The bacterial cell wall plays a key role in viability and is an important drug target. The cell wall is made of elongated polymers that are cross-linked to one another to form a load-bearing mesh. An alternative cell wall cross-linking mechanism used by the l,d-transpeptidase YcbB has been implicated in the stress-regulated roles of β-lactam resistance, outer membrane defect rescue, and typhoid toxin release. The role for this stress-linked cross-linking in the context of a host infection was unclear.
KEYWORDS: l,d-transpeptidase; bacterial infection; carbapenem; structural biology
ABSTRACT
The bacterial cell wall plays a key role in viability and is an important drug target. The cell wall is made of elongated polymers that are cross-linked to one another to form a load-bearing mesh. An alternative cell wall cross-linking mechanism used by the l,d-transpeptidase YcbB has been implicated in the stress-regulated roles of β-lactam resistance, outer membrane defect rescue, and typhoid toxin release. The role for this stress-linked cross-linking in the context of a host infection was unclear. Here, we resolve the crystallographic structures of both Salmonella Typhi YcbB and Citrobacter rodentium YcbB acylated with ertapenem that delineate the conserved structural characteristics of YcbB. In parallel, we show that the general involvement of YcbB in peptidoglycan reinforcement under conditions of bacterial outer envelope stress does not play a significant role in acute infections of mice by C. rodentium and S. Typhimurium. Cumulatively, in this work we provide a foundation for the development of novel YcbB-specific antibacterial therapeutics to assist in treatment of increasingly drug-resistant S. Typhi infections.
INTRODUCTION
The biosynthesis of the bacterial cell wall has long been a focus of antibacterial strategies both from competing species (1) and in drug development (2, 3). This is exemplified by the widespread use of β-lactam antibiotics, which inhibit the final stages of the pathway. The targeting of cell wall biosynthesis is an excellent strategy due to the importance of the bacterial cell wall in bacterial viability in natural environs and the unique nature of the enzymes and substrates involved. The bacterial cell wall consists of peptidoglycan (PG)—extended glycan strands of alternating β-1,4-linked N-acetylglucosamine–N-acetyl-muramic acid which are cross-linked into a net-like mesh via short peptide segments covalently attached at the C3-OH position of the muramic acid moiety (4).
PG is synthesized through a multistage pathway transitioning from the initial steps in the bacterial cytosol with the formation of soluble precursors, to the inner leaflet of the cytoplasmic membrane where these precursors are assembled onto lipid carriers (terminating in formation of lipid II building blocks), to the outer leaflet of the cytoplasmic membrane where polymerization and cross-linking of the PG occur through the activity of penicillin binding proteins (PBPs) and l,d-transpeptidases (4, 5).
PBPs are split into two major classes, class A and class B (aPBP and bPBP, respectively). The aPBPs perform both a glycosyltransferase activity which polymerizes the flipped lipid II molecules at the outer leaflet into a growing PG strand and a d,d-transpeptidase activity which catalyzes serine-mediated acylation and cleavage of a d-Ala4-d-Ala5 peptide bond of the acyl donor and subsequent transfer of the penultimate d-Ala4 carbonyl to the primary amine of a diaminopimelic acid (DAP) residue on the acceptor. This results in a d-Ala4donor-DAP3acceptor cross-link necessary for cell wall strength and integrity. The bPBPs perform only the latter d,d-transpeptidase activity (5, 6) but typically do so in complex with a glycosyltransferase-bearing partner, such as a SEDS (shape, elongation, division, and sporulation) protein or an aPBP, to create a functional PG “synthase.” The d,d-transpeptidase activity of both aPBPs and bPBPs can be blocked by β-lactam antibiotics, which act as a substrate mimetic of the donor strand peptide bond. Inhibition of the crucial transpeptidation step in PG biosynthesis results in destabilization of the cell wall and, ultimately, in cell death (7–9).
Despite the successful use of β-lactam antibiotics in the clinic, the use and abuse of these potent antibiotics have led to the emergence of numerous bacterial resistance mechanisms. These include the widely disseminated and well-characterized serine and metallo-β-lactamase classes of β-lactamases and drug efflux pumps, as well as many others (10). One more recently discovered mechanism was the capability of bypass of d,d-transpeptidation through the use of an alternative cross-linking mechanism—l,d-transpeptidation (11, 12).
It has been shown that, in Escherichia coli, an l,d-transpeptidase (Ldt) known as YcbB (and, alternatively, LdtD) can compensate for inhibition of PBPs by β-lactam antibiotics with remarkably few other factors. Upon upregulation of alarmone [(p)ppGpp], YcbB, a carboxypeptidase (PBP5), and an aPBP (PBP1b) can form an alternative synthase complex and produce mature PG in a β-lactam-resistant manner (12, 13). Ldts, such as YcbB, cross-link PG using an l,d-transpeptidation mechanism to form a DAP3donor-DAP3acceptor cross-link between a tetrapeptide-containing donor and a tetra- or tripeptide acceptor (12, 14). l,d-Transpeptidases, including YcbB, are not efficaciously inactivated by β-lactams, with the exception of the members of the carbapenem subclass. The thioester-containing l,d-transpeptidase–β-lactam adduct is slow to form for both penicillins and cephalosporins. Additionally, the acylation that does occur is swiftly followed by hydrolysis. Together, these attributes prevent full inactivation of the enzyme (15).
In addition to this, YcbB has been implicated in rescue of outer membrane defects (16), as well as in the release of typhoid toxin in Salmonella enterica serovar Typhi (17). A general trend in these scenarios is the presence of bacterial outer envelope stress, which can stimulate the well-known Cpx stress response. Indeed, in E. coli, the ycbB gene was previously shown to be under the control of the Cpx response transcription factor CpxR (18, 19). Additionally, it has been proposed that l,d-transpeptidation may play a key role in PG maintenance in intracellular pathogens (20). It is possible that YcbB remodeling could act to promote an optimal equilibrium between the inherent physical traits of the d,d-cross-linked PG layer and those of the adjacent outer membrane (21), mediating proper maintenance of overall outer envelope integrity under stressful conditions.
It has been postulated that anti-typhoid toxin immunization or therapeutics may benefit the treatment of Salmonella Typhi (22), a host-restricted pathogen responsible for 10.9 million cases of typhoid fever (23) and ∼200,000 deaths every year (22). PG editing by YcbB has been linked to the release of this toxin (17), and inhibition of YcbB in S. Typhi may prove efficacious in reducing the severity of S. Typhi infection. Beyond this, it remains unknown whether these stress-regulated proteins play a broader role in the virulence of the bacterial pathogens in which they exist. YcbB is found in a few clinically relevant Gram-negative pathogens, such as enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), and S. enterica (including, but not limited to, the serovar Typhi), so it was of interest to decipher the importance of its role in infection outside the context of typhoid toxin release.
Fortunately, there are mouse models for both EPEC and S. Typhi infection, in C. rodentium (24) and S. enterica serovar Typhimurium (25), respectively. S. Typhimurium provokes typhoid-like symptoms in mice while not producing typhoid toxin, allowing one to probe the impact of YcbB in virulence outside the context of typhoid toxin’s role in Salmonella infection. Additionally, using an S. Typhimurium mouse infection model with mice pretreated with streptomycin, one can model Salmonella gastroenteritis.
Recently, we solved the structure of the E. coli meropenem-YcbB acyl-enzyme complex via X-ray crystallography (13). The structure was seen to consist of a novel tridomain architecture which was quite distinct from those of previously characterized l,d-transpeptidases (13). There was also an interesting addition of a capping subdomain on the otherwise canonical Ldt catalytic domain. We proposed that the capping subdomain would hinge relative to the catalytic domain during the formation of l,d-cross-links, in order to fully facilitate the cross-linking mechanism and release of cross-linked PG. Despite the insight gleaned from this structure, many additional issues arose regarding the novel features observed.
Here, we pursued characterization of YcbB from both Salmonella Typhi and Citrobacter rodentium at the atomic level, resulting in resolution of the crystallographic structures of both S. Typhi YcbB and C. rodentium YcbB acylated with ertapenem. From these data, we delineated the conserved structural characteristics and domain architecture of YcbB across a variety of Gram-negative pathogens and determined the breadth of conservation across their catalytic domains. In addition, we observed reorientations in the capping subdomain which provide further insights into the conformational space that these subdomains sample in the greater mechanistic role of these enzymes. In tandem, we show that the general involvement of YcbB in PG reinforcement under conditions of bacterial outer envelope stress does not play a significant role in acute infections of mice by C. rodentium and S. Typhimurium. Cumulatively, in this work we provide a foundation for the development of novel YcbB-specific antibacterial therapeutics to assist in treatment of increasingly drug-resistant S. Typhi infections (26).
RESULTS AND DISCUSSION
Structures of Salmonella Typhi and Citrobacter rodentium YcbB.
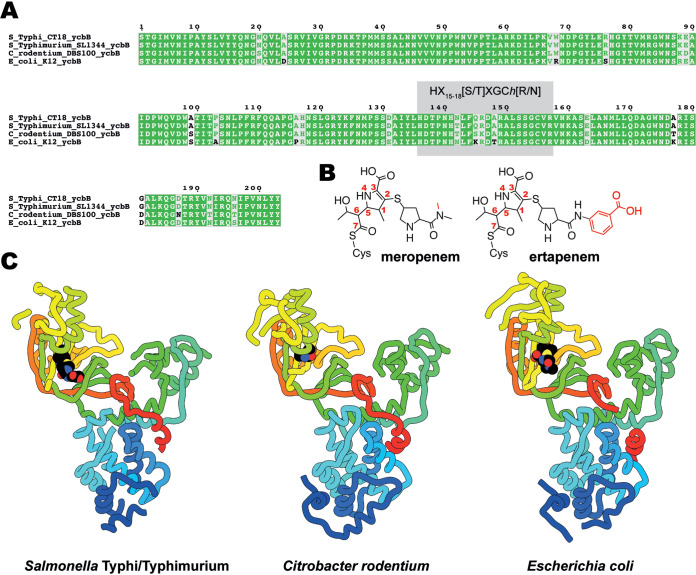
The structures of YcbB from both C. rodentium and S. Typhi/Typhimurium were pursued to provide insight into the structural conservation of the relevant Ldts. It should be noted that YcbB is 98% to 100% conserved among Salmonella Typhi and Typhimurium serovars. The S. enterica Typhi/Typhimurium YcbB studied here is 98.5% identical to YcbB of the clinically relevant multidrug-resistant CT18 strain, with no residues differing in the active site and only one differing within the catalytic domain (Fig. 1A). Among Salmonella, Escherichia, and Citrobacter YcbBs, there is between 78 and 84% identity.
FIG 1.
Multiple-sequence alignment and structures of YcbB. (A) Alignment of amino acid sequences from the catalytic domains of YcbB from Salmonella Typhi CT18, Salmonella Typhimurium SL1344, Citrobacter rodentium DBS100, and Escherichia coli K-12. HX15-18[S/T]XGCh[R/N] (where X represents any residue and h is any hydrophobic residue) is highlighted in gray. (B) Chemical diagrams of meropenem and ertapenem postacylation by YcbB. Differences between meropenem and ertapenem are indicated in red. (C) The structures of S. Typhi/Typhimurium and C. rodentium YcbB are shown in ribbon representation, colored in rainbow from the N terminus (blue) to the C terminus (red), in comparison to the previously solved E. coli YcbB (PDB ID 6NTW [13]) represented in the same scheme. Acylated drugs (ertapenem for S. Typhi/Typhimurium and meropenem for E. coli) are represented by spherical atoms, colored in black and by heteroatom.
Ertapenem was chosen for cocrystallization after extensive screening performed with a small library of commercially available and clinically utilized carbapenem antibiotics. Interestingly, for both S. Typhi YcbB and C. rodentium YcbB, cocrystals with ertapenem were the only crystals obtained. This is in contrast to E. coli YcbB, where crystals were obtained with a variety of carbapenem antibiotics, with a complex with meropenem providing the best ultimate resolution (13). Both ertapenem and meropenem are of current use in the treatment of bacterial infection, particularly in β-lactam-resistant cases (27). The two molecules are closely related, differing only at the terminal amide group of the carbapenem R2 substituent, with an additional methyl group on the amide in meropenem and the addition of a benzoic acid group in ertapenem (Fig. 1B). Cocrystals of S. Typhi YcbB, generated in the presence of 1 mM ertapenem (Fig. 1B) at pH 8.5, displayed P312 symmetry with unit cell dimensions of a = 75.3 Å, b = 75.3 Å, and c = 194.4 Å and a diffraction resolution of 3.6 Å. Extensive optimization efforts to improve on this resolution failed. Crystals of C. rodentium YcbB, generated in the presence of 1 mM ertapenem at pH 6.5, displayed C2221 symmetry with unit cell dimensions of a = 90.0 Å, b = 117.7 Å, and c = 125.2 Å and a diffraction resolution of 2.6 Å. There is one molecule of YcbB in the asymmetric unit of each crystal form. The structure solution was phased by molecular replacement using E. coli YcbB (13), with side chains retained, split into two pieces, and placed sequentially (84% and 78% sequence identity of the S. Typhi and C. rodentium variants with that of E. coli, respectively). The dimensions of both the C. rodentium YcbB and S. Typhi YcbB are approximately 75 by 75 by 40 Å, similar to those of E. coli YcbB. The overall architectures of the three currently characterized YcbB enzymes (Fig. 1C) are also similar, featuring the same distinct tridomain format. The well-ordered central catalytic domain, with the YcbB-specific substrate capping subdomain, proximal scaffold domain, and PG domain, is conserved across all three YcbBs. The backbone root mean square deviations (RMSD) among the three are 2.1 Å across the entirety of the modeled protein (452 residues), 1.6 Å excluding of the capping loop subdomain region (across 396 residues—excluding residues 420 to 493 per S. Typhi numbering), and 1.0 Å across 372 common trimmed residues.
Conservation of YcbB active-site architecture.
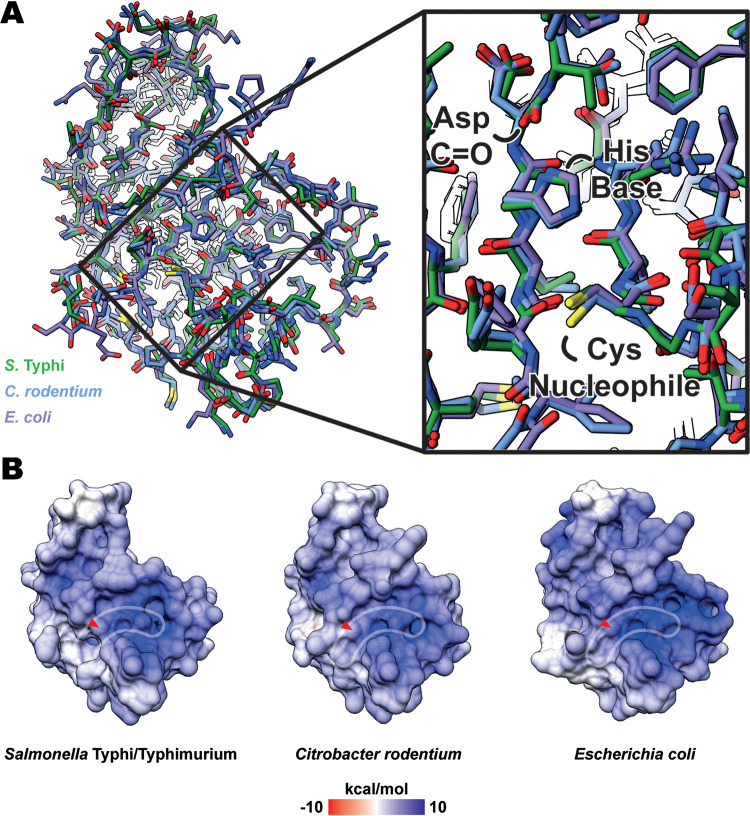
The catalytic domains of S. Typhi and C. rodentium YcbB show both sequence conservation (Fig. 1A; see also Fig. S1 in the supplemental material) and structural conservation (Fig. 1C; see also Fig. 2), with the canonical l,d-transpeptidase folds harboring extended electropositive (Fig. 2B) active-site clefts with the conserved active-site motif (28) (HX15-18[S/T]XGCh[R/N], where X represents any residue and h is any hydrophobic residue), as seen in the E. coli YcbB (13).
FIG 2.
Molecular architecture and electrostatic surface potential of YcbB catalytic domain and active site. (A) An overlay of the catalytic domain of various YcbB homologues (in their carbapenem-acylated states, with carbapenem atoms removed for visualization), showing structural conservation of residues across the domain. S. Typhi/Typhimurium is represented in green, C. rodentium is represented in blue, and E. coli (PDBID 6NTW [13]) is represented in purple. (B) The electrostatic surface potential of catalytic domains from various YcbB homologues, showing conservation of the extended electropositive active site (outlined in white) across species. Placement of the catalytic cysteine is noted with red arrows.
Despite the differences in substrates and substrate stabilization among the three YcbB homologues, there are remarkably few differences in either the peptide backbone or residue placement in the active site among the three (Fig. 2A). There is excellent structural conservation of the nucleophilic cysteine, the histidine general base which activates it, and the main-chain nitrogen atoms of the catalytic cysteine and adjacent tyrosine which form the oxyanion hole to stabilize the negatively charged transition state. In addition, many of the positively charged residues in and around the active site are well conserved, resulting in common positive electrostatic surface distributions among the three homologues (Fig. 2B). On the basis of the characteristics of the conserved PG substrates and subsequent conserved structural elements of the catalytic domain, the broad ability of these YcbB homologues to be acylated by carbapenem ligands is understandable. It would therefore potentially pose a challenge to develop inhibitory molecules that are wholly specific to the YcbB of a single species.
Acylation of YcbB by ertapenem and meropenem.
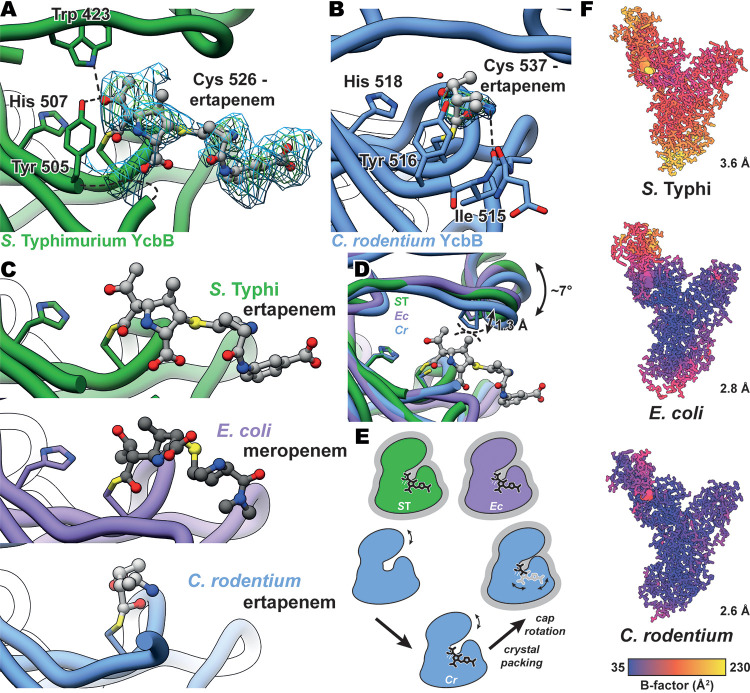
With the new structures of YcbB from S. Typhi and C. rodentium characterized here, we gain additional insights into the acylation of these YcbB proteins by carbapenem antibiotics. Among all three species’ structures of YcbB, there is a lack of extensive hydrogen bonding networks seen between carbapenems and the various conserved motifs (SXXK, SXN, KTG) in PBPs (Fig. 3A and B).
FIG 3.
Carbapenem acylation of YcbB and movement of the capping loop subdomain. (A) mFo-DFc simulated annealing omit map for ertapenem-acylated Cys526 of S. Typhi YcbB, contoured at 2 and 2.5 σ and indicated in blue and green, respectively. S. Typhi YcbB is in green, ertapenem is in light gray, and selected residues and ertapenem are colored by heteroatom. Hydrogen bonding between ertapenem, Tyr507, and Trp423 is represented with dashed lines. Some poorly resolved density is seen for residues 502 to 504, which are represented with a curved dashed line. (B) mFo-DFc simulated annealing omit map for the ertapenem-acylated Cys537 of C. rodentium YcbB, contoured at 2 and 2.5 σ and indicated in blue and green, respectively. C. rodentium YcbB is indicated in blue, and the remainder is colored as described for panel A. Hydrogen bonding between ertapenem and the backbone carbonyl of Ile515 is represented with dashed lines. (C) Comparisons of ertapenem-S. Typhi YcbB (gray/green), ertapenem-C. rodentium YcbB (gray/blue), and meropenem-E. coli YcbB (gray/purple—PDB ID 6NTW [13]). Acylated catalytic cystine and catalytic histidine residues are shown. Similar orientations of the active-site residues are seen among all three homologues, while the general positions of both ertapenem and meropenem are conserved among the S. Typhi and E. coli structures, with the benzoic acid extension protruding away from the active site in the ertapenem-S. Typhi YcbB structure. (D) An overlay of S. Typhi (ST), E. coli (Ec), and C. rodentium (Cr) YcbB structures, highlighting capping loop rearrangements between the three. Modest rearrangements of the capping loop are seen to occlude the stability of ertapenem in the crystal structure of C. rodentium YcbB, likely through the steric interference shown in dashed lines between an overlay of ertapenem and C. rodentium YcbB. (E) A schematic representation of the capping loop rearrangement and the impact on the acylation state during crystallization. Representations of states with structures are additionally outlined in gray. (F) B-factors of S. Typhi, E. coli, and C. rodentium YcbB structures, highlighting the increased levels of B-factors of the capping loop within each model and the increased overall levels of B-factors scaling with resolution (plotted in UCSF Chimera [42]).
In the case of the S. Typhi ertapenem-YcbB structure, we see well-resolved density for the ligand as it acylates the catalytic Cys526 (Fig. 3A). The ertapenem is positioned similarly to the meropenem in the E. coli YcbB structure (13), sitting in the donor side of the active site (Fig. 3C). The conserved core of carbapenem in particular plays the same role in mediating the only hydrogen bonding present between drug and the active site, with the ethyl-alcohol group on C6 hydrogen bonding with both the Tyr505 of the active-site Ldt motif and the conserved Trp423 of the capping loop (Fig. 3A). The proline-rich insertion centered at Pro426 appears to provide a structural ridge that prevents significant interaction with nearby electrostatic residues, as also seen in the E. coli YcbB structure. This observed lack of complex hydrogen bonding in favor of increased hydrophobic interactions is observed both in the E. coli YcbB structure and in those of many non-YcbB Ldts (13, 29).
One interesting feature of the ertapenem acylation of the S. Typhi YcbB is the presumed destabilization of the loop consisting of residues 502 to 504, which are not well resolved and could not be modeled reliably in the electron density. We see a similar region of poor density on the same loop in the higher-resolution C. rodentium ertapenem-YcbB structure, with poorly resolved density and higher-temperature factors for residues 511 to 513, though only residue 512 could not be reliably modeled. In the case of the S. Typhi YcbB, the poorer density and presumed destabilization of this loop could in part be due to the crystal packing, as Tyr561 from an adjacent YcbB in the crystal lattice packs against residues 499 and 500 just prior to the region of poor density. No such packing is seen in the C. rodentium YcbB structure, leading one to postulate that some of this destabilization could indeed be due to the ertapenem acylation.
Beyond the conserved carbapenem core, the ertapenem extends outward from the active site. In particular, the additional benzoic acid extension on ertapenem protrudes into a solvent channel and interacts with a neighboring YcbB in the crystal lattice. This interaction stabilizes the extended ertapenem molecule, which likely would not be well resolved to its terminus in the absence of these contacts. In the context of native YcbB in the periplasm, the acylation of YcbB by ertapenem would likely provide little additional contact in comparison to that of a shorter carbapenem such as meropenem (Fig. 1B).
Conformational plasticity of YcbB capping loop subdomain.
In the structure of the ertapenem-C. rodentium YcbB acyl-enzyme complex, we see poor density for the bulk of the ertapenem-acylated catalytic cysteine (Fig. 3B). Due to the high sequence similarity among YcbB from C. rodentium, YcbB from S. Typhi, and YcbB from E. coli and to the acylated nature of the ligand, this observation is perhaps surprising. On the basis of the well-resolved nature of the catalytic cysteine, which is observed to be rotated away from its conjugate base, His518, we propose that the C. rodentium YcbB is indeed in an acylated state (6, 13). Upon further observation, it is apparent that the capping loop is positioned in the lowest, most active site-occluding position in this structure (Fig. 3D). As we have previously proposed that these capping loop subdomains are likely motile in solution (13), it is possible that the ertapenem formed key stabilizing interactions with C. rodentium YcbB in solution, prior to crystallization. In solution, there is likely an equilibrium between a stabilized acylated ertapenem and a lowered capping loop state with a destabilized ertapenem. The former stabilized state is likely favored in this equilibrium (Fig. 3E). During the crystallization process, this equilibrium could have been shifted toward the observed destabilized state during the crystallization process, with only the portion of the acylated ertapenem closest to the C7 acylation being stabilized, as observed in the density. We see minimal density beyond the acylation site, with some density for part of the secondary ring of the carbapenem, due in part to a hydrogen bond between N4 and the backbone carbonyl of Ile515, similar to the interaction seen with the backbone carbonyl of Ala505 in the E. coli YcbB structure.
With the structures provided here, we begin to see the potential dynamics of the capping loop subdomain. Rotation of the latter is seen about the previously proposed hinge region (13), with a modest rotation of ∼7° (measured by the angle formed between the backbone nitrogen atoms of Pro429, Pro424, and Ile 506, per E. coli numbering). Due to the conserved acylated and crystallized nature of these three YcbB structures, we see relatively similar slices of the potentially far greater landscape of motion that this capping loop can undergo. In all three structures observed to date, there is no potential pathway for the exit of the cross-linked substrate of the respective YcbBs, again alluding to the presence of an additional unobserved, further open state for the capping loop such as we have previously proposed (13).
Exploration of YcbB as a virulence factor.
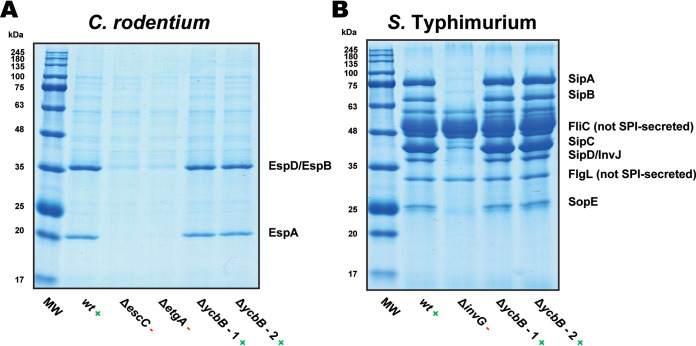
In parallel with our structural studies, we wished to further probe the role of YcbB as a virulence factor. Both EPEC and S. Typhi and their respective mouse models are known to depend on the proper functioning of a type three secretion system (T3SS) to persist within the host in a virulent state (30). The assembly of a functional T3SS in the bacterial membranes and cell wall requires PG remodeling. Therefore, it was of interest to first assay the role of YcbB in the ability of these bacteria to secrete effectors via the T3SS. YcbB knockout strains were generated for both C. rodentium (CrΔycbB) and S. Typhimurium (STmΔycbB) and tested for secretion activity. Neither YcbB knockout strain showed any appreciable loss of T3SS secretion function in this established assay (31) (Fig. 4).
FIG 4.
Secreted-protein profiles from ΔycbB mutants of C. rodentium and S. Typhimurium. (A) Secreted-protein profiles from C. rodentium strain DBS100, which utilizes a LEE (locus of enterocyte effacement)-encoded T3SS. The wild-type (wt) strain was used as a positive control, and ΔescC and ΔetgA mutant strains were used as negative controls. The ΔycbB strain is seen to retain LEE T3SS activity. (B) Secreted-protein profiles from S. Typhimurium strain SL1344, which utilizes an SPI-1 (Salmonella pathogenicity island 1)-encoded T3SS. The wild-type (wt) strain was used as a positive control, and the ΔinvG mutant strain was used as a negative control. The ΔycbB strain is seen to retain SPI-1 T3SS activity. The two ΔycbB lanes in each of panels A and B represent two independent isolates, which should be genetically identical. Lanes MW are molecular weight markers.
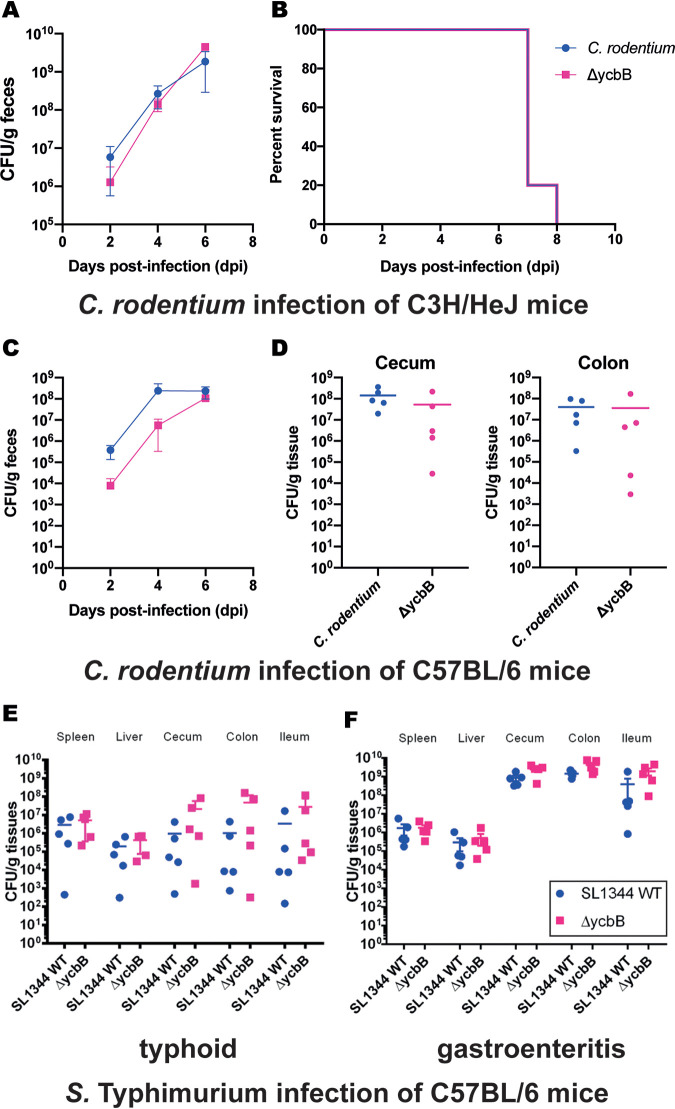
Once it was determined that the role of YcbB in a host environment was independent of the T3SS, mouse infection trials were conducted to see if the more fundamental role of YcbB in strengthening the cell wall under outer envelope stress conditions would be required in an in vivo infection setting. In the first C. rodentium mouse infection model, fecal bacterial shedding postinfection was monitored, as well as the survival of the infected C3H/HeJ mice. Similar levels of bacterial shedding were observed in wild-type C. rodentium-infected and strain ΔycbB-infected mice as the disease progressed (Fig. 5A). The survival curves seen with the two strains were likewise identical, with all mice reaching a humane endpoint within 8 days postinfection (Fig. 5B).
FIG 5.
Infection and colonization of mice by C. rodentium and S. Typhimurium. (A) Bacterial shedding in feces of C3H/HeJ mice (n = 5) were subjected to gavage with ∼3 × 108 CFU of C. rodentium wild-type DBS100 (blue) and ΔycbB mutant (pink). (B) Survival curve of C. rodentium DBS100-infected mice, treated as described for panel A. Mice were assessed daily for weight loss and clinical symptoms, and upon reaching the humane endpoint, mice were euthanized, and this time point was taken as the time of death. (C) Bacterial shedding in feces of C57BL/6 mice (n = 5) were subjected to gavage with ∼3 × 108 CFU of the C. rodentium wild-type strain (blue) and ΔycbB mutant (pink). (D) C. rodentium wild-type DBS100 (blue) and ΔycbB mutant (pink) colonization of C57BL/6 cecum (left panel) and colon (right panel) (n = 5) 6 days postinfection with ∼3 × 108 CFU of bacteria by oral gavage. (E) S. enterica serovar Typhimurium SL1344 wild-type (blue) and ΔycbB mutant (pink) colonization of C57BL/6 mouse organs (n = 5) 3 days postinfection with ∼5 × 107 CFU of bacteria by oral gavage to generate typhoid symptoms. (F) S. enterica serovar Typhimurium SL1344 wild-type (blue) and ΔycbB mutant (pink) colonization of C57BL/6 mouse organs (n = 5) 3 days postinfection with ∼5 × 107 CFU of bacteria by oral gavage. The mice were pretreated with 20 mg streptomycin 1 day before S. Typhimurium infection to generate gastroenteritis symptoms.
As C3H/HeJ mice are quite susceptible to infection by C. rodentium, a second C. rodentium mouse infection model was tested with the infection of more-resilient C57BL/6 mice. As in the C3H/HeJ model, fecal bacterial shedding postinfection was monitored (Fig. 5C). Modest differences were observed at 2 and 4 days postinfection, with the ΔycbB strain-infected mice showing less fecal shedding. By day 6, the levels of shedding in the ΔycbB strain-infected mice had reached the level observed in the wild-type-infected mice. The C57BL/6 mice were not observed to succumb fully to C. rodentium infection; therefore, levels of C. rodentium colonization and attachment in the cecum and colon were determined 6 days postinfection to terminate these experiments (Fig. 5D). Decreases in colonization and attachment of the cecum and colon were observed for the ΔycbB mutant, though the data did not reach statistical significance.
In the S. Typhimurium model of typhoid infection, levels of colonization of C57BL/6 mouse organs were determined at 3 days postinfection. Similar levels of Salmonella were found in wild-type S. Typhimurium-infected and ΔycbB strain-infected mice in all organs assayed (spleen, liver, cecum, colon, and ileum) (Fig. 5E). Likewise, in the S. Typhimurium model of gastroenteritis, levels of colonization of organs from C57BL/6 mice pretreated with streptomycin at 3 days postinfection were determined. Similar levels of Salmonella were found in the wild-type S. Typhimurium-infected mice and the ΔycbB strain-infected mice in all organs assayed (spleen, liver, cecum, colon, and ileum), with the increases in the colonization of the cecum, colon, and ileum expected of the gastroenteritis model holding true for both strains (Fig. 5F).
From the results seen in these mouse models, we conclude that the general role of YcbB in maintenance of the PG layer under conditions of bacterial outer envelope stress is not a contributing factor in the ability of either C. rodentium or S. Typhimurium to establish acute infections in mice.
Implications in development of anti-YcbB therapeutics.
Here, we have shown that YcbB did not act as a general virulence factor in a number of mouse models of bacterial infection. While this does not preclude the possibility that the general role of YcbB in bacterial outer envelope stress response could play a role in EPEC, EHEC, or nontyphoidal Salmonella infections, we believe that such a scenario is unlikely, given the conserved mechanisms of bacterial infections and host responses between the mouse models used and human infections. This leaves the previously discovered role of YcbB in the release of typhoid toxin in S. Typhi (17) as the most attractive role of YcbB from a therapeutic standpoint.
Despite the wealth of research regarding the role of typhoid toxin in the virulence and physiology of S. Typhi, this toxin was discovered only relatively recently (32) within the broader context of research on typhoid infection, which is one of the oldest human diseases on written record. In the decade and a half since that discovery, our understanding of the role of this toxin in the divergent symptoms of infection by S. Typhi in comparison to the majority of other S. enterica serovars, which are limited to causing gastroenteritis, has greatly increased (23, 33–35). Despite the considerable evidence supporting the idea of a role of the typhoid toxin in the pathogenesis of S. Typhi, a recent human challenge study of typhoid infection found little to no role for the toxin in the early stages of S. Typhi infection (36). Due to the limitations imposed on the severity of inflicted infections, for ethical reasons, the authors of that study noted that the typhoid toxin may play a role in more-severe cases of infection or in the development of persistent infection through modulation of the immune response. In addition, those authors noted that the study population was generally Westernized and likely had little prior immune priming, such as may be seen in populations where S. Typhi infection is endemic. Clearly, there is a need to further dissect the role of typhoid toxin in the context of human infection. This provides a further rationale for the development and use of anti-YcbB therapeutic agents (in addition to the existing, albeit nonspecific, anti-YcbB carbapenem antibiotics), which could be used to help dissect the intricacies in the role of the typhoid toxin during infection.
With the structural insights provided in this work, we begin to see trends in the inhibition of these YcbBs. As first seen in the E. coli structure (13), there are remarkably few factors which stabilize these carbapenems in the active site of each YcbB. This leads to the possibility of further design of evolved carbapenem antibiotics which can additionally extend into the active site of YcbB, as has been done with the Ldt enzymes from Mycobacterium tuberculosis (37). In the S. Typhi structure of YcbB, which is acylated with ertapenem, we see additional extension of the C2 group away from the donor site of the enzyme, providing little to no benefit to the inhibitor (Fig. 3). The possibility of extension at the C3 carboxylic acid group could potentially maximize the interaction of the inhibitor with the donor site, while extension at the C5 ethyl alcohol group could possibly be exploited to begin to explore additional inhibition of the adjacent acceptor site.
In addition to the insights provided in this work with respect to the potential evolution of carbapenem antibiotics which would enable further exploration of the active site of YcbB, the expanded understanding of the motion and steric hindrances of the capping loop will be of benefit to the development of novel anti-YcbB agents. There are two potential routes that could be pursued for the development of evolved carbapenems and other anti-YcbB agents. First, one could attempt to avoid interaction with the lower face of the capping loop, perhaps through removal of the C1 methyl group of the various carbapenems, to remove the steric hindrance proposed to have occurred during crystal formation in the C. rodentium YcbB structure. As the capping loop can likely hinge and occlude only so far before steric intervention by the residues along the edge of the active site, removal of the C1 methyl group may reduce binding interference between the capping loop and the drug, resulting in a more favorable binding event. Alternatively, now that there is further understanding of the hinge point for the rotation and movement of the capping loop, one could explore extension of the C1 methyl group to form specific contacts with the lower face of the capping loop. This could provide additional chemical space to explore in the evolution of carbapenem antibiotics to inhibit YcbB.
In this work, we have probed the role of YcbB in the infection of mice by C. rodentium and S. Typhimurium. We show that the general involvement of YcbB in PG reinforcement under conditions of bacterial outer envelope stress does not play a significant role in these models and therefore likely does not play a profound role in E. coli or Salmonella infection of humans. Further exploration is needed to determine if the circumstances where YcbB mediates β-lactam resistance could occur in an infection model. This results in the limited, yet highly specific, potential for anti-YcbB therapeutic agents in treatment of drug-resistant S. Typhi infections. We have determined the structures of both Salmonella Typhi and C. rodentium YcbB and from them observed the conserved structural components of YcbB across a variety of Gram-negative pathogens. Cumulatively, in this work we provide a foundation for the development of novel YcbB-specific antibacterial therapeutics to assist in treatment of S. Typhi infection.
MATERIALS AND METHODS
Cloning and protein expression.
The coding regions of Salmonella Typhimurium and C. rodentium YcbB without their signal peptides (residues 31 and 48 onward, respectively) were cloned into expression vector pET28a with a thrombin cleavable, N-terminal His tag. Expression constructs were transformed into E. coli BL21(DE3) for expression. Cells were cultured in ZYP-5052 autoinduction media for 4 h at 37°C followed by overnight protein expression at 25°C. Cells were pelleted and stored at −80°C until required.
Protein purification.
For purification of S. Typhimurium and C. rodentium YcbB, cell pellets were resuspended in lysis buffer (20 mM HEPES [pH 8.0], 300 mM NaCl, 10% glycerol) and lysed by processing twice with a homogenizer (Avestin) (15 kPa). Cellular debris was pelleted by centrifugation at 125,000 × g for 1 h. The resultant supernatant was loaded onto 10 ml of nickel-nitrilotriacetic acid (Ni-NTA) Superflow resin (Qiagen) and washed with 65 mM imidazole–buffer A (20 mM HEPES [pH 8.0], 300 mM NaCl), and the protein was eluted with 300 mM imidazole–buffer A. One 1-U volume of thrombin was added per mg of protein to remove the N-terminal His tag overnight at 4°C. Samples were purified further by size exclusion chromatography (SEC) with a Superdex 200 column (GE Lifesciences) equilibrated in buffer B (20 mM HEPES [pH 8.0], 150 mM NaCl). Fractions containing purified protein were pooled and concentrated to 9 mg/ml for S. Typhimurium and 8.8 mg/ml for C. rodentium. Protein was frozen rapidly in liquid nitrogen and stored at –80°C until required.
X-ray crystallography and structure determination.
S. Typhi YcbB was crystallized at 20°C by sitting-drop vapor diffusion using 1 μl protein solution (9 mg/ml purified protein–buffer A) and 1 μl of mother liquor (0.1 M Tris [pH 8.5], 0.18 M MgCl2, 17% polyethylene glycol [PEG] 8000, 2% ethanol) with the addition of 1 mM ertapenem (Millipore Sigma). C. rodentium YcbB was crystallized at 20°C by sitting-drop vapor diffusion using 0.2 μl protein solution (8.8 mg/ml purified protein–buffer A) and 0.1 μl of mother liquor (0.16 M calcium acetate, 0.08 M sodium cacodylate [pH 6.5[], 14% [vol/vol] PEG 8000, 20% [vol/vol] glycerol) with the addition of 1 mM ertapenem. Data for S. Typhi ertapenem-YcbB and C. rodentium ertapenem-YcbB were collected on Advanced Photon Source beamline 23-ID-B and Canadian Light Source beamline 08B1-1, respectively. All data sets were processed with XDS (38). For both YcbB data sets reported here, the structures were solved by molecular replacement using Phaser (39). The previously reported structure of E. coli YcbB (PDB identifier [ID] 6NTW [13]) was divided into the following two parts: (i) its capping loop subdomain (residues 424 to 493) and (ii) the remainder of the protein (all modeled residues minus residues 422 to 495). The second part was placed first by manual methods followed by the capping subdomain. For phasing, all atom models were used with no side chain modification, due to the high levels of homology. These structures were then refined using Phenix (40) and Coot (41).
For S. Typhi YcbB, the portions of the N-terminal face of the scaffolding domain and portions of the capping loop opposite the active site were manually trimmed from the phased model, as they could not reliably be fit into the density. The resulting model starts at residue 72; has the unmodeled regions at residues 82 to 82, 107 to 108, 179 to 180, 201 to 204, 213 to 215 (lower portion of the scaffolding domain), 267 to 311, 331 (PG binding domain—disordered loop observed in E. coli YcbB), 451 to 456, 461 to 463, 480, 485 to 490, and 502 to 503 (capping loop, opposite the active site); and ends at residue 609. The model has a MolProbity score of 2.69 and statistics that are in line with deposited models at this resolution (see Fig. S2A in the supplemental material). In addition, the interpretations that we make from this structure are appropriate for the resolution and observed density, with the key observations centering on the global architecture and structural conservation in comparison to the homologous structures of higher resolutions.
For C. rodentium YcbB, the modeled residues are in line with the regions modeled for E. coli YcbB. The resulting model starts at residue 72; has the unmodeled regions at residues 288 to 322 (PG binding domain—disordered loop observed in E. coli YcbB), 512 (loop below active site), and 466 to 467 (capping loop, opposite the active site); and ends at residue 622. The model has a MolProbity score of 2.26 and statistics that are in line with deposited models at this resolution (Fig. S2B). See Table S1 in the supplemental material for data collection and refinement statistics.
Generation of ycbB knockout strains.
The sacB gene-based allelic exchange method and suicide vector pRE112 were used to generate in-frame deletion mutants of ycbB in both S. enterica serovar Typhimurium strain SL1344 and C. rodentium strain DBS100. Two DNA fragments flanking ∼1 kb upstream and downstream of the coding region of ycbB were generated by PCR using the following primers: TATAAGGTACCATAGCGTGCGGTTAAACGTCAACG (forward primer for upstream S. Typhimurium; KpnI site underlined) and ATATTGAATTCCCCTTGCCCCCTGTTTTCG (reverse primer for upstream S. Typhimurium; EcoRI site underlined); TATAAGAATTCATGAAGAAGTTCTGGTAAATATGTTGTCC (forward primer for downstream S. Typhimurium; EcoRI site underlined) and ATATTGAGCTCCGTGAGTAAAATTTGCATCAACG (reverse primer for downstream S. Typhimurium; SacI site underlined); TATAAGAGCTCACGCCAGCAAGCAGCGATACC (forward primer for upstream C. rodentium; SacI site underlined) and ATATTGAATTCTATAGAAAGGCCAGAACCCTGTTGCC (reverse primer for upstream C. rodentium; EcoRI site underlined); and TATAAGAATTCATGAAGACGTTCTGGTCATTATGG (forward primer for downstream C. rodentium; EcoRI site underlined) and ATATTGGTACCGTAACCAGAACTCATCTTCTTTTTCC (reverse primer for C. rodentium; KpnI site underlined). After digestion with KpnI/EcoRI or SacI/EcoRI, respectively, the DNA fragment pairs were subjected to gel purification and cloned into KpnI/SacI-digested pRE112 in a three-way ligation, generating pRE112-ΔST_ycbB or pRE112-ΔCr_ycbB. These suicide vectors were transformed into E. coli strain MFDpir by electroporation and introduced into S. Typhimurium strain SL1344 or C. rodentium strain DBS100 by conjugation. After sucrose selection, colonies resistant to sucrose and sensitive to chloramphenicol were screened for ycbB deletion by PCR.
Secretion assays.
For the type III secretion assay performed for C. rodentium, C. rodentium strains were grown overnight in LB broth at 37°C in a shaker at 225 rpm. The overnight cultures were diluted 1:40 into 3 ml of prewarmed Dulbecco’s modified Eagle’s medium (DMEM) (HyClone) supplemented with 4,500 mg/liter glucose, 4 mM l-glutamine, and 110 mg/liter sodium pyruvate in a 6-well tissue culture plate (Corning Inc.) and grown statically at 37°C for 6 h in a tissue culture incubator containing 5% (vol/vol) CO2 to induce type III secretion.
For Salmonella pathogenicity island 1 (SPI-1) secretion assays for S. enterica serovar Typhimurium, Salmonella strains were grown overnight in LB broth containing 100 μg/ml of streptomycin sulfate at 37°C in a shaker at 225 rpm. The cultures were diluted 1:100 into 4 ml of fresh LB with 100 μg/ml of streptomycin sulfate and grown under the same conditions for 6 h to induce SPI-1 type III secretion.
The bacterial cultures for either C. rodentium or S. Typhimurium were then centrifuged at 16,100 × g for 10 min to pellet the bacteria. The culture supernatant was collected and passed through a Millex-GV 0.22-μm-pore-size filter unit (Millipore) to remove any remaining bacteria, and the secreted proteins were precipitated with trichloroacetic acid (TCA) at a final concentration of 10% (vol/vol). The secreted proteins were then collected by centrifugation at 16,100 × g for 30 min, and the protein pellet was dried in air and dissolved in SDS-PAGE sample buffer, with the residual TCA neutralized with 0.5 μl of saturated Tris. The amount of the sample buffer used to resuspend the bacterial pellet or dissolve the precipitated proteins was normalized according to the A600 values of the cultures to ensure equal loading of the samples. The secreted proteins were analyzed in SDS–12% PAGE and stained with Coomassie blue G250.
C3H/HeJ mouse infections by C. rodentium wild-type strain DBS100 and its isogenic ycbB deletion mutant, the ΔycbB strain.
All mouse experiments were approved by the University of British Columbia (UBC) Animal Care Committee and performed in strict accordance with the guidelines of the Canadian Council on Animal Care and the UBC Animal Care Committee. Six‐week‐old female C3H/HeJ mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in a specific-pathogen‐free facility at UBC. Groups of 5 mice were subjected to oral gavage with ∼3 × 108 CFU of bacteria in 100 μl of overnight cultures of C. rodentium DBS100 grown in LB at 37°C and 225 rpm. Bacterial shedding in stool specimens was monitored by plating dilutions of fecal samples on MacConkey agar every 2 days throughout the infection. Mice were assessed daily for weight loss and clinical symptoms, and, upon reaching the humane endpoint (typically weight loss of 20%, or moribund signs of bloody diarrhea, severe hunching, and/or rectal prolapse), mice were euthanized by isoflurane anesthesia followed by carbon dioxide inhalation, and this time point was taken as the time of death.
C57BL/6 mouse infections by C. rodentium wild-type strain DBS100 and its isogenic ycbB deletion mutant, the ΔycbB strain.
All mouse experiments were approved by the UBC Animal Care Committee and performed in strict accordance with the guidelines of the Canadian Council on Animal Care and the UBC Animal Care Committee. Five‐week‐old female C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in a specific-pathogen‐free facility at UBC. Groups of 5 mice were subjected to oral gavage with ∼3 × 108 CFU of bacteria in 100 μl of overnight cultures of C. rodentium DBS100 grown in LB at 37°C and 225 rpm. Bacterial shedding in stool specimens was monitored by plating dilutions of fecal samples on MacConkey agar every 2 days throughout the infection. Mice were euthanized 6 days postinfection by anesthesia with isoflurane followed by CO2 asphyxiation. Bacterial colonization and attachment to mouse cecum and colon were monitored by plating on MacConkey agar.
C57BL/6 mouse infections by S. enterica serovar Typhimurium and its isogenic ycbB deletion mutant, the ΔycbB strain.
All mouse experiments were approved by the UBC Animal Care Committee and performed in accordance with the ethical requirements of the Canadian Council on Animal Care and the UBC Animal Care Committee. Five-week-old female C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in a specific-pathogen‐free facility at UBC. Two models of Salmonella infections were used, the typhoid model and the gastroenteritis model, which differ in that each mouse was given 20 mg of streptomycin by oral gavage 24 h prior to Salmonella infection in the gastroenteritis model only. S. enterica serovar SL1344 strains were grown overnight in LB containing 100 μl of streptomycin at 37°C and 225 rpm and diluted 10-fold in phosphate-buffered saline (PBS). The mice were then infected with 100 μl of the diluted bacterial cultures containing ∼5 × 107 CFU of bacteria. Mice were euthanized 3 days postinfection by anesthesia with isoflurane followed by CO2 asphyxiation. Mouse organs (cecum, colon, ileum, spleen, and liver) were dissected and collected in 1 ml of sterile PBS and homogenized in a FastPrep homogenizer (MP Biochemicals). Serial dilutions in PBS were plated in LB agar plates containing 100 μg/ml of streptomycin for Salmonella CFU enumeration.
Data availability.
Atomic coordinates for the Salmonella Typhi YcbB-ertapenem and C. rodentium YcbB-ertapenem models have been deposited in the Protein Data Bank with accession codes 7KGN and 7KGM, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jan Burian for thoughtful discussion and advice on microbial genetics.
Research described in this work was performed using beamline 08ID-1 at the Canadian Light Source and beamline 23-ID-B at the Advanced Photon Source. The Canadian Light Source is supported by the Canada Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada; the University of Saskatchewan; the Government of Saskatchewan; Western Economic Diversification Canada; the National Research Council Canada; and the Canadian Institutes of Health Research (CIHR). This research also used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. This work was funded by operating grants to N.C.J.S. from Joint Programming Initiative on Antimicrobial Resistance (JPIAMR)-CIJR and the Howard Hughes International Senior Scholar program and by operating grants to B.B.F. from CIHR. We also acknowledge infrastructure funding from the Canadian Foundation of Innovation and British Columbia Knowledge Development Fund. N.A.C. holds an NSERC PGS D Award, B.B.F. is the UBC Peter Wall Distinguished Professor, and N.C.J.S. is a Tier I Canada Research Chair in Antibiotic Discovery.
N.A.C. contributed to conceptualization, methodology, investigation, analysis, writing (original draft), review, and editing. A.S.-P., S.E.W., and T.B. performed the mouse infections and analyzed the data, W.D. contributed to supervision, methodology, investigation, analysis, and editing. G.C. contributed to investigation, review, and editing. M.V. contributed to investigation. B.B.F. contributed to supervision and funding acquisition. N.C.J.S. contributed to conceptualization, supervision, writing (review and editing), and funding acquisition.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Aoki H, Sakai HI, Kohsaka M, Konomi T, Hosoda J, Iguchi E, Imanaka H, Kubochi Y. 1976. Nocardicin a, a new monocyclic β-lactam antibiotic. I. Discovery, isolation and characterization. J Antibiot (Tokyo) 29:492–500. doi: 10.7164/antibiotics.29.492. [DOI] [PubMed] [Google Scholar]
- 2.Sabe VT, Tolufashe GF, Ibeji CU, Maseko SB, Govender T, Maguire GEM, Lamichhane G, Honarparvar B, Kruger HG. 2019. Identification of potent L,D-transpeptidase 5 inhibitors for Mycobacterium tuberculosis as potential anti-TB leads: virtual screening and molecular dynamics simulations. J Mol Model 25:328. doi: 10.1007/s00894-019-4196-z. [DOI] [PubMed] [Google Scholar]
- 3.Decuyper L, Deketelaere S, Vanparys L, Jukič M, Sosič I, Sauvage E, Amoroso AM, Verlaine O, Joris B, Gobec S, D'hooghe M. 2018. In silico design and enantioselective synthesis of functionalized monocyclic 3-amino-1-carboxymethyl-β-lactams as inhibitors of penicillin-binding proteins of resistant bacteria. Chemistry 24:15254–15266. doi: 10.1002/chem.201801868. [DOI] [PubMed] [Google Scholar]
- 4.Caveney NA, Li FK, Strynadka NC. 2018. Enzyme structures of the bacterial peptidoglycan and wall teichoic acid biogenesis pathways. Curr Opin Struct Biol 53:45–58. doi: 10.1016/j.sbi.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Sobhanifar S, King DT, Strynadka NCJ. 2013. Fortifying the wall: synthesis, regulation and degradation of bacterial peptidoglycan. Curr Opin Struct Biol 23:695–703. doi: 10.1016/j.sbi.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 6.King DT, Wasney GA, Nosella M, Fong A, Strynadka NCJ. 2017. Structural insights into inhibition of Escherichia coli penicillin-binding protein 1B. J Biol Chem 292:979–993. doi: 10.1074/jbc.M116.718403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming A 1929. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzæ. Br J Exp Pathol 10:226–236. [Google Scholar]
- 8.Liu HH, Tomasz A. 1985. Penicillin tolerance in multiply drug-resistant natural isolates of streptococcus pneumoniae. J Infect Dis 152:365–372. doi: 10.1093/infdis/152.2.365. [DOI] [PubMed] [Google Scholar]
- 9.Tomasz A 1986. Penicillin-binding proteins and the antibacterial effectiveness of β-lactam antibiotics. Rev Infect Dis 8(Suppl 3):S260–S278. doi: 10.1093/clinids/8.supplement_3.s260. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Nishino K, Roberts MC, Tolmasky M, Aminov RI, Zhang L. 2015. Mechanisms of antibiotic resistance. Front Microbiol 6:34. doi: 10.3389/fmicb.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M. 2005. A novel peptidoglycan cross-linking enzyme for a β-lactam-resistant transpeptidation pathway. J Biol Chem 280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- 12.Hugonnet J-E, Mengin-Lecreulx D, Monton A, den Blaauwen T, Carbonnelle E, Veckerlé C, Brun YV, van Nieuwenhze M, Bouchier C, Tu K, Rice LB, Arthur M. 2016. Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. Elife 5:e19469. doi: 10.7554/eLife.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caveney NA, Caballero G, Voedts H, Niciforovic A, Worrall LJ, Vuckovic M, Fonvielle M, Hugonnet J-E, Arthur M, Strynadka NCJ. 2019. Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli. Nat Commun 10:1849. doi: 10.1038/s41467-019-09507-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erdemli SB, Gupta R, Bishai WR, Lamichhane G, Amzel LM, Bianchet MA. 2012. Targeting the cell wall of Mycobacterium tuberculosis: structure and mechanism of L,D-transpeptidase 2. Structure 20:2103–2115. doi: 10.1016/j.str.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triboulet S, Dubée V, Lecoq L, Bougault C, Mainardi J-L, Rice LB, Ethève-Quelquejeu M, Gutmann L, Marie A, Dubost L, Hugonnet J-E, Simorre J-P, Arthur M. 2013. Kinetic features of L,D-transpeptidase inactivation critical for β-lactam antibacterial activity. PLoS One 8:e67831. doi: 10.1371/journal.pone.0067831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morè N, Martorana AM, Biboy J, Otten C, Winkle M, Serrano CKG, Montón Silva A, Atkinson L, Yau H, Breukink E, den Blaauwen T, Vollmer W, Polissi A. 2019. Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. mBio 10:e02729-18. doi: 10.1128/mBio.02729-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiger T, Pazos M, Lara-Tejero M, Vollmer W, Galán JE. 2018. Peptidoglycan editing by a specific ld-transpeptidase controls the muramidase-dependent secretion of typhoid toxin. Nat Microbiol 3:1243–1254. doi: 10.1038/s41564-018-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernal-Cabas M, Ayala JA, Raivio TL. 2015. The Cpx envelope stress response modifies peptidoglycan cross-linking via the l,d-transpeptidase LdtD and the novel protein YgaU. J Bacteriol 197:603–614. doi: 10.1128/JB.02449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delhaye A, Collet JF, Laloux G. 2016. Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio 7:e00047-16. doi: 10.1128/mBio.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-del Portillo F 2020. Building peptidoglycan inside eukaryotic cells: a view from symbiotic and pathogenic bacteria. Mol Microbiol 113:613–626. doi: 10.1111/mmi.14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC. 2018. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559:617–621. doi: 10.1038/s41586-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galán JE 2016. Typhoid toxin provides a window into typhoid fever and the biology of Salmonella typhi. Proc Natl Acad Sci U S A 113:6338–6344. doi: 10.1073/pnas.1606335113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanaway JD, Reiner RC, Blacker BF, Goldberg EM, Khalil IA, Troeger CE, Andrews JR, Bhutta ZA, Crump JA, Im J, Marks F, Mintz E, Park SE, Zaidi AKM, Abebe Z, Abejie AN, Adedeji IA, Ali BA, Amare AT, Atalay HT, Avokpaho EFGA, Bacha U, Barac A, Bedi N, Berhane A, Browne AJ, Chirinos JL, Chitheer A, Dolecek C, El Sayed Zaki M, Eshrati B, Foreman KJ, Gemechu A, Gupta R, Hailu GB, Henok A, Hibstu DT, Hoang CL, Ilesanmi OS, Iyer VJ, Kahsay A, Kasaeian A, Kassa TD, Khan EA, Khang YH, Magdy Abd El Razek H, Melku M, Mengistu DT, et al. . 2019. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis 19:369–381. doi: 10.1016/S1473-3099(18)30685-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogt SL, Scholz R, Peng Y, Guest RL, Scott NE, Woodward SE, Foster LJ, Raivio TL, Finlay BB. 2019. Characterization of the Citrobacter rodentium Cpx regulon and its role in host infection. Mol Microbiol 111:700–716. doi: 10.1111/mmi.14182. [DOI] [PubMed] [Google Scholar]
- 25.Ehrhardt K, Steck N, Kappelhoff R, Stein S, Rieder F, Gordon IO, Boyle EC, Braubach P, Overall CM, Finlay BB, Grassl GA. 2019. Persistent Salmonella enterica serovar typhimurium infection induces protease expression during intestinal fibrosis. Inflamm Bowel Dis 25:1629–1643. doi: 10.1093/ibd/izz070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Browne AJ, Kashef Hamadani BH, Kumaran EAP, Rao P, Longbottom J, Harriss E, Moore CE, Dunachie S, Basnyat B, Baker S, Lopez AD, Day NPJ, Hay SI, Dolecek C. 2020. Drug-resistant enteric fever worldwide, 1990 to 2018: a systematic review and meta-analysis. BMC Med 18:1. doi: 10.1186/s12916-019-1443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nørgaard SM, Jensen CS, Aalestrup J, Vandenbroucke-Grauls CMJE, De Boer MGJ, Pedersen AB. 2019. Choice of therapeutic interventions and outcomes for the treatment of infections caused by multidrug-resistant gram-negative pathogens: a systematic review. Antimicrob Resist Infect Control 8:170. doi: 10.1186/s13756-019-0624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianchet MA, Pan YH, Basta LAB, Saavedra H, Lloyd EP, Kumar P, Mattoo R, Townsend CA, Lamichhane G. 2017. Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2by biapenem and tebipenem. BMC Biochem 18:8. doi: 10.1186/s12858-017-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gokulan K, Khare S, Cerniglia CE, Foley SL, Varughese KI. 2018. Structure and inhibitor specificity of L,D-transpeptidase (LdtMt2) from Mycobacterium tuberculosis and antibiotic resistance: calcium binding promotes dimer formation. AAPS J 20:44. doi: 10.1208/s12248-018-0193-x. [DOI] [PubMed] [Google Scholar]
- 30.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, Strynadka NCJ, Finlay BB. 2017. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol 15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 31.Worrall LJ, Hong C, Vuckovic M, Deng W, Bergeron JRC, Majewski DD, Huang RK, Spreter T, Finlay BB, Yu Z, Strynadka NCJ. 2016. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature 540:597–601. doi: 10.1038/nature20576. [DOI] [PubMed] [Google Scholar]
- 32.Haghjoo E, Galán JE. 2004. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A 101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song J, Gao X, Galán JE. 2013. Structure and function of the Salmonella Typhi chimaeric A 2 B 5 typhoid toxin. Nature 499:350–354. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Bel Belluz L, Guidi R, Pateras IS, Levi L, Mihaljevic B, Rouf SF, Wrande M, Candela M, Turroni S, Nastasi C, Consolandi C, Peano C, Tebaldi T, Viero G, Gorgoulis VG, Krejsgaard T, Rhen M, Frisan T. 2016. The typhoid toxin promotes host survival and the establishment of a persistent asymptomatic infection. PLoS Pathog 12:e1005528. doi: 10.1371/journal.ppat.1005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang YA, Lee S, Zhao J, Thompson AJ, McBride R, Tsogtbaatar B, Paulson JC, Nussinov R, Deng L, Song J. 2018. In vivo tropism of Salmonella Typhi toxin to cells expressing a multiantennal glycan receptor. Nat Microbiol 3:155–163. doi: 10.1038/s41564-017-0076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibani MM, Jones E, Barton A, Jin C, Meek J, Camara S, Galal U, Heinz E, Rosenberg-Hasson Y, Obermoser G, Jones C, Campbell D, Black C, Thomaides-Brears H, Darlow C, Dold C, Silva-Reyes L, Blackwell L, Lara-Tejero M, Jiao X, Stack G, Blohmke CJ, Hill J, Angus B, Dougan G, Galán J, Pollard AJ. 2019. Investigation of the role of typhoid toxin in acute typhoid fever in a human challenge model. Nat Med 25:1082–1088. doi: 10.1038/s41591-019-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar P, Kaushik A, Lloyd EP, Li S-G, Mattoo R, Ammerman NC, Bell DT, Perryman AL, Zandi TA, Ekins S, Ginell SL, Townsend CA, Freundlich JS, Lamichhane G. 2017. Non-classical transpeptidases yield insight into new antibacterials. Nat Chem Biol 13:54–61. doi: 10.1038/nchembio.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates for the Salmonella Typhi YcbB-ertapenem and C. rodentium YcbB-ertapenem models have been deposited in the Protein Data Bank with accession codes 7KGN and 7KGM, respectively.