, so it was used because there is no genetic basis in this data set for clustering isolates lacking shared alleles.
Phylogenetics.
For the likelihood tree, nucleotide sequences from each cluster of orthologous groups (COG) in the core genome were aligned using MAFFT (version 7.427) with “—adjustdirection” to find reverse complements of genes when necessary to ensure that genes were oriented in the same direction (63). The alignments were then concatenated with a custom Python script. The best likelihood tree as well as the 100 bootstrap trees were found using raxmlHPC-PTHREADS (version 8.2.9) rapid bootstrapping with a general time reversal rate categories (GTRCAT) model of nucleotide substitution and rate heterogeneity (64).
For the distance tree, the assembly of paired-end reads was done with ABySS 1.5.2 using a k value of 64 (65) and as used previously (28, 29). Pairwise distances were determined using the Meier-Kolthoff method as reported previously (66) and implemented locally using PanCake (29). A distance tree was inferred using Mega7 with neighbor joining (67, 68) and visualized using MATLAB software (MathWorks, Natick, MA, USA).
Genome variation was determined using total genomic distance with a k-mer (31-mer) approach as a method to use the entire genome to determine relatedness between isolates as previously described (56). Population partitioning using nucleotide k-mers (PopPUNK) was used to determine related genomic clusters based on the whole-genome distance of the Vibrio isolates used in this study. PopPUNK uses genomic distance with variable-length k-mer comparisons, enabling the analysis of divergence in core and accessory gene contents within the same analysis to visualize a network-like vision of genome diversity (69).
Ortholog discovery and annotation.
Open reading frames were found using Prodigal 2.6.3 (70). The core genome was identified by first running all-against-all pairwise reciprocal BLAST (71), finding pairwise reciprocal best hits (PRBHs) requiring an E value of <1 × 10−45 and a length of at least 80 nucleotides, and then grouping these into COGs (72). COGs were found with a custom Python script that first identified preclusters from a sorted list of all PRBHs by clustering pairs sharing the first element and then recursively looked for and grouped preclusters that overlapped by at least two gene identifications. Clusters overlapping by only one gene identification remained as separate clusters. In these cases, one was removed, with a preference for keeping the larger cluster. The core genome was then assembled from COGs containing every isolate exactly once.
COGs were annotated by BLAST comparisons to isolates AQ3810 and RIMD_2210633 (downloaded from https://theseed.org [73]) and with InterProScan (74) using the RESTful Web service and Python 3.6. The InterPro annotations (identifiers starting with “IPR”) were used. In both cases, annotations with E values of <1 × 10−45 were accepted. For T3SS genes, SEED annotations were preferentially used over InterPro annotations because they were presumed to be more species specific and refined, but if InterPro annotated a gene as being T3SS related, it was used unless the SEED annotation indicated that it was flagellar. TDH family gene clusters were annotated with InterPro.
Data availability.
All accession numbers and associated metadata are provided in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported in part by an appointment of C. H. M. Lüdeke and J. J. Miller to the Research Participation Program at the Center for Food Safety and Applied Nutrition administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
REFERENCES
- 1.Jones JL. 2014. Vibrio: introduction, including Vibrio parahaemolyticus, Vibrio vulnificus, and other Vibrio species, p 691–698. In Batt CA, Tortorello ML (ed), Encyclopedia of food microbiology, vol 3 Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 2.Iwamoto M, Ayers T, Mahon BE, Swerdlow DL. 2010. Epidemiology of seafood-associated infections in the United States. Clin Microbiol Rev 23:399–411. doi: 10.1128/CMR.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker-Austin C, Trinanes JA, Taylor NGH, Hartnell R, Siitonen A, Martinez-Urtaza J. 2013. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat Clim Chang 3:73–77. doi: 10.1038/nclimate1628. [DOI] [Google Scholar]
- 4.CDC. 2016. Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet surveillance report for 2014 (final report). US Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 5.Powell JL. 1999. Vibrio species. Clin Lab Med 19:537–552. doi: 10.1016/S0272-2712(18)30103-3. [DOI] [PubMed] [Google Scholar]
- 6.Lynch T, Livingstone S, Buenaventura E, Lutter E, Fedwick J, Buret AG, Graham D, DeVinney R. 2005. Vibrio parahaemolyticus disruption of epithelial cell tight junctions occurs independently of toxin production. Infect Immun 73:1275–1283. doi: 10.1128/IAI.73.3.1275-1283.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park KS, Ono T, Rokuda M, Jang MH, Iida T, Honda T. 2004. Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol Immunol 48:313–318. doi: 10.1111/j.1348-0421.2004.tb03512.x. [DOI] [PubMed] [Google Scholar]
- 8.Broberg CA, Zhang L, Gonzalez H, Laskowski-Arce MA, Orth K. 2010. A Vibrio effector protein is an inositol phosphatase and disrupts host cell membrane integrity. Science 329:1660–1662. doi: 10.1126/science.1192850. [DOI] [PubMed] [Google Scholar]
- 9.Jesser KJ, Valdivia-Granda W, Jones JL, Noble RT. 2019. Clustering of Vibrio parahaemolyticus isolates using MLST and whole-genome phylogenetics and protein motif fingerprinting. Front Public Health 7:66. doi: 10.3389/fpubh.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones JL, Ludeke CH, Bowers JC, Garrett N, Fischer M, Parsons MB, Bopp CA, DePaola A. 2012. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J Clin Microbiol 50:2343–2352. doi: 10.1128/JCM.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burdette DL, Yarbrough ML, Orth K. 2009. Not without cause: Vibrio parahaemolyticus induces acute autophagy and cell death. Autophagy 5:100–102. doi: 10.4161/auto.5.1.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caburlotto G, Lleo MM, Hilton T, Huq A, Colwell RR, Kaper JB. 2010. Effect on human cells of environmental Vibrio parahaemolyticus strains carrying type III secretion system 2. Infect Immun 78:3280–3287. doi: 10.1128/IAI.00050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiou CS, Hsu SY, Chiu SI, Wang TK, Chao CS. 2000. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J Clin Microbiol 38:4621–4625. doi: 10.1128/JCM.38.12.4621-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton AE, Garrett N, Stroika SG, Halpin JL, Turnsek M, Mody RK, Centers for Disease Control and Prevention. 2014. Increase in Vibrio parahaemolyticus infections associated with consumption of Atlantic Coast shellfish—2013. MMWR Morb Mortal Wkly Rep 63:335–336. [PMC free article] [PubMed] [Google Scholar]
- 15.Turner JW, Paranjpye RN, Landis ED, Biryukov SV, Gonzalez-Escalona N, Nilsson WB, Strom MS. 2013. Population structure of clinical and environmental Vibrio parahaemolyticus from the Pacific Northwest coast of the United States. PLoS One 8:e55726. doi: 10.1371/journal.pone.0055726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CDC. 2016. Cholera and other Vibrio illness surveillance (COVIS), summary data, 2014. CDC, Atlanta, GA: https://www.cdc.gov/nationalsurveillance/pdfs/covis-annual-summary-2014-508c.pdf. Accessed 27 October 2020. [Google Scholar]
- 17.Xu F, Ilyas S, Hall JA, Jones SH, Cooper VS, Whistler CA. 2015. Genetic characterization of clinical and environmental Vibrio parahaemolyticus from the Northeast USA reveals emerging resident and non-indigenous pathogen lineages. Front Microbiol 6:272. doi: 10.3389/fmicb.2015.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePaola A, Jones JL, Woods J, Burkhardt W III, Calci KR, Krantz JA, Bowers JC, Kasturi K, Byars RH, Jacobs E, Williams-Hill D, Nabe K. 2010. Bacterial and viral pathogens in live oysters: 2007 United States market survey. Appl Environ Microbiol 76:2754–2768. doi: 10.1128/AEM.02590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePaola A, Nordstrom JL, Bowers JC, Wells JG, Cook DW. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl Environ Microbiol 69:1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson CN, Flowers AR, Noriea NF III, Zimmerman AM, Bowers JC, DePaola A, Grimes DJ. 2010. Relationships between environmental factors and pathogenic vibrios in the Northern Gulf of Mexico. Appl Environ Microbiol 76:7076–7084. doi: 10.1128/AEM.00697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus L-A, DePaola A. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J Bacteriol 190:2831–2840. doi: 10.1128/JB.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harth-Chu E, Espejo RT, Christen R, Guzman CA, Hofle MG. 2009. Multiple-locus variable-number tandem-repeat analysis for clonal identification of Vibrio parahaemolyticus isolates by using capillary electrophoresis. Appl Environ Microbiol 75:4079–4088. doi: 10.1128/AEM.02729-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludeke CH, Fischer M, LaFon P, Cooper K, Jones JL. 2014. Suitability of the molecular subtyping methods intergenic spacer region, direct genome restriction analysis, and pulsed-field gel electrophoresis for clinical and environmental Vibrio parahaemolyticus isolates. Foodborne Pathog Dis 11:520–528. doi: 10.1089/fpd.2013.1728. [DOI] [PubMed] [Google Scholar]
- 24.Lüdeke CHM, Kong N, Weimer BC, Fischer M, Jones JL. 2015. Complete genome sequences of a clinical isolate and an environmental isolate of Vibrio parahaemolyticus. Genome Announc 3:e00216-15. doi: 10.1128/genomeA.00216-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haendiges J, Timme R, Allard M, Myers RA, Payne J, Brown EW, Evans P, Gonzalez-Escalona N. 2014. Draft genome sequences of clinical Vibrio parahaemolyticus strains isolated in Maryland (2010 to 2013). Genome Announc 2:e00776-14. doi: 10.1128/genomeA.00776-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haendiges J, Timme R, Allard MW, Myers RA, Brown EW, Gonzalez-Escalona N. 2015. Characterization of Vibrio parahaemolyticus clinical strains from Maryland (2012-2013) and comparisons to a locally and globally diverse V. parahaemolyticus strains by whole-genome sequence analysis. Front Microbiol 6:125. doi: 10.3389/fmicb.2015.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazen TH, Lafon PC, Garrett NM, Lowe TM, Silberger DJ, Rowe LA, Frace M, Parsons MB, Bopp CA, Rasko DA, Sobecky PA. 2015. Insights into the environmental reservoir of pathogenic Vibrio parahaemolyticus using comparative genomics. Front Microbiol 6:204. doi: 10.3389/fmicb.2015.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taff CC, Weis AM, Wheeler S, Hinton MG, Weimer BC, Barker CM, Jones M, Logsdon R, Smith WA, Boyce WM, Townsend AK. 2016. Influence of host ecology and behavior on Campylobacter jejuni prevalence and environmental contamination risk in a synanthropic wild bird species. Appl Environ Microbiol 82:4811–4820. doi: 10.1128/AEM.01456-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weis AM, Storey DB, Taff CC, Townsend AK, Huang BC, Kong NT, Clothier KA, Spinner A, Byrne BA, Weimer BC. 2016. Genomic comparison of Campylobacter spp. and their potential for zoonotic transmission between birds, primates, and livestock. Appl Environ Microbiol 82:7165–7175. doi: 10.1128/AEM.01746-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui Y, Yang X, Didelot X, Guo C, Li D, Yan Y, Zhang Y, Yuan Y, Yang H, Wang J, Wang J, Song Y, Zhou D, Falush D, Yang R. 2015. Epidemic clones, oceanic gene pools, and Eco-LD in the free living marine pathogen Vibrio parahaemolyticus. Mol Biol Evol 32:1396–1410. doi: 10.1093/molbev/msv009. [DOI] [PubMed] [Google Scholar]
- 31.Yang C, Pei X, Wu Y, Yan L, Yan Y, Song Y, Coyle NM, Martinez-Urtaza J, Quince C, Hu Q, Jiang M, Feil E, Yang D, Song Y, Zhou D, Yang R, Falush D, Cui Y. 2019. Recent mixing of Vibrio parahaemolyticus populations. ISME J 13:2578–2588. doi: 10.1038/s41396-019-0461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiden MCJ. 2006. Multilocus sequence typing of bacteria. Annu Rev Microbiol 60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Stine OC, Badger JH, Gil AI, Nair GB, Nishibuchi M, Fouts DE. 2011. Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence. BMC Genomics 12:294. doi: 10.1186/1471-2164-12-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raszl SM, Froelich BA, Vieira CRW, Blackwood AD, Noble RT. 2016. Vibrio parahaemolyticus and Vibrio vulnificus in South America: water, seafood and human infections. J Appl Microbiol 121:1201–1222. doi: 10.1111/jam.13246. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez-Escalona N, Gavilan RG, Brown EW, Martinez-Urtaza J. 2015. Transoceanic spreading of pathogenic strains of Vibrio parahaemolyticus with distinctive genetic signatures in the recA gene. PLoS One 10:e0117485. doi: 10.1371/journal.pone.0117485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornelis GR. 2000. Molecular and cell biology aspects of plague. Proc Natl Acad Sci U S A 97:8778–8783. doi: 10.1073/pnas.97.16.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H. 2003. Phage as agents of lateral gene transfer. Curr Opin Microbiol 6:417–424. doi: 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 38.Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, DeMaere MZ, Ting L, Ertan H, Johnson J, Ferriera S, Lapidus A, Anderson I, Kyrpides N, Munk AC, Detter C, Han CS, Brown MV, Robb FT, Kjelleberg S, Cavicchioli R. 2009. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci U S A 106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pao SS, Paulsen IT, Saier MH Jr.. 1998. Major facilitator superfamily. Microbiol Mol Biol Rev 62:1–34. doi: 10.1128/MMBR.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giraud MF, Naismith JH. 2000. The rhamnose pathway. Curr Opin Struct Biol 10:687–696. doi: 10.1016/S0959-440X(00)00145-7. [DOI] [PubMed] [Google Scholar]
- 41.Ma B, Simala-Grant JL, Taylor DE. 2006. Fucosylation in prokaryotes and eukaryotes. Glycobiology 16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 42.Wang G, Boulton PG, Chan NWC, Palcic MM, Taylor DE. 1999. Novel Helicobacter pylori alpha1,2-fucosyltransferase, a key enzyme in the synthesis of Lewis antigens. Microbiology 145(Part 11):3245–3253. doi: 10.1099/00221287-145-11-3245. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, Kaur I, Niesel DW, Seetharamaiah GS, Peterson JW, Prabhakar BS, Klimpel GR. 1997. Lipoprotein from Yersinia enterocolitica contains epitopes that cross-react with the human thyrotropin receptor. J Immunol 158:1976–1983. [PubMed] [Google Scholar]
- 44.Ganesan B, Dobrowolski P, Weimer BC. 2006. Identification of the leucine-to-2-methylbutyric acid catabolic pathway of Lactococcus lactis. Appl Environ Microbiol 72:4264–4273. doi: 10.1128/AEM.00448-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rottig A, Steinbuchel A. 2013. Acyltransferases in bacteria. Microbiol Mol Biol Rev 77:277–321. doi: 10.1128/MMBR.00010-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. 2004. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol 186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rontani JF, Bonin PC, Volkman JK. 1999. Production of wax esters during aerobic growth of marine bacteria on isoprenoid compounds. Appl Environ Microbiol 65:221–230. doi: 10.1128/AEM.65.1.221-230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong HC, Wang P. 2004. Induction of viable but nonculturable state in Vibrio parahaemolyticus and its susceptibility to environmental stresses. J Appl Microbiol 96:359–366. doi: 10.1046/j.1365-2672.2004.02166.x. [DOI] [PubMed] [Google Scholar]
- 49.McLaughlin JB, DePaola A, Bopp CA, Martinek KA, Napolilli NP, Allison CG, Murray SL, Thompson EC, Bird MM, Middaugh JP. 2005. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N Engl J Med 353:1463–1470. doi: 10.1056/NEJMoa051594. [DOI] [PubMed] [Google Scholar]
- 50.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. 2011. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog 7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gengenbacher M, Kaufmann SHE. 2012. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev 36:514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeannotte R, Lee E, Kong N, Ng W, Weimer BC. 2014. High-throughput analysis of foodborne bacterial genomic DNA using Agilent 2200 TapeStation and genomic DNA ScreenTape system. Agilent Technologies, Santa Clara, CA. [Google Scholar]
- 53.Weimer BC. 2017. 100K Pathogen Genome Project. Genome Announc 5:e00594-17. doi: 10.1128/genomeA.00594-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong N, Davis M, Arabyan N, Huang BC, Weis AM, Chen P, Thao K, Ng W, Chin N, Foutouhi S, Foutouhi A, Kaufman J, Xie Y, Storey DB, Weimer BC. 2017. Draft genome sequences of 1,183 Salmonella strains from the 100K Pathogen Genome Project. Genome Announc 5:e00518-17. doi: 10.1128/genomeA.00518-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bandoy DJDR, Weimer BC. 2019. Biological machine learning combined with bacterial population genomics reveals common and rare allelic variants of genes to cause disease. bioRxiv 739540 10.1101/739540. [DOI] [PMC free article] [PubMed]
- 57.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 58.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. 2018. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:238. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falush D, Stephens M, Pritchard JK. 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT, p 39–64. In Posada D. (ed), Bioinformatics for DNA sequence analysis, 1st ed Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 64.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 69.Lees JA, Harris SR, Tonkin-Hill G, Gladstone RA, Lo SW, Weiser JN, Corander J, Bentley SD, Croucher NJ. 2019. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res 29:304–316. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 73.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All accession numbers and associated metadata are provided in Table 1.