Abstract
Most cells of the body contain molecular clocks, but the requirement of peripheral clocks for rhythmicity, and their effects on physiology, are not well understood. Here we show that deletion of core clock components REV-ERBα and β in adult mouse hepatocytes disrupted diurnal rhythms of a subset of liver genes and altered the diurnal rhythm of de novo lipogenesis. Liver function is also influenced by non-hepatocytic cells, and the loss of hepatocyte REV-ERBs remodeled the rhythmic transcriptomes and metabolomes of multiple cell types within the liver. Finally, alteration of food availability demonstrated the hierarchy of the cell-intrinsic hepatocyte clock mechanism and the feeding environment. Together, these studies reveal previously unsuspected roles of the hepatocyte clock in the physiological coordination of nutritional signals and cell-cell communication controlling rhythmic metabolism.
One Sentence Summary
The hepatocyte clock and food availability control gene expression and metabolism in multiple cell types in the liver.
Biological rhythms are intricately involved in sleeping/waking, feeding/fasting, and activity/rest phenomena, and are essential to maintain physiological homeostasis (1). The mammalian core clock includes transcriptional activators BMAL1/CLOCK and transcriptional repressors REV-ERBα and β that function in interlocked transcriptional feedback loops (2). Central clocks in the suprachiasmatic nucleus (SCN) are believed to synchronize clocks in peripheral tissues (3), and dyssynchrony of this system is associated with metabolic dysfunction (4, 5). Nevertheless, major questions remain as to how the environment and genetic factors control the clocks in peripheral tissues, and whether communication exists between clocks in different cell types within an organ.
To dissect the cell-autonomous and non-autonomous regulation of diurnal rhythms in peripheral tissues, we focused on the liver, a metabolic hub (6). REV-ERBα and β were specifically deleted in hepatocytes (HepDKO) by injecting the AAV8-TBG-CRE virus into adult REV-ERBα/β floxed mice. This model excludes developmental effects and potential confounding due to direct manipulation of the clock in other tissues (7, 8). Expression of both REV-ERBα and β was nearly undetectable after 2 weeks, even at ZT10 when REV-ERBs were highly expressed at the mRNA (Fig. 1A) and protein level (Fig. S1). REV-ERBs physiologically repress Bmal1 and Npas2 in a circadian manner (9, 10), and both of these genes were constitutively derepressed in the REV-ERB HepDKO (Fig. 1B). Other core clock genes also demonstrated reduced rhythmicity (Fig. 1B).
Figure 1. Disruption of REV-ERBα and β in hepatocytes remodels the liver diurnal rhythmic transcriptome and lipid metabolism.
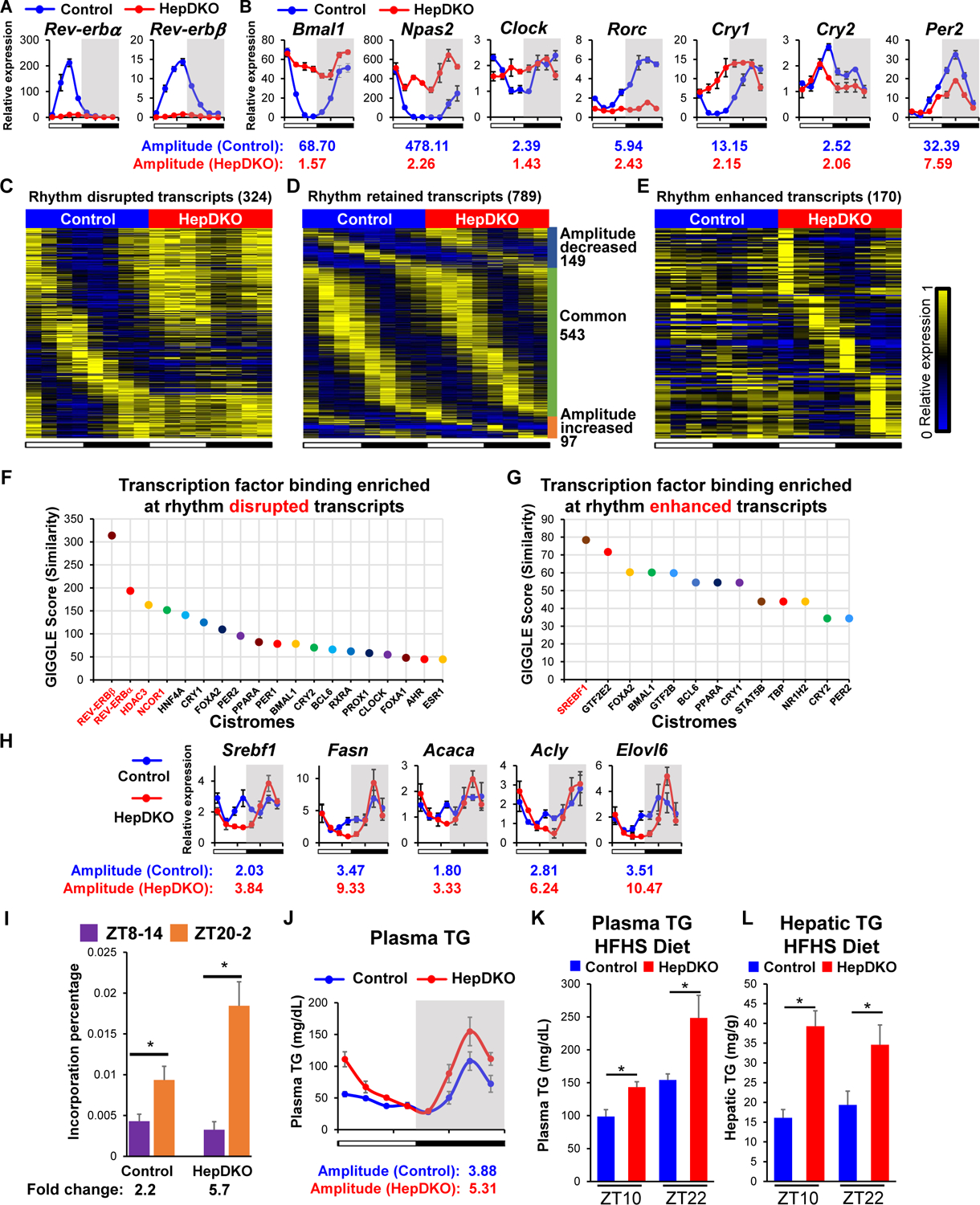
(A–B) Relative mRNA expression of Rev-erbα and Rev-erbβ (A) and REV-ERBs target genes (B) in control and HepDKO livers. (C–E) Heat map of the relative expression of rhythm disrupted (C), retained (D), and enhanced (E) transcripts in control and HepDKO livers. The color bar indicates the scale used to show the expression of transcripts across eight time points, with the highest expression normalized to 1. JTK_CYCLE, adjusted p < 0.01, 21 ≤ period (t) ≤ 24 hr, peak to trough ratio > 2 (n = 3 per time point). (F–G) Transcription factor binding similarity screening on rhythm disrupted (F) and enhanced (G) transcripts based on all published liver cistromes from CistromeDB (11). (H) Relative mRNA expression of Srebf1 and its target genes in control and HepDKO livers (n = 4–6 per time point). (I) Incorporation of deuterated water into liver fatty acids was measured in mice 6 hours after oral gavage of D2O either at ZT8 or at ZT20. Data are presented as mean ± SEM. *p < 0.05 in Student’s t-test (n = 6 mice per group). (J) Serum triglyceride measurements in control and HepDKO mice. (K–L) Serum triglyceride (K) and hepatic triglyceride (L) measurements in HFHS-fed control and HepDKO mice. Data are presented as mean ± SEM (n = 4–6 per time point).
We next examined the effect of REV-ERB HepDKO on the liver rhythmic transcriptome. Two weeks after AAV treatment, RNA-seq performed on livers harvested every 3 hours revealed the attenuation of the rhythmicity of a large group of transcripts that were highly rhythmic in the controls, including genes involved in diurnal rhythm pathways such as Bmal1, Npas2 and Clock (Fig. 1C, S2A and Table S1A). This fits the prevailing hypothesis that REV-ERBs are major controllers of the clock and suggests that the rhythmic expression of these genes depends on the intrinsic core clock feedback loop. Many genes, however, maintained diurnal rhythmicity in the absence of REV-ERBs (Fig. 1D, S2B and Table S1B). These included ~170 genes, enriched for lipid metabolism, that showed enhanced rhythmic amplitudes (Fig. 1E, S2C and Table S1C). KEGG and Gene Set Enrichment Analysis (GSEA) indicate that rhythmic transcripts regulated by REV-ERBs were involved in circadian rhythms, hormone secretion, and lipid metabolism (Fig. S2A–S2D). These results indicated an unexpected rhythmic transcriptomic reprogramming in the liver upon the depletion of REV-ERBs in adult hepatocytes. Importantly, rhythmic locomotor activity (Fig. S3A), feeding (Fig. S3B), and plasma insulin levels (Fig. S3C) were not much affected in REV-ERB HepDKO mice, indicating that disruption of the hepatocyte clock did not affect diurnal activity and feeding responses and excluding the possibility that the remodeling of the liver rhythmic transcriptome was due to changes in behavior.
Rhythmic expression of REV-ERB directly regulates many target genes by binding to ROR/REV-ERB-response element (RORE), where it represses transcription both by recruiting corepressors and by competing with ROR nuclear receptors, as well as by tethering to liver factors (11). ROR targets represented a high percentage of rhythm-disrupted but not enhanced transcripts (Fig. S3D), suggesting that REV-ERB’s direct binding was more relevant to the rhythmically disrupted transcripts. This was confirmed in livers expressing REV-ERBα DNA binding domain deficient mutant and lacking REV-ERBβ (Fig. S3E)(12).
To explore the transcriptional mechanism underlying rhythmic disruption in hepatocytes upon REV-ERB HepDKO, we used CistromeDB (13) to perform transcription factor (TF) binding similarity screening based on all published liver cistromes. Reassuringly, REV-ERBs and their corepressors HDAC3 and NCOR1 were the top TFs bound near genes whose diurnal rhythm was disrupted by REV-ERB HepDKO (Fig. 1F and Table S2A). The binding sites of BMAL1, PER2, and CRY1 were enriched in rhythm retained transcripts, suggesting that systemic signals drive the rhythmic expression of these genes via these core clock genes (14) (Fig. S3F and Table S2B).
Interestingly, SREBF1 was the most enriched TF near genes whose diurnal rhythms were induced by the loss of REV-ERBs (Fig. 1G and Table S2C) although there is no REV-ERBα binding site near Srebf1. Indeed, the rhythmic expression of Srebf1 was enhanced upon REV-ERB HepDKO, as was that of many of its target genes involved in de novo lipogenesis (DNL) (Fig. 1H), which is consistent with a previous REV-ERBα whole-body knockout mouse model (15). Enhanced diurnal rhythmic expression of Srebf1 was also observed in livers from reverse phase feeding (RPF) Cry1−/−, Cry2−/− mice (16), suggesting a general role of core clock TF repressors in maintaining the homeostasis of hepatic lipid metabolism. The physiological significance of this finding was assessed by directly measuring DNL using deuterated water as a tracer. Consistent with the new rhythm of Srebf1 and the DNL pathway, the normal rhythm of DNL was markedly amplified in the livers of the REV-ERB HepDKO mice (Fig. 1I). This was accompanied by an increase in the amplitude of plasma triglyceride rhythms, both on normal chow (Fig. 1J) as well as on high fat/high sucrose (HFHS) diet (Fig. 1K). Consistent with the increase in DNL, liver TG concentration was also increased in the livers of the HFHS-fed mice (Fig. 1L). Thus, REV-ERBs in hepatocytes are required to maintain lipid metabolism homeostasis.
Although hepatocytes are the most abundant cell type in the liver, the organ is composed of many other cell types that have critical roles in metabolic diseases (17–19). To better understand the effects of hepatocyte clock disruption, we performed single nucleus sequencing (sNuc-Seq) on livers harvested at ZT8, when REV-ERBs are highly expressed, from control and HepDKO mice. sNuc-Seq avoided skewing the results against lipid-laden hepatocytes that may be lost due to lysis or size exclusion during single-cell isolation, and ~3000 genes were detected per nucleus. Based on cell-specific markers (Fig. S4A and S4B), populations of hepatocytes, endothelial cells (ECs), Kupffer cells (KCs), stellate cells, and immune cells were clearly distinguishable, as were the subpopulations of hepatocytes corresponding to the previously defined markers of zonation (20) (Fig. 2A). As expected, many changes in gene expression were observed between control and HepDKO hepatocytes (Fig. 2B) with ~ 2/3 of the changes being common to hepatocytes in different zones (Fig. S4C). The percentage of different cell populations in the liver was largely unchanged (Fig. S4D), but, interestingly, gene expression in non-hepatocyte cells in the REV-ERB HepDKO livers was markedly altered, with the largest number of changes observed in ECs (Fig. 2B). Considerable changes were also noted in KCs, which are liver-resident macrophages that have critical roles in the liver (17). Together, these two cell types were selected for more detailed studies.
Figure 2. Hepatocyte REV-ERBs control non-hepatocytic diurnal rhythmic transcriptome.
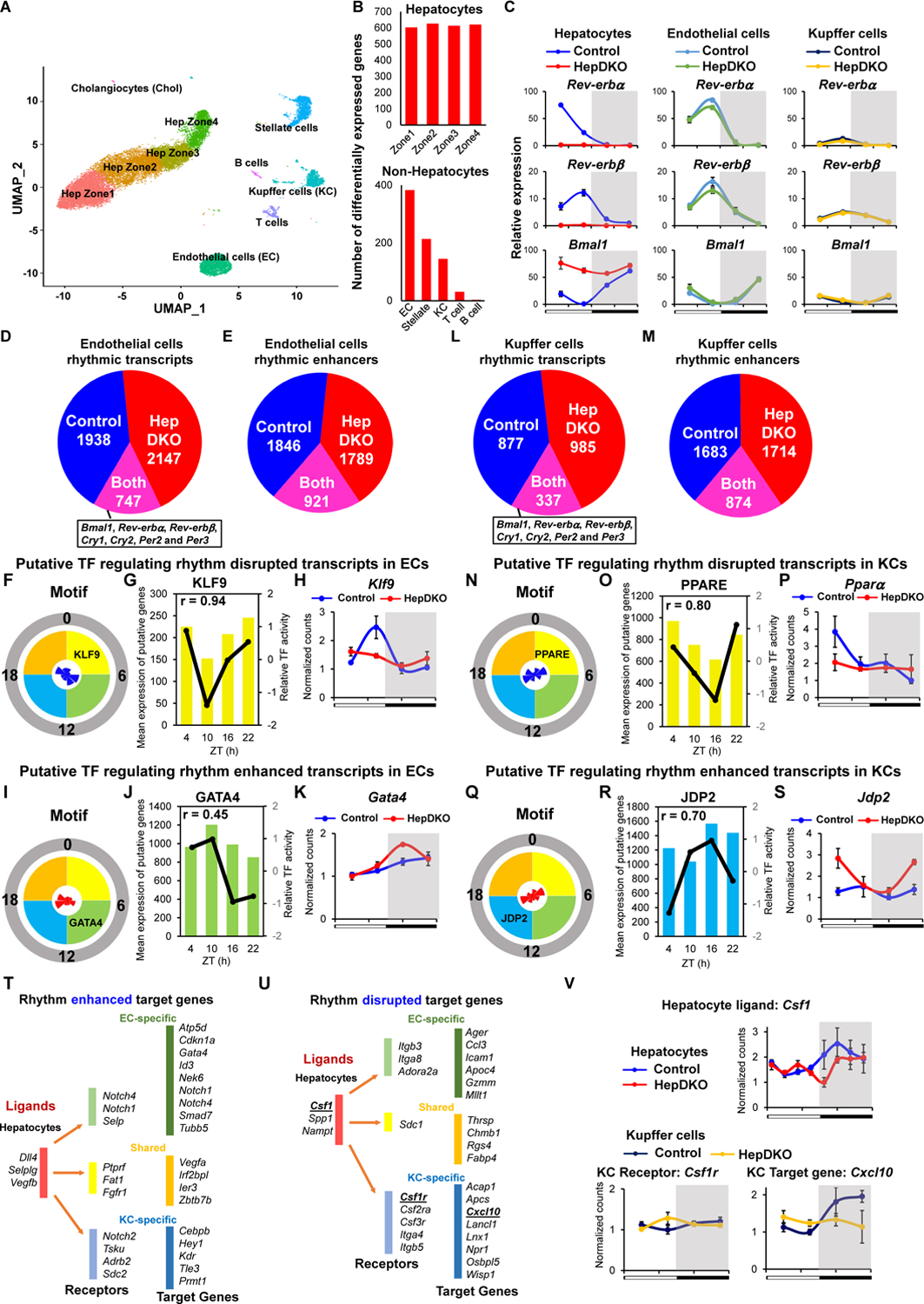
(A) UMAP visualization of liver cell clusters based on 18,239 single-cell transcriptomes. (B) The number of differentially expressed transcripts in hepatocytes (upper panel) or non-hepatocytes (lower panel) upon REV-ERBs HepDKO. (C) Relative mRNA expression of Rev-erbα, Rev-erbβ and Bmal1 in isolated hepatocytes, endothelial cells and Kupffer cells from control and HepDKO livers. (D and E) Identification of diurnal rhythmic transcripts (D) and enhancers (E) in isolated endothelial cells from control and HepDKO livers. JTK_CYCLE (Hughes et al., 2010), adjusted p ≤ 0.05, 21 ≤ period (t) ≤ 24 hr, peak to trough ratio > 1.5. (F and I) Rose diagrams show the prevalence of rhythmic transcripts in each phase group, and motifs enriched at sites of rhythmic enhancers, which were correlated with rhythm disrupted (F) and enhanced (I) transcripts and enhancers from IMAGE in isolated ECs. (G and J) Correlation of mean expression of putative target genes and relative TF transcription activity in four phase groups in isolated ECs from control (G) and HepDKO (J) livers. In each plot, the bars represent the mean expression of putative TF target genes of each phase, and the black line represents the predicted TF relative transcription activity. Correlation coefficient r shows the strength of the relationship between the mean expression of putative TF target genes and relative transcription activity. (H and K) Expression level (normalized read counts) of Klf9 (H) and Gata4 (K) in isolated ECs from control and HepDKO livers. (L and M) Identification of diurnal rhythmic transcripts (L) and enhancers (M) in isolated Kupffer cells from control and HepDKO livers. (N and Q) Rose diagrams show the prevalence of rhythmic transcript in each phase group, and motifs enriched at sites of rhythmic enhancers, which were correlated with rhythm disrupted (N) and enhanced (Q) transcripts and enhancers from IMAGE in isolated KCs. (O and R) Correlation of mean expression of putative target genes and relative TF transcription activity in four phase groups in isolated KCs from control (O) and HepDKO (R) livers. In each plot, the bars represent the mean expression of putative TF target genes of each phase, and the black line represents the predicted TF relative transcription activity. Correlation coefficient r shows the strength of the relationship between the mean expression of putative TF target genes and relative transcription activity. (P and S) Expression level (normalized read counts) of Pparα (P) and Jbp2 (S) in isolated KCs from control and HepDKO livers. Data are presented as mean ± SEM (n = 4 per time point). (T and U) Ligand-receptor interaction analysis. Top 3 putative ligands from hepatocytes affect receptors (in ECs and KCs) for the regulation of rhythm enhanced (T) and disrupted (U) transcripts in ECs and KCs. (V) Examples of rhythm disrupted ligand (Csf1) from hepatocytes, receptor (Csf1r) and rhythm disrupted target gene (Cxcl10) in Kupffer cells.
To quantify whole cell transcriptomes with greater depth than possibly done by sNuc-Seq, we performed diurnal rhythmic transcriptomics on ECs and KCs isolated every 6 hours, 2 weeks after hepatocyte-specific deletion of REV-ERBs. The deletion of Rev-erbα and β along with their constitutively induced repression target Bmal1 were confirmed in isolated hepatocytes. Rev-erbα/β gene expression was virtually unchanged in the ECs and KCs from the HepDKO livers, although the amplitude of Rev-erbα/β rhythms were muted in KCs (Fig. 2C). The relative expression of lineage specific markers Stab2 (ECs) and Csf1r (KCs) confirmed the specificity of the cell populations (Fig. S4E).
Remarkably, despite the physiologically rhythmic expression of the core clock genes, the diurnal rhythmic transcriptomes were extensively remodeled in ECs (Fig. 2D, S5A and Table S3). These results indicated that disruption of the hepatocyte clock was communicated to the ECs. In addition, we quantified enhancer RNA (eRNA) expression in isolated ECs by mapping RNA-seq reads to intergenic regions of open chromatin determined by Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) (21), which identified a widespread reprogramming of rhythmic enhancers (Fig. 2E, S5B and Table S3).
We next used Integrated Analysis of Motif Activity and Gene Expression (IMAGE) (22) to ascertain sequence motifs enriched at sites of rhythmic enhancers associated with rhythmic genes in order to identify potential TFs with corresponding binding preferences and diurnal rhythmicity. These putative factors that were potentially responsible for rhythm disrupted and enhanced enhancers and transcripts were identified (Table S4). For example, Kruppel-like Factor 9 (KLF9), a ubiquitous regulator of oxidative stress (23), was identified as one of the putative TFs responsible for the loss of rhythmic enhancers associated with lost rhythmic genes that peaked between ZT0–6 (Fig. S5C and 2F), and there was a positive correlation between KLF9 transcription activity and its putative target gene expression (Fig. 2G). Indeed, the expression of Klf9 was rhythmic in control cells but not in ECs from HepDKO livers (Fig. 2H). Conversely, gained rhythmic enhancers peaking between ZT6–12 (Fig. S5D and Fig. 2I) were enriched for the GATA binding motif (Fig. 2J), corresponding to a gained rhythmic expression of Gata4, a known regulator of the hepatic microvasculature (24) (Fig. 2K).
Similarly, the KC rhythmic transcriptome was extensively reprogrammed in REV-ERB HepDKO livers (Fig. 2L, S5E and Table S5), and this was associated with both the loss and gain of rhythmic enhancers (Fig. 2M, S5F and Table S5). The factors identified as potentially responsible for rhythmic disrupted and enhanced enhancers and transcripts are listed in Table S4. As an example, the PPAR-binding motif was enriched at sites of ZT0–6 rhythmic enhancers that decreased in KCs of the HepDKO livers (Fig. S5G and 2N) and was associated with the highest transcriptional activity in this phase (Fig. 2O). Consistent with this, the expression of Pparα, a regulator of the macrophage inflammatory response (25), was also rhythmic, peaking between ZT0–6 in control cells but not in KCs isolated from HepDKO livers (Fig. 2P). By contrast, the motif of Jun dimerization protein 2 (JDP2) was enriched in REV-ERB HepDKO-specific enhancers whose activity peaked at ZT12–18 (Fig. S5H and 2Q), and also had the highest predicted transcriptional activity in this phase (Fig. 2R). The phase of the gained rhythmic expression of JDP2 was antiphase to its transcriptional activity (Fig. 2S), consistent with its transcriptional repression function (26). Moreover, comparative analysis of rhythmic remodeled transcripts between hepatocytes, ECs, and KCs revealed little overlap between different cell types, indicating a cell-type specific response to loss of REV-ERB in hepatocytes (Fig. S6A–S6C).
To uncover potential signals from hepatocytes lacking REV-ERBs to other cell types we used NicheNet (27) to identify ligand-receptor pairs in which the ligand was altered in HepDKO hepatocytes, and the receptor was expressed in ECs or KCs, and the downstream genes exhibited enhanced (Fig. 2T and S6D) or disrupted (Fig. 2U and S6E) rhythms. For example, the colony stimulating factor 1 gene Csf1 lost rhythmicity in HepDKO hepatocytes (Fig. 2V). The CSF1 receptor was expressed in KCs and, while it was not rhythmically expressed, downstream genes of the CSF1 signaling pathways such as Cxcl10 (28) lost rhythmicity (Fig. 2V). These results demonstrate how disruption of the hepatocyte clock could lead to new diurnal rhythms of gene expression in surrounding non-hepatocytic cells. Note that this analysis does not incorporate post-transcriptional regulation of predicted ligands and receptors that were not regulated at the transcript level (Table S6) (29).
To understand the impact of HepDKO-induced diurnal rhythm remodeling on non-hepatocytic cells, we performed GSEA on the rhythmic transcriptomes of ECs and KCs. Lipid metabolism related pathways were found to be enriched in both ECs and KCs (Fig. S7A and S7B). This rhythm remodeling may be regulated not only via mapped ligand-receptor pairs but also via metabolites from hepatocytes, since we observed rhythmic metabolome reprogramming in isolated hepatocytes in the liver upon the depletion of REV-ERBs (Fig. S7C, S7D and Table S7). Consistently, mouse phenotype enrichment analysis (30) indicates that phenotypes most enriched in altered rhythmic transcripts of both ECs and KCs from HepDKO livers were related to homeostasis and metabolism (Fig. S7E and S7F).
To test this prediction, we performed diurnal rhythmic metabolomic profiling, identifying many metabolites whose diurnal rhythms were disrupted or enhanced in ECs and KCs from HepDKO livers (Fig. S7G and Table S7). Integrated analysis of rhythm-remodeled transcripts and metabolites by MetaboAnalyst (31) revealed a number of significantly impacted metabolic pathways. In ECs, multiple rhythmic metabolic pathways were disrupted, including glutathione metabolism (Fig. 3A and Fig. S8), as illustrated by expression of the Gpx1 gene and glutathione disulfide (Fig. 3B). Other pathways exhibited enhanced diurnal rhythmicity, including glucose metabolism and its conversion into hexosamines (Fig. 3C), as illustrated by the gained rhythm of Pfkl gene expression and UDP-N-acetyl-glucosamine levels (Fig. 3D). These changes likely impact the function of ECs, which rely on glycolysis for energy production, with the hexosamine pathway controlling nitric oxide (NO) production and angiogenesis (32). In KCs, the correlated rhythmic disrupted transcripts and metabolites were related to lipid metabolism (Fig. 3E), exemplified by Enpp6 gene expression and C22:4 levels (Fig. 3F) (33, 34), whereas rhythm enhanced pathways included one-carbon metabolism (Fig. 3G) regulated by the Dhfr gene (Fig. 3H). Together, the cell-type specific rhythm remodeling in non-hepatocytic cells upon the loss of hepatocyte REV-ERBs identifies a previously unknown, coordinated response to hepatocyte clock disturbance.
Figure 3. Hepatocyte REV-ERBs regulate non-hepatocytic diurnal rhythmic metabolic process.
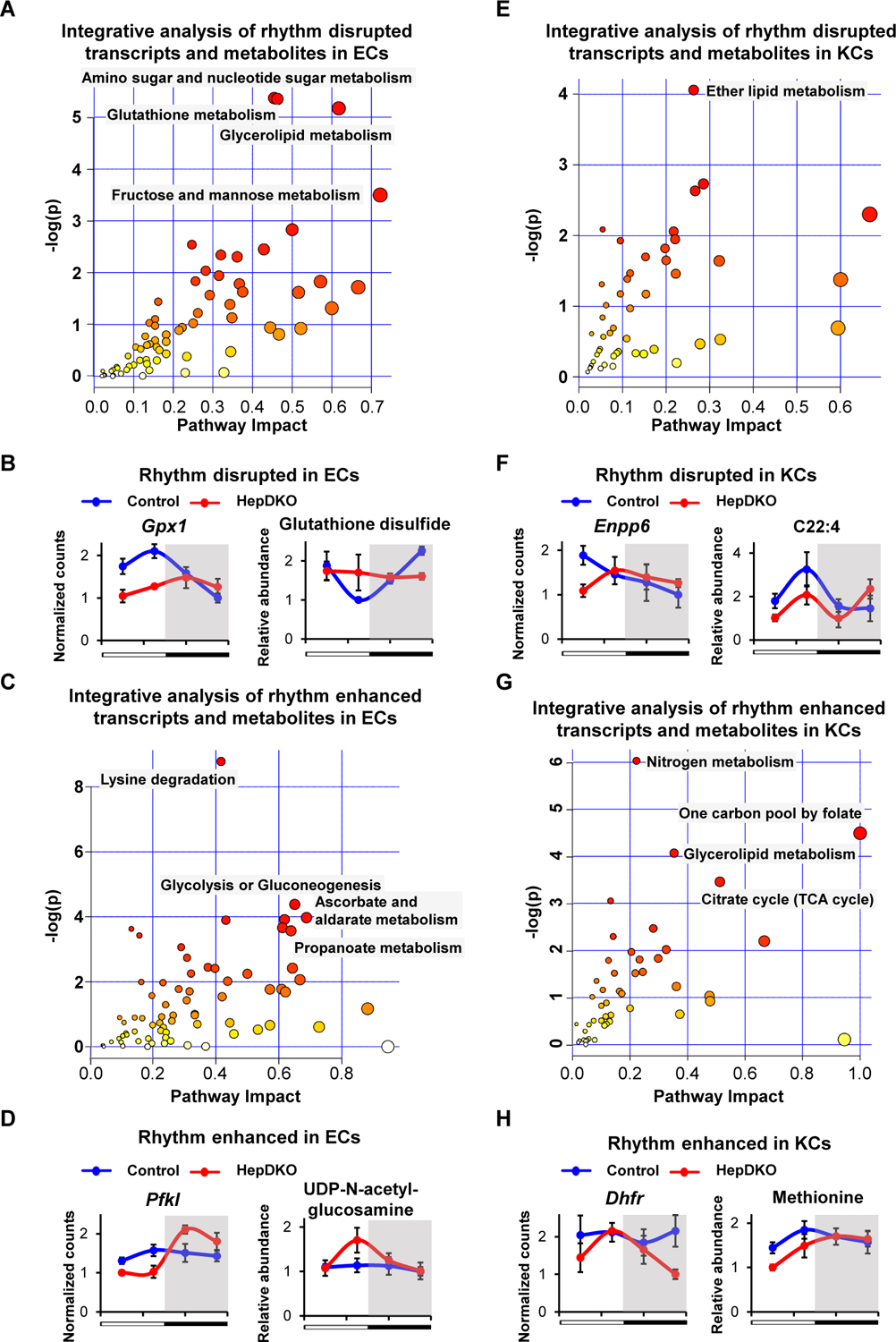
(A and C) Metabolic pathway analysis integrating the enrichment of genes and metabolites in rhythm disrupted (A) and enhanced (C) transcripts and metabolites in isolated ECs. (B and D) Examples of rhythm disrupted (B) and enhanced (D) metabolites and related transcripts in ECs upon REV-ERBs DKO in hepatocytes. (E and G) Metabolic pathway analysis integrating the enrichment of genes and metabolites in rhythm disrupted (E) and enhanced (G) transcripts and metabolites in isolated KCs. (F and H) Examples of rhythm disrupted (F) and enhanced (H) metabolites and related transcripts in KCs upon REV-ERBs DKO in hepatocytes. Pathways were considered significant if p < 0.01 using Hypergeometric Test. Metabolites and transcripts data are presented as mean ± SEM (n = 3–4 per time point).
Although light/dark cycles act as zeitgebers to entrain behavioral rhythms via the central rhythmic oscillator in the suprachiasmatic nucleus of the hypothalamus, feeding/fasting cycles are important synchronizers of peripheral clocks (35–37), and time-restricted feeding uncouples liver rhythms from behavioral rhythms (35). Having demonstrated the role of the hepatocyte clock in controlling cell-autonomous and non-cell autonomous rhythms in the liver, we therefore considered its role in the response to nutrition by performing diurnal rhythmic transcriptomic analysis on mice subjected to three weeks of reverse phase feeding (RPF), in which food was available only during the light phase (Fig. 4A). As expected based on previous work (35), RPF of control mice led to a 12-hour phase shift in the rhythms of core clock genes such as Rev-erbα and Rev-erbβ (Fig. 4B). Indeed, transcriptomic analysis indicated that nearly all rhythmic transcripts exhibited a 12-hour phase shift in the livers of control mice under RPF (Fig. 4C) suggesting a dominant role of feeding on rhythmic phase regulation.
Figure 4. Control of liver diurnal rhythms by the hepactocyte clock and feeding.
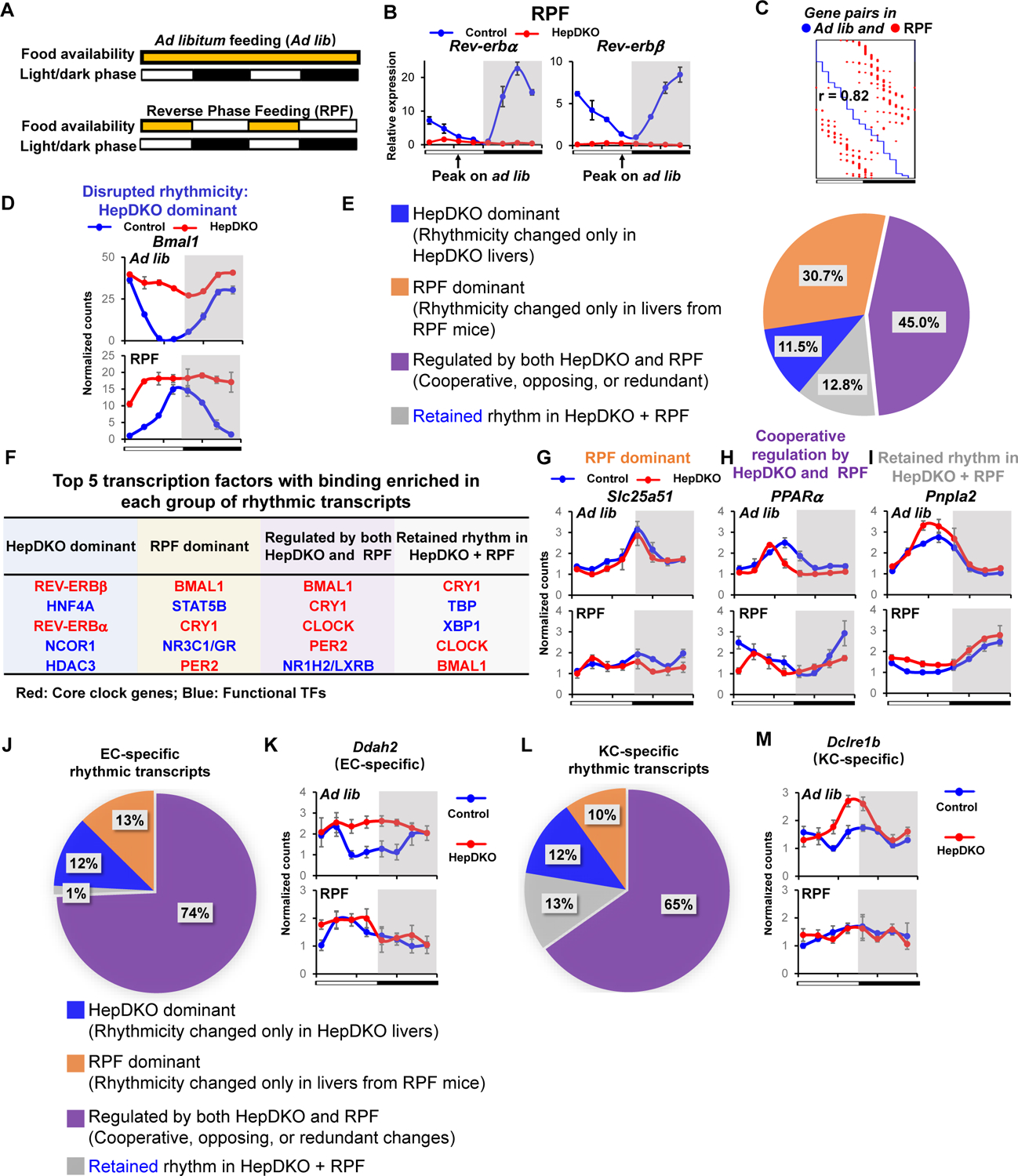
(A) Diagram of feeding schedule of Ad libitum feeding (Ad lib) and reverse phase feeding (RPF). (B) Relative expression of Rev-erbα and Rev-erbβ in control and HepDKO livers from RPF mice. (C) Phase correlation of hepatic rhythmic transcripts between ad lib and RPF mice. Each row has a unique pair of hepatic rhythmic transcripts in ad lib mice (blue dot) and RPF mice (red dot). (D) Expression level (normalized read counts) of Bmal1 in control and HepDKO livers from ad lib and RPF mice. (E) Identification of rhythmic transcripts that were dominantly regulated by HepDKO (blue) or RPF (orange), regulated by both HepDKO and RPF (purple), and retained in HepDKO+RPF (grey). (F) Top 5 TFs from four groups in (E) identified from transcription factor binding similarity screening based on all published liver cistromes from CistromeDB (11). (G-I) Expression level (normalized read counts) of Slc25a51, Pparα and Pnpla2 in control and HepDKO livers from ad lib and RPF mice. (J and L) Identification of EC-specific (J) and KC-specific (L) rhythmic transcripts that were dominantly regulated by HepDKO (blue) or RPF (orange), regulated by both HepDKO and RPF (purple), and retained in HepDKO+RPF (grey). (K and M) Expression level (normalized read counts) of EC-specific gene Ddah2 (K) and KC-specific gene Dclre1b (M) in control and HepDKO livers from ad lib and RPF mice. Data are presented as mean ± SEM (n = 3 per time point).
The rhythm of the core clock gene Bmal1 was also phase-shifted by ~12 hours under RPF in control livers. In contrast, in the livers of REV-ERB HepDKO mice, Bmal1 expression was constitutive, robust, and non-rhythmic both under RPF and ad libitum (ad lib) feeding (Fig. 4D), indicating cell-autonomous clock regulation of the hepatocyte endogenous clock by REV-ERBs. Since most rhythmic genes were phase-shifted ~12 hours by RPF, we assessed changes in rhythmicity using a classification that integrated amplitude (fold change of peak/trough > 2), period (between 21–24 hours) and adjusted p value (< 0.01) from the JKT algorithm (38). This analysis identified 4 categories of rhythmic genes that we refer to as “HepDKO-dominant” (rhythmicity of transcripts is changed only in HepDKO livers), “RPF-dominant” (rhythmicity of transcripts is changed only in livers from RPF mice), “regulated by both HepDKO and RPF” (including cooperative, redundant, or opposing changes), and “retained rhythm in of HepDKO and RPF” (rhythmicity unchanged in HepDKO+RPF).
11.5% of all rhythmic transcripts were HepDKO-dominant (Fig. 4E, S9A and Table S8A) and, based on TF binding similarity screening analysis, this group of rhythmic transcripts was likely directly regulated by REV-ERB and its corepressor complexes (Fig. 4F). RPF-dominant transcripts represented 30.7% of rhythmic transcripts (Fig. 4E and Table S8B), implying non-cell autonomous regulation by feeding. For example, the diurnal rhythmicity of Slc25a51 was indistinguishable in control and HepDKO livers from ad lib fed mice but disrupted in both control/RPF and HepDKO/RPF livers (Fig. 4G). Binding sites for STAT and GR TFs were enriched near these genes (Fig. 4F) (39, 40).
45% of rhythmic transcripts were regulated by both HepDKO and RPF, either cooperatively, oppositely, or redundantly (Fig. 4E and S9B). Binding sites enriched near these genes included those of lipid-regulating LXR (Fig. 4F), whose activation was reported to be rhythmically enhanced in livers of REV-ERBα whole-body knockout mice (15). Cooperative changes were exemplified by the Pparα gene (Fig. 4H and Table S8C). Note that these results largely reflect hepatocytes, whose Pparα expression pattern was different from that shown for KCs. By contrast, the HepDKO-induced diurnal rhythmic enhancement of Phf8 was negated by RPF while the rhythmic disrupting effect by RPF on Ccnd1 was counteracted by HepDKO (Fig. S9B and S9C). The cooperative and opposing effects on rhythmicity demonstrate interdependence of the hepatocyte clock and feeding. However, for genes termed “redundant” the separate effects of HepDKO and RPF on rhythmicity were similar to each other and to the combination (e.g. Srebf1) (Fig. S9B and S9D).
In the final group of rhythmic transcripts, although the phase was dependent on food entrainment, the rhythmicity per se was retained in both HepDKO and RPF, suggesting that this was controlled by other signals independent of the intrinsic clock and feeding (Fig. 4E, 4I and Table S8D). Interestingly, although the rhythmic mRNA expression of core clock genes Bmal1, Cry1 and Per2 was attenuated upon REV-ERB depletion, the binding sites were still enriched in these non-intrinsic rhythmic transcripts (Fig. 4F), suggesting that systemic signals drive the rhythmic transcription activity of these TFs (14, 41).
Finally, we sought to determine the extent to which the hepatocyte clock and feeding/fasting cycles control diurnal rhythms in non-hepatocytes. We defined the EC-specific rhythmic genes based on the RNA-seq data from ECs isolated from the HepDKO livers and then determined their rhythmic expression during RPF, both in control and REV-ERB HepDKO livers. Remarkably, ~74% of rhythmic genes (Fig. 4J and Table S9A) were regulated by both HepDKO and RPF, with enrichment for genes regulating NO synthesis (Fig. S9E), including EC-specific Ddah2 (42) (Fig. 4K). Similarly, in KCs, ~65% of cell-specific rhythmic genes were regulated by both HepDKO and RPF (Fig. 4L and Table S9B), with enrichment for genes regulating histone-serine phosphorylation (Fig. S9F), including Dclre1b, which regulates DNA repair (43) and whose rhythmic expression was KC-specific in the liver (Fig. 4M). Thus, non-autonomous signals resulting from feeding and communication from hepatocytes play vital roles in the rhythmic gene expression of non-hepatocytic cells in the liver.
In sum, our studies shed new light on the physiological importance and function of peripheral clocks, whose existence was originally established in vitro (44–46). We demonstrate that some but not all hepatocyte diurnal rhythms are controlled by the core clock in a cell-autonomous manner in vivo. Moreover, the enhanced diurnal rhythms upon REV-ERB deletion (e.g. DNL genes) suggests that the clock not only anticipates daily environment changes but also buffers against certain fluctuations. Previous studies manipulating the liver clock found that it was not essential for weight loss due to food restriction during the normal feeding period (47) or behavioral diurnal rhythms for which the light/dark cycle acts as a zeitgeber (7). However, when feeding is restricted to the light phase, it becomes the predominant hepatocytic zeitgeber for liver (35), and our studies demonstrate the hierarchy and interdependence of feeding and the cell-autonomous clock for diurnal rhythmic hepatocyte gene expression. Moreover, rhythmic gene expression and metabolism in non-hepatocytic cells in the liver are highly influenced both by the hepatocyte clock as well as feeding. These findings are likely to apply to peripheral clocks in other cell types.
Supplementary Material
Acknowledgments
We thank Hannah J. Richter, Kun Zhu, Yuxiang Zhang, Yong Hoon Kim and other members of the Lazar lab for technical support and valuable discussions, particularly during the revision of this manuscript while sheltered in place during the Coronavirus pandemic. We thank Jesper Grud Skat Madsen and Susanne Mandrup for advice on using the IMAGE program.
Funding:
We thank the Functional Genomics Core and the Viral Vector Core of the Penn Diabetes Research Center (P30 DK19525) for next-generation sequencing and virus preparation, respectively. We also acknowledge the Penn DRC Metabolomics Core at Princeton (DK19525). This work was supported by the JPB Foundation (M.A.L.) and the Cox Medical Research Institute (M.A.L.) as well as by National Institutes of Health grants (R01-DK045586, M.A.L.; and F32DK116519 D.G.). C.J. and W.X. were supported by American Diabetes Association Training Grant (1-17-PDF-076 C.J. and 1-18-PDF-132 W.X.). P.D. was supported by a post-doctoral fellowship from the American Heart Association (20POST35210738).
Footnotes
Competing interests: J.D.R. is consultant for Pfizer and scientific advisor to C.R.R., and M.A.L. is consultant to Pfizer, Novartis, Madrigal Pharmaceuticals, and Calico.
Data and materials availability: The GEO accession number for the RNA-seq, ATAC-seq, and sNuc-seq data reported in this paper is GSE143528. M.A.L. obtained REV-ERBβ floxed under a material transfer agreement with the Centre Europeen de Recherche en Biologie et Medecine.
References
- 1.Bass J, Takahashi JS, Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi JS, Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welsh DK, Takahashi JS, Kay SA, Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 72, 551–577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green CB, Takahashi JS, Bass J, The meter of metabolism. Cell 134, 728–742 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panda S, Circadian physiology of metabolism. Science 354, 1008–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trefts E, Gannon M, Wasserman DH, The liver. Curr Biol 27, R1147–R1151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koronowski KB et al. , Defining the Independence of the Liver Circadian Clock. Cell 177, 1448–1462 e1414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunger MK et al. , Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crumbley C, Wang Y, Kojetin DJ, Burris TP, Characterization of the core mammalian clock component, NPAS2, as a REV-ERBalpha/RORalpha target gene. J Biol Chem 285, 35386–35392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillaumond F, Dardente H, Giguere V, Cermakian N, Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms 20, 391–403 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y et al. , GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science 348, 1488–1492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho H et al. , Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 485, 123–127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng R et al. , Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res 47, D729–D735 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U, System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS biology 5, e34 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Martelot G et al. , REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS biology 7, e1000181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vollmers C et al. , Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proceedings of the National Academy of Sciences of the United States of America 106, 21453–21458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krenkel O, Tacke F, Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 17, 306–321 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Xu M et al. , LECT2, a Ligand for Tie1, Plays a Crucial Role in Liver Fibrogenesis. Cell 178, 1478–1492 e1420 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Xiong X et al. , Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol Cell 75, 644–660 e645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halpern KB et al. , Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z et al. , Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nature communications 10, 4562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madsen JGS et al. , Integrated analysis of motif activity and gene expression changes of transcription factors. Genome Res 28, 243–255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zucker SN et al. , Nrf2 amplifies oxidative stress via induction of Klf9. Mol Cell 53, 916–928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geraud C et al. , GATA4-dependent organ-specific endothelial differentiation controls liver development and embryonic hematopoiesis. The Journal of clinical investigation 127, 1099–1114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brocker CN et al. , Hepatocyte-specific PPARA expression exclusively promotes agonist-induced cell proliferation without influence from nonparenchymal cells. Am J Physiol Gastrointest Liver Physiol 312, G283–G299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin C et al. , JDP2, a repressor of AP-1, recruits a histone deacetylase 3 complex to inhibit the retinoic acid-induced differentiation of F9 cells. Mol Cell Biol 22, 4815–4826 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonnardel J et al. , Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 51, 638–654 e639 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett T et al. , NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 41, D991–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atger F et al. , Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proceedings of the National Academy of Sciences of the United States of America 112, E6579–6588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng MP, Liao BY, modPhEA: model organism Phenotype Enrichment Analysis of eukaryotic gene sets. Bioinformatics 33, 3505–3507 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Chong J, Wishart DS, Xia J, Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr Protoc Bioinformatics 68, e86 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Stapor P, Wang X, Goveia J, Moens S, Carmeliet P, Angiogenesis revisited-role and therapeutic potential of targeting endothelial metabolism. J Cell Sci 127, 4331–4341 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Norata GD et al. , The Cellular and Molecular Basis of Translational Immunometabolism. Immunity 43, 421–434 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Wahli W, Michalik L, PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab 23, 351–363 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Damiola F et al. , Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14, 2950–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenwell BJ et al. , Rhythmic Food Intake Drives Rhythmic Gene Expression More Potently than the Hepatic Circadian Clock in Mice. Cell Rep 27, 649–657 e645 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Mange F et al. , Diurnal regulation of RNA polymerase III transcription is under the control of both the feeding-fasting response and the circadian clock. Genome Res 27, 973–984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes ME et al. , Guidelines for Genome-Scale Analysis of Biological Rhythms. J Biol Rhythms 32, 380–393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalvisa A et al. , Insulin signaling and reduced glucocorticoid receptor activity attenuate postprandial gene expression in liver. PLoS biology 16, e2006249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quagliarini F et al. , Cistromic Reprogramming of the Diurnal Glucocorticoid Hormone Response by High-Fat Diet. Mol Cell 76, 531–545 e535 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDearmon EL et al. , Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 314, 1304–1308 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pope AJ, Karrupiah K, Kearns PN, Xia Y, Cardounel AJ, Role of dimethylarginine dimethylaminohydrolases in the regulation of endothelial nitric oxide production. J Biol Chem 284, 35338–35347 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye J et al. , TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142, 230–242 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Nagoshi E et al. , Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Ollinger R et al. , Dynamics of the circadian clock protein PERIOD2 in living cells. J Cell Sci 127, 4322–4328 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Dunlap JC, Molecular bases for circadian clocks. Cell 96, 271–290 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Chaix A, Lin T, Le HD, Chang MW, Panda S, Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell metabolism 29, 303–319 e304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dierickx P et al. , SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc Natl Acad Sci U S A 116, 12147–12152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan D, Factor D, Liu Y, Wang Z, Kao HY, The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene 32, 3819–3828 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su X, Lu W, Rabinowitz JD, Metabolite Spectral Accuracy on Orbitraps. Anal Chem 89, 5940–5948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz S et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes ME, Hogenesch JB, Kornacker K, JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25, 372–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guan D et al. , Diet-Induced Circadian Enhancer Remodeling Synchronizes Opposing Hepatic Lipid Metabolic Processes. Cell 174, 831–842 e812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu G, Wang LG, Han Y, He QY, clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corces MR et al. , Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–21 29 29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Browaeys R, Saelens W, Saeys Y, NicheNet: modeling intercellular communication by linking ligands to target genes. Nature methods 17, 159–162 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Gu Z, Gu L, Eils R, Schlesner M, Brors B, circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Madsen JGS et al. , Integrated analysis of motif activity and gene expression changes of transcription factors. Genome research, (2017). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


