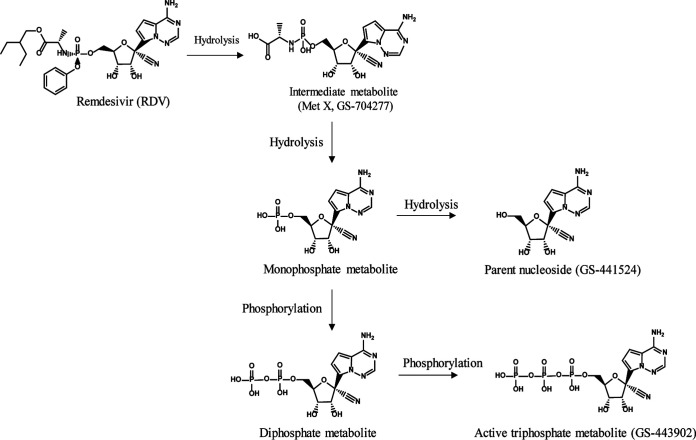
Remdesivir (RDV, GS-5734), the first FDA-approved antiviral for the treatment of COVID-19, is a single diastereomer monophosphoramidate prodrug of an adenosine analogue. It is intracellularly metabolized into the active triphosphate form, which in turn acts as a potent and selective inhibitor of multiple viral RNA polymerases.
KEYWORDS: COVID-19, SARS-CoV-2, antiviral agents, mitochondria, mitochondrial respiration, nucleoside analogs, remdesivir, toxicity
ABSTRACT
Remdesivir (RDV, GS-5734), the first FDA-approved antiviral for the treatment of COVID-19, is a single diastereomer monophosphoramidate prodrug of an adenosine analogue. It is intracellularly metabolized into the active triphosphate form, which in turn acts as a potent and selective inhibitor of multiple viral RNA polymerases. RDV has broad-spectrum activity against members of the coronavirus family, such as SARS-CoV-2, SARS-CoV, and MERS-CoV, as well as filoviruses and paramyxoviruses. To assess the potential for off-target toxicity, RDV was evaluated in a set of cellular and biochemical assays. Cytotoxicity was evaluated in a set of relevant human cell lines and primary cells. In addition, RDV was evaluated for mitochondrial toxicity under aerobic and anaerobic metabolic conditions, and for the effects on mitochondrial DNA content, mitochondrial protein synthesis, cellular respiration, and induction of reactive oxygen species. Last, the active 5′-triphosphate metabolite of RDV, GS-443902, was evaluated for potential interaction with human DNA and RNA polymerases. Among all of the human cells tested under 5 to 14 days of continuous exposure, the 50% cytotoxic concentration (CC50) values of RDV ranged from 1.7 to >20 μM, resulting in selectivity indices (SI, CC50/EC50) from >170 to 20,000, with respect to RDV anti-SARS-CoV-2 activity (50% effective concentration [EC50] of 9.9 nM in human airway epithelial cells). Overall, the cellular and biochemical assays demonstrated a low potential for RDV to elicit off-target toxicity, including mitochondria-specific toxicity, consistent with the reported clinical safety profile.
INTRODUCTION
Nucleoside/tide analogs have played a key role in the treatment of viral infections caused by DNA viruses such as herpesvirus and hepatitis B virus, as well as RNA viruses such as human immunodeficiency virus (HIV) and hepatitis C virus (HCV) (1, 2). Studies over the past several decades have significantly expanded our knowledge on the potential off-target effects for this class of compounds, including inhibition of host nucleic acid polymerases such as human mitochondrial DNA polymerase γ (POL γ) (3) and mitochondrial RNA polymerase (POLRMT) (4, 5), as well as perturbation of nucleotide metabolism, mitochondrial respiration, and deoxynucleoside triphosphate/nucleoside triphosphate (dNTP/NTP) pools (6). Importantly, suitable methods to evaluate the potential for these types of toxicities are currently available.
Remdesivir (RDV, GS-5734) is a single diastereomer monophosphoramidate prodrug of an adenosine analog (Fig. 1). Remdesivir has broad-spectrum activity against coronaviruses (SARS-CoV-2, SARS-CoV, and MERS-CoV) (7, 8), filoviruses (Ebola virus [EBOV] and Marburg virus) (9, 10), and paramyxoviruses (respiratory syncytial virus [RSV], Nipah virus, and Hendra virus) (11, 12). RDV was the first FDA-approved antiviral for the treatment of COVID-19. Its safety has been evaluated in two phase I studies in healthy volunteers (13) and multiple phase III studies in Ebola-infected (14) and SARS-CoV-2-infected patients (15–17). These studies demonstrated RDV as an effective antiviral agent with a favorable benefit/risk profile. RDV is intracellularly metabolized to its active triphosphate form (GS-443902) (Fig. 1), which in turn acts as a potent and selective inhibitor of multiple viral RNA polymerases. Additional metabolites are formed both intracellularly and systemically in plasma, including alanine metabolite (GS-704277), parent nucleoside (GS-441524), and mono- and diphosphate metabolites. To assess the potential off-target toxicity of RDV, the drug and its metabolites, including parent nucleoside analog (GS-441524), intermediate metabolite MetX (GS-704277), and active triphosphate metabolite (GS-443902) (Fig. 1), were characterized in a broad panel of cellular and biochemical assays.
FIG 1.
Structures of RDV and major metabolites.
RESULTS
Evaluation of cytotoxicity.
Cytotoxicity in human cell lines and primary cells. The cytotoxicity of RDV and GS-441524 was tested in four immortalized human cell lines (laryngeal, hepatoma, prostate, and lymphoblastoid transformed cell lines) and in seven primary human cell types, including primary human hepatocytes (PHH), primary renal proximal tubule epithelial cells (RPTECs), quiescent and stimulated human peripheral blood mononuclear cells (PBMCs), and human hematopoietic progenitor cells, including erythroid, myeloid, and megakaryoid progenitors (Table 1). These cells were chosen for their reported high sensitivity to or potential organ toxicity associated with nucleoside/tide analogs (6). ATP level was used as an indication of cell viability. After 5 to 14 days of continuous exposure to RDV, the CC50 values ranged from 1.7 to > 20 μM, resulting in selectivity indices (SI, CC50/EC50) from >170 to 20,000 with respect to RDV anti-SARS-CoV-2 activity (EC50 of 9.9 nM in human airway epithelial cells [HAE]) (18). Among the four cell lines tested, the MT-4 cell line was the most sensitive toward RDV and GS-441524, consistent with reports using other nucleoside/tide analogues (5, 19). Among the seven primary cell types tested, PHH showed the lowest CC50 toward RDV, suggesting that PHHs are susceptible to RDV-mediated toxicity.
TABLE 1.
In vitro cytotoxicity of RDV and GS-441524 in human cell lines and primary human cells after a 5 to 14 day treatment
| Cell type | CC50 (μM)a,b |
||
|---|---|---|---|
| RDV (SI)c | GS-441524 | Positive controld | |
| Cell lines | |||
| HEp-2 | 6.0 ± 1.5 (600) | >100 | 0.53 ± 0.10 |
| HepG2 | 3.7 ± 0.2 (370) | >100 | 0.73 ± 0.01 |
| PC-3 | 8.9 ± 1.6 (890) | >100 | 0.52 ± 0.11 |
| MT-4 | 1.7 ± 0.4 (170) | 69 ± 26 | 0.12 ± 0.03 |
| Primary cells | |||
| PHH | 2.5 ± 0.6 (250) | >100 | 1.60 ± 0.01 |
| RPTEC | 12.9 ± 6.2 (12,900) | >100 | 0.85 ± 0.01 |
| Quiescent PBMC | >20 (>20,000) | >100 | 5.05 ± 0.01 |
| Stimulated PBMC | 14.8 ± 5.8 (14,800) | >100 | 1.10 ± 0.01 |
| Erythroid progenitors | 8.5 ± 1.9 (850) | 13.9 ± 1.3 | 3.2 ± 1.6 |
| Myeloid progenitors | 5.1 ± 2.3 (510) | 11.7 ± 9.6e | 2.2 ± 0.6 |
| Megakaryoid progenitors | 4.9 ± 2.5 (490) | 9.6 ± 2.4 | 2.3 ± 1.7 |
All CC50 values represent the average ± SD of three or more independent experiments. Cells from three different donors were tested in the primary cell assays. The compounds were refreshed every other day during the 5-day PHH study.
Treament duration: all studies were 5 days except studies for the three human hematopoietic progenitor cells, where an 11- to 14-day treatment was used.
Selectivity index (SI) = CC50/EC50, where the EC50 value of 10 nM against SARS-CoV-2 was used (18).
Positive control: puromycin was used in all assays except the three human hematopoietic progenitor cells, where 5-fluorouracil was used as a positive control. CC50 values of control compounds are consistent with historical values.
The myeloid CC50 values for GS-441524 tested against cells from three different donors were 6.37, 22.70, and 5.93 μM, respectively.
The parent nucleoside GS-441524 showed no cytotoxicity at up to 100 μM in any of the cell lines tested, except in MT-4 (CC50 = 69 ± 26 μM). Among the primary human cells, GS-441524 showed no toxicity at the highest concentration tested (100 μM), except in the three human hematopoietic progenitor cells (CC50 = 9.6 to 13.9 μM after 11 to 14 days of exposure).
Effect on production of reactive oxygen species. Reactive oxygen species (ROS), including peroxides, superoxides, and hydroxyl radicals, are natural by-products of normal cell metabolism and have important roles in cell signaling and homeostasis. However, excessive production of ROS can be induced by drugs and may result in cellular damage and cell death (20). In this study, we evaluated the potential effects of RDV on ROS generation in HepG2 cells, a human hepatic cell line. After a 24-h incubation, RDV-treated cells showed no significant increase in ROS levels at concentrations up to 50 μM, and a 60% increase at 100 μM (P = 0.042 compared with the DMSO control) accompanied by a parallel 60% decrease in cell viability, indicating that ROS is unlikely to be the driver of cytotoxicity (Fig. 2). In comparison, the control compound menadione showed a >3-fold increase in ROS levels after a 30-minute incubation.
FIG 2.
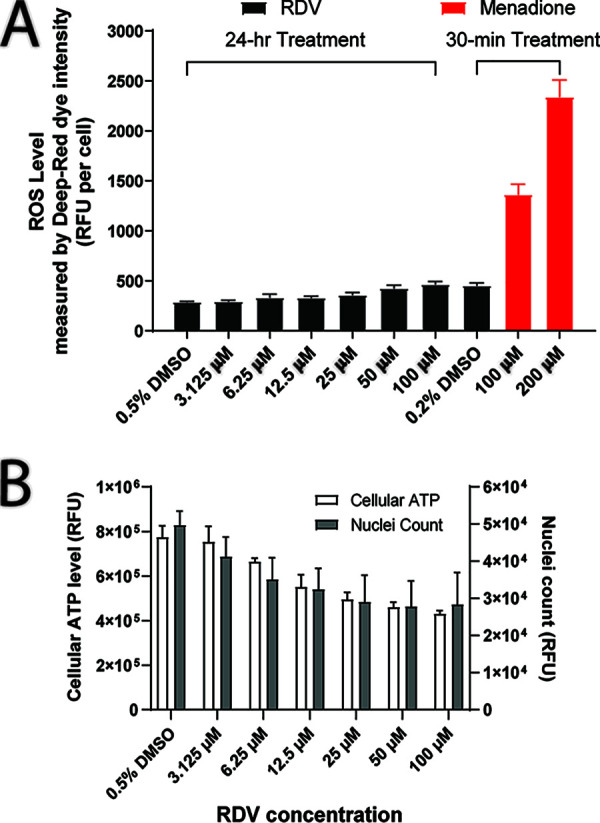
Effect of RDV on cellular ROS, ATP, and cell count after 24 h of treatment in HepG2 cells. (A) Treatment of 3.1 to 50 μM RDV showed no effect on ROS, while 100 μM RDV caused a 60% increase in ROS (P = 0.042). The positive control menadione showed a >3-fold increase in ROS levels after a 30-min incubation. (B) A concentration-dependent cytotoxicity was observed during the 24-h incubation of RDV, especially for concentrations of ≥12.5 μM.
Evaluation of mitochondrial toxicity.
Cytotoxicity under aerobic metabolic conditions. Some nucleoside analogues have the potential to affect mitochondrial functions via diverse mechanisms. One approach to assess mitochondrial function is a comparison of effects on cell viability in the presence of glucose-favoring glycolysis (i.e., anaerobic metabolism) and galactose-favoring oxidative phosphorylation (i.e., aerobic metabolism). The latter condition may sensitize cells to compounds affecting mitochondrial functions (21). However, the predictability of this assay has not been validated for nucleoside/tide analogues (6). Using intracellular ATP quantification as a readout for cell viability, the effects of glucose and galactose on the cytotoxicity of RDV and GS-441524 were assessed in the HepG2 hepatoma cell line, which was previously identified as a suitable model for testing compounds under aerobic conditions (Table 2) (21). In addition, a similar study was done using the PC-3 prostate-derived cell line, a model of rapidly proliferating cells. The CC50 values of RDV in HepG2 cells were 3.7 and 11.1 μM in the presence of glucose and galactose, respectively (Table 2), indicating that aerobic conditions did not enhance the cytotoxicity of RDV. In contrast, aerobic conditions in PC-3 cells enhanced RDV cytotoxicity by 6-fold (CC50 = 1.4 in galactose culture versus 8.9 μM in glucose culture). The divergent results from HepG2 and PC-3 cells suggest that the observed effects are cell line dependent.
TABLE 2.
In vitro cytotoxicity of RDV and GS-441524 under anaerobic and aerobic metabolic conditions after 5-day treatment in HepG2 and PC-3 cells
| Compound | 5-day CC50 (μM)a |
|||
|---|---|---|---|---|
| HepG2 cells |
PC-3 cells |
|||
| Anaerobic (glucose) | Aerobic (galactose) | Anaerobic (glucose) | Aerobic (galactose) | |
| RDV | 3.7 ± 0.2 | 11.1 ± 1.2 | 8.9 ± 1.6 | 1.4 ± 0.1 |
| GS-441524 | >100 | >100 | >100 | >100 |
| Puromycin | 0.73 ± 0.01 | 0.96 ± 0.13 | 0.52 ± 0.11 | 0.48 ± 0.01 |
All CC50 values represent the average ± SD of three or more independent experiments.
The parent nucleoside GS-441524 did not show any cytotoxicity in either PC-3 or HepG2 cells at the highest concentrations tested (100 μM), irrespective of the metabolic conditions (Table 2). Puromycin, used as general cytotoxic control, exhibited similar cytotoxicity in both cell types in the presence of glucose or galactose. The lack of a known nucleoside mitotoxin prevented us from using one as a positive control in this study.
Effect on mitochondrial DNA. The potential effect of RDV and GS-441524 on mitochondrial DNA (mtDNA) was assessed in vitro by quantitative real-time PCR (qPCR) analysis following continual treatment of HepG2 cells for 10 days. Dideoxycytidine (ddC), a known inhibitor of mtDNA replication, was used as a positive control. HepG2 cells treated with RDV showed a lack of dose response, indicating the lack of a specific effect on mtDNA synthesis. The effect of the parent nucleoside GS-441524 on mtDNA content was overall minimal (Table S1 in the supplemental material).
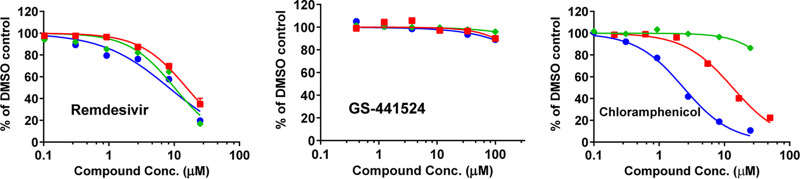
Effect on mitochondrial protein synthesis. The effect of RDV and GS-441524 on mitochondrial protein synthesis was assessed following a 5-day incubation with PC-3 cells. This particular cell model was chosen for its prior successful use in the studies of mitochondrial toxins (6). The selective effect of a compound on mitochondrial protein synthesis was determined by quantification of the level of cytochrome oxidase subunit 1 (COX-1, encoded by mtDNA) and succinate dehydrogenase A (SDH-A, encoded by nuclear DNA) (5, 19). RDV affected the levels of COX-1 and SDH-A to a similar extent, with CC50 values of 8.9 and 8.6 μM, respectively (Table 3, Fig. 3). These effects manifested in the same range of concentrations as the cytotoxicity measured by cellular ATP levels, indicating a lack of any selective effect of RDV on mitochondrial protein synthesis. GS-441524 showed no effect on protein synthesis up to the highest concentration tested (100 μM) (Table 3, Fig. 3). Chloramphenicol was used as a positive control, and its specific effect on mitochondrial protein synthesis was consistent with published data (5, 19).
TABLE 3.
In vitro effect of RDV and GS-441524 on mitochondrial proteosynthesis after 5-day treatment in PC-3
| Compound | Mitochondrial and cellular protein synthesis 5-day CC50 (μM)a |
||
|---|---|---|---|
| COX-1 | SDH-A | Cellular ATP | |
| RDV | 8.9 ± 1.1 | 8.6 ± 1.3 | 11.3 ± 3.3 |
| GS-441524 | >100 | >100 | >100 |
| Chloramphenicolb | 2.6 ± 0.6 | >25 | 14.1 ± 3.6 |
CC50 values represent the average ± SD of three or more independent experiments.
Positive control.
FIG 3.
Effects of RDV and its parent nucleoside GS-441524 on mitochondrial protein synthesis after 5 days of treatment in PC-3 cells. The effects on COX-1 levels (● blue, solid lines), SDH-A (♦ green, solid lines), and ATP levels (■ red, solid lines) are shown as % of the DMSO control. RDV showed nonselective inhibition of COX-1, SDH-A, and ATP levels, while GS-441524 showed no effect up to 100 μM. The positive control chloramphenicol, a known mitochondrial toxin, specifically inhibited COX-1 synthesis.
Effect on mitochondrial respiration. RDV and GS-441524 were further evaluated for their effects on mitochondrial spare respiratory capacity in the human cell lines PC-3 and HepG2 and the primary cells PHH and RPTEC by measuring the rate of oxygen consumption (OCR) using a Seahorse Extracellular Flux Analyzer (5, 22–24). Overall, there was a lack of a specific effect on cellular respiration among the four cellular systems tested. The cellular respiration of RDV-treated cells decreased in parallel with decreases in ATP level and total DNA level in HepG2, PHH, and RPTEC, indicating a lack of specific inhibition of cellular respiration (Table 4, Fig. 4). The most profound effect of RDV on spare mitochondrial respiratory capacity was seen in PC-3 cells, where RDV inhibited mitochondrial spare respiratory capacity with a CC50 value (CC50 = 2.5 μM) lower than those for the inhibition of ATP level and total DNA (CC50 = 24.0 and 12.5 μM, respectively). Among all four cell systems, GS-441524 showed no effect on mitochondrial respiration at the highest concentration tested (100 μM) (Table 4, Fig. S1). In contrast, the positive control chloramphenicol showed specific inhibition of mitochondrial respiration and minimal impact on ATP level and total DNA (Fig. S2).
TABLE 4.
In vitro effect of RDV and its metabolites GS-704277 and GS-441524 on mitochondrial respiration after 3-day treatment in PC-3, HepG2, PHH, and RPTECs
| Compound | Cell model | 3-day CC50 (μM)a |
||
|---|---|---|---|---|
| Spare respiration | Total DNA content | Cellular ATP | ||
| RDV | PC-3 | 2.5 ± 0.1 | 12.5 ± 0.7 | 24.0 ± 1.4 |
| HepG2 | 10.6 ± 0.1 | 6.3 ± 0.9 | 7.9 ± 0.1 | |
| PHH | 7.6 ± 1.9 | 13.4 ± 1.7 | 7.8 ± 2.2 | |
| RPTECs | 7.3 ± 2.7 | 14.3 ± 3.3 | 16.9 ± 4.1 | |
| GS-441524 | PC-3 | >100 | >100 | >100 |
| HepG2 | >100 | >100 | >100 | |
| PHH | >100 | >100 | >100 | |
| RPTECs | >100 | >100 | >100 | |
| GS-704277 | PHH | >100 | >100 | >100 |
| Control chloramphenical | PC-3 | 4.8 ± 1.3 | >50 | >50 |
| Control phenformin | PHH | 1.9 ± 0.5 | 22.1 ± 1.9 | 9.7 ± 4.5 |
CC50 values represent the average ± SD from three or more independent experiments.
FIG 4.
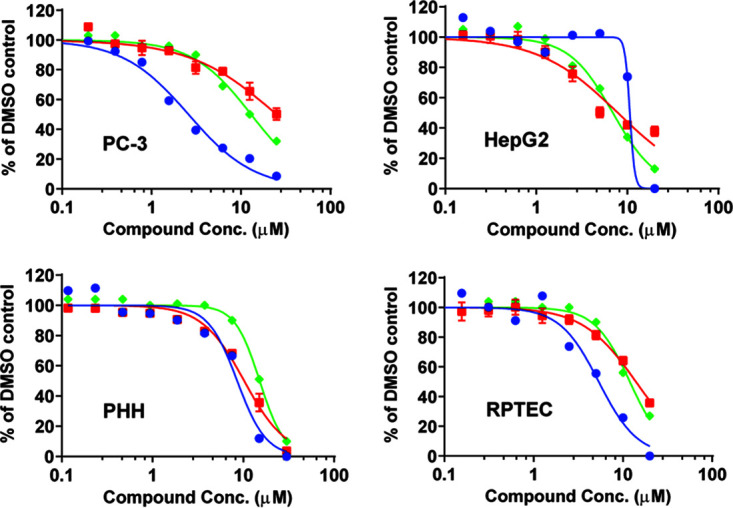
Effects of RDV on mitochondrial respiration (spare respiratory capacity) (● blue, solid lines), ATP levels (■ red, solid lines), and total DNA (♦ green, solid lines) after a 3-day treatment in PC-3, HepG2, PHH, and RPTEC cells. The spare respiratory capacity was normalized by cell numbers. Overall, RDV showed simultaneous inhibition of mitochondrial respiration, ATP levels, and total DNA except in PC-3 cells, where a mitochondria-specific inhibition was observed.
GS-704277, a major intermediate RDV metabolite, was evaluated for its effect on mitochondrial spare respiratory capacity, ATP levels, and total DNA in PHH after both an acute 4-h treatment and a chronic 3-day incubation, the default condition in the experiments described in the previous paragraph. After the 4-h incubation, RDV-treated PHH showed parallel decreases in mitochondrial spare respiratory capacity (21% to 27%) and cellular ATP levels (17% to 31%), indicating a lack of mitochondria-specific toxicity (Fig. S3). After a 3-day treatment with RDV, PHH showed parallel decreases in mitochondrial spare respiratory capacity, ATP levels, and total DNA, with CC50 values of 7.6, 7.8, and 13.4 μM, respectively, indicating general toxicity instead of specific mitochondrial toxicity (Table 4, Fig. 4). Neither of the two major systemic metabolites of RDV, GS-704277 nor GS-441524, exhibited any effects on mitochondrial spare respiratory capacity, ATP levels, or total DNA at the highest concentration tested (100 μM) after either a 4-h or a 3-day treatment (Table 4, Fig. S1, Fig. S3, Fig. S4).
Profiling of the active triphosphate metabolites.
Formation of active 5′-triphosphate metabolite GS-443902 in cells. Formation of GS-443902 from RDV and GS-441524 was measured in HEp-2, PC-3, and PHH cells after 24 h of continuous incubation with the compounds. As shown in Fig. 5 and Table S2, RDV formed 43 ± 6, 153 ± 16, and 45 ± 23 pmol/106 cells of GS-443902 in HEp-2, PC-3, and PHH cells, respectively. In contrast, GS-441524 formed 7.8- to 23-fold less active metabolites in these cells, at 5.5 ± 1.1, 6.6 ± 4.9, and 1.9 ± 1.6 pmol/106 cells, respectively.
FIG 5.
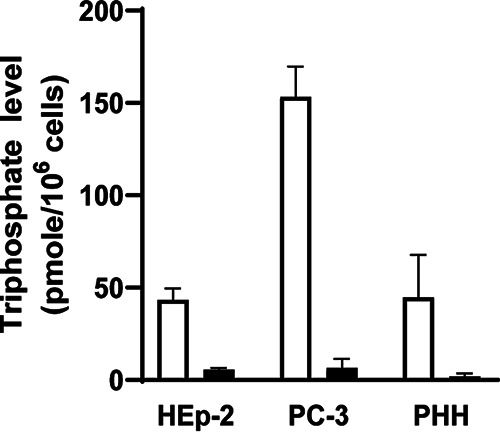
Metabolism of RDV and GS-441524 in HEp-2, PC-3, and PHH cells. The active 5′-triphosphate levels were measured after 24 h of continuous incubation of each compound. RDV-treated cells (open bars) formed 7.8-, 23-, and 23-fold higher triphosphate levels than GS-441524-treated (filled bars) in HEp-2, PC-3, and PHH cells, respectively.
Interaction with host RNA and DNA polymerases. The active triphosphate metabolite GS-443902 was tested in multiple biochemical assays to assess its interaction with key human DNA and RNA polymerases. The enzymatic activities of human DNA polymerases α and β, as well as that of RNA polymerase II, were unaffected by GS-443902 up to 200 μM, the highest concentration tested (Table 5). In addition, GS-443902 was tested for its incorporation into nucleic acids by host mitochondrial DNA and RNA polymerases using a single nucleotide incorporation assay. GS-443902 was a poor substrate of mtDNA polymerase γ, with no detectable incorporation when tested under the supratherapeutic concentration of 50 μM (versus a maximum drug concentration in serum [Cmax] of 6 μM at a 150-mg clinical dose) (13). It was also a poor substrate for POLRMT, with a rate of incorporation equal to 5.8% relative to ATP when tested under the supratherapeutic concentration of 500 μM (Table 6). This result contrasts with the significantly higher incorporation rates of 92% and 112%, for the triphosphate forms of BMS-986094 and balapiravir, respectively, two anti-HCV nucleosides associated with clinical toxicity (5). Together, these data further support the hypothesis that RDV has low potential to induce mitochondrial toxicity.
TABLE 5.
Inhibition of host DNA and RNA polymerases by the active triphosphate metabolite GS-443902
| Compound | IC50 (μM)a |
||||
|---|---|---|---|---|---|
| DNA pol α | DNA pol β | DNA pol γ | RNA pol II | POLRMT pol | |
| GS-443902 | >200 | >200 | >200 | >200 | >200 |
| Positive Control | Aphidicolin 4.7 ± 3.3 | 3′dTTP 1.9 ± 0.8 | 3′dTTP 1.2 ± 0.6 | α-amanitin 0.0035 ± 0.0015 | 3′deoxy GTP 4.2 ± 1.4 |
IC50 values represent the average ± SD from three independent experiments.
TABLE 6.
Relative rate of incorporation of the active triphosphate metabolite GS-443902 by human mitochondrial DNA and RNA polymerases
| Clinical compound | Nucleotide triphosphate | Rate of incorporation (% of natural dNTP or NTP)a |
|
|---|---|---|---|
| DNA polymerase γ | mtRNA polymerase | ||
| RDV | GS-443902 | 0% | 5.8% ± 1.4% |
| Decitabine | 5-aza-2′-dCTP | 79% ± 8% | ND |
| BMS-986094 | 2’CMe-GTP | ND | 92% ± 33% |
| Balapiravir/RG1626 | 4’CN-CTP | ND | 112% ± 10% |
The rate of single nucleotide incorporation was measured in the presence of 50 μM nucleotide analog for DNA Pol γ and 500 μM for POLRMT and expressed as the % of the natural dNTP or NTP incorporation at the same concentration. Data are presented as the average ± SD from three or more independent experiments. ND, not done.
Molecular target screen.
In addition to the aforementioned known off-targets of nucleoside/tide anlaogues, the RDV-containing diastereomeric mixture GS-466547 and the parent nucleoside GS-441524 were screened against a panel of 87 targets consisting of receptors, ion channels, transporters, and enzymes involved in a wide range of biological processes (Table S3). At 10 μM, none of the compounds showed detectable interaction with any of the 87 targets tested.
DISCUSSION
RDV is a single diastereomer monophosphoramidate prodrug of an adenosine analog with broad-spectrum antiviral activity. Using a prodrug as a way to deliver an active molecule may potentially result in intracellular accumulation of the less cell-permeable metabolites, depending on the activity of the enzymes participating in the metabolism of a particular drug and its conversion into the active form. Therefore, detailed examination of the functional consequences of exposure to high doses is critical for the evaluation of the drug safety.
This study presents a detailed characterization of RDV in a panel of in vitro cytotoxicity and off-target screening assays, undertaken to identify potential safety and toxicity liabilities. In this report, the general cytotoxicity of RDV measured as 50% cytotoxic concentration (CC50) following 5 to 14 days of continuous exposure to the drug ranged from 1.7 μM to >20 μM in selected human cell lines and primary human cells. It is worth noting that the RDV exposure in these cell culture studies was significantly higher than the systemic exposure to RDV with the repeated dosing of 150 mg in humans (plasma Cmax = 2,720 ng/ml = 4.5 μM; t1/2 = 1.11 h) (13). The parent nucleoside and major systemic metabolite GS-441524 showed less cytotoxicity than RDV. The differences in toxicity between these two compounds are likely due to the significantly lower level of active triphosphate in GS-441524-treated cells than in RDV-treated cells, as observed in this study as well as other studies (18). However, RDV and GS-441524 showed similar levels of toxicity in three hematopoietic progenitor cells. Cell-dependent cytotoxicity has been widely observed for nucleoside/tide analogs, which can be affected by the duration of compound exposure (5 day versus 14 day), compound permeability, intracellular activation, and differential cellular characteristics, including replication rate and bioenergetic status (5).
Among the primary cells tested, PHH showed the highest sensitivity to RDV, possibly due to high cellular permeability and efficient intracellular metabolism, leading to high levels of the triphosphate metabolite. This observation could shed light on the liver enzyme elevations observed in healthy volunteers treated with repeated doses of RDV (13). In contrast, the two main RDV metabolites in plasma, GS-704277 and GS-441524, are unlikely to contribute to clinical liver toxicity due to their low permeability and ineffective intracellular metabolism or limited systemic exposure. In two phase I studies, RDV was evaluated in healthy subjects with single-dose intravenous (i.v.) administration (across the dose range of 3 to 225 mg; n = 78 RDV and 18 placebo) or multiple doses of 150 mg once daily for 7 or 14 days (n = 16 RDV and 8 placebo) (13). Overall, RDV was well tolerated in both studies, and all adverse events were grade 1 or 2 in severity. In the repeat-dose study, reversible, treatment-emergent grade 1 or 2 alanine aminotransferase (ALT) (50% RDV, 13% placebo) and aspartate aminotransferase (AST) (44% RDV, 0% placebo) elevations were observed. Because transaminase elevations have been observed in patients with COVID-19 (25), the potential of RDV to exacerbate transaminase elevations in patients with COVID-19 may be difficult to discern. This has been supported by multiple RDV clinical trials. First, an open-label study (n = 397 RDV) without a placebo control arm showed little difference between 5-day and 10-day RDV treatment durations for adverse events of elevations in ALT (6% and 8%, respectively) or AST (5% and 7%, respectively) (15). Second, in a study of RDV in 584 hospitalized patients with moderate COVID-19, patients were randomized at a 1:1:1 ratio to receive 10-day RDV, 5-day RDV, or standard-of-care treatment; there was little difference between the three arms for adverse events of elevations in ALT (32%, 34%, and 39%, respectively) or AST (32%, 32%, and 33%, respectively). Grade 3 or 4 adverse events were reported less frequently for patients who received RDV (ALT: 3%, 2%, and 8%, respectively; AST: 1%, 3%, and 6%, respectively) (17). Finally, in the recently completed double-blind study of RDV versus placebo in 1,062 hospitalized patients with COVID-19, adverse events of increased amino transferase levels, including ALT, AST, or both, were reported in 6% of subjects treated with RDV for up to 10 days compared with 11% of subjects in the placebo group, indicating lack of specific drug-induced liver enzyme elevation (16). All of these reports consistently support minimal contribution of RDV to transaminase elevations in patients with COVID-19.
To understand whether the cytotoxicity is associated with mitochondrial toxicity, we measured the effects of RDV on mitochondrial DNA content, protein level, and cellular respiration. In HepG2 cells, RDV showed a lack of a specific dose-dependent effect on mtDNA synthesis. Similarly, RDV did not show specific inhibition of a mitochondrial DNA-encoded protein (CC50 of 8.9 μM) over a nuclear DNA-encoded protein (CC50 of 8.6 μM) in PC-3 cells. Additional biochemical assays showed the active triphosphate metabolite GS-443902 was a poor substrate for mitochondrial DNA and RNA polymerases, even when tested at supraphysiological concentrations of 50 and 500 μM, respectively. A recent paper reported the effects of RDV on cellular toxicity through a transcriptomic analysis, and reported effects of RDV on mitochondrial function (26). However, it is not clear whether the results were due to specific mitochondrial toxicity or general cellular toxicity. In addition, the relation between specific gene expression and mitochondrial toxicity has not been fully validated.
Taken together, we conclude that RDV has low potential for the off-target toxicities described for other nucleoside analogs, including mitochondrial toxicity. Furthermore, neither RDV nor its systemic metabolites induced ROS formation in vitro or interacted with any of the 87 targets in the molecular screen. Consistent with the clinical observations of drug-related elevations of liver transaminases following multiple doses of RDV, primary hepatocytes showed high in vitro susceptibility to RDV. In clinical settings of COVID-19 treatment in hospitalized patients, the risk associated with possible RDV-related liver enzyme elevations is substantially lower than its established benefits in hospitalized COVID-19 patients.
MATERIALS AND METHODS
Reagents.
Remdesivir (RDV, GS-5734), its parent nucleoside GS-441524, metabolite intermediate GS-704277 (Met X), and 5′-triphosphorylated metabolite GS-443902 were synthesized by Gilead Sciences, Inc. (Foster City, CA). Nucleotide triphosphates (TP) used as positive controls, including decitabine-TP (5-aza-2′-deoxyCTP), 2′CMe-GTP (active metabolite of BMS-986094), 4′-CN-CTP (active metabolite of Balapiravir/RG1626) were synthesized by Gilead Sciences, Inc. 3′-Deoxy ATP, 3′-deoxy GTP, 3′-deoxy CTP, and 3′-deoxy UTP were purchased from TriLink BioTechnologies (San Diego, CA). Control compounds for CC50 assays, such as dideoxycytidine (ddC), puromycin, chloramphenicol, menadione, aphidicolin, and α-amanitin, were purchased from Sigma-Aldrich (St. Louis, MO). All radioactively labeled nucleoside triphosphates (NTPs) were purchased from PerkinElmer (Shelton, CT).
Cell lines and primary cells.
The following cell lines were obtained from the indicated sources: HEp-2 (laryngeal carcinoma; ATCC), HepG2 (hepatoblastoma; ATCC), PC-3 (prostate metastatic carcinoma; ATCC), and MT-4 (human T-cell leukemia virus 1 [HTLV-1]-transformed human T lymphoblastoid cells; NIH AIDS Research and Reference Reagent Program). Cryopreserved primary human renal proximal tubule epithelial cells (RPTECs) were obtained from LifeLine Cell Technology (Frederick, MD) and isolated from the tissue of human kidney. Freshly isolated primary human hepatocytes (PHH) were from Bioreclamation IVT (Westbury, NY) or Life Technologies (Carlsbad, CA). Human primary blood mononuclear cells (PBMC) were isolated from human buffy coats, which were obtained from healthy volunteers (Stanford Blood Bank, Palo Alto, CA).
Cell cultures.
The HepG2 galactose-adapted cell line was established by culturing HepG2 cells in glucose-free Dulbecco’s modified Eagle medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, 100 units/ml streptomycin, 2 mM glutamine, 1 mM sodium pyruvate, and 10 mM galactose for 3 weeks prior to using in the assays. The PC-3 galactose-adapted cell line was established by culturing PC-3 cells in glucose-free Kaighn’s F12 medium supplemented with 10% FBS, 100 units/ml penicillin, 100 units/ml streptomycin, and 7 mM galactose for 3 weeks prior to using in the assays. The cells were passaged twice per week to maintain subconfluent densities. The cell culture procedures for RPTEC and PHH cells followed the vendor’s recommendations. PBMCs were isolated from human buffy coats using standard Ficoll separation, stimulated as described elsewhere (19), and tested at both quiescent and stimulated stages. The stimulated PBMCs were obtained from quiescent PBMCs stimulated with 10 units per ml of recombinant human interleukin 2 (hIL-2) and 1 μg/ml phytohemagglutinin P (PHA-P) for 48 h prior to drug treatment. Normal human primary bone marrow (BM) light-density cells were from three different lots obtained from AllCells (Emeryville, CA) or Lonza (Walkersville, MD). All primary human cells were cultured according to the manufacturer’s protocols. All cells were cultured at 37°C in a 5% CO2 incubator with 90% humidity, unless noted otherwise.
General cytotoxicity and cell viability.
Cytotoxicity in human cell-lines, PHH, RPTEC, and PBMC. Cells were treated with compounds for 4 h to 14 days depending on assay type. Cultures of freshly isolated PHH cells required fresh medium every 24 to 48 h, so compounds and medium were refreshed on days 0, 2, and 4 or days 0, 1, and 3 during the 5-day culture. After the incubation period, cell viability was measured by the addition of CellTiter Glo viability reagents (Promega, Madison, WI). The luminescence signal was quantified on an EnVision or Victor luminescence plate reader (Perkin-Elmer, Waltham, MA) after the cells were incubated with the reagents for 10 min at room temperature. The compound concentration that caused a 50% decrease in the luminescence signal (CC50), a measure of toxicity, was calculated by nonlinear regression using a sigmoidal dose-response (variable slope) equation as described in the Data Analysis section.
Cytotoxicity in human hematopoietic progenitor cells. The effects of the compounds on the proliferation of human erythroid, myeloid, and megakaryoid progenitors were tested in MethoCult84434, a methylcellulose-based colony assay conducted by StemCell Technology (Vancouver, Canada) (27). After an 11- to 14-day culture, the hematopoietic progenitor colonies (CFU-E, BFU-E, CFU-GM, and CFU-GEMM [E, erythroid; BFU, burstforming unit; GM, granulocyte/macrophage; GEMM, multilineage progenitors]) were enumerated and the CC50 values were calculated as described in the Data Analysis section.
Detection of intracellular reactive oxygen species level.
HepG2 cells were seeded at densities of 12 × 103 cells per well in 96-well plates, with a final volume of 160 μl per well. The cells were incubated overnight at 37°C in a 5% CO2 and 90% humidity incubator with Eagle’s minimum essential medium (EMEM) supplemented with 10% FBS (HyClone, Logan, UT), 100 units/ml penicillin, and 100 μg/ml streptomycin (P/S) (Gibco, Carlsbad, CA). On the next day (day 1), cells in 42 out of the 96 wells per plate were treated with RDV for 24 h. RDV was added to the cell culture plate directly using an HP D300 dispenser (Hewlett-Packard, Palo Alto, CA) with a starting concentration of 100 μM, with 2-fold dilutions to generate a total of 6 concentration points with six replicates for each concentration and 6 wells without RDV as negative controls. On the day of the reactive oxygen species (ROS) assay (day 2), 18 wells out of the remaining untreated 54 wells on the original 96-well plate were incubated with either 0 μM, 100 μM, or 200 μM menadione (Sigma-Aldrich, St. Louis, MO) for 30 min, with 6 replicates for each treatment. A mixture of CellROX Deep Red reagent and NucBlue Hoechst 33342 was added to the cells treated with menadione, RDV, and dimethyl sulfoxide (DMSO). The final DMSO concentration was 0.5% in all wells. The cell culture plate was incubated in a 37°C incubator for 30 min. The cell culture plate was then washed 3 to 4 times with 200 μl/well of 1× phosphate-buffered saline (PBS) buffer without Ca2+ and Mg2+ (Corning, Corning, New York), followed by a final addition of 100 μl of 1× PBS in each well. The assay plate was scanned using a Cellomics ARRAYSCAN VTI (Thermo Fisher Scientific, Waltham, MA) high content screening instrument with two channels. One channel measured the intensity of CellROX Deep Red using excitation/emission at 640/665 nm, while the other channel measured the signal intensity from NucBlue/Hoechst 3334 using excitation/emission at 360/460 nm. The ATP level was measured using CellTiter Glo viability reagents (Promega, Madison, WI) in cells treated with RDV for 24 h in a parallel experiment.
Data analysis
of calculation of CC50 and IC50 values. The CC50 values were defined as the concentration causing 50% decrease in cell viability, DNA level, protein level, or spare respiratory capacity, in comparison to the DMSO control. The 50% inhibitory concentration (IC50) values were defined as the concentration causing a 50% decrease in product formation in the biochemical assays. Data were analyzed using GraphPad Prism 8.0 (La Jolla, CA). The CC50 and IC50 values were calculated by nonlinear regression analysis using sigmoidal dose-response (variable slope) equation (four parameters logistic equation):
| (1) |
| (2) |
where X is the log of the concentration of the test compound and Y is the response. The CC50 or IC50 values were calculated as an average of three or more independent experiments.
Data analysis of measuring mitochondrial DNA. The relative amount of mitochondrial DNA (mtDNA) in treated samples was determined using a relative quantification method based upon the threshold cycle (2−ΔΔCT) formula (28). The amount of mtDNA in compound-treated samples relative to the DMSO-treated controls (% mtDNA) was calculated based on the following formulas:
where CT, cyt b and CT, β-actin represent the cycle threshold values for the amplification of cytochrome b and β-actin, respectively, as determined by the computational analysis of amplification curves using the ABI Prism software. The final results are presented as the mean % mtDNA ± SD (standard deviation) from two independent experiments, each performed in triplicate. The 2−ΔΔCT method was validated for cytochrome b and β-actin genes by determining the ΔCT values for amplification reaction mixtures containing various amounts of total cellular DNA. Minimal differences were observed in the ΔCT values in samples containing 5 to 40 ng of total cellular DNA, indicating that neither the amplification nor detection efficiencies of cytochrome b and β-actin were affected by the amount of DNA template within the dilution range relevant for the quantitative analysis performed (Table S5 in the supplemental material).
Measuring mitochondrial DNA.
mtDNA was generated by real-time PCR from total DNA isolated from HepG2 cells using the QIAamp DNA minikit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Real-time PCRs were performed using TaqMan universal mastermix (Applied Biosystems, Foster City, CA) in an ABI Prism 7900HT Fast real-time PCR system (Applied Biosystems). Quantification of mtDNA was achieved by amplification of a fragment of the mitochondrial specific cytochrome b gene using the primers and probe. The oligonucleotide sequences of the primer and probe used were cytochrome b forward 5′-CCTTCCACCCTTACTACACAATCAA-3′; cytochrome b reverse 5′-GGTCTGGTGAGAATAGTGTTAATGTCA-3′; and cytochrome b FAM-ACGCCCTCGGCTTAC-BHQ1. Chromosomal DNA was quantified by the amplification of a fragment of the β-actin gene using a β-actin Assay-on-Demand kit (Applied Biosystems). Quantification of mitochondrial and chromosomal DNA were performed independently using approximately HepG2 cell culture and compounds’ treatment was based on a previously published method (19, 29).
Mitochondrial protein synthesis assay.
PC-3 cells were treated with the compounds for 5 days and analyzed with the MitoTox MitoBiogenesis in-cell enzyme-linked immunosorbent assay (ELISA) kit (MitoSciences/Abcam, Eugene, Oregon) as described previously (5, 19). The assay uses quantitative immunocytochemistry to measure the protein levels of nuclear DNA-encoded succinate dehydrogenase (SDH-A; complex II) and mitochondrial DNA-encoded cytochrome c oxidase (COX-1; complex IV) in cultured cells.
Mitochondrial respiration in cell lines and primary cells.
Mitochondrial respiration was monitored by measuring the rate of oxygen consumption (OCR) of the cells after 4-h or 3-day treatments with the compounds, using a Seahorse extracellular flux analyzer (XFe-96) based on published protocols (5, 22–24) and modified from a previously published method (5). The OCR signals were normalized by cell numbers using DNA content, which method was based on a previously published method (5). The PC-3, HepG2, RPTEC, and PHH cells were seeded at optimized densities of 5 × 103, 5 × 103, 7.5 × 103, and 25 × 103 cells per well, respectively, in XF 96-well plates (Seahorse Bioscience, North Billerica, MA) with a final volume of 160 μl per well. The cells were incubated with compounds for 3 days, except for PHH, which received compound treatment for 4 h or 3 days. Compounds were added to the assay plate directly using an HP D300 dispenser (Hewlett-Packard, Palo Alto, CA) with 2-fold dilutions to give a total of 9 concentration points with six replicates. On the day of the assay, the cell culture medium was replaced with XF assay medium (pH 7.4) containing 10 mM glucose and 1 mM freshly prepared pyruvate. The mitochondrial respiration was monitored by measuring the rate of oxygen consumption (OCR) on a Seahorse Extracellular Flux (XFe-96) analyzer based on published protocols (5, 22–24). All concentrations were final after mixing unless noted otherwise. Multiple parameters were measured after the sequential injection of the ATP synthase inhibitor oligomycin (2 μM for all cell models), the uncoupler FCCP (0.25 μM for PC-3, 0.5 μM for HepG2 and PHH, and 1.5 μM for RPTEC), and a mixture of the mitochondrial complex I inhibitor rotenone and the complex III inhibitor antimycin A (0.5 μM each for all cell models) (Mito Stress Test kit, Seahorse Biosciences) (22–24). The spare respiratory capacity was obtained by subtracting the rate of basal respiration from the rate of maximal respiration and normalized to the cell number obtained from DNA level measurements. The data reported for each treatment are the average of the results from six replicates. The methods of detection of measuring ATP level in parallel experiments was based on a previously published method (5).
Biochemical assays.
DNA and RNA templates and primers. The DNA polymerase alpha and gamma inhibition assays utilized a 78-mer DNA template and a 19-mer DNA primer: D78 5′-ACACATGATACTACGAATTTTATGCTTCCAATGCCTTACAGTTCTCTAGCGGTGGCGCCCGAACA GGGACCTGAAAGC-3′ and D19 5′-GTCCCTGTTCGGGCGCCAC-3′ were purchased from Integrated DNA Technologies (Coralville, IA). Activated fish sperm DNA was purchased from USB/Affymetrix (Santa Clara, CA) and used as a template for the DNA polymerase beta inhibition assay. The DNA template used in the RNA POLII inhibition assay was a 1,188-bp restriction fragment containing the cytomegalovirus (CMV) immediate early promoter (Promega, Madison, WI). For single nucleotide incorporation, a DNA 19-mer primer and a DNA 36-mer template were used for POL γ–catalyzed reactions, whereas an RNA 12-mer primer and a DNA 18-mer template were used for POLRMT-catalyzed reactions (Table S6). All primers were 5′-labeled with [γ-32P]ATP (3,000 Ci/mmol) and T4-kinase (New England BioLabs, Ipswich, MA). In brief, a mixture containing 100 μM primer, 0.4 unit/μl T4-kinase, 1× T4-kinase reaction buffer, and 4 μCi/μl (0.4 μM) [γ-32P]ATP was incubated at 37°C for 60 to 120 min, followed by heat inactivation at 65°C for 5 to 10 min. 5′-32P-labeled R12/D18 were annealed at a 1:1.1 molar ratio at a concentration of 10 μM in a solution of 10 mM Tris-HCl (pH 8.0) and 0.1 mM EDTA using a thermocycler where samples were heated for 1 min at 90°C and cooled to 10°C at a rate of 5°C per minute. 5′-32P-labeled D18/D36 were annealed at a 1:1.1 molar ratio at 10 μM in a solution containing 10 mM Tris-HCl (pH 8.0) and 0.1 mM EDTA. Samples were heated at 90°C for 10 min, cooled at 50°C for 30 min, and placed on ice for 10 min.
Enzymatic inhibition assays. Human DNA polymerase alpha, isolated from HeLa cell extracts, was from CHIMERx (Madison, WI). Recombinant human DNA polymerase beta, expressed in Escherichia coli, was a gift from Zucai Suo at The Ohio State University. Recombinant human DNA polymerase gamma (including both the large subunit and the small subunit) was cloned, expressed, and purified from insect cells by Gilead Sciences (Foster City, CA) (19). RNA PolII was purchased as part of the HeLaScribe nuclear extract in vitro transcription system kit from Promega (Madison, WI). The recombinant human POLRMT and the transcription factors mitochondrial transcription factor A (mtTFA) and B2 (mtTFB2) were purchased from Enzymax (Lexington, KY). The inhibition of DNA polymerases alpha, beta, and gamma, POLRMT, and PolII has been described previously in detail (4, 19, 30).
Single nucleotide incorporation assay by human mitochondrial DNA Pol γ. With all concentrations given as the final concentration, 1.2 nM DNA Pol γ large subunit and 3.4 nM Pol γ accessory subunit were preincubated on ice for 5 min and added to a reaction mixture containing 50 mM Tris-HCl (pH 8.0), 2 mM dithiothreitol (DTT), 0.2 mg/ml bovine serum albumin (BSA), 200 nM D19/D36 p/t, and 10 mM MgCl2. Reactions were heated for 37°C and initiated by addition of 50 μM of natural deoxynucleoside triphosphate (dNTP) or analogs, plus 50 μM dGTP (for T or C template) or dCTP (for A or G template). At 0, 0.5, 2, 5, 10, 30, and 60 min, 10 μl of the reaction mixture was removed and quenched with10 μl of gel loading buffer containing 100 mM EDTA, 80% formamide, and 1% bromophenol blue, and heated for 65°C for 5 min. The samples were run on a 20% polyacrylamide gel (8 M urea) and the gel was exposed to a phosphorimager screen. The substrate and the incorporation products, D19 and D20-28, were quantified using a Typhoon Trio Imager and Image Quant TL software.
Single nucleotide incorporation assay by human mitochondrial POLRMT. The assay was modified based on a published report (31). With all concentrations given as final concentrations, a mixture of MTCN buffer (50 mM MES [morpholineethanesulfonic acid], 25 mM Tris-HCl, 25 mM CAPS [N-cyclohexyl-3-aminopropanesulfonic acid], and 50 mM NaCl, pH 7.5), 200 nM 5′-32P-labeled R12/D18, 10 mM MgCl2, 1 mM DTT, and 376 nM POLRMT (Enzymax, Lexington, KY) was preincubated at 30°C for 1 min. The reaction was started by addition of 500 μM natural NTP or NTP analogs. At 0.17, 0.5, 2, 5, 10, and 30 min, 3 μl of the reaction mixture was removed and quenched with10 μl of gel loading buffer containing 100 mM EDTA, 80% formamide, and 1% bromophenol blue, and heated for 65°C for 5 min. The samples were run on a 20% polyacrylamide gel (8 M urea) and the gel was processed and analyzed in the same way as described above.
Molecular target screen of the RDV-containing diastereomeric mixture GS-466547 and parent nucleoside GS-441524.
RDV-containing diastereometric mixture GS-466547 and parent nucleoside GS-441524 were tested at 10 μM in a SafetyScreen87 Panel (Eurofins Panlabs Taiwan, Ltd.) (Table S3). Effects of compounds were evaluated along with a known receptor ligand or substrate. Positive results were defined as >50% inhibition of ligand binding.
Determination of GS-441524 and its phosphorylated metabolites in PC-3 and PHH cells.
HEp-2, PC-3, or PHH cells were seeded at 0.20 × 106, 0.44 × 106, and 0.88 × 106 cells/well, respectively, in a 12-well plate. On the next day, HEp-2, PC-3, or PHH cells were treated with either 1 μM RDV or GS-441524, in duplicate, or 0.01% DMSO in eight replicates. After a 24-h treatment, cells were washed twice with ice-cold 0.9% NaCl and then treated with 500 μl of dry ice-cold extraction buffer (0.1% potassium hydroxide and 67 mM ethylenediamine tetraacetic acid in 70% methanol, containing 0.5 μM chloro-ATP as an internal standard). The above solution was vortexed for 5 min, then centrifuged at 20,000 × g for 10 min. Supernatant was transferred to clean 1.5-ml Eppendorf vials and loaded onto a centrifuging evaporator. Once dry, samples were reconstituted with 80 μl of 1 mM ammonium phosphate buffer (pH 7.0) and transferred to high-pressure liquid chromatography (HPLC) vials, where a 10 μl injection was used for analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Standard calibration curves for HEp-2, PC-3, or PHH were constructed based on pmol of compound per sample with known cell numbers in each sample. The standard curve was prepared by spiking an appropriate amount of GS-441524, GS-441524-MP, GS-441524-DP, and GS-443902 solution, prepared in water, into blank HEp-2, PC-3, or PHH matrix, with serial dilutions to complete the calibration standard curve. Detailed HPLC and mass spectrometry parameters are provided in the Materials and Methods section of the supplemental material.
Determination of cell volume.
Measurements of the cell volume for various cell types were conducted using confocal microscope Leica SP8 (Leica Microsystems GmbH, Wetzlar, Germany) and image analysis software Imaris (Bitplane, Zürich, Switzerland). The intracellular space was labeled with a fluorescent dye, Calcein AM (1 to 10 μg/ml, Thermo Fisher Scientific, Waltham, MA, USA), which loads into live cells with active esterases. The nuclei were labeled with DNA-binding dye Hoechst 33342 (16 to 32 μM, Thermo Fisher Scientific, Waltham, MA, USA) to facilitate separation and correct counting of adjacent cells. The labeling was conducted by incubating the cells in a culture medium containing the dyes at 37°C and 5% CO2 for at least 15 min. The series of images (Z-stacks) were collected using a 63× objective with numerical aperture of 1.4 at different focal planes to enable computational reconstruction of three-dimensional objects with complex shape and estimation of their volume. Calcein was excited with the white laser at 488 nm and the fluorescence signal was detected in the range of 498 to 600 nm using a Leica HyD hybrid detector. The imaging was performed at 37°C to avoid temperature-related changes in shape of the cells. The estimation of the cell volume was verified using the fluorescent beads of known diameter by comparison of the values obtained after calculations based on either the diameter or 3-D reconstruction. This revealed that only minor correction was needed to account for elongation of the point spread function in vertical direction for the objective with high (1.4) numerical aperture, unlike for the objective with low (0.4) numerical aperture. In addition, the results of confocal imaging were confirmed using a cell counter (Cellometer K2, Nexcelom Bioscience, Lawrence, MA, USA) with imaging capabilities, allowing for determining the diameter of the cells in suspension after trypsinization.
Statistical analysis.
The differences between compound-treated groups and DMSO-treated groups were analyzed using ordinary one-way ANOVA (Sidak’s multiple-comparison test) (GraphPad Prism 8.1) and P values less than 0.05 were defined as statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
Becky Norquist provided medical writing assistance on behalf of Gilead. All authors are employees of Gilead Sciences, Inc. except O.B. and M.P. who were previously employed by Gilead Sciences, Inc. All authors may hold stock or stock options in Gilead Sciences, Inc.
The studies were fully sponsored by Gilead Sciences, Inc.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.De Clercq E 2011. A 40-year journey in search of selective antiviral chemotherapy. Annu Rev Pharmacol Toxicol 51:1–24. doi: 10.1146/annurev-pharmtox-010510-100228. [DOI] [PubMed] [Google Scholar]
- 2.Asselah T, Marcellin P, Schinazi RF. 2018. Treatment of hepatitis C virus infection with direct-acting antiviral agents: 100% cure? Liver Int 38 Suppl 1:7–13. doi: 10.1111/liv.13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson AA, Ray AS, Hanes J, Suo Z, Colacino JM, Anderson KS, Johnson KA. 2001. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J Biol Chem 276:40847–40857. doi: 10.1074/jbc.M106743200. [DOI] [PubMed] [Google Scholar]
- 4.Arnold JJ, Sharma SD, Feng JY, Ray AS, Smidansky ED, Kireeva ML, Cho A, Perry J, Vela JE, Park Y, Xu Y, Tian Y, Babusis D, Barauskus O, Peterson BR, Gnatt A, Kashlev M, Zhong W, Cameron CE. 2012. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog 8:e1003030. doi: 10.1371/journal.ppat.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng JY, Xu Y, Barauskas O, Perry JK, Ahmadyar S, Stepan G, Yu H, Babusis D, Park Y, McCutcheon K, Perron M, Schultz BE, Sakowicz R, Ray AS. 2016. Role of mitochondrial RNA polymerase in the toxicity of nucleotide inhibitors of hepatitis C virus. Antimicrob Agents Chemother 60:806–817. doi: 10.1128/AAC.01922-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng JY 2018. Addressing the selectivity and toxicity of antiviral nucleosides. Antivir Chem Chemother 26:2040206618758524–2040206618758528. doi: 10.1177/2040206618758524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, Park Y, Babusis D, Clarke MO, Mackman RL, Spahn JE, Palmiotti CA, Siegel D, Ray AS, Cihlar T, Jordan R, Denison MR, Baric RS. 2017. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheahan TP, Sims AC, Leist SR, Schafer A, Won J, Brown AJ, Montgomery SA, Hogg A, Babusis D, Clarke MO, Spahn JE, Bauer L, Sellers S, Porter D, Feng JY, Cihlar T, Jordan R, Denison MR, Baric RS. 2020. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun 11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porter DP, Weidner JM, Gomba L, Bannister R, Blair C, Jordan R, Wells J, Wetzel K, Garza N, Van Tongeren S, Donnelly G, Steffens J, Moreau A, Bearss J, Lee E, Bavari S, Cihlar T, Warren TK. 2020. Remdesivir (GS-5734) is efficacious in cynomolgus macaques infected with Marburg virus. J Infect Dis 222:1894–1901. doi: 10.1093/infdis/jiaa290. [DOI] [PubMed] [Google Scholar]
- 10.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N, Strickley R, Wells J, Stuthman KS, Van Tongeren SA, Garza NL, Donnelly G, Shurtleff AC, Retterer CJ, Gharaibeh D, Zamani R, Kenny T, Eaton BP, Grimes E, Welch LS, Gomba L, Wilhelmsen CL, Nichols DK, Nuss JE, Nagle ER, Kugelman JR, Palacios G, Doerffler E, Neville S, Carra E, Clarke MO, Zhang L, Lew W, Ross B, Wang Q, Chun K, Wolfe L, Babusis D, Park Y, Stray KM, Trancheva I, Feng JY, Barauskas O, Xu Y, Wong P, et al. 2016. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan PC, Liu C, Raynaud P, Lo MK, Spiropoulou CF, Symons JA, Beigelman L, Deval J. 2018. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog 14:e1006889. doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo MK, Feldmann F, Gary JM, Jordan R, Bannister R, Cronin J, Patel NR, Klena JD, Nichol ST, Cihlar T, Zaki SR, Feldmann H, Spiropoulou CF, de Wit E. 2019. Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci Transl Med 11:eaau9242. doi: 10.1126/scitranslmed.aau9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humeniuk R, Mathias A, Cao H, Osinusi A, Shen G, Chng E, Ling J, Vu A, German P. 2020. Safety, tolerability, and pharmacokinetics of remdesivir, an antiviral for treatment of COVID-19, in healthy subjects. Clin Transl Sci 13:896–906. doi: 10.1111/cts.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulangu S, Dodd LE, Davey RT, Tshiani Mbaya O, Proschan M, Mukadi D, Lusakibanza Manzo M, Nzolo D, Tshomba Oloma A, Ibanda A, Ali R, Coulibaly S, Levine AC, Grais R, Diaz J, Lane HC, Muyembe-Tamfum J-J, Sivahera B, Camara M, Kojan R, Walker R, Dighero-Kemp B, Cao H, Mukumbayi P, Mbala-Kingebeni P, Ahuka S, Albert S, Bonnett T, Crozier I, Duvenhage M, Proffitt C, Teitelbaum M, Moench T, Aboulhab J, Barrett K, Cahill K, Cone K, Eckes R, Hensley L, Herpin B, Higgs E, Ledgerwood J, Pierson J, Smolskis M, Sow Y, Tierney J, Sivapalasingam S, Holman W, Gettinger N, Vallée D, PALM Consortium Study Team, et al. 2019. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med 381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldman JD, Lye DCB, Hui DS, Marks KM, Bruno R, Montejano R, Spinner CD, Galli M, Ahn MY, Nahass RG, Chen YS, SenGupta D, Hyland RH, Phil D, Osinusi AO, Cao H, Blair C, Wei X, Gaggar A, Brainard DM, Towner WJ, Munoz J, Mullane KM, Marty FM, Tashima KT, Diaz G, Subramanian A. 2020. Remdesivir for 5 or 10 days in patients with severe Covid-19. N Eng J Med 383:1827–1837. doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh M, Ruiz-Palacios GM, Benfield T, Fatkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC. 2020. Remdesivir for the treatment of Covid-19—final report. N Engl J Med 383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spinner CD, Gottlieb RL, Criner GJ, Arribas Lopez JR, Cattelan AM, Soriano Viladomiu A, Ogbuagu O, Malhotra P, Mullane KM, Castagna A, Chai LYA, Roestenberg M, Tsang OTY, Bernasconi E, Le Turnier P, Chang SC, SenGupta D, Hyland RH, Osinusi AO, Cao H, Blair C, Wang H, Gaggar A, Brainard DM, McPhail MJ, Bhagani S, Ahn MY, Sanyal AJ, Huhn G, Marty FM, GS-US-540–5774 Investigators. 2020. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. JAMA 324:1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruijssers AJ, George AS, Schäfer A, Leist SR, Gralinksi LE, Dinnon KH, Yount BL, Agostini ML, Stevens LJ, Chappell JD, Lu X, Hughes TM, Gully K, Martinez DR, Brown AJ, Graham RL, Perry JK, Du Pont V, Pitts J, Ma B, Babusis D, Murakami E, Feng JY, Bilello JP, Porter DP, Cihlar T, Baric RS, Denison MR, Sheahan TP. 2020. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep 32:107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng JY, Cheng G, Perry J, Barauskas O, Xu Y, Fenaux M, Eng S, Tirunagari N, Peng B, Yu M, Tian Y, Lee YJ, Stepan G, Lagpacan LL, Jin D, Hung M, Ku KS, Han B, Kitrinos K, Perron M, Birkus G, Wong KA, Zhong W, Kim CU, Carey A, Cho A, Ray AS. 2014. Inhibition of hepatitis C virus replication by GS-6620, a potent C-nucleoside monophosphate prodrug. Antimicrob Agents Chemother 58:1930–1942. doi: 10.1128/AAC.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shuhendler AJ, Pu K, Cui L, Uetrecht JP, Rao J. 2014. Real-time imaging of oxidative and nitrosative stress in the liver of live animals for drug-toxicity testing. Nat Biotechnol 32:373–380. doi: 10.1038/nbt.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. 2007. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol Sci 97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- 22.Beeson CC, Beeson GC, Schnellmann RG. 2010. A high-throughput respirometric assay for mitochondrial biogenesis and toxicity. Anal Biochem 404:75–81. doi: 10.1016/j.ab.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadanaciva S, Rana P, Beeson GC, Chen D, Ferrick DA, Beeson CC, Will Y. 2012. Assessment of drug-induced mitochondrial dysfunction via altered cellular respiration and acidification measured in a 96-well platform. J Bioenerg Biomembr 44:421–437. doi: 10.1007/s10863-012-9446-z. [DOI] [PubMed] [Google Scholar]
- 24.Dranka BP, Benavides GA, Diers AR, Giordano S, Zelickson BR, Reily C, Zou L, Chatham JC, Hill BG, Zhang J, Landar A, Darley-Usmar VM. 2011. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic Biol Med 51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C, Shi L, Wang FS. 2020. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol 5:428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akinci E, Cha M, Lin L, Yeo G, Hamilton CM, Donahue JC, Bermudez-Cabrera CH, Zanetti CL, Chen M, Barkal AS, Khowpinitchai B, Chu N, Velimirovic M, Jodhani R, Fife DJ, Sovrovic M, Cole AP, Davey AR, Cassa AC, Sherwood IR. 2020. Elucidation of remdesivir cytotoxicity pathways through genome-wide CRISPR-Cas9 screening and transcriptomics. BioRxiv doi: 10.1101/2020.08.27.270819. [DOI]
- 27.Pessina A, Albella B, Bayo M, Bueren J, Brantom P, Casati S, Croera C, Gagliardi G, Foti P, Parchment R, Parent-Massin D, Schoeters G, Sibiril Y, Van Den Heuvel R, Gribaldo L. 2003. Application of the CFU-GM assay to predict acute drug-induced neutropenia: an international blind trial to validate a prediction model for the maximum tolerated dose (MTD) of myelosuppressive xenobiotics. Toxicol Sci 75:355–367. doi: 10.1093/toxsci/kfg188. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Birkus G, Hitchcock MJM, Cihlar T. 2002. Assessment of mitochondrial toxicity in human cells treated with tenofovir: comparison with other nucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 46:716–723. doi: 10.1128/aac.46.3.716-723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke MO, Mackman R, Byun D, Hui H, Barauskas O, Birkus G, Chun BK, Doerffler E, Feng J, Karki K, Lee G, Perron M, Siegel D, Swaminathan S, Lee W. 2015. Discovery of beta-d-2′-deoxy-2′-alpha-fluoro-4′-alpha-cyano-5-aza-7,9-dideaza adenosine as a potent nucleoside inhibitor of respiratory syncytial virus with excellent selectivity over mitochondrial RNA and DNA polymerases. Bioorg Med Chem Lett 25:2484–2487. doi: 10.1016/j.bmcl.2015.04.073. [DOI] [PubMed] [Google Scholar]
- 31.Smidansky ED, Arnold JJ, Reynolds SL, Cameron CE. 2011. Human mitochondrial RNA polymerase: evaluation of the single-nucleotide-addition cycle on synthetic RNA/DNA scaffolds. Biochemistry 50:5016–5032. doi: 10.1021/bi200350d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.