Two multidrug-resistant (MDR) mcr-1-harboring Klebsiella pneumoniae isolates from patients with urinary tract infections and one MDR Klebsiella quasipneumoniae isolate from a patient with bloodstream infection were identified to carry tmexCD1-toprJ1. The addition of the efflux pump inhibitor reduced the tigecycline MIC against all three isolates by 8- to 16-fold. pKQBSI104-1 was transferred from K. quasipneumoniae to Escherichia coli J53 via conjugation.
KEYWORDS: Klebsiella spp., efflux pump, tmexCD1-toprJ1, tigecycline, antimicrobial resistance
ABSTRACT
Two multidrug-resistant (MDR) mcr-1-harboring Klebsiella pneumoniae isolates from patients with urinary tract infections and one MDR Klebsiella quasipneumoniae isolate from a patient with bloodstream infection were identified to carry tmexCD1-toprJ1. The addition of the efflux pump inhibitor reduced the tigecycline MIC against all three isolates by 8- to 16-fold. pKQBSI104-1 was transferred from K. quasipneumoniae to Escherichia coli J53 via conjugation. The tmexCD1-toprJ1-carrying plasmids pKP15ZE495-1 (102,569 bp) and pKQBSI104-1 (121,996 bp) were completely sequenced and analyzed.
INTRODUCTION
Klebsiella pneumoniae is a clinically important species that causes nosocomial infections such as septicemia, pneumonia, urinary tract infection, surgical site infection, and soft tissue infection (1, 2). In comparison, Klebsiella quasipneumoniae is an emerging pathogenic species that is frequently detected at infection sites in humans with bloodstream infection (BSI) (3). However, K. quasipneumoniae and K. pneumoniae are indistinguishable via standard laboratory methods and can be discriminated by whole-genome sequencing (WGS) (4, 5). The treatment of these pathogens is significantly challenging for clinicians due to the rapid development of antimicrobial resistance (AMR) associated with Klebsiella strains.
Tigecycline is considered one of the last-resort treatments against human infections caused by multidrug-resistant (MDR) Enterobacteriaceae (6). The mechanism of tigecycline resistance has been associated with the overexpression of nonspecific active efflux pumps or mutations within the drug-binding sites in the ribosome (7). To date, there have been reports of plasmid-mediated tigecycline resistance determinants, including mutations in the plasmid-mediated efflux pump gene tet(A) (8) and carriage of the tet(X3) and tet(X4) genes (6, 9). Recently, a novel plasmid-harbored resistance-nodulation-division (RND) efflux pump gene cluster, tmexCD1-toprJ1, which confers multidrug resistance, including resistance to tigecycline, was identified from MDR K. pneumoniae isolates from chickens and patients in China (10, 11). However, since the tmexCD1-toprJ1 genes were rarely found in human clinical isolates (10), the characteristics of tmexCD1-toprJ1-harboring plasmids from clinical isolates are not fully understood. Here, we identified three tmexCD1-toprJ1-positive Klebsiella isolates from inpatients and two tmexCD1-toprJ1-harboring plasmids, which were identified from an mcr-1-harboring K. pneumoniae and an MDR K. quasipneumoniae isolate, respectively.
From routine monitoring of clinical Enterobacterales isolates conferring tigecycline resistance, we identified 23 tigecycline-resistant isolates, including 22 Klebsiella species isolates and 1 Enterobacter cloacae isolate. By PCR using specific primers (see Table S1 in the supplemental material), 3 of the 23 isolates carried tmexCD1-toprJ1. These three isolates (KQBSI104, KP15ZE215, and KP15ZE495) were retrospectively collected from two Chinese hospitals from 2012 to 2017. Strains KP15ZE215 and KP15ZE495 were isolated from the same hospital. The sequence type 3447 (ST3447) K. pneumoniae strain KP15ZE215 was recovered in July 2012 from urine samples of a 22-year-old woman who was hospitalized for 21 days because of a urinary tract infection. The ST3447 K. pneumoniae strain KP15ZE495 was isolated in June 2015 from urine samples of a 49-year-old woman who was suffering from urinary tract infection and hospitalized for 8 days. The K. quasipneumoniae strain KQBSI104 was isolated in 2017 from the blood samples of a 25-year-old female patient who suffered from BSI (Table 1).
TABLE 1.
Characteristics of the isolatesa
| Characteristic | Value |
||||
|---|---|---|---|---|---|
| KP15ZE215 | KP15ZE495 | KQBSI104 | Transconjugant | E. coli J53Azr | |
| Source | Human | Human | Human | ||
| Isolation site | Urine | Urine | Blood | ||
| Yr of isolation | 2012 | 2015 | 2017 | ||
| MLST type | 3447 | 3447 | |||
| Plasmid replicon types | FIB, FII, FIA, X4 | FIB, FII, FIA, X4 | FIB, FII, HI1B | ||
| MIC (MIC with NMP) (μg/ml) | |||||
| Tigecycline (+NMP) | 32 (4) | 32 (4) | 16 (1) | 4 (0.25) | 0.25 (0.125) |
| Tetracycline (+NMP) | 64 (16) | 64 (16) | >128 (32) | 128 (32) | ≤1 (≤1) |
| Chloramphenicol (+NMP) | 64 (16) | 64 (16) | >128 (64) | 128 (32) | 8 (4) |
| Ciprofloxacin (+NMP) | >256 (256) | >256 (256) | 32 (16) | 32 (16) | ≤0.03 (≤0.03) |
| Colistin (+NMP) | 64 (64) | 64 (64) | 1 (≤0.25) | 0.25 (≤0.25) | 0.25 (0.25) |
| Aztreonam (+NMP) | 128 (64) | 128 (128) | 64 (64) | 16 (16) | 0.25 (0.25) |
| Cefepime (+NMP) | 128 (64) | 128 (64) | 32 (32) | 32 (32) | ≤0.5 (≤0.5) |
| Ceftazidime (+NMP) | 64 (32) | 64 (32) | 32 (32) | 32 (32) | ≤1 (≤1) |
| Gentamicin (+NMP) | >256 (>256) | >256 (>256) | >256 (>256) | >256 (>256) | 1 (2) |
| Amikacin (+NMP) | >256 (>256) | >256 (>256) | >256 (>256) | >256 (>256) | 1 (2) |
| Cefotaxime (+NMP) | 128 (64) | 128 (128) | 64 (64) | 16 (32) | ≤1 (≤1) |
| Trimethoprim-sulfamethoxazole (+NMP) | 2 (0.5) | 2 (0.5) | >16 (>16) | >16 (>16) | ≤0.25 (≤0.25) |
| Imipenem | 1 | 2 | 0.5 | 0.5 | 0.5 |
| Meropenem | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
MLST, multilocus sequence typing. Note that 1-(1-naphthylmethyl)-piperazine (NMP) was added at a concentration of 100 μg/ml.
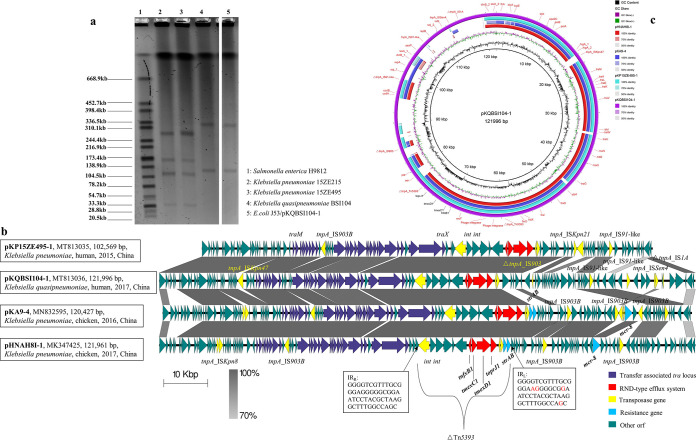
S1 nuclease digestion and pulsed-field gel electrophoresis (S1-PFGE) were used to identify the number and size of the plasmids in tmexCD1-toprJ1-positive isolates (12). The results showed that both KP15ZE215 and KP15ZE495 carried four plasmids with sizes ranging from ∼33 kb to ∼280 kb, while strain KQBSI104 harbored two plasmids with sizes of ∼120 kb and ∼320 kb. The tmexCD1-toprJ1-carrying plasmid (pKQBSI104-1) was successfully transferred from the donor strain KQBSI104 into an Escherichia coli J53 strain via conjugation experiments (13). However, the tmexCD1-toprJ1 plasmids in KP15ZE215 and KP15ZE495 failed to be transferred into a recipient cell via conjugation, transformation, or electroporation. Furthermore, S1-PFGE confirmed that the other plasmid within KQBSI104 was cotransferred with pKQBSI104-1 to the recipient strain (Fig. 1a).
FIG 1.
Characteristics of tmexCD1-toprJ1-positive isolates and associated plasmids. (a) S1-PFGE map of K. pneumoniae KP15ZE215, K. pneumoniae KP15ZE495, K. quasipneumoniae KQBSI104, and E. coli transconjugant J53/pKQBSI104-1. (b) Detailed comparison of linear maps of four tmexCD1-toprJ1-carrying plasmids. Two recently identified plasmids, pHNAH8I-1 (GenBank accession number MK347425) and pKA9-4 (GenBank accession number MN832595), were used as references. Dark-gray shading indicates homologous regions. Arrows show the direction of transcription of open reading frames (ORFs). Genes, mobile elements, and other features are colored based on functional classification. The tmexCD1-toprJ1 and resistance genes are marked. The locations of SNPs in IRL of Tn5393 are marked in red. The figure is drawn to scale. (c) Circular genetic map of four tmexCD1-toprJ1-carrying plasmids, pHNAH8I-1 (GenBank accession number MK347425), pKA9-4 (GenBank accession number MN832595), pKP15ZE495-1 (GenBank accession number MT813035), and pKQBSI104-1 (GenBank accession number MT813036). The map was drawn using BLAST Ring Image Generator (BRIG).
Susceptibilities to tetracycline, chloramphenicol, ciprofloxacin, aztreonam, cefepime, ceftazidime, gentamicin, amikacin, cefotaxime, trimethoprim-sulfamethoxazole, imipenem, and meropenem were determined for KQBSI104, KP15ZE215, and KP15ZE495 by the agar dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (14). In addition, the MICs of tigecycline and colistin were determined by a broth dilution method according to the guidelines of EUCAST. MIC determinations were also carried out with a fixed concentration (100 μg/ml) of the efflux pump inhibitor 1-(1-naphthylmethyl)-piperazine (NMP) against those antibiotics among all the tmexCD1-toprJ1-positive strains. E. coli ATCC 25922, E. coli EC600, and E. coli J53 were used as control strains for efflux pump inhibition (Table S2). From the MIC results, all isolates tested were determined to be MDR (Table 1). With the exceptions of trimethoprim-sulfamethoxazole and carbapenems, strains KP15ZE215 and KP15ZE495 were resistant to all the tested antimicrobial agents, whereas strain KQBSI104 exhibited resistance to all antibiotics except for colistin and carbapenems. KP15ZE215, KP15ZE495, and KQBSI104 showed resistance to tigecycline with MICs of 32 μg/ml, 32 μg/ml, and 16 μg/ml, respectively. In the presence of the efflux pump inhibitor NMP, there was a significant reduction in the isolates’ MICs of tigecycline, tetracycline, and chloramphenicol. The addition of NMP reduced the tigecycline MICs in all three isolates by 8- to 16-fold. The resistance to tigecycline was conferred with an MIC of 4 μg/ml in the E. coli J53 transconjugant strain (Table 1). This was in agreement with a previous study showing that the transfer of a tmexCD1-toprJ1-carrying plasmid leads to an 8- to 32-fold increase of the tigecycline MIC (10).
After extracting the genomic DNA and plasmids of the tmexCD1-toprJ1-positive isolates, we constructed DNA libraries with 350-bp paired-end fragments and performed sequencing using an Illumina HiSeq 2000 platform. The sequencing reads were assembled into contigs using SPAdes version 3.10 (15). To determine the complete sequences of tmexCD1-toprJ1 plasmids, we combined the sequencing data from the genomic DNA and the plasmids and closed predicted gaps within the sequences by PCR and Sanger sequencing using primers listed in Table S3. Acquired antibiotic resistance genes (ARGs) were identified in the ResFinder database (16). Insertion sequence (IS) elements were confirmed by searching in ISFinder. We found that both strains KP15ZE215 and KP15ZE495 carried the plasmid-mediated colistin resistance gene mcr-1.1 (Table S4). Furthermore, considering that these two isolates have the same STs, plasmid components, and resistome, we conclude that these two isolates are clonally related. These data indicate the potential dissemination of tmexCD1-toprJ1-positive K. pneumoniae and extend the period of its isolation from 2014 to 2018 (10, 11) to 2012 to 2015. We obtained complete sequences of tmexCD1-toprJ1-harboring plasmids for KP15ZE495 and KQBSI104, named pKP15ZE495-1 (102,569 bp) and pKQBSI104-1 (121,996 bp), respectively. pKQBSI104-1 contains FIB and FII replicons and harbors 134 open reading frames (ORFs), while pKP15ZE495-1 contains FIA and FII replicons and harbors 117 ORFs (Fig. 1b). In pKP15ZE495-1 and pKQBSI104-1, the tmexCD1-toprJ1 gene cluster (6,489 bp) was colocated with two genes encoding site-specific integrases (int) and two hypothetical protein-encoding genes. This 15,261-bp sequence was inserted into the transposon Tn5393 at bp +226 within tnpA_Tn5393. A BLAST comparison of plasmids found in this study with two recently identified tmexCD1-toprJ1-harboring plasmids, pHNAH8I-1 (GenBank accession number MK347425) (10) and pKA9-4 (GenBank accession number MN832595) (11), was then performed. They shared sequence identities across the backbone module, while significant differences were observed in the regions of rearrangement associated with multiple IS elements (Fig. 1c). The tmexCD1-toprJ1 gene cluster in plasmids from this study showed 100% nucleotide identity to those in pHNAH8I-1 and pKA9-4. Among these four plasmids, the tmexCD1-toprJ1 genes were inserted into tnpA_Tn5393 at the same position. The intact inverted repeat right (IRR) of Tn5393 was found in all four plasmids, while the inverted repeat left (IRL) sequence was found only in pHNAH8I-1 and pKA9-4, with a difference of four single nucleotide polymorphisms (SNPs) compared to Tn5393 IRL. Additionally, pHNAH8I-1 and pKA9-4 harbored the colistin resistance gene mcr-8 and the streptomycin resistance genes strA and strB, which were absent in the plasmids isolated in this study (Fig. 1b). Furthermore, an intact IS903B element (1,057 bp; IS5 family) was located within the tra locus in pHNAH8I-1, pKA9-4, and pKP15ZE495-1. However, an intact ISKpn47 element (IS256 family) was found only in pKQBSI104-1, causing a 1,317-bp difference compared to the other three plasmids. Compared to our plasmids, pHNAH8I-1 and pKA9-4 harbored an intact ISKpn8 element (1,443 bp; IS3 family), which was located close to the psiAB genes. These data indicate that tmexCD1-toprJ1-harboring plasmids are capable of integrating ARGs to form MDR plasmids.
In conclusion, we identified tmexCD1-toprJ1-carrying plasmid pKQBSI104-1 in an MDR clinical K. quasipneumoniae isolate and pKP15ZE495-1 in an mcr-1.1-harboring clinical K. pneumoniae isolate. The transferability of a tmexCD1-toprJ1-carrying plasmid and its potential to form MDR plasmids lead to difficulties of treatment options for MDR Klebsiella infections, representing a threat to public health.
Data availability.
The nucleotide sequences of pKP15ZE495-1 and pKQBSI104-1 have been submitted to GenBank and assigned accession numbers MT813035 and MT813036, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant numbers 81722030, 81830103, and 81902123), the National Key Research and Development Program (grant number 2017ZX10302301), the Guangdong Natural Science Foundation (grant number 2017A030306012), the Project of High-Level Health Teams of Zhuhai in 2018 (The Innovation Team for Antimicrobial Resistance and Clinical Infection), the 111 Project (grant number B12003), the Open Project of the Key Laboratory of Tropical Disease Control (Sun Yat-sen University), the Ministry of Education (grant numbers 2020kfkt04 and 2020kfkt07), the China Postdoctoral Science Foundation (grant number 2019M653192), and the Science, Technology, & Innovation Commission of Shenzhen Municipality (JCYJ20190807151601699).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Herridge WP, Shibu P, O’Shea J, Brook TC, Hoyles L. 2020. Bacteriophages of Klebsiella spp., their diversity and potential therapeutic uses. J Med Microbiol 69:176–194. doi: 10.1099/jmm.0.001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee W-H, Choi H-I, Hong S-W, Kim K-S, Gho YS, Jeon SG. 2015. Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Exp Mol Med 47:e183. doi: 10.1038/emm.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imai K, Ishibashi N, Kodana M, Tarumoto N, Sakai J, Kawamura T, Takeuchi S, Taji Y, Ebihara Y, Ikebuchi K, Murakami T, Maeda T, Mitsutake K, Maesaki S. 2019. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japan, 2014–2017. BMC Infect Dis 19:946. doi: 10.1186/s12879-019-4498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long SW, Linson SE, Saavedra MO, Cantu C, Davis JJ, Brettin T, Olsen RJ. 2017. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2:e00290-17. doi: 10.1128/mSphereDirect.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, Shen Y, Wang G, Sun L, Lei L, Lv Z, Li Y, Pang M, Wang L, Sun Q, Fu Y, Song H, Hao Y, Shen Z, Wang S, Chen G, Wu C, Shen J, Wang Y. 2019. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol 4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, Cai Y, Liu X, Bai N, Liang B, Wang R. 2013. The emergence of clinical resistance to tigecycline. Int J Antimicrob Agents 41:110–116. doi: 10.1016/j.ijantimicag.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Du XX, He F, Shi QC, Zhao F, Xu J, Fu Y, Yu YS. 2018. The rapid emergence of tigecycline resistance in blaKPC-2 harboring Klebsiella pneumoniae, as mediated in vivo by mutation in tetA during tigecycline treatment. Front Microbiol 9:648. doi: 10.3389/fmicb.2018.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsberg KJ, Patel S, Wencewicz TA, Dantas G. 2015. The tetracycline destructases: a novel family of tetracycline-inactivating enzymes. Chem Biol 22:888–897. doi: 10.1016/j.chembiol.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv L, Wan M, Wang C, Gao X, Yang Q, Partridge SR, Wang Y, Zong Z, Doi Y, Shen J, Jia P, Song Q, Zhang Q, Yang J, Huang X, Wang M, Liu J-H. 2020. Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. mBio 11:e02930-19. doi: 10.1128/mBio.02930-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun S, Gao H, Liu Y, Jin L, Wang R, Wang X, Wang Q, Yin Y, Zhang Y, Wang H. 2020. Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae. Emerg Microbes Infect 9:1102–1113. doi: 10.1080/22221751.2020.1768805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirichote P, Hasman H, Pulsrikarn C, Schonheyder HC, Samulioniene J, Pornruangmong S, Bangtrakulnonth A, Aarestrup FM, Hendriksen RS. 2010. Molecular characterization of extended-spectrum cephalosporinase-producing Salmonella enterica serovar Choleraesuis isolates from patients in Thailand and Denmark. J Clin Microbiol 48:883–888. doi: 10.1128/JCM.01792-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang YC, Tang HL, Liao YC, Chiou CS, Chen YT, Chiang MK, Lu MC, Lai YC. 2019. Cocarriage of distinct blaKPC-2 and blaOXA-48 plasmids in a single sequence type 11 carbapenem-resistant Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 63:e02282-18. doi: 10.1128/AAC.02282-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; 28th informational supplement. M100-S28 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequences of pKP15ZE495-1 and pKQBSI104-1 have been submitted to GenBank and assigned accession numbers MT813035 and MT813036, respectively.