The Mycobacterium tuberculosis drug discovery effort has generated a substantial number of new/repurposed drugs for therapy for this pathogen. The arrival of these drugs is welcome, but another layer of difficulty has emerged.
KEYWORDS: Mycobacterium tuberculosis, combination therapy, metabolic state, Greco model
ABSTRACT
The Mycobacterium tuberculosis drug discovery effort has generated a substantial number of new/repurposed drugs for therapy for this pathogen. The arrival of these drugs is welcome, but another layer of difficulty has emerged. Single agent therapy is insufficient for patients with late-stage tuberculosis because of resistance emergence. To achieve our therapeutic ends, it is requisite to identify optimal combination regimens. These regimens go through a lengthy and expensive evaluative process. If we have a modest group of 6 to 8 new or repurposed agents, this translates into 15 to 28 possible 2-drug combinations. There is neither time nor resources to give an extensive evaluation for all combinations. We sought a screening procedure that would identify combinations that had a high likelihood of achieving good bacterial burden decline. We examined pretomanid, moxifloxacin, linezolid, and bedaquiline in log-phase growth, acid-phase growth, and nonreplicative persister (NRP) phase in the Greco interaction model. We employed the interaction term α and the calculated bacterial burden decline as metrics to rank different regimens in different metabolic states. No relationship was found between α and bacterial kill. We chose bacterial kill as the prime metric. The combination of pretomanid plus moxifloxacin emerged as the clear frontrunner, as the largest bacterial declines were seen in log phase and acid phase with this regimen and it was second best in NRP phase. Bedaquiline also produced good kill. This screening process may identify optimal combinations that can be further evaluated in both the hollow-fiber infection model and in animal models of Mycobacterium tuberculosis infection.
TEXT
New agents for the therapy of Mycobacterium tuberculosis have been slow in development. Thankfully, in the last several years, several new (e.g., pretomanid [PMD], delamanid, bedaquiline [BDQ]) or repurposed agents (e.g., linezolid [LZD], moxifloxacin [MXF], levofloxacin, clofazimine) have become available for clinical trial evaluation. Multiple other new agents are in the preclinical or early (phase I) clinical phase of development (e.g., TBAJ-587, TBI-223, SPR-720, TBAJ-876).
Therapy for M. tuberculosis is almost always multiagent and of long duration. The multiagent nature of therapy is mostly due to the high rate of resistance when monotherapy is employed. It is also due to high between-subject variance in important pharmacokinetic (PK) parameters, such as clearance, differences in penetration to effect sites between drugs (pharmacokinetic siloing), and the prolonged nature of therapy increasing the risk of nonadherence (1–5).
Multiple-agent therapy may also increase the rate of kill for the organism, providing a possibility of shortening therapy. It is important to realize that M. tuberculosis may exist in multiple physiologic states in the infected patient. The most commonly studied are log-phase M. tuberculosis, acid-phase M. tuberculosis, and nonreplicative persister (NRP)-phase M. tuberculosis. Optimally, a multidrug regimen will generate good bacterial kill and suppress resistance for all physiological states at exposures that do not drive concentration-related toxicities.
Unfortunately, just having a combination of agents is not a guarantee of a successful regimen. Our group has had experience with two relatively recent combination chemotherapy regimens. These were evaluated in the hollow-fiber infection model (HFIM), an in vitro system that was recently qualified as an M. tuberculosis drug development tool by the European Medicines Agency (EMA) (6).
In the first regimen, we evaluated moxifloxacin plus rifampin (7). In this evaluation, the combination shut off resistance emergence when the organisms were in log phase. However, when the NRP phase was examined, the rate of bacterial kill was significantly decreased with the combination therapy (antagonism for rate of bacterial kill). The clinical importance of this observation was identified in a randomized trial by Gillespie et al. (8), where the regimen with moxifloxacin plus rifampin failed to attain its clinical endpoint of achieving shortening of therapy duration in spite of clearing the sputum significantly faster. This implies but does not absolutely prove that the antagonism seen for the NRP-phase organisms had an effect on the rate of clearance at the end of therapy and may have resulted in failure to attain the endpoint.
In the second regimen, we examined the combination of linezolid plus rifampin (9). Here, the combination did not guarantee resistance suppression. In the 9 arms where combination therapy was evaluated, 7 had emergence of resistance to one drug or the other or both.
With the advent of new drugs, we felt an important issue was to identify a screening process so that promising combinations could be focused upon with more resource- and time-intensive investigations, such as HFIM evaluations and investigations in animal models of infection and, ultimately, clinical trials. If, for example, we have 6 or 8 new agents, all possible combinations would be 15 to 28 in number. The time, effort, and money required for traditional evaluation of all possible regimens would be daunting. Therefore, we decided to look at combination therapy with a modified in vitro checkerboard assay in which quantitative culture values of all single and combination drug concentrations are mathematically analyzed. We did not focus on resistance suppression, as this was meant to be a screening exercise. The aim was to identify the most promising regimens for further, more in-depth study. We chose to examine the agents pretomanid, moxifloxacin, bedaquiline, and linezolid. We evaluated these agents for M. tuberculosis strain H37Rv in the log-phase and acid-phase metabolic states and for M. tuberculosis 18b, a streptomycin-resistant streptomycin auxotroph that exists in an NRP metabolic state when it is streptomycin starved (10). For statistical evaluation, we used the Greco combination therapy model (11).
RESULTS
MIC values for H37Rv.
MIC values for pretomanid, moxifloxacin, linezolid, and bedaquiline were 0.125, 0.25, 1.0, and 0.0625 mg/liter. The lack of growth of streptomycin-starved strain 18b (NRP phase) made it impossible to determine the MIC of this isolate in this metabolic state.
Greco model interaction analysis.
We examined 2-drug combinations of 4 agents (pretomanid, moxifloxacin, bedaquiline, linezolid) in a modified, quantitative plate assay for three metabolic states (log phase, acid phase, and NRP phase). Since pretomanid is unstable in medium, the broth microdilution MICs were read at 14 days as recommended for susceptibility testing of this drug (12). The plates were read at 14 days, and quantitative bacterial counts were determined for each well. Each combination regimen for each physiological state was performed in duplicate. Each replicate was analyzed alone, and then both replicates were analyzed together. The Greco model (11) was employed to analyze all of the observations simultaneously as we have previously described (10, 13). The parameter estimates of the model, including the α interaction parameter for each of the combinations and for each metabolic state, are reported in Table 1. The range of α values was large (see Tables 1 and 2). Two of the analyses (PMD/BDQ in the NRP phase and LZD/PMD in the acid phase) showed negative values. The 95% confidence interval (CI) for PMD/BDQ crossed zero and was recorded to be additive, while LZD/PMD was recorded as statistically significantly antagonistic (the upper bound of the 95% confidence interval was still negative). In one other instance (PMD/MXF in NRP state), the α value was positive and the lower bound of the 95% CI remained positive, indicating synergy. The rest of the evaluations were deemed additive.
TABLE 1.
Greco model parameters for all drugs and physiologic statesa
| Drug and physiologic state | ECON log10 (CFU/ml) | EC50,1 (mg/liter) | m1 value | EC50,2 (mg/liter) | m2 value | α value | 95% CIb |
|---|---|---|---|---|---|---|---|
| PMD+MXF | |||||||
| Log phase | 7.24 | 0.0659 | 2.97 | 0.0663 | 15.6 | 4.59 × 10−5 | −0.70–0.70 |
| Acid phase | 6.14 | 0.118 | 5.50 | 0.0932 | 5.34 | 1.84 × 10−4 | −0.215–0.215 |
| NRP phase | 4.40 | 0.421 | 2.01 | 0.556 | 1.40 | 1.60 | 0.281–2.91 |
| LZD+BDQ | |||||||
| Log phase | 7.84 | 4.24 | 0.962 | 0.286 | 2.17 | 9.05 × 10−6 | −0.729–0.729 |
| Acid phase | 9.10 | 2.10 | 0.414 | 0.164 | 0.338 | 1.06 | −0.457–2.59 |
| NRP phase | 6.31 | 2.57 | 1.96 | 0.688 | 1.21 | 0.392 | −0.103–0.886 |
| PMD+LZD | |||||||
| Log phase | 6.92 | 3.08 | 1.12 | 0.118 | 0.766 | 0.256 | −0.495–1.01 |
| Acid phase | 3.77 | 2.91 | 6.02 | 0.424 | 2.42 | −0.535 | −0.723 to −0.347 |
| NRP phase | 2.77 | 0.267 | 2.46 | 0.814 | 7.34 | 0.808 | −0.0881–1.70 |
| PMD+BDQ | |||||||
| Log phase | 4.45 | 0.0318 | 1.76 | 0.0367 | 3.85 | 1.62 × 10−7 | −0.534–0.534 |
| Acid phase | 3.50 | 0.370 | 0.978 | 2.25 | 0.764 | 36.9 | −3.37–77.1 |
| NRP phase | 2.76 | 0.585 | 2.09 | 0.461 | 2.22 | −0.110 | −1.01–0.789 |
ECON, number of colonies at baseline; EC50,1, concentration of drug 1 that provides 50% of maximal activity; m1, Hill’s constant for drug 1; EC50,2, concentration of drug 2 that provides 50% of maximal activity; m2, Hill’s constant for drug 2; α, interaction parameter; 95% CI, 95% confidence interval for α; PMD, pretomanid; MXF, moxifloxacin; LZD, linezolid; BDQ, bedaquiline.
Bolded confidence intervals are significantly synergistic (PMD+MXF, NRP phase) or antagonistic (PMD+LZD, acid phase).
TABLE 2.
α interaction values for each regimen and the amount of M. tuberculosis bacterial kill for each regimen in three metabolic states as determined by simulation
| Metabolic state and drug regimen | α value | Type of interactiona | Regimen | M. tuberculosis kill (start/end colony counts) |
|---|---|---|---|---|
| Log phase | ||||
| LZD/PMD | 0.256 | ADD | PMD/MXF | 7.00/0.000 |
| PMD/MXF | 4.59 × 10−5 | ADD | PMD/BDQ | 7.00/0.010 |
| LZD/BDQ | 9.05 × 10−6 | ADD | LZD/BDQ | 7.00/0.530 |
| PMD/BDQ | 1.62 × 10−7 | ADD | LZD/PMD | 7.00/1.060 |
| Acid phase | ||||
| PMD/BDQ | 36.9 | ADD | PMD/MXF | 7.00/0.010 |
| LZD/BDQ | 1.06 | ADD | PMD/BDQ | 7.00/0.250 |
| PMD/MXF | 1.84 × 10−4 | ADD | LZD/PMD | 7.00/0.720 |
| LZD/PMD | −0.535 | ANTAG | LZD/BDQ | 7.00/2.38 |
| NRP phase | ||||
| PMD/MXF | 1.60 | SYN | LZD/PMD | 7.00/0.020 |
| LZD/PMD | 0.808 | ADD | PMD/MXF | 7.00/0.190 |
| LZD/BDQ | 0.392 | ADD | PMD/BDQ | 7.00/0.480 |
| PMD/BDQ | −0.110 | ADD | LZD/BDQ | 7.00/2.560 |
SYN, synergistic; ADD, additive; ANTAG, antagonistic.
As the raw data demonstrated differing declines in bacterial burden among the regimens, we wished to examine some measure of bacterial load decline in order to have a ranking of regimens other than just α values. We employed the simulation module of the ADAPT software (version 5) into which the Greco model had been implemented. We employed pharmacokinetic data from the literature (14–18). Predicted bacterial burden decline was calculated relative to an initial condition of a 7.0 log10 (CFU/ml). The α values and the predicted bacterial burden declines are listed in Table 2. While there was a wide range of α values, the calculated bacterial burden decline had a more foreshortened range.
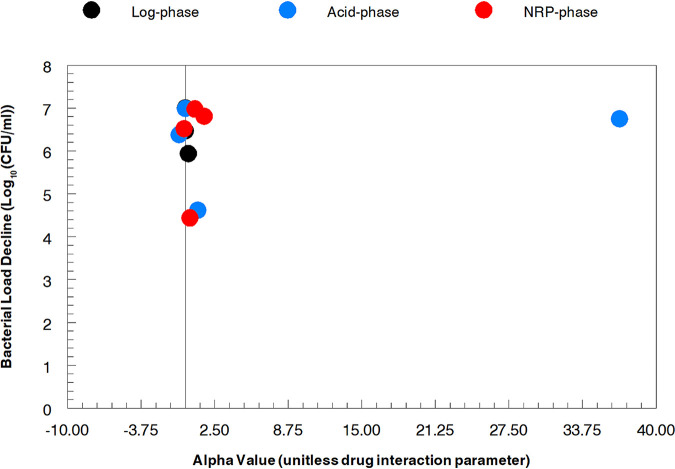
The plotted α values in relationship to the predicted bacterial burden declines for each drug combination and M. tuberculosis metabolic state are displayed in Fig. 1. As may be seen, there is no discernible relationship between the two metrics. Because the α values are employed along with the 50% inhibitory concentration (IC50) values and Hill’s constants for each drug as well as the dose for each agent and its pharmacokinetics to generate a log decline, we chose to employ this metric for rank ordering of regimens. Examining Table 2, the combination of pretomanid plus moxifloxacin generated the largest drop in bacterial load for log-phase and acid-phase organisms. For NRP-phase organisms, this distinction belonged to the combination of linezolid plus pretomanid where the computed bacterial burden went from 7.00 log10 (CFU/ml) at baseline to 0.02 log10 (CFU/ml) at the end of the evaluation. However, the combination of pretomanid plus moxifloxacin was a very close second best, declining to 0.19 log10 (CFU/ml) at the end of the experiment. All of the evaluated combinations performed reasonably well with the highest residual burdens at the end of therapy being 1.06 log10 (CFU/ml) for log phase (linezolid plus pretomanid), 2.38 log10 (CFU/ml) for acid phase (linezolid plus bedaquiline), and 2.56 log10 (CFU/ml) for NRP-phase (linezolid plus bedaquiline). In terms of absolute decline from baseline, the combination of pretomanid plus moxifloxacin caused calculated decreases of 7.00, 6.90, and 6.81 log10 (CFU/ml), respectively, for log-phase, acid-phase, and NRP-phase organisms. Pretomanid plus bedaquiline caused declines of 6.90, 6.75, and 6.52 log10 (CFU/ml). Given the integrated metric of bacterial load decline, we came to the conclusion that the combination of pretomanid plus moxifloxacin was the most promising 2-drug regimen followed very closely by pretomanid plus bedaquiline.
FIG 1.
Lack of correlation between the value of the interaction parameter α and the calculated M. tuberculosis bacterial burden decline.
DISCUSSION
The need for multidrug combination chemotherapy for infections is relatively uncommon. Examples include the therapy for tuberculosis and nontuberculous mycobacteria, some patients with ventilator-associated bacterial pneumonia, some infective endocarditis patients (e.g., enterococcal endocarditis), and some viral diseases, such as HIV and hepatitis C. A common thread here is that the pathogen burden is high, and therefore, it is highly likely that less-susceptible subpopulations are already extant at therapy initiation. However, when required, it is critical that the combinations chosen are optimal. The determinants of optimal combination therapy are straightforward. We would like to have combinations that rapidly kill the pathogen (or in the case of viruses, suppress viral turnover) and also lower the probability of amplification of less-susceptible subpopulations. The third determinant of optimal therapy, which we will not address here is that the regimen chosen is not associated with concentration-driven toxicities for the patient.
In the investigations described here, we have chosen M. tuberculosis as the pathogen. As noted previously, we are fortunate that drug discovery for M. tuberculosis therapy has markedly accelerated in the past 5 to 10 years. As the number of agents available for evaluation grows, the number of possible 2-drug regimens grows substantially.
Again, over this same time frame, we have developed evaluation methodologies that include preclinical systems, such as the HFIM, multiple murine models, and also nonhuman primate models of drug effect (19–25). These evaluative approaches are expensive and time-consuming. If one has even a modest number of new chemical entities such as 8 (the following website has a large number of preclinical- and clinical-phase candidates: https://www.newtbdrugs.org/meetings/2019-wgnd-annual-meeting), this leads to 28 possible 2-drug regimens. There are not resources or time to make an encyclopedic evaluation of such a large number of possible combinations.
The focus of this manuscript was to identify a relatively rapid screening procedure to allow focus on the most promising regimens. A relatively large number of possible 2-drug combinations would need to be “funneled down” to the most promising regimens. It should be noted that we did not focus on resistance suppression but rather used predicted bacterial burden reduction as the metric for rank ordering different 2-drug regimens. We felt that more thorough evaluations, such as in the HFIM, would be more appropriate for evaluation of resistance suppression after the number of regimens was more limited.
From Table 2, it is clear that the combination of pretomanid plus moxifloxacin was a leading contender, with pretomanid plus bedaquiline a very close second. The bacterial load decline was excellent for all three metabolic states studied. The real question is whether this regimen lived up to its potential after it passed through the screening process.
Recently, we had studied the pretomanid plus moxifloxacin combination in a static time-kill assay (26). We did not perform this evaluation in the HFIM model, as the physicochemical properties of pretomanid and bedaquiline (with bedaquiline added to pretomanid plus moxifloxacin to generate a 3-drug regimen) made achievement of a desired concentration-time profile over a 28-day experiment difficult. In this time-kill study, we checked glucose weekly, checked drug concentrations by liquid chromatography-tandem mass spectrometry (LC-MS/MS) thrice weekly, and completely changed medium weekly to assure that no issues with drug degradation would skew results.
The combination of pretomanid plus moxifloxacin against H37Rv in log phase demonstrated excellent activity. The baseline burden was 7.45 log10 (CFU/ml). There were nine combination therapy arms. Three drug exposures were studied for each agent (maximum concentration of drug in serum [Cmax], average concentration of drug in serum [Cavg], and minimum concentration of drug in serum [Cmin]), resulting in the nine combination therapy arms. For arms with any combination of Cmax or Cavg, there was reduction of the bacterial burden to below detection by day 21. This was also the case for arms with Cmin of pretomanid with either Cmax or Cavg of moxifloxacin. Also evaluated was the impact of the regimen on less-susceptible subpopulations. In all combinations, all less-susceptible populations were reduced to undetectable by day 7.
This experiment validated the promise of the pretomanid plus moxifloxacin combination against log-phase organisms with regard to rate of bacterial load reduction. In addition, it also showed great promise by rapidly shutting down all amplification of less-susceptible populations.
We also wished to look at addition of a third drug in this time-kill study, and on the basis of this screening procedure, we chose bedaquiline. In this evaluation, the enhanced combination reduced the 7.45 log10 (CFU/ml) baseline burden to undetectable by 14 days instead of 21 days. We believe this adds a layer of believability to the ability of the screening procedure to identify promising 2-drug combinations.
What are the weaknesses of the data presented here? The first is that the screening procedure only takes bacterial burden reduction into account and does not touch upon resistance suppression. The second is that the validation procedure (26) has, to date, only been done with log-phase organisms. However, acid-phase and NRP-phase evaluations are in process. Finally, the validation process was performed in a static system. In the future, this validation should be performed in the HFIM and the murine system to provide the exposure dynamics inherent in these systems. Another issue is that we employed the plasma concentration-time profile for the calculation of the predicted bacterial burden reduction. Some of the drugs in this investigation (e.g., bedaquiline) have very prolonged pharmacokinetic profiles because of distribution to peripheral compartments, especially effect compartments where the organisms are located. We recognize that concentration-time profiles at the effect site would be more informative, but we are unaware of data indicating full effect site profiles. We, therefore, chose to use the human plasma concentration-time profiles. In the future, we believe that effect compartment concentration-time profiles will provide greater precision for this combination therapy screening procedure.
In summary, we have set forth a screening procedure that allows relatively rapid evaluation of 2-drug combination therapy, in this case for M. tuberculosis. However, the approach is flexible enough to be employed in other combination therapy settings. Rapid evaluation with hierarchical ranking will allow the scientific community to focus resources and time on the most promising regimens. The hope is that identification of better combination therapies for multiple metabolic states will result in faster and more complete bacterial kill. As the total burden declines, the likelihood of resistant subpopulation amplification declines. The more rapid decline may result in shorter duration therapy, which again, may reduce resistance because of improved regimen adherence. Hopefully, as new drugs come on line, we can retain them for a longer period through use of optimized combination regimens.
MATERIALS AND METHODS
Bacterium and generation of metabolic phases.
M. tuberculosis strains H37Rv (ATCC 27294) and 18b were used. M. tuberculosis 18b is a clinical isolate that is a streptomycin-resistant streptomycin auxotroph (kindly provided by Stewart Cole, Global Health Institute, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland). It requires at least 10 mg/liter of streptomycin to be added to agar and broth medium for it to propagate in log phase. In streptomycin-free medium, it converts to the NRP phase and reverts back to log phase when streptomycin is added back to the medium (10). Stocks of the bacterium were stored at −80°C. For experiments using log-phase H37Rv, an aliquot of the stock culture was thawed and incubated at 37°C at 5% CO2 with shaking in Middlebrook 7H9 broth supplemented with 10% albumin, dextrose, and catalase (ADC) and 0.05% Tween 80 (tuberculosis [TB] broth) for 7 to 10 days to achieve log-phase growth. To generate log phase for M. tuberculosis strain 18b, the same procedure was followed, except streptomycin 100 mg/liter was added to the medium. To transition M. tuberculosis 18b to an NRP state, log-phase 18b grown in streptomycin-containing medium was washed thrice by centrifugation with phosphate-buffered saline (PBS) containing 0.05% Tween 80 and resuspended in streptomycin-free TB broth at pH 7 (neutral pH environment). Acid-phase bacteria were generated by transferring 100 µl of log-phase H37Rv to 40 ml of TB broth at pH 6. The culture was incubated at 37°C at 5% CO2 for 7 to 10 days before they were used in susceptibility testing or in the modified quantitative checkerboard assay (8 by 9 matrix of concentrations of drugs alone and in combination for PMD/MXF and PMD/BDQ; 8 by 8 matrix of concentration of drugs alone and in combination for LZD/PMD; 8 by 7 or 8 by 9 for LZD/BDQ) studies.
Drugs.
Linezolid solution (600 mg/300 ml) was purchased from TEVA Pharmaceuticals (North Wales, PA), while pharmaceutical bedaquiline and moxifloxacin was purchased from BOC Sciences (Shirley, NY); both compounds were stored according to the manufacturers’ instructions. Bedaquiline was dissolved in dimethyl sulfoxide (DMSO). Linezolid was dissolved with sterile water. Streptomycin was purchased from Sigma-Aldrich (St. Louis, MO) and was dissolved in sterile water. Pretomanid was graciously supplied by the Global Alliance for TB Drug Development. Moxifloxacin was dissolved in sterile water; pretomanid was dissolved in dimethyl sulfoxide (DMSO). Subsequent dilutions were performed in TB medium. When employed, the final concentration of DMSO was 0.5%.
MIC determination.
Broth microdilution MIC values (12, 27) of all drugs were determined for the H37Rv strain at log phase and acid phase and for the 18b strain at log phase. The final bacterial inoculum added to round-bottom 96-well dilution plates was 1 × 104 CFU/well (100-µl wells; 105 CFU/ml). The bacteria in the different metabolic phases were prepared as described earlier in TB broth. The bacterial suspensions were added to wells containing geometric 2-fold dilutions of all drugs. For studies with acid-phase M. tuberculosis, the medium was adjusted to pH 6. After 14 days of incubation at 37°C at 5% CO2, the broth MICs were read. Initially, MICs were read at 14 and 21 days. There were no differences noted for MXF, LZD, or BDQ. PMD MIC values became unreadable at day 21 because of resistance emergence. The MIC was defined as the lowest concentration that resulted in no visible growth. The susceptibility studies for both drugs were performed using polystyrene 96-well plates and dilution tubes to minimize nonspecific drug binding by BDQ.
In vitro drug interaction studies in the plate system.
Each bacterial metabolic state (H37Rv in log phase and acid phase and 18b in log phase and NRP phase at neutral [pH 7] environments) was prepared in TB broth, with adjustment of the medium pH to 6 to generate M. tuberculosis in the acid-phase metabolic state. At time zero, the bacterial suspensions were inoculated at 104 CFU/well (105 CFU/ml) for BDQ plus LZD. For any PMD-containing combination, the initial inoculum was 103 CFU/well (104 CFU/ml) because we wished to decrease the probability of amplification of a less-susceptible population, particularly in the PMD-alone wells. All experiments were carried out in 96-well round-bottom microdilution plates (Falcon, Corning, NY), containing an 8 by 8 or 9 by 8 matrix consisting of no drug or serial 2-fold increments of all drugs (BDQ, LZD, PMD, MXF) alone and in all possible 2-drug combinations. The checkerboard studies were performed using polystyrene 96-well plates and dilution tubes to minimize the nonspecific binding reported for BDQ. Since antibiotic MICs cannot be determined for M. tuberculosis strain 18b in NRP phase, the middle concentration of drugs in the range of concentrations evaluated singly and in combination in the checkerboard experiments of the other metabolic states studied was the value for the middle concentration for strain 18b. Log-phase and acid-phase phenotypes were incubated for 14 days. The M. tuberculosis suspensions were washed twice with normal saline to remove drug carryover and then quantitatively plated on 7H10 agar supplemented with 10% oleic acid-ADC (OADC). The cultures were incubated at 37°C at 5% CO2 for 4 weeks before the colonies were enumerated. For all studies with 18b, the agar used for the quantitative cultures was also supplemented with 100 mg/liter of streptomycin.
All plate assays were performed in duplicate and analyzed as below simultaneously.
Mathematical model.
The quantitative culture counts obtained from the combination regimens for each metabolic state (log, acid, and NRP phases) were analyzed with the ADAPT software (version 5) (28) using maximum likelihood estimation in the ID application. Data were modeled by the universal response surface approach (URSA) equation of Greco and colleagues (11).
where drug1 and drug2 are the drug concentrations for each agent studied in a particular 2-drug regimen, IC50,1 and IC50,2 are the concentrations of the drugs for which the effect is half maximal, m1 and m2 are Hill’s constants, ECON is the effect for the control, α is the interaction parameter, and E is the fractional effect.
The use of the Greco model enabled us to distinguish between the presence of additivity, synergy, and antagonism in a quantitative manner by evaluating the α value and its associated confidence interval. Additivity is declared if α and its 95% confidence interval include zero. Synergy is declared if α and its 95% confidence interval are positive and do not include zero. Antagonism is declared if α and the 95% confidence interval are negative and do not include zero.
Simulations of the concentration-time profiles were performed using the SIM application in ADAPT (version 5) (28). The concentration-time profile was linked to the simulation module containing the Greco equation with the specific parameter values for the 2-drug combination of interest. The ECON was fixed to 7.0 log10 (CFU/ml). Thus, all bacterial load declines are relative to this value. The specific pharmacokinetic parameter values were taken from the literature (14–18).
The literature PK values were inserted into the simulation module of ADAPT 5, which then employed these values to generate simulated drug concentrations at hourly intervals. These values were then ported to the Greco model to simulate the microbiological effect. The ending values were then subtracted from a baseline value of 7.00 log10 (CFU/ml) to estimate the decline in bacterial numbers. The file was set up to simulate the decline at the end of the experiment.
ACKNOWLEDGMENTS
This work was supported by P01AI0123036 and R01AI121430 from NIAID as well as P41EB001978 from NIBIB and the Alfred Mann Institute for Biomedical Engineering at USC.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Selkon JB, Devadatta S, Kulkarni KG, Mitchison DA, Narayana AS, Nair CN, Ramachandran K. 1964. The emergence of isoniazid-resistant cultures in patients with pulmonary tuberculosis during treatment with isoniazid alone or isoniazid plus PAS. Bull World Health Organ 31:273–294. [PMC free article] [PubMed] [Google Scholar]
- 2.Wasserman S, Denti P, Brust JCM, Abdelwahab M, Hlungulu S, Wiesner L, Norman J, Sirgel FA, Warren RM, Esmail A, Dheda K, Gandhi NR, Meintjes G, Maartens G. 2019. Linezolid pharmacokinetics in South African patients with drug-resistant tuberculosis and a high prevalence of HIV coinfection. Antimicrob Agents Chemother 63:e02164-18. doi: 10.1128/AAC.02164-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alghamdi WA, Al-Shaer MH, An G, Alsultan A, Kipiani M, Barbakadze K, Mikiashvili L, Ashkin D, Griffithg DE, Cegielski JP, Kempker RR, Peloquin CA. 2020. Population pharmacokinetics of linezolid in tuberculosis patients: dosing regimens simulation and target attainment analysis. Antimicrob Agents Chemother 64:e01174-20. doi: 10.1128/AAC.01174-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarathy JP, Zuccotto F, Hsinpin H, Sandberg L, Via LE, Marriner GA, Masquelin T, Wyatt P, Ray P, Dartois V. 2016. Prediction of drug penetration in tuberculosis lesions. ACS Infect Dis 2:552–563. doi: 10.1021/acsinfecdis.6b00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1993. Approaches to improving adherence to antituberculosis therapy–South Carolina and New York, 1986–1991. MMWR Morb Mortal Wkly Rep 42:74–75. [PubMed] [Google Scholar]
- 6.Cavaleri M, Manolis E. 2015. Hollow fiber system model for tuberculosis: The European Medicines Agency experience. Clin Infect Dis 61:S1–S4. doi: 10.1093/cid/civ484. [DOI] [PubMed] [Google Scholar]
- 7.Drusano GL, Sgambati N, Eichas A, Brown DL, Kulawy R, Louie A. 2010. The Combination of rifampin plus moxifloxacin is synergistic for suppression of resistance but antagonistic for cell kill of Mycobacterium tuberculosis as determined in a hollow-fiber infection model. mBio 1:e00139-10. doi: 10.1128/mBio.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillespie SH, Crook AM, McHugh TD, Mendel CM, Meredith SK, Murray SR, Pappas F, Phillips PPJ, Nunn AJ, REMoxTB Consortium . 2014. Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. N Engl J Med 371:1577–1587. doi: 10.1056/NEJMoa1407426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drusano GL, Neely M, Van Guilder M, Schumitzky A, Brown D, Fikes S, Peloquin C, Louie A. 2014. Analysis of combination drug therapy to develop regimens with shortened duration of treatment for tuberculosis. PLoS One 9:e101311. doi: 10.1371/journal.pone.0101311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Miranda Silva C, Hajihosseini A, Myrick J, Nole J, Louie A, Schmidt S, Drusano GL. 2018. Effect of moxifloxacin plus pretomanid against Mycobacterium tuberculosis in log phase, acid phase and nonreplicating-persister phase in an in vitro assay. Antimicrob Agents Chemother 63:e01695-18. doi: 10.1128/AAC.01695-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greco WR, Bravo G, Parsons JC. 1995. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev 47:331–385. [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2011. Susceptibility testing for mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard—2nd ed CLSI document A24-A Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 13.De Miranda Silva C, Hajihosseini A, Myrick J, Nole J, Louie A, Schmidt S, Drusano GL. 2018. Effect of linezolid plus bedaquiline against Mycobacterium tuberculosis in log phase, acid phase, and nonreplicating-persister phase in an in vitro assay. Antimicrob Agents Chemother 62:e00856-18. doi: 10.1128/AAC.00856-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Food and Drug Administration. 2019. Pretomanid. Food and Drug Administration, Washington, DC: https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/212862s000lbl.pdf. [Google Scholar]
- 15.McLeay SC, Vis P, van Heeswijk RP, Green B. 2014. Population pharmacokinetics of bedaquiline (TMC207), a novel antituberculosis drug. Antimicrob Agents Chemother 58:5315–5324. doi: 10.1128/AAC.01418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stalker DJ, Jungbluth GL, Hopkins NK, Batts DH. 2003. Pharmacokinetics and tolerance of single- and multiple-dose oral or intravenous linezolid, an oxazolidinone antibiotic, in healthy volunteers. J Antimicrob Chemother 51:1239–1246. doi: 10.1093/jac/dkg180. [DOI] [PubMed] [Google Scholar]
- 17.Food and Drug Administration. Avelox tablets. Final draft package insert. Food and Drug Administration, Washington, DC: https://www.accessdata.fda.gov/drugsatfda_docs/label/1999/21085lbl.pdf. [Google Scholar]
- 18.Öbrink-Hansen K, Hardlei TF, Brock B, Jensen-Fangel S, Thomsen MK, Petersen E, Kreilgaard M. 2015. Moxifloxacin pharmacokinetic profile and efficacy evaluation in empiric treatment of community-acquired pneumonia. Antimicrob Agents Chemother 59:2398–2404. doi: 10.1128/AAC.04659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gumbo T, Louie A, Liu W, Ambrose PG, Bhavnani SM, Brown D, Drusano GL. 2007. Isoniazid’s bactericidal activity ceases because of the emergence of resistance, not depletion of Mycobacterium tuberculosis in the log phase of growth. J Infect Dis 195:194–201. doi: 10.1086/510247. [DOI] [PubMed] [Google Scholar]
- 20.Gumbo T, Louie A, Deziel MR, Liu W, Parsons LM, Salfinger M, Drusano GL. 2007. Concentration-dependent Mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrob Agents Chemother 51:3781–3788. doi: 10.1128/AAC.01533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louie A, Duncanson B, Myrick J, Maynard M, Nole J, Brown D, Schmidt S, Neely M, Scanga CA, Peloquin C, Drusano GL. 2018. Activity of moxifloxacin against Mycobacterium tuberculosis in acid phase and nonreplicative-persister phenotype phase in a hollow-fiber infection model. Antimicrob Agents Chemother 62:e01470-18. doi: 10.1128/AAC.01470-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drusano GL, Sgambati N, Eichas A, Brown D, Kulawy R, Louie A. 2011. Effect of administering moxifloxacin plus rifampin against Mycobacterium tuberculosis 7 of 7 Days versus 5 of 7 days in an in vitro pharmacodynamic system. mBio 2:e00108-11. doi: 10.1128/mBio.00108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams KN, Stover CK, Zhu T, Tasneen R, Tyagi S, Grosset JH, Nuermberger E. 2009. Promising antituberculosis activity of the oxazolidinone PNU 100480 relative to that of linezolid in a murine model. Antimicrob Agents Chemother 53:1314–1319. doi: 10.1128/AAC.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramnik I, Dietrich WF, Demant P, Bloom BR. 2000. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc Natl Acad Sci USA 97:8560–8565. doi: 10.1073/pnas.150227197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman MT, Chen RY, Lee M, Lin PL, Dodd LE, Maiello P, Via LE, Kim Y, Marriner G, Dartois V, Scanga C, Janssen C, Wang J, Klein E, Cho SN, Barry CE, III, Flynn JL. 2014. PET/CT imaging reveals a therapeutic response to oxazolidinones in macaques and humans with tuberculosis. Sci Transl Med 6:265ra167. doi: 10.1126/scitranslmed.3009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drusano GL, Neely MN, Kim S, Yamada WM, Schmidt S, Duncanson B, Nole J, Mtchedlidze N, Peloquin CA, Louie A. 2020. Building optimal three-drug combination chemotherapy regimens. Antimicrob Agents Chemother 64:e01610-20. doi: 10.1128/AAC.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sala C, Dhar N, Hartkoorn RC, Zhang M, Ha YH, Schneider P, Cole ST. 2010. Simple model for testing drugs against nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 54:4150–4158. doi: 10.1128/AAC.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Argenio DZ, Schumitzky A, Wang X. 2009. ADAPT 5 user’s guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource, Los Angeles, CA. [Google Scholar]