Recently, a complete genome sequence of Mycoplasma bovirhinis HAZ141_2 was published showing the presence of a 54-kB prophage-like region. Bioinformatic analysis revealed that this region has a more than 40% GC content and a chimeric organization with three structural elements—a prophage continuous region, a restriction-modification cassette, and a highly transmittable aadE-sat4-aphA-3 gene cluster found in both Gram-positive and Gram-negative bacteria.
KEYWORDS: Mycoplasma bovirhinis, aadE-sat4-aphA-3 gene cluster, aminoglycosides, horizontal gene transfer, kanamycin and neomycin resistance, prophage
ABSTRACT
Recently, a complete genome sequence of Mycoplasma bovirhinis HAZ141_2 was published showing the presence of a 54-kB prophage-like region. Bioinformatic analysis revealed that this region has a more than 40% GC content and a chimeric organization with three structural elements—a prophage continuous region, a restriction-modification cassette, and a highly transmittable aadE-sat4-aphA-3 gene cluster found in both Gram-positive and Gram-negative bacteria. It is known that aadE confers resistance to streptomycin, sat4 governs resistance to streptothricin/nourseothricin, and aphA-3 is responsible for resistance to kanamycin and structurally related antibiotics. An aadE-like (aadE*) gene of strain HAZ141_2 encodes a 228-amino acid (aa) polypeptide whose carboxy-terminal domain (positions 44 to 206) is almost identical to that of a functional 302-aa AadE (positions 140 to 302). Transcription analysis of the aadE*-sat4-aphA-3 genes showed their cotranscription in M. bovirhinis HAZ141_2. Moreover, a common promoter for aadE*-sat4-aphA-3 was mapped upstream of aadE* using 5′ rapid amplification of cDNA ends analysis. Determination of MICs to aminoglycosides and nourseothricin revealed that M. bovirhinis HAZ141_2 is highly resistant to kanamycin and neomycin (≥512 μg/ml). However, MICs to streptomycin (64 μg/ml) and nourseothricin (16 to 32 μg/ml) were similar to those identified in the prophageless M. bovirhinis type strain PG43 and Israeli field isolate 316981. We cloned the aadE*-sat4-aphA-3 genes into a low-copy-number vector and transferred them into antibiotic-sensitive Escherichia coli cells. While the obtained E. coli transformants were highly resistant to kanamycin, neomycin, and nourseothricin (MICs, ≥256 μg/ml), there were no changes in MICs to streptomycin, suggesting a functional defect of the aadE*.
INTRODUCTION
Aminoglycosides are broad-spectrum bactericidal antibiotics, which are primarily used to treat infections caused by Gram-negative aerobic bacilli, staphylococci and some other Gram-positive bacteria, and Mycobacterium and Mycoplasma spp. (1, 2). Based on the identity of the aminocyclitol moiety, aminoglycosides are grouped into 4,6-disubstitued 2-deoxystreptamine (DOS)-containing kanamycin, tobramycin, amikacin, gentamicin, and others; 4,5-disubstitued DOS-containing neomycin, paromomycin, lividomycin, and others; and a group of compounds with alternative core rings or organization, such as streptomycin, spectinomycin, apramycin, and hygromycin B (3). Aminoglycosides impair protein synthesis by binding to different sites in the 16S rRNA of the 30S ribosomal subunit and to some ribosomal proteins (4). However, the drug binding site of 2-DOS aminoglycosides is composed mainly of nucleotides located within the decoding region A site of bacterial helix 44 of the 30S subunit (5). In addition, some 2-DOS aminoglycosides have a secondary binding site in helix 69 of the 50S subunit (6). Acquired resistance to aminoglycosides can arise via a number of various mechanisms, including modification of the ribosomal target, antibiotic uptake and accumulation, efflux of antibiotic, and inactivation of the drugs by aminoglycoside-modifying enzymes (7).
In the members of the class Mollicutes, a large group of wall-less bacteria (8), the main mechanisms of acquired resistance described until now are target modification with point mutation(s) and ribosome protection by the tet(M) determinant (9–11). A single study also reported that in ureaplasmas, macrolide resistance can be achieved through ribosomal methylation mediated by ermB and through drug efflux mediated by msrA, msrB, or msrD (12), but these data have not been confirmed by any other group. In addition, in Mycoplasma hominis mutants selected in vitro on ethidium bromide and showing increased MICs to ciprofloxacin, an active efflux mechanism has been demonstrated (13). Regarding aminoglycosides, it was shown previously that point mutations at positions 912 and 1192 (Escherichia coli numbering) of 16S rRNAs were associated with decreased susceptibility to streptomycin and spectinomycin, respectively, in several mycoplasmas (14, 15). However, to the best of our knowledge, no studies describing mechanisms of resistance to kanamycin and neomycin in mycoplasmas have been published.
Mycoplasma bovirhinis is a species that is frequently isolated from the upper and lower respiratory tracts of both healthy and diseased cattle and buffaloes all over the world, and it is often coisolated with other bacterial or mycoplasmal pathogens (16). Recently, complete genomes of three M. bovirhinis strains were released in databases, namely, the type strain PG43 (NCTC 10118/ATCC 27748; GenBank accession no. LR214972.1), the Japanese isolate HAZ141_2 (AP018135.1 [17]), and the Chinese isolate GS01 (CP024049.1 [18]). Initial reports describing genomes of the two latter strains mentioned a 53.5-kb DNA sequence uniquely present in HAZ141_2 (17, 18). Chen et al. (18) noticed that the HAZ141_2 strain-specific genomic region, besides bacteriophage-like DNA, contains three open reading frames (ORFs) resembling the following known genes: aadE, encoding 6′ adenyltransferase [AAD(6′)] conferring resistance to streptomycin, sat4, encoding streptothricin N-acetyltransferase conferring resistance to streptothricin/nourseothricin, and aphA-3, encoding a 3′ phosphotransferase [APH(3′)-III] conferring resistance to kanamycin/neomycin/amikacin. This information caught our attention and motivated the analysis of these genes never seen before in Mollicutes.
The aadE-sat4-aphA-3 genes are one of highly transmittable and disseminated clusters found mainly on the plasmids in both Gram-positive (e.g., Enterococcus spp., Streptococcus spp., Staphylococcus aureus, etc.,) and Gram-negative bacteria (e.g., Campylobacter species). Indeed, two genes, aadE and aphA-3, were described as a part of the composite S. aureus transposon Tn5405 (19). Later, the same group identified a pseudogene (truncated ORF) within Tn5405 located between aadE and aphA-3 (20), which shared a strong similarity to sat4 described in Gram-negative Campylobacter coli (21). A comprehensive analysis of 50 S. aureus strains carrying the addE-sat4-aphA-3 locus revealed that only 14 strains contained an intact sat4 (22). The obvious transmittable character of the aadE-sat4-aphA-3 cluster became apparent after the discovery of Streptococcus phages carrying these genes (23). Since genes encoding resistance to aminoglycosides and strepthotricin were identified for the first time in Mollicutes, the aim of this study was the preliminary characterization of these genes and analysis of their phenotypic expression in both M. bovirhinis HAZ141_2 and in antibiotic-susceptible Escherichia coli cells where the genes were cloned on a low-copy-number vector.
RESULTS
Characterization of the M. bovirhinis HAZ141_2 prophage-like region.
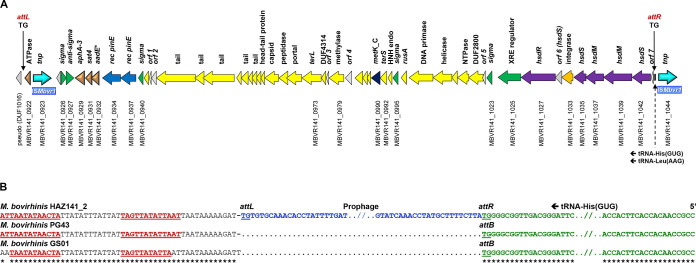
Our in silico analysis of the M. bovirhinis HAZ141_2 prophage-like region (Fig. 1A) revealed its putative integration sites, attR and attL, to be a dinucleotide (TG) located within a 3′ end of the tRNA-His gene (MBVR141_t0068; nucleotides [nt] 924505 to 924506) and within an intergenic region (nt 870582– to 870583), respectively (Fig. 1B). Moreover, this strain HAZ141_2-specific region was inserted precisely between a 3′ end of the tRNA-His gene and its putative transcriptional terminator, whose stable stem-loop structure (ΔG, 8.3 kcal/mol) is formed by two perfect 13-bp inverted repeats (IR), ATTAATATAACTA (nt 870531 to 870569; Fig. 1B). An identical terminator-like sequence was found downstream of orthologous tRNA-His genes in both prophageless genomes, PG43 and GS01, suggesting a remarkable expression variability of this gene in strain HAZ141_2.
FIG 1.
Genomic organization of M. bovirhinis HAZ141_2 prophage and its integration sites. (A) Schematic representation of the prophage-like genomic region. Arrows represent ORFs, with the arrowheads indicating the direction of transcription. The ORFs are color coded based on the putative function of the predicted encoded proteins. Brown, antibiotic resistance genes from the aadE*-sat4-aphA-3 gene cluster; different shades of blue, ISMbvr1 encoding transposases and PinE-related recombinases; different shades of yellow, phage-associated genes; purple, restriction-modification enzymes; green, transcriptional regulation; gray, not annotated, hypothetical or unknown proteins. Only ORFs discussed in the text are tagged. The attL and attR sites are indicated by a vertical black line, and their sequences (TG) are presented. Schematic representation of the prophage-like genomic region prepared according to a scale using ISMbvr1 (2,010 bp) as a scale bar. (B) Analysis of the putative integration sites of the M. bovirhinis HAZ141_2 prophage. Putative integration sites attL and attR as predicted in M. bovirhinis HAZ141_2 are shown. The prophage integrated into a putative 2-bp target sequence (TG) at the 3′ end of the tRNA-His (anticodon GUG) gene (MBVR141_t0068); its duplication is present in the genome of strain HAZ141_2, but not in the M. bovirhinis PG43 and GS01 strains. Perfect inverted repeats of a putative transcriptional terminator of the tRNA-His gene are shown in red bold and underlined. Asterisks mark identical nucleotides.
An entire prophage-like region (including an attP site) was estimated to be 53,923 bp with 40.7% GC content, which is much higher than the average GC content of mycoplasmal genomes (28 to 29%) and M. bovirhinis HAZ141_2 (28.24% [17]). The codon usage in the M. bovirhinis HAZ141_2 prophage-like region is similar to that of Firmicutes bacteria and their phages (https://www.kazusa.or.jp/codon/). For example, analysis of prophage ORFs showed that tryptophan (Trp) residues are mainly encoded by the TGG codon, while in Mycoplasma species, Trp is encoded by either TGG or TGA codons (24, 25), suggesting a relatively recent acquisition of this region by M. bovirhinis. Remarkably, translation of TGA as Trp leads to some extension of translated polypeptides (e.g., MBVR141_0932, MBVR141_0995, MBVR141_0999, etc.; see below). The influence of such extension on protein stability and putative function is unknown.
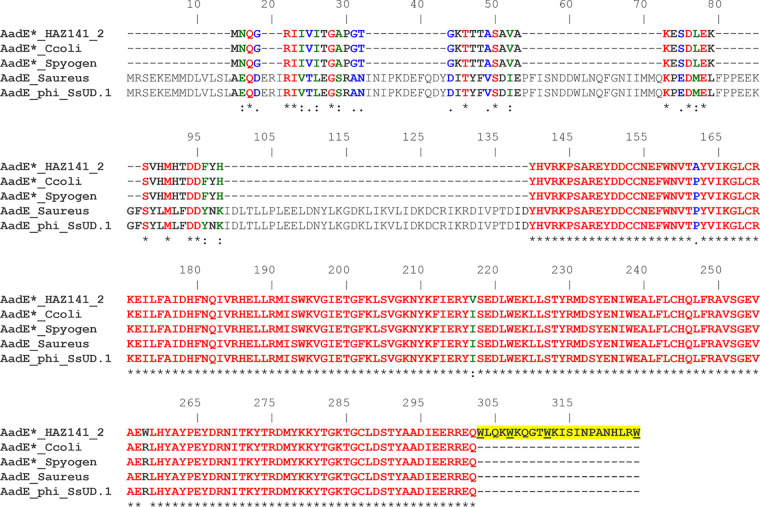
Genome sequence analysis revealed 55 ORFs (48 were annotated in previous publications [17, 18] and 7 were newly annotated as ORFs 1 to 7), of which 52 were transcribed in the same direction and 3 (MBOV141_0923, 0926, and 0927) were transcribed in the opposite direction (Fig. 1A; see Table S1 in the supplemental material). The M. bovirhinis HAZ141_2 prophage-like region has a chimeric structural organization. The 5′ end of the prophage consists of a highly transmittable aadE*-sat4-aphA-3 gene cluster (nt 875016 to 877062). aadE* (MBVR141_0932) encodes a protein whose C terminus shares a strong similarity to 6′ adenyltransferase [AAD(6′)] enzyme AadE, conferring resistance to streptomycin. The M. bovirhinis AadE* deduced amino acid sequence contains 228 residues, which is nearly 25% less than a length of an enzymatically active AadE (302 amino acids [aa]; Fig. 2). Three features of the M. bovirhinis AadE* should be mentioned as follows. First, the amino (N)-terminal part of the M. bovirhinis AadE* (residues 1 to 43) does not show any similarity to amino acid sequences of known AadE enzymes with experimentally validated activity. Second, the carboxy (C)-terminal residues of AadE* (residues 44 to 206) are almost identical to the C terminus of a classic 6′ adenyltransferase AadE (residues 140 to 302 of a 302-aa polypeptide). Third, AadE* has a 22-aa C-terminal elongation (residues 207 to 228), as a result of a specific usage of the TGA translation stop codon by mycoplasmas; this sequence is absent from any other enzymatically studied AadE proteins (Fig. 2). The next gene of the cluster is sat4 (MBVR141_0931), which encodes a 180-aa protein with 100% identity to streptothricin acetyltransferase Sat4, conferring resistance to aminoglycoside glycopeptide antibiotics of the streptothricin class in multiple Gram-positive and Gram-negative bacterial species (Table S1). In M. bovirhinis, there is a 70-bp overlap between the 3′ end of an extended aadE* and the 5′ end of sat4. Next to sat4 is aphA-3, encoding a 264-aa protein showing 100% identity to 3′ phosphotransferase [APH(3′)-III], conferring resistance to kanamycin as well as to other structurally related aminoglycosides (e.g., neomycin and amikacin) in both Gram-positive and Gram-negative bacteria (Table S1). The aadE*-sat4-aphA-3 gene cluster is bordered downstream by two genes encoding a putative alternative sigma factor resembling YlaD (MBVR141_0926) and its cognate membrane-associated negative regulator (an antisigma, MBVR141_0927), while two genes coding for putative serine site-specific recombinases (MBVR141_0934 and 0937) are located upstream of the cluster (Fig. 1A and Table S1).
FIG 2.
Amino acid sequence alignment of the M. bovirhinis strain HAZ141_2 AadE*-like protein with homologs from other bacterial species. The comparison includes AadE*_HAZ141_2 (MBVR141_0932, GenBank accession no. AP018135.1, 228 aa), Mycoplasma bovirhinis strain HAZ141_2; AadE*_Ccoli (accession no. KJ610809.1, 206 aa), Campylobacter coli strain SH-CCD11C145; AadE*_Spyogen (SD89_05780, accession no. CP010449.1, 206 aa), Streptococcus pyogenes strain NGAS322; AadE_Saureus (SA268_2518, accession no. AII57085.1, 302 aa), Staphylococcus aureus strain SA268; AadE_phi_SsUD1 (accession no. CBR26932.1, Orf62, 302 aa), Streptococcus suis phage phi-SsUD.1. The alignment length is 324 aa residues; the protein sequences were aligned using CLUSTAL W (70). Identical amino acids (53.39%) are shown in red and marked with an asterisk (*); strongly similar amino acids (2.78%) are shown in green and marked with a colon (:); weakly similar amino acids (2.47%) are shown in blue and marked with a dot (.); different amino acids (41.36%) are indicated in black. A 22-aa C-terminal extension of AadE*_HAZ141_2 is highlighted in yellow, and four Trp (W) residues encoded by TGA codons are underlined.
The central part of the M. bovirhinis HAZ141 prophage-like region contains multiple orfs encoding tail- and capsid-associated components as well as both subunits of terminase, TerL (MBVR141_0973) and TerS (MBVR141_0992), an essential enzyme of bacteriophages responsible for packaging genomic DNA into proheads of future virions (Fig. 1A and Table S1). The 3′ end of the prophage consists of 9 genes (from MBVR141_1025 to MBVR141_1042 and two putative nonannotated genes, orf6, resembling hsdS, and orf7). Six out of those nine genes encode type I restriction and modification (RM)-related proteins. Finally, an insertion-like sequence, which we designated ISMbvr1 (MBVR141_0923), is located at the 5′ end of the prophage-like region, while an additional copy of ISMbvr1 (which encodes transposase MBVR141_1044) flanks the 3′ end of the prophage (Fig. 1A).
In vitro susceptibility of M. bovirhinis strains to aminoglycosides and nourseothricin.
Since the presence of the aadE*-sat4-aphA-3 gene cluster, conferring resistance to aminoglycosides and strepthotricin in other bacteria, has been identified in Mollicutes for the first time, we tested their phenotypic expression in M. bovirhinis HAZ141_2 by determining the MICs. The results of in vitro susceptibility data of three M. bovirhinis strains to the aminoglycosides gentamicin, kanamycin, neomycin, streptomycin, and spectinomycin, as well as to nourseothricin, are presented in Table 1. While the MICs to gentamicin, spectinomycin, streptomycin, and nourseothricin were compatible among the strains, M. bovirhinis HAZ141_2 demonstrated high-level MICs to kanamycin and neomycin (≥512 μg/ml), which were at least 32 to 64 and 8 times higher, respectively, than those in M. bovirhinis 316981 and M. bovirhinis PG43 (Table 1).
TABLE 1.
In vitro susceptibility to aminoglycosides and nourseothricin of M. bovirhinis strains used in this study
| MIC (μg/ml)a |
||||||
|---|---|---|---|---|---|---|
| Strain | Km | Gm | Nm | Sp | Sm | NTC |
| PG43 | 8 | 4 | 64 | 4 | 32 | 16–32 |
| 316981 | 16 | 8 | 64 | 8 | 32 | 16–32 |
| HAZ141-2 | ≥512 | 8 | ≥512 | 8 | 64 | 16–32 |
Km, kanamycin; Gm, gentamicin; Nm, neomycin; Sp, spectinomycin; Sm, streptomycin; NTC, nourseothricin.
The prophage-borne aadE*-sat4-aphA-3 gene cluster expressed in E. coli provides resistance to kanamycin, neomycin, and nourseothricin, but not to streptomycin.
Since we showed a significant difference in susceptibility to kanamycin and neomycin between M. bovirhinis HAZ141_2 and two other M. bovirhinis strains (Table 1), we first checked if there are any mutations present in the 16S rRNA-encoding genes (rrs) of strain HAZ141_2 in comparison to the type strain, PG43. The sequence analysis of 3 alleles of the 16S rRNA-encoding gene revealed that 7 positions differed between two strains, namely, 91, 138, 178, 494, 847, 987, and 1290 (E. coli numbering [Fig. S1]). None of the substitutions detected in M. bovirhinis HAZ141_2 were located within the helix 44 of 16S rRNA, which is known as a binding site for 2-DOS-containing aminoglycosides (5). In addition, since some 2-DOS aminoglycosides may have a secondary binding site in the ribosome—helix 69 of the 50S subunit (6)—the sequences of 23S rRNA-encoding genes (rrl) were compared between M. bovirhinis strains PG43 and HAZ141_2. No nucleotide difference was found at positions 1906 to 1930 (E. coli numbering) between two M. bovirhinis strains or between them and E. coli (data not shown).
Then, since the aadE*-sat4-aphA-3 genes were identified only in M. bovirhinis HAZ141_2, we wanted to test whether this cluster was, indeed, active and might confer resistance to kanamycin and neomycin as well as to streptomycin and nourseothricin in a heterologous host. To that end, the aadE*-sat4-aphA-3 cluster was cloned into the low-copy-number plasmid vector pACYC184 derivative and introduced by transformation into E. coli JM109 (see Materials and Methods). No increase in MIC values to gentamicin, spectinomycin, or streptomycin was observed in recombinant clone pAC10 in comparison to host E. coli JM109 and E. coli transformants carrying an empty pACYC184 plasmid vector (Table 2). In contrast, the MIC values for neomycin and kanamycin were at least 128-fold higher in E. coli JM109 transformants carrying pAC10 than in E. coli JM109 and the plasmid-containing E. coli JM109 recipient, respectively (Table 2), and were comparable to that demonstrated in M. bovirhinis HAZ141_2 (Table 1). In addition, in E. coli JM109, the prophage-borne aadE*-sat4-aphA-3 gene cluster provided resistance to nourseothricin, with the MIC value of ≥256 μg/ml in pAC10 in comparison to 2 μg/ml in E. coli JM109 and in its transformant carrying only the pACYC184 plasmid (Table 2). Notably, the MIC value for nourseothricin was at least 8-fold less in M. bovirhinis HAZ141_2 than in E. coli containing pAC10 (Tables 1 and 2).
TABLE 2.
In vitro susceptibility to aminoglycosides and nourseothricin of the E. coli JM109 recombinant plasmid pAC10 carrying the aadE*-sat4-aphA3 gene cluster
| MIC (μg/ml)a |
||||||
|---|---|---|---|---|---|---|
| Strain | Km | Gm | Nm | Sp | Sm | NTC |
| JM109 | 2 | 1 | 2 | 16 | 4 | 2 |
| JM0109 (pACYC184) | 2 | 1 | 2 | 16 | 4 | 2 |
| JM109 (pAC10) | ≥256 | 1 | ≥256 | 16 | 4 | ≥256 |
Km, kanamycin; Gm, gentamicin; Nm, neomycin; Sp, spectinomycin; Sm, streptomycin; NTC, nourseothricin.
Transcription analysis of the M. bovirhinis HAZ141_2 aadE*-sat4-aphA-3 genes confirms their expression and cotranscription.
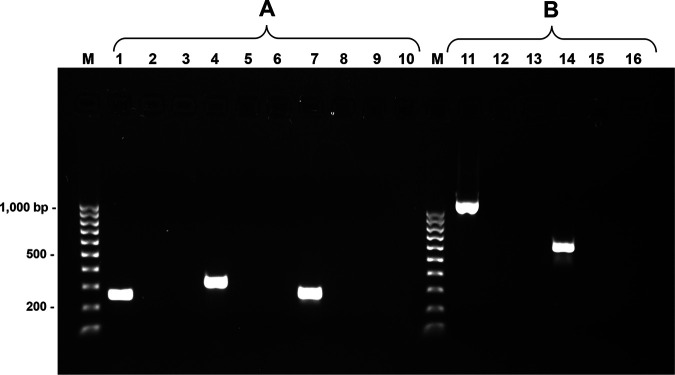
Regardless of the presence of aadE* and sat4 in M. bovirhinis HAZ141_2, no significant difference in MICs to streptomycin and nourseothricin has been identified among the 3 M. bovirhinis strains tested in this study. In addition, no increase in MICs to streptomycin was evident in recombinant plasmid pAC10 (Tables 1 and 2). To explore this issue, reverse transcription PCR (RT-PCR) analysis was performed on total RNA of M. bovirhinis HAZ141_2 using primers complementary to the aadE*, sat4, and aphA-3 genes (Table S3). The results of RT-PCR showed expression of all three genes (Fig. 3A). Moreover, an RT-semiquantitative PCR analysis of each individual gene revealed the same pattern of expression (Fig. S2). In addition, RT-PCR analysis performed on cDNA, obtained with the aphA3-R2 primer, complementary to the aphA-3 gene, revealed the cotranscription of the aadE*, sat4, and aphA-3 genes in M. bovirhinis HAZ141_2 (Fig. 3B).
FIG 3.
Transcription analysis of the M. bovirhinis HAZ141_2 aadE*-sat4-aphA-3 genes. (A) RT-PCR analysis of the aadE*, sat4, and aphA-3 genes. Agarose gel electrophoresis of RT-PCR products. cDNAs, synthesized with either aadE-R1, sat4-R1, or aphA3-R2 primers complementary to the aadE*, sat4, and aphA-3 genes, respectively, were subjected to PCR amplifications to determine the expression of each individual gene. The PCR products were obtained using the aadE-F2 and aadE-R2 (lane 1), sat4-F1 and sat4-R1 (lane 4), and aphA3-F1 and aphA3-R2 (lane 7) primers. The control cDNAs (lanes 2, 5, and 8) were without RT in the reaction. Negative PCR controls are presented in lanes 3, 6, and 9. (B) Coexpression of the M. bovirhinis HAZ141_2 aadE*-sat4-aphA-3 genes. PCR amplifications of the cDNA, obtained with the aphA3-R2 primer complementary to the aphA-3 gene, were produced to determine cotranscription of the aadE*, sat4, and aphA-3 genes. The amplification products were obtained using the aadE-F3 and aphA3-R1 (lane 11) and the aadE-F3 and sat-R2 (lane 14) primers. The control samples (lanes 12 and 15) were those without RT in the reaction. Negative PCR controls are presented in lanes 13 and 16. The 100-bp ladder (Bio-Rad, CA, USA) is shown as M.
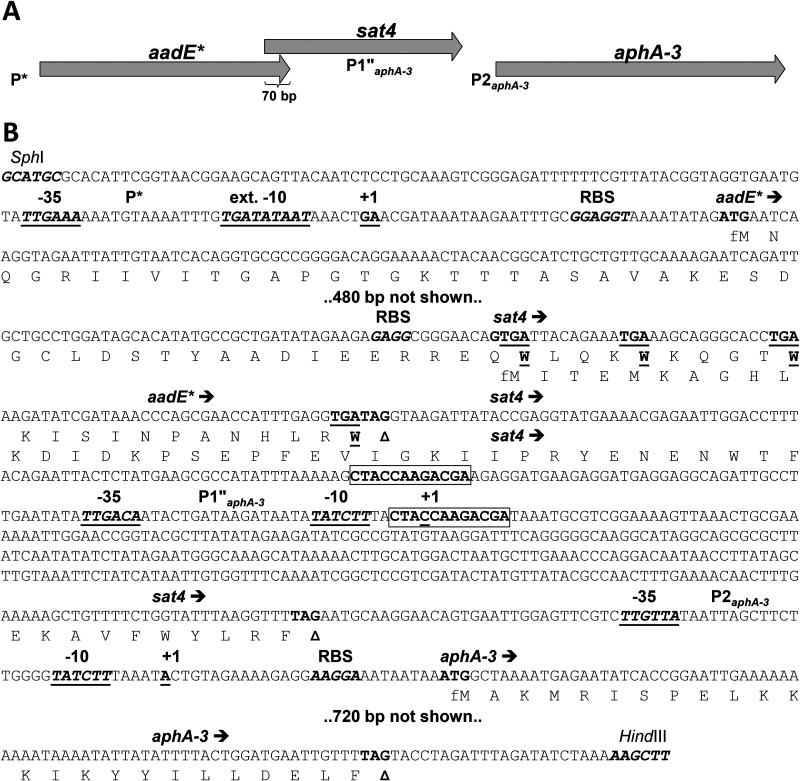
In silico analysis of the DNA region upstream of the aphA-3 gene revealed two putative promoters, one resembling both P1 and P1′, described in literature (in this study designated P1″), and another identical to P2 (Fig. 4) that was previously characterized (26–28). The M. bovirhinis P2 promoter is located within an intergenic region between the sat4 and aphA-3 genes and is totally identical to the sequence of P2, which was experimentally validated first on plasmid pJH1 of the Gram-positive Enterococcus faecalis (28) and on plasmid pIP1433 of the Gram-negative Campylobacter coli (27). The P1 promoter of aphA-3 was identified within the encoding region of sat4 of C. coli plasmid pIP1433 (27), and it was flanked by two 12-bp direct repeats (DRs) like those found in the corresponding region of the M. bovirhinis HAZ141_2 sat4 (Fig. 4 and Fig. S3). In contrast to the P1 promoter of aphA-3 in plasmid pIP1433, which contained a tandemly repeated 7-bp sequence, ATAATAT, located between the –35 and –10 elements, the M. bovirhinis HAZ141_2 P1″ promoter possessed only one ATAATAT repeat resembling the experimentally determined P1′ promoter in the pJH1 and pLG2 plasmids of E. faecalis (28, 29). An identical to P1″ promoter was previously described in strepthotricin- and kanamycin-resistant C. coli BE/G4 (21). It is noteworthy that a deletion of one of the ATAATAT repeats changed the –10 element of P1 from the optimal consensus TATAAT sequence to TATCTT (Fig. 4 and Fig. S3) and restored an intact ORF of the sat4 gene that was truncated in plasmid pIP1433.
FIG 4.
Genomic organization of the aadE*-sat4-aphA-3 gene cluster of M. bovirhinis HAZ141_2 and its regulatory sites and features. (A) Schematic representation of the aadE*-sat4-aphA-3 cluster. Gene designations: aadE*, a partial homolog of aadE encoding streptomycin aminoglycoside 6-adenyltransferase (see the text for other details); sat4 encoding streptothricin acetyltransferase; aphA-3 encoding aminoglycoside O-phosphotransferase APH(3′)-IIIa (neomycin-kanamycin phosphotransferase type III). Promoters are designated P*, P1″aphA-3, and P2aphA-3. (B) Partial nucleotide sequence of the aadE*-sat4-aphA-3 gene cluster (two hidden DNA stretches are indicated with the labels “480 bp not shown” and “720 bp not shown”). Deduced amino acid sequences of corresponding gene products are shown only for C- and N-terminal regions; fMet (formyl-methionine) is used for the first translated residue of each polypeptide. C-terminal tryptophane (W) residues of AadE* encoded by TGA (UGA) codons are shown as underlined boldface letters. The 5′- and 3′-terminal restriction sites, SphI (designed in this study) and HindIII (authentic), respectively, are those used for cloning and expression of the cluster in E. coli. Promoter elements are shown with −35 and −10 or ext. −10 (extended −10) above the underlined boldface italicized letters. Two 12-bp direct repeats flanking promoter P1″ are shown as boxed boldface letters. The experimentally validated two-nucleotide transcription start site is shown with +1 above the underlined boldface nucleotides. Predicted ribosome-binding sites are shown with RBS above the boldface italicized letters. Arrows indicate the positions and directions of transcription of the genes. Start and stop codons are shown in bold, with the latter marked with a triangle.
Using 5′ rapid amplification of cDNA ends (5′-RACE) analysis, we identified a promoter upstream of the aadE* gene with a transcriptional start site (TSS) represented by two nucleotides, A and G, located 35 and 36 nucleotides upstream of the ATG start codon of the aadE* gene, respectively. The –35 site, TTGAAA, and an extended –10 Pribnow box, TGaTATAAT, (30) were also mapped (Fig. 4 and Fig. S4). Notably, the TTS was identified on cDNA obtained with either aphA-3, sat4, or aadE* complementary primers (see Table S2), supporting cotranscription of these genes demonstrated by RT-PCR (Fig. 3).
DISCUSSION
Bacteriophages are intracellular parasites of bacteria that are highly prevalent in nature (31) and can be categorized by their life cycle as either lytic or lysogenic. In the latter scenario, the phage genome (prophage) is usually integrated into the bacterial genome and transmitted vertically during replication. Prophages play an important role in bacterial evolution, population shaping, virulence, and genetic transfer through horizontal gene transfer (HGT). Indeed, many prophages carrying virulence as well as fitness factors (32, 33) influence bacterial traits and allow the bacteria to access new environmental niches (34–36). Recently, a unique 53.5-kb chromosomal insertion was identified in the genome of M. bovirhinis HAZ141_2 (17). Later, Chen et al. (18) reported that this region is missing in the genome of strain GS01 and provided its initial analysis. Many ORFs of the 53.5-kb region resembled genes of bacterial phages. Our in silico analysis deduced a more precise size of the prophage-like region, which is 53,523 bp, including a copy of a novel insertion element, ISMbvr1 (2,010 bp) (Fig. 1 and Table S1). To the best of our knowledge, it is the biggest prophage-like genomic region ever identified in Mollicutes. Its high GC content in comparison to the rest of the M. bovirhinis HAZ141_2 genome, as well as the almost sole use of the TGG codon (rather than TAG) to encode Trp, suggests the recent exogenous acquisition of this prophage by mycoplasma. BLAST analysis of amino acid sequences of proteins encoded by the M. bovirhinis HAZ141_2 prophage-like region revealed that most of the homologous polypeptides were detected in species of Firmicutes (Gram-positive), and several homologous ORFs were associated with both Gram-positive and Gram-negative bacteria (Table S1).
The M. bovirhinis HAZ141_2 prophage-like region has a modular organization similar to that of Streptococcus suis phage phi-SsUD.1 (23), with the phage-specific genes located in the middle of this region, while the aminoglycoside/streptothricin resistance gene cluster aadE*-sat4-aphA-3 and the stretch of 6 genes encoding type I RM enzymes were identified at the 5′ and 3′ ends of the prophage, respectively (Fig. 1). The aadE-sat4-aphA-3 gene cluster, characterized so far in different Gram-positive and Gram-negative bacteria (22, 37–39), has never been described in Mollicutes (as of 20 September 2020). However, our BLASTN analysis identified the aadE-sat4-aphA-3 locus in the recently published genome of the Acholeplasmatales human gut isolate UBA11505 (GenBank accession number DPOU01000014.1), but its neither presence nor its function were not discussed or mentioned by Parks et al. (40). Moreover, the aadE gene (GenBank accession number HCX08069.1) of the Acholeplasmatales isolate encodes a full-size 302-aa AadE-like polypeptide, suggesting another gene-transferring source of this cluster.
The aadE*-sat4-aphA-3 gene cluster confers high-level resistance to kanamycin and neomycin in M. bovirhinis HAZ141_2 and to kanamycin, neomycin, and nourseothricin in E. coli JM109 (pAC10) cells, while no expression of resistance to streptomycin was evident in either host system (Tables 1 and 2). The obtained phenotypic discrepancy between MIC values to nourseothricin in M. bovirhinis HAZ141_2 (16 to 32 μg/ml) and in E. coli transformants carrying pAC10 (≥256 μg/ml) can be explained, at least partially, by the fact that in M. bovirhinis HAZ141_2, the extended 3′ end of aadE* extensively (70 nucleotides) overlaps with a 5′ end of sat4 and impairs the translational expression of the latter (Fig. 4). Moreover, a GC-rich GTG as a start codon for sat4 might be an additional reason for translational silencing of this gene in mycoplasma cells. In addition, no significant difference in MICs to streptomycin was found among three M. bovirhinis strains (Table 1), and low MIC to streptomycin was identified in the recombinant E. coli pAC10 clone (Table 2). Actually, M. bovirhinis HAZ141_2 prophage-encoded AadE* is a chimeric protein with truncated N-terminal and extended C-terminal parts (Fig. 2). These changes, especially a lack of the regular AadE N-terminal part, apparently resulted in a loss of function, i.e., in a failure to confer resistance to streptomycin in either M. bovirhinis HAZ141_2 or E. coli (Tables 1 and 2). The presence of a similar aadE*-like gene encoding a 206-aa polypeptide that did not provide resistance to streptomycin was previously identified on a chromosome of C. coli (41) and on conjugative plasmids from Camplylobacter jejuni (39) and C. coli (42).
Despite the fact that acquired resistance to kanamycin and neomycin mainly arises via inactivation of the drugs by aminoglycoside-modifying enzymes, the modifications of the aminoglycoside binding site (helix 44) located within the 16S rRNA by point mutations or methylation have also been reported (43–47). We did not identify any nucleotide differences within the primary and secondary 2-DOS aminoglycoside binding sites (4, 6, 48) comparing rrs and rrl genes, respectively, of M. bovirhinis HAZ141_2 and PG43 (Fig. S1).
Our analysis revealed the high identity (>99%) of the M. bovirhinis aadE*-sat4-aphA-3 region to the genome of Streptococcus pyogenes strain NGAS322 (CP010449.1). S. pyogenes is almost exclusively restricted to humans, but several reports described identification of this pathogen from different mammals, including dogs, rabbits, sheep (49, 50), and even hedgehog (Erinaceus europaeus [51]), indicating the ability of S. pyogenes to colonize and infect these animals. One may hypothesize that aadE*-sat4-aphA-3 cluster, as a part of some yet unknown Streptococcus phage, was transferred directly to M. bovirhinis HAZ141_2. Indeed, Palmieri et al. (23) described the S. suis phage phi-SsUD.1, which carries a related aadE-sat4-aphA-3 cluster that encodes an intact 302-aa AadE. Another possible hypothesis is that this locus was first inserted in a prophage-like region present in other bacteria, whose sequences do still are not present in GenBank, and then transferred to M. bovirhinis. Such putative gene transfer could illustrate the exchange between evolutionarily distanced bacterial species and demonstrates the ability of HGT in M. bovirhinis. In any scenario, the M. bovirhinis-specific element ISMbvr1 (which encodes transposase MBVR141_0923) was then inserted between MBVR141_0922 and MBVR141_0925 (Fig. 1).
The distribution of the aadE*-sat4-aphA-3 gene cluster among other M. bovirhinis strains is not known, and our preliminary attempts to detect this cluster by PCR in a cohort of strains isolated in Austria (n = 17) and Italy (n = 32) were not successful (data not shown). It might be that the prevalence of M. bovirhinis isolates carrying the aadE*-sat4-aphA-3 gene cluster is low and/or restricted to the particular geographic region. The presence of M. bovirhinis with high MICs to kanamycin (>100 μg/ml) was previously reported from Japan (52, 53). In addition, Mycoplasma bovis strains with MICs of ≥64 μg/ml to kanamycin were also identified in Japan (52–54). Unfortunately, it was impossible to compare kanamycin susceptibility of bovine mycoplasmas isolated in Japan and that in other countries since kanamycin is usually not included in the panel of antibiotics tested. Moreover, the susceptibility of M. bovirhinis isolates was almost solely tested in Japan.
The 3′-terminal module of the M. bovirhinis HAZ141_2 prophage-like region contains a cluster of 6 genes encoding type I RM enzymes (Fig. 1). Methylation is crucial in many aspects of cell life, such as metabolism, gene regulation, DNA replication and repair, and epigenetic programming, and it is often used as a defense system against invading foreign DNA (55). Therefore, such abundance of RMs within the prophage may limit other parasitic DNA from integrating into a prophage or/and into a chromosome of prophage-occupied mycoplasma. We can speculate that RMs may also help the prophage and themselves to establish a stable symbiosis between AT-rich mycoplasma genomes and GC-rich prophage regions or protect the prophage DNA from an auto-immune degradation. Moreover, RMs are known to cause genomic rearrangements by promoting homologous or site-specific recombination, generating bacterial diversity and regulating gene expression, for example, by phase variation as previously described in the case of hsdS genes in Mycoplasma pulmonis (56). The site-specific XerC family recombinase (MBVR141_1033) encoded by the RM cluster probably generates such recombinations and rearrangements (Fig. 1). In addition to the type I RM system, an ORF similar to S-adenosylmethionine synthetase MetK was identified in the M. bovirhinis HAZ141_2 prophage region (MBVR141_0990). The metK-like genes were annotated in certain genomes of Firmicutes viruses, including S. suis phage phi-SsUD.1 (23), Staphylococcus phage UPMK_1 (GenBank accession no. MG543995.1), Faecalibacterium phage FP_Brigit (GenBank accession no. MG711465.1), etc. The primary function of orphan MTases is to actively methylate phage DNA and protect it from multiple host-encoded restriction endonucleases (57), but more pleotropic regulatory roles have also been proposed (58, 59). We can also speculate that methyltransferases may play an additional role in conferring resistance to aminoglycosides in M. bovirhinis, as has been shown for other bacteria (60–62).
Conclusion.
Our results revealed for the first time that Mycoplasma species can stably maintain a large composite GC-rich prophage-like genomic island that carries a highly conserved aadE-sat4-aphA-3-like gene cluster previously characterized in certain Gram-positive and Gram-negative pathogens. This cluster is expressed in mycoplasma cells conferring resistance to the aminoglycosides kanamycin and neomycin, but not to streptomycin and nourseothricin, although the M. bovirhinis HAZ141_2 sat4 gene encoding streptothricin acetyltransferase is intact, and it does provide resistance to nourseothricin in E. coli recombinant clones. The expression of the aadE-sat4-aphA-3 gene cluster in kanamycin/neomycin- and streptothricin-sensitive M. bovirhinis strain or other ruminant mycoplasma species would provide the final demonstration of the drug resistance phenotype conferred by this prophage-like genomic region. In addition, we suggest that other Mycoplasma species associated with both human and animal diseases be monitored for a presence of this transmittable gene cluster.
MATERIALS AND METHODS
Mycoplasma bovirhinis strains and growth conditions.
M. bovirhinis strain HAZ141_2, isolated from the nasal discharge of a coughing calf in Japan in 2008 (17), was kindly provided by Eiji Hata (National Institute of Animal Health, National Agriculture and Food Research Organization, Tsukuba, Ibaraki, Japan). M. bovirhinis strain 316981 was isolated from the lungs of cattle in Israel in 2018, and the M. bovirhinis PG43 type strain (NCTC 10118) was purchased from the National Collection of Type Cultures (NCTC; Public Health England, UK). All three isolates were propagated at 37°C and 5% CO2 in modified Friis (FF) broth or agar medium (63). Stock cultures were aliquoted and maintained at –80°C. For each stock, the number of CFU per ml of was determined by performing serial 10-fold dilutions in FF broth and by plating each dilution on agar in triplicates.
Genomic DNA extraction and PCR amplification.
M. bovirhinis genomic DNA was extracted from 10 ml logarithmic-phase broth cultures using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The primers were developed and commercially synthesized (Sigma-Aldrich, Rehovot, Israel) based on the nucleotide sequence of the M. bovirhinis strain HAZ141_2 genome (GenBank accession no. AP018135.1 [17]). The nucleotide sequences and locations of the oligonucleotide primers are given in Table S2. When needed, primers used in the PCRs or primers complementary to the internal sequences of the amplicons were used to complete the sequence of the PCR products (data not shown). PCRs were carried out in 50-μl volumes containing 50 to 100 ng of template DNA, 1 μl of Phire Hot Start II DNA polymerase (Thermo Fisher Scientific, Waltham, MA), 5× Phire reaction buffer, 1 μl of 10 mM deoxynucleoside triphosphate (dNTP), and 0.4 μM each primer. PCR amplifications were carried out in a C1000 Touch thermal cycler (Bio-Rad, CA). PCR amplicons were purified using a PureLink quick PCR purification kit (Invitrogen, CA, USA). PCR amplicons used for cloning were extracted from the gels and purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany). Sequencing was performed at the DNA Sequencing Unit of the Hebrew University (Jerusalem, Israel).
Enzymes and antibiotics.
Restriction enzymes (SphI and HindIII) and T4 ligase were purchased from New England Biolabs (NEB) (MA, USA) and Promega, Inc., (Madison, USA), respectively, and used according to the manufacturer’s recommendations. Antibiotics, chloramphenicol, gentamicin sulfate, kanamycin sulfate, neomycin trisulfate, spectinomycin hydrochloride, and streptomycin sulfate were purchased from Sigma (Sigma-Aldrich, Rehovot, Israel), and nourseothricin sulfate was purchased from ENCO Diagnostics Ltd. (Israel).
Cloning and transformation.
To clone aadE*-sat4-aphA-3 genes from M. bovirhinis HAZ141-2, forward primer aadE-F1-SphI containing the SphI site and reverse primer down-aphA3-R1-HindIII containing the HindIII site were used for PCR amplification of the genes (Table S2). The PCR amplification program was as follows: 35 cycles of denaturation at 98°C for 30 s, primer annealing at 58°C for 30 s, and extension at 72°C for 2 min. The resulting PCR fragment was purified, sequenced, cut with HindIII and SphI, and cloned into a derivative of the low-copy-number pACYC184 plasmid vector (gifted from M. Kolot, Tel Aviv University) cut with the same enzymes. The pACYC184 derivative was obtained by PCR using the pACYC184-F1-HindIII and pACYC184-R1-SphI primers (Table S2). The PCR amplification program was as follows: 35 cycles of denaturation at 98°C for 30s, primer annealing at 59°C for 30 s, and extension at 72°C for 2 min. The pACYC184 derivative product was 2,280 bp long and contained the p15A origin of replication as well as the cat gene encoding a chloramphenicol acetyltransferase responsible for chloramphenicol resistance (CmR). The recombinant clones and the pACYC184 plasmid itself were transformed into competent cells of E. coli strain JM109 (Promega Inc., Madison, USA). The transformants were plated on Luria-Bertani broth (LB) plates containing chloramphenicol (15 μg/ml). DNA of the recombinant plasmids was isolated using a PureLink quick plasmid miniprep kit (Invitrogen, CA, USA) according to the manufacturer’s instructions. The resulting recombinant plasmid pAC10 was completely sequenced on both strands (HU; Jerusalem, Israel).
Antimicrobial susceptibility testing.
The In vitro susceptibility of M. bovirhinis strains to gentamicin, kanamycin, neomycin, spectinomycin, streptomycin, and nourseothricin was determined using the agar dilution method as previously described (64) with some modifications. Briefly, 2-fold dilutions of antimicrobials from 0.5 to 1,024 μg/ml were incorporated onto the FF agar plates, and 2.5 μl of each isolate, containing 1 × 105 to 1 × 106 CFU, was spotted onto the agar. Plates were incubated at 37°C with 5% CO2 for 3 days. The procedure was repeated independently three times.
In vitro susceptibility of the host E. coli strain JM109 and that of its transformants carrying either the empty pACYC184 vector or the recombinant clone pAC10 containing the aadE*-sat4-aphA-3 gene cluster from M. bovirhinis HAZ141_2 was tested using the agar dilution method following instructions of the European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) (65). Briefly, overnight-grown cultures of E. coli JM109 and recombinant clones were diluted with LB broth to give 107 CFU/ml. The volume of the suspension drop was 2.5 μl, and it contained 104 CFU of E. coli. Bacterial growth of inoculates was determined after incubation at 37°C for 20 h.
RNA isolation, reverse transcription (RT), and RT-PCR.
Total RNA was extracted from a mid-logarithmic-phase culture of M. bovirhinis strain HAZ141-2 using the RNeasy minikit (Qiagen, Hilden, Germany). The elimination of contaminating genomic DNA was performed during RNA isolation as specified by the manufacturer. In addition, after extraction, the RNA was treated with DNase I (NEB, USA) to remove any contaminated chromosomal DNA and then cleaned up using an RNA mini-prep column (Zymo Research, USA). The concentration of total RNA was determined using a NanoDrop system, and a ratio of about 2.0 was considered a good indication of purity. RT was performed with the RevertAid Moloney murine leukemia virus reverse transcriptase (M-MuLV; Promega, Inc., Madison, USA) using the aadE-R1, sat4-R1, and aphA3-R2 primers specific to the aadE*, sat4, and aphA-3 genes, respectively (Table S2). Briefly, about 1 μg total RNA and 20 μM oligonucleotide primer at the final volume of 15 μl were added to a microtube, incubated for 5 min at 70°C, and cooled on ice. dNTP (10 mM), M-MuLV buffer X1, RNase inhibitor (25 u; NEB, USA), and 200 u M-MuLV enzyme were added to the cocktail (25 μl final volume) and incubated for 60 min at 42°C. To inactivate the enzyme, the reaction was then incubated for 10 min at 70°C. Synthesized cDNA was stored at –20°C. For each RT-PCR, the negative control containing all reagent, except the M-MuLV enzyme, was included. The resultant cDNA products were subjected to PCRs using MyTaq DNA polymerase (Bioline) and different primers specific to the aadE*, sat4, and aphA-3 genes (Table S2). The PCR amplification program was as follows: 30 cycles of denaturation at 95°C for 15 s, primer annealing at 56°C for 15 s, and extension at 72°C for 45 s.
Rapid amplification of 5′ cDNA ends (5′-RACE) analysis.
The 5′-RACE analysis was performed as previously described by Amram et al. (66) using a 2nd-generation 5′/3′-RACE kit (Roche Diagnostics GmbH, Mannheim, Germany) and according to the manufacturer’s instructions. Briefly, total cellular RNA was extracted from mid-logarithmic-phase cultures of M. bovirhinis strain HAZ141-2 using the High Pure RNA isolation kit (Roche Diagnostics GmbH, Mannheim, Germany). The cDNA was obtained using 1 μg of DNA-free RNA of M. bovirhinis strain HAZ141-2 with either aadE-R1, sat4-R1, or aphA3-R2 primers complementary to the sequences of the aadE*, sat4, and aphA3 genes, respectively (Table S2). In the first 5′-RACE PCR, the cDNA products with a poly(A) tail were subjected to PCR using the forward oligonucleotide(dT) anchor primer provided with the kit and either aadE*-specific aadE-R2, sat4-specific sat4-R3, or aphA3-specific aphA3-R1 reverse primers (Table S2). In the nested 5′-RACE PCRs, an aliquot from the first 5′-RACE PCRs was amplified using the forward PCR anchor primer, provided with the kit, and the reverse aadE_R3-specific primer (Table S2). The final PCR products were purified using the MEGA quick-spin PCR and agarose gel extraction system (iNtRON Biotechnology, South Korea) and sequenced.
Computational analysis.
BLAST analysis of protein and nucleotide sequences was performed using the NCBI server (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The different functional domains were identified using the Pfam protein family database (http://pfam.xfam.org/), the Integrated Resource of Protein Domains (InterPro) (http://www.ebi.ac.uk/interpro/), and the database of protein families and domains PROSITE (https://prosite.expasy.org/). Primary DNA sequence analyses (GC content, DRs, dyad symmetries, etc.) were performed with either Clone Manager 9, professional edition, software (Scientific & Educational Software, Durham, NC) or DNASTAR software version 5.06/5.51, 2003 (Lasergene, Inc., Madison, WI). DNA promoter motif searches were performed with the Pattern Locator program (67) (http://www.cmbl.uga.edu/software/patloc.html). The search was carried out using the general motif for RpoD (SigA)-dependent bacterial promoters, which is TTGACA in the –35 element and TATAAT in the –10 element (or Pribnow box), with a spacer of 16 to 19 nucleotides between –35 and –10 (68, 69). Multiple sequence alignments were performed with the PRABI Lyon Gerland public servers for DNA sequences (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalwan.html) and for amino acid sequences (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html).
Data availability.
The data supporting the findings of this study are available within the paper and its supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the receipt of M. bovirhinis strain HAZ141_2 from Eiji Hata (National Institute of Animal Health, National Agriculture and Food Research Organization, Tsukuba, Ibaraki, Japan). We also acknowledge the receipt of DNAs of M. bovirhinis field strains from Joachim Spergser (Division of Clinical Microbiology and Infection Biology, Institute of Bacteriology, Mycology and Hygiene, University of Veterinary Medicine Vienna, Austria) and Salvatore Catania (Istituto Zooprofilattico Sperimentale delle Venezie, viale Dell’Universitä, Legnaro, Italy).
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Yao J, Moellering R. 2007. Antibacterial agents, p 1077–1113. In Murray P, Baron E, Jorgensen J, Landry M, Pfaller M (ed), Manual of clinical microbiology. ASM Press, Washington, DC. [Google Scholar]
- 2.Ho YI, Chan CY, Cheng AF. 1997. In-vitro activities of aminoglycoside-aminocyclitols against mycobacteria. J Antimicrob Chemother 40:27–32. doi: 10.1093/jac/40.1.27. [DOI] [PubMed] [Google Scholar]
- 3.Bryskier A 2005. Antibiotics and antibacterial agents: classifications and structure—activity relationships, p 13–38. In Bryskier A (ed), Antimicrobial agents. ASM Press, Washington, DC. [Google Scholar]
- 4.Puglisi JD, Blanchard SC, Dahlquist KD, Eason RG, Fourmy D, Lynch SR, Recht MI, Yoshizawa S. 2000. Aminoglycoside antibiotics and decoding, p 419–430. In Garrett RA, Douthwaite SR, Liljas A, Matheson AT, Moore PB, Noller HF (ed), The ribosome: structure, functions, antibiotics, and cellular interactions. ASM Press, Washington, DC. [Google Scholar]
- 5.Moazed D, Noller HF. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 6.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Doudna Cate JH. 2007. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol 14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 7.Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev 16:430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Razin S, Yogev D, Naot Y. 1998. Molecular biology and pathogenesis of mycoplasmas. Microbiol Mol Biol Rev 62:1094–1156. doi: 10.1128/MMBR.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bébéar CM, Kempf I. 2005. Antimicrobial therapy and antimicrobial resistance, p 535–568. In Blanchard A, Browning GF (ed), Mycoplasmas: molecular biology, pathogenicity and strategies for control. Horizon Bioscience, Norwich, UK. [Google Scholar]
- 10.Waites KB, Lysnyansky I, Bébéar CM. 2014. Emerging antimicrobial resistance in mycoplasmas of humans and animals, p 289–322. In Browning GF, Citti C (ed), Mollicutes: molecular biology and pathogenesis. Caister Academic Press, Norfolk, England. [Google Scholar]
- 11.Gautier-Bouchardon AV 2018. Antimicrobial resistance in Mycoplasma spp, p 425–446. In Schwarz S, Cavaco L, Shen J (ed), Antimicrobial resistance in bacteria from livestock and companion animals. ASM Press, Washington, DC. doi: 10.1128/microbiolspec.ARBA-0030-2018. [DOI] [Google Scholar]
- 12.Lu C, Ye T, Zhu G, Feng P, Ma H, Lu R, Lai W. 2010. Phenotypic and genetic characteristics of macrolide and lincosamide resistant Ureaplasma urealyticum isolated in Guangzhou, China. Curr Microbiol 61:44–49. doi: 10.1007/s00284-009-9574-9. [DOI] [PubMed] [Google Scholar]
- 13.Raherison S, Gonzalez P, Renaudin H, Charron A, Bebear C, Bebear CM. 2002. Evidence of active efflux in resistance to ciprofloxacin and to ethidium bromide by Mycoplasma hominis. Antimicrob Agents Chemother 46:672–679. doi: 10.1128/aac.46.3.672-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konigsson MH, Bolske G, Johansson KE. 2002. Intraspecific variation in the 16S rRNA gene sequences of Mycoplasma agalactiae and Mycoplasma bovis strains. Vet Microbiol 85:209–220. doi: 10.1016/s0378-1135(01)00517-x. [DOI] [PubMed] [Google Scholar]
- 15.Sulyok KM, Kreizinger Z, Wehmann E, Lysnyansky I, Bányai K, Marton S, Jerzsele Á, Rónai Z, Turcsányi I, Makrai L, Jánosi S, Nagy SÁ, Gyuranecz M. 2017. Mutations associated with decreased susceptibility to seven antimicrobial families in field and laboratory-derived Mycoplasma bovis strains. Antimicrob Agents Chemother 61:e01983-16. doi: 10.1128/AAC.01983-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholas RAJ, Ayling R, McAuliffe L. 2009. Mycoplasma Diseases of Ruminants. CABI, Wallingford, UK. [Google Scholar]
- 17.Hata E, Nagai K, Murakami K. 2017. Complete genome sequence of Mycoplasma bovirhinis strain HAZ141_2 from bovine nasal discharge in Japan. Genome Announc 5:e01000-17. doi: 10.1128/genomeA.01000-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, Hao H, Zhao P, Liu Y, Chu Y. 2018. Genome-wide analysis of Mycoplasma bovirhinis GS01 reveals potential virulence factors and phylogenetic relationships. G3 (Bethesda) 8:1417–1424. doi: 10.1534/g3.118.200018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derbise A, Dyke KG, el Solh N. 1996. Characterization of a Staphylococcus aureus transposon, Tn5405, located within Tn5404 and carrying the aminoglycoside resistance genes, aphA-3 and aadE. Plasmid 35:174–188. doi: 10.1006/plas.1996.0020. [DOI] [PubMed] [Google Scholar]
- 20.Derbise A, de Cespedes G, el Solh N. 1997. Nucleotide sequence of the Staphylococcus aureus transposon, Tn5405, carrying aminoglycosides resistance genes. J Basic Microbiol 37:379–384. doi: 10.1002/jobm.3620370511. [DOI] [PubMed] [Google Scholar]
- 21.Jacob J, Evers S, Bischoff K, Carlier C, Courvalin P. 1994. Characterization of the sat4 gene encoding a streptothricin acetyltransferase in Campylobacter coli BE/G4. FEMS Microbiol Lett 120:13–17. doi: 10.1111/j.1574-6968.1994.tb07000.x. [DOI] [PubMed] [Google Scholar]
- 22.Derbise A, Aubert S, El Solh N. 1997. Mapping the regions carrying the three contiguous antibiotic resistance genes aadE, sat4, and aphA-3 in the genomes of staphylococci. Antimicrob Agents Chemother 41:1024–1032. doi: 10.1128/AAC.41.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmieri C, Princivalli MS, Brenciani A, Varaldo PE, Facinelli B. 2011. Different genetic elements carrying the tet(W) gene in two human clinical isolates of Streptococcus suis. Antimicrob Agents Chemother 55:631–636. doi: 10.1128/AAC.00965-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inamine JM, Ho KC, Loechel S, Hu PC. 1990. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J Bacteriol 172:504–506. doi: 10.1128/jb.172.1.504-506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamao F, Muto A, Kawauchi Y, Iwami M, Iwagami S, Azumi Y, Osawa S. 1985. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A 82:2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caillaud F, Trieu-Cuot P, Carlier C, Courvalin P. 1987. Nucleotide sequence of the kanamycin resistance determinant of the pneumococcal transposon Tn1545: evolutionary relationships and transcriptional analysis of aphA-3 genes. Mol Gen Genet 207:509–513. doi: 10.1007/BF00331623. [DOI] [PubMed] [Google Scholar]
- 27.Trieu-Cuot P, Gerbaud G, Lambert T, Courvalin P. 1985. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J 4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trieu-Cuot P, Klier A, Courvalin P. 1985. DNA sequences specifying the transcription of the streptococcal kanamycin resistance gene in Escherichia coli and Bacillus subtilis. Mol Gen Genet 198:348–352. doi: 10.1007/BF00383017. [DOI] [PubMed] [Google Scholar]
- 29.Laverde Gomez JA, Hendrickx AP, Willems RJ, Top J, Sava I, Huebner J, Witte W, Werner G. 2011. Intra- and interspecies genomic transfer of the Enterococcus faecalis pathogenicity island. PLoS One 6:e16720. doi: 10.1371/journal.pone.0016720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campagne S, Marsh ME, Capitani G, Vorholt JA, Allain FH. 2014. Structural basis for -10 promoter element melting by environmentally induced sigma factors. Nat Struct Mol Biol 21:269–276. doi: 10.1038/nsmb.2777. [DOI] [PubMed] [Google Scholar]
- 31.Brussow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108:13–16. doi: 10.1016/S0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 32.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, Penades JR, Ingmer H. 2016. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun 7:13333. doi: 10.1038/ncomms13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fillol-Salom A, Alsaadi A, Sousa JAM, Zhong L, Foster KR, Rocha EPC, Penades JR, Ingmer H, Haaber J. 2019. Bacteriophages benefit from generalized transduction. PLoS Pathog 15:e1007888. doi: 10.1371/journal.ppat.1007888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brussow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Gomez C, Blanco-Picazo P, Brown-Jaque M, Quiros P, Rodriguez-Rubio L, Cerda-Cuellar M, Muniesa M. 2019. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci Rep 9:13281. doi: 10.1038/s41598-019-49898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lekunberri I, Subirats J, Borrego CM, Balcazar JL. 2017. Exploring the contribution of bacteriophages to antibiotic resistance. Environ Pollut 220:981–984. doi: 10.1016/j.envpol.2016.11.059. [DOI] [PubMed] [Google Scholar]
- 37.Boerlin P, Burnens AP, Frey J, Kuhnert P, Nicolet J. 2001. Molecular epidemiology and genetic linkage of macrolide and aminoglycoside resistance in Staphylococcus intermedius of canine origin. Vet Microbiol 79:155–169. doi: 10.1016/s0378-1135(00)00347-3. [DOI] [PubMed] [Google Scholar]
- 38.Werner G, Hildebrandt B, Witte W. 2001. Aminoglycoside-streptothricin resistance gene cluster aadE-sat4-aphA-3 disseminated among multiresistant isolates of Enterococcus faecium. Antimicrob Agents Chemother 45:3267–3269. doi: 10.1128/AAC.45.11.3267-3269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nirdnoy W, Mason CJ, Guerry P. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob Agents Chemother 49:2454–2459. doi: 10.1128/AAC.49.6.2454-2459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 41.Qin S, Wang Y, Zhang Q, Chen X, Shen Z, Deng F, Wu C, Shen J. 2012. Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob Agents Chemother 56:5332–5339. doi: 10.1128/AAC.00809-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Mukherjee S, Hoffmann M, Kotewicz ML, Young S, Abbott J, Luo Y, Davidson MK, Allard M, McDermott P, Zhao S. 2013. Whole-genome sequencing of gentamicin-resistant Campylobacter coli isolated from U.S. retail meats reveals novel plasmid-mediated aminoglycoside resistance genes. Antimicrob Agents Chemother 57:5398–5405. doi: 10.1128/AAC.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki Y, Katsukawa C, Tamaru A, Abe C, Makino M, Mizuguchi Y, Taniguchi H. 1998. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J Clin Microbiol 36:1220–1225. doi: 10.1128/JCM.36.5.1220-1225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Criswell D, Tobiason VL, Lodmell JS, Samuels DS. 2006. Mutations conferring aminoglycoside and spectinomycin resistance in Borrelia burgdorferi. Antimicrob Agents Chemother 50:445–452. doi: 10.1128/AAC.50.2.445-452.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Recht MI, Douthwaite S, Puglisi JD. 1999. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J 18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Recht MI, Puglisi JD. 2001. Aminoglycoside resistance with homogeneous and heterogeneous populations of antibiotic-resistant ribosomes. Antimicrob Agents Chemother 45:2414–2419. doi: 10.1128/aac.45.9.2414-2419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, Shibata N, Ike Y, Arakawa Y. 2007. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 51:4401–4409. doi: 10.1128/AAC.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.François B, Russell RJM, Murray JB, Aboul-Ela F, Masquida B, Vicens Q, Westhof E. 2005. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res 33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vela AI, Villalon P, Saez-Nieto JA, Chacon G, Dominguez L, Fernandez-Garayzabal JF. 2017. Characterization of Streptococcus pyogenes from animal clinical specimens, Spain. Emerg Infect Dis 23:2013–2016. doi: 10.3201/eid2312.151146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willard MD, Berridge B, Braniecki A, Bouley D. 1998. Possible antibiotic-associated colitis in a dog. J Am Vet Med Assoc 213:1775–1779, 1753–1754. [PubMed] [Google Scholar]
- 51.Franklinos LH, Efstratiou A, Macgregor SK, John SK, Hopkins T, Cunningham AA, Lawson B. 2015. Streptococcus pyogenes infection in a free-living European hedgehog (Erinaceus europaeus). Ecohealth 12:689–692. doi: 10.1007/s10393-015-1051-2. [DOI] [PubMed] [Google Scholar]
- 52.Hirose K, Kobayashi H, Ito N, Kawasaki Y, Zako M, Kotani K, Ogawa H, Sato H. 2003. Isolation of mycoplasmas from nasal swabs of calves affected with respiratory diseases and antimicrobial susceptibility of their isolates. J Vet Med B Infect Dis Vet Public Health 50:347–351. doi: 10.1046/j.1439-0450.2003.00681.x. [DOI] [PubMed] [Google Scholar]
- 53.Uemura R, Sueyoshi M, Nagatomo H. 2010. Antimicrobial susceptibilities of four species of mycoplasma isolated in 2008 and 2009 from cattle in Japan. J Vet Med Sci 72:1661–1663. doi: 10.1292/jvms.10-0165. [DOI] [PubMed] [Google Scholar]
- 54.Kawai K, Higuchi H, Iwano H, Iwakuma A, Onda K, Sato R, Hayashi T, Nagahata H, Oshida T. 2014. Antimicrobial susceptibilities of mycoplasma isolated from bovine mastitis in Japan. Anim Sci J 85:96–99. doi: 10.1111/asj.12144. [DOI] [PubMed] [Google Scholar]
- 55.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dybvig K, Sitaraman R, French CT. 1998. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc Natl Acad Sci U S A 95:13923–13928. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy J, Mahony J, Ainsworth S, Nauta A, van Sinderen D. 2013. Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl Environ Microbiol 79:7547–7555. doi: 10.1128/AEM.02229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sternberg N, Coulby J. 1990. Cleavage of the bacteriophage P1 packaging site (pac) is regulated by adenine methylation. Proc Natl Acad Sci U S A 87:8070–8074. doi: 10.1073/pnas.87.20.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Łobocka MB, Rose DJ, Plunkett G III, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. 2004. Genome of bacteriophage P1. J Bacteriol 186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson J, Skeggs PA, Cundliffe E. 1985. Methylation of 16S ribosomal RNA and resistance to the aminoglycoside antibiotics gentamicin and kanamycin determined by DNA from the gentamicin-producer, Micromonospora purpurea. Mol Gen Genet 201:168–173. doi: 10.1007/BF00425655. [DOI] [PubMed] [Google Scholar]
- 61.Beauclerk AA, Cundliffe E. 1987. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J Mol Biol 193:661–671. doi: 10.1016/0022-2836(87)90349-4. [DOI] [PubMed] [Google Scholar]
- 62.Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. 1999. Aminoglycosides: activity and resistance. Antimicrob Agents Chemother 43:727–737. doi: 10.1128/AAC.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friis NF 1975. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare. Nordisk Veterinaermedicin 27:333–339. [PubMed] [Google Scholar]
- 64.Amram E, Mikula I, Schnee C, Ayling RD, Nicholas RA, Rosales RS, Harrus S, Lysnyansky I. 2015. 16S rRNA gene mutations associated with decreased susceptibility to tetracycline in Mycoplasma bovis. Antimicrob Agents Chemother 59:796–802. doi: 10.1128/AAC.03876-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.European Committee for Antimicrobial Susceptibility Testing of the European Society of Clinical M, Infectious D. 2000. EUCAST Definitive Document E.DEF 3.1, June 2000: determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin Microbiol Infect 6:509–515. doi: 10.1046/j.1469-0691.2000.00142.x. [DOI] [PubMed] [Google Scholar]
- 66.Amram E, Borovok I, Nachum-Biala Y, Ayling R, Lerner U, Harrus S, Lysnyansky I. 2016. High prevalence of diverse insertion sequences within the rRNA operons of Mycoplasma bovis. Appl Environ Microbiol 82:6386–6394. doi: 10.1128/AEM.01628-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mrazek J, Xie S. 2006. Pattern locator: a new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics 22:3099–3100. doi: 10.1093/bioinformatics/btl551. [DOI] [PubMed] [Google Scholar]
- 68.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. 2007. Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res 35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feklistov A, Darst SA. 2011. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell 147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the paper and its supplemental material.