SUMMARY
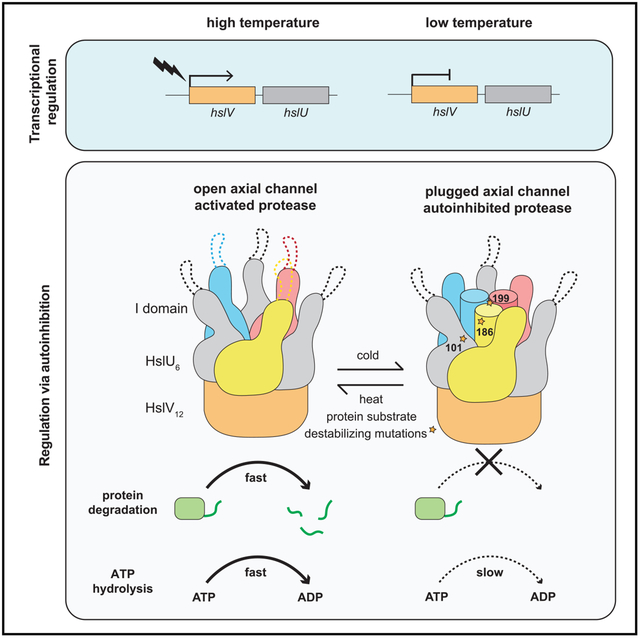
At low temperatures, protein degradation by the AAA+ HslUV protease is very slow. New crystal structures reveal that residues in the intermediate domain of the HslU6 unfoldase can plug its axial channel, blocking productive substrate binding and subsequent unfolding, translocation, and degradation by the HslV12 peptidase. Biochemical experiments with wild-type and mutant enzymes support a model in which heat-induced melting of this autoinhibitory plug activates HslUV proteolysis.
Graphical Abstract
In Brief
Baytshtok et al. demonstrate that the activity of HslUV, a AAA+ heat shock protease, is regulated by thermal melting of its autoinhibitory axial plug, which activates ATP hydrolysis, substrate binding, and energy-dependent proteolysis and ensures that robust protein degradation by HslUV occurs only at elevated temperatures in the cell.
INTRODUCTION
By degrading incomplete, unfolded, or unneeded proteins, intracellular AAA+ proteases serve important roles in protein-quality control, often removing proteins damaged by heat or other destabilizing stresses (Sauer and Baker, 2011). HslUV consists of one or two AAA+ HslU6 ring hexamers and the double-ring HslV12 peptidase (Figure 1A; Rohrwild et al., 1997; Sousa et al., 2000; Wang et al., 2001). In Escherichia coli, expression of the HslU and HslV enzymes increases ~10-fold at high temperatures (Rohrwild et al., 1996), and HslUV proteolysis is substantially faster at high than low temperatures in vitro (Burton et al., 2005). ATP-dependent proteolysis requires HslU6 to bind the degrons of target proteins in its axial channel and then unfold and translocate the substrate into the proteolytic chamber of HslV12 (Sundar et al., 2012). To reach the axial channel of the HslU hexamer, substrates must transit a funnel-like structure formed by segments of its intermediate (I) domain (Figures 1A and 1B; Sousa et al., 2000; Wang et al., 2001). Both the I-domain and this funnel-like structure are unique to the HslU subfamily of AAA+ unfoldases (Sauer and Baker, 2011). An I-domain L199Q mutation enhances degradation of some protein substrates and the rate of ATP hydrolysis (Baytshtok et al., 2016), possibly by relief of autoinhibition. However, the molecular mechanism by which Leu199 and surrounding residues affect proteolysis and ATP hydrolysis is unknown, as amino acids 177–212 are disordered in known structures (Bochtler et al., 2000; Sousa et al., 2000; Wang et al., 2001). Here, we present evidence for a model in which HslU hexamers are autoinhibited at low temperatures by a trimeric I-domain plug that blocks the axial channel but melts at high temperatures to activate proteolysis.
Figure 1. HslUV with open and closed axial channels.
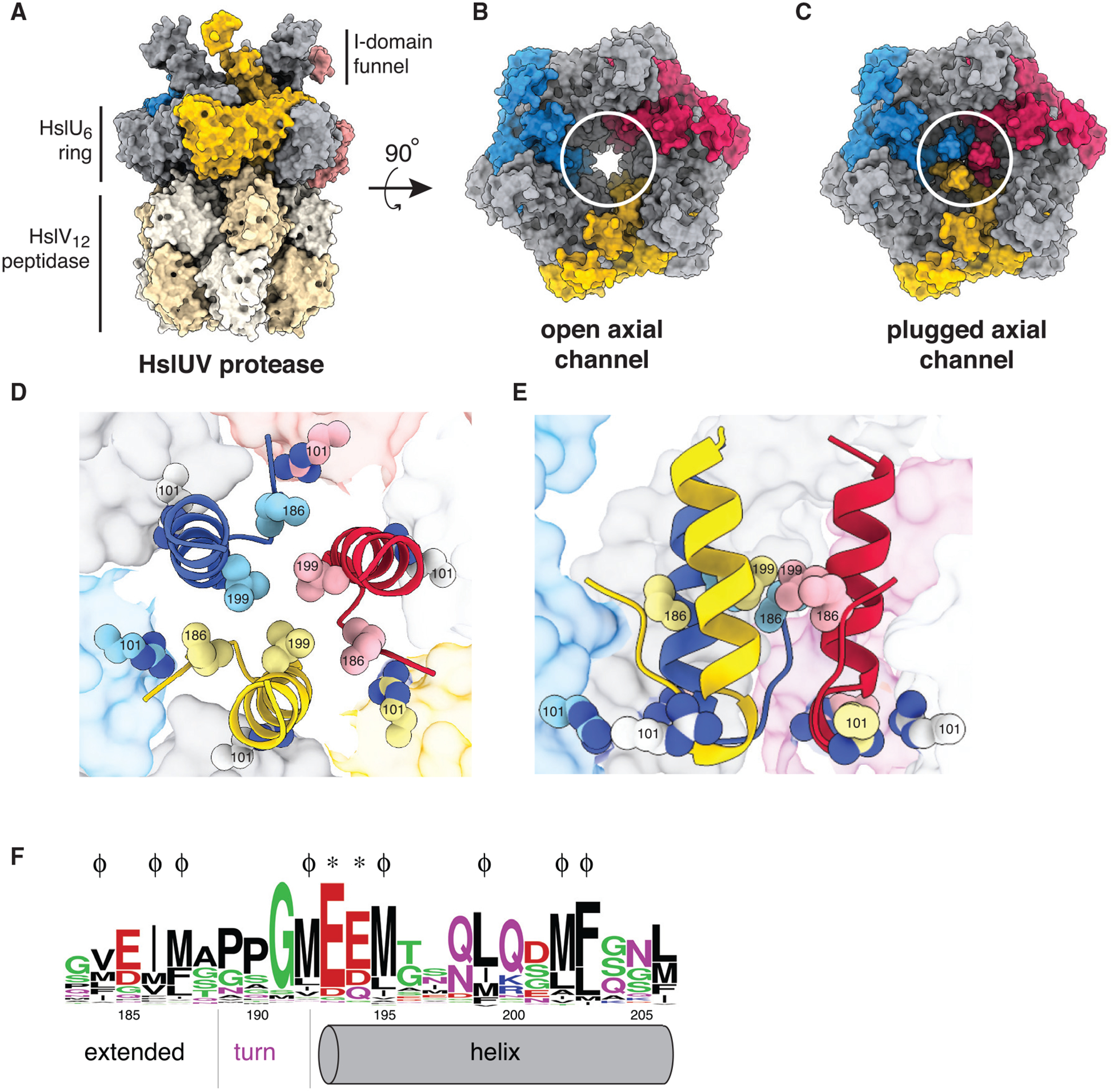
(A) Surface-representation side view of the HslUV protease (PDB: 5JI3).
(B) Surface-representation top view of open-channel HslUV structure (PDB: 5JI3).
(C) Surface-representation top view of plugged-channel HslUV structure (PDB: 6PXI).
(D) Top view of axial plug (PDB: 6PXI) in cartoon representation. The side chains of Arg101, Ile186, and Leu199 are shown as spheres. The axial channel of HslU is shown in surface representation.
(E) Cutaway side view of axial plug (PDB: 6PXI) in cartoon representation with Arg101, Ile186, and Leu199 shown as spheres.
(F) WebLogo representation of conservation of plug residues. ϕ indicates positions of hydrophobic residues (Val184, Ile186, Met187, Met192, Met195, Leu199, Met202, and Phe203) that engage in packing within the plug; asterisk (*) indicates the positions of negatively charged residues (Glu193 and Glu194) that appear to form favorable electrostatic interactions with Arg101 and Lys293 in the axial channel of the HslU hexamer.
RESULTS AND DISCUSSION
Autoinhibited structures of HslU and HslUV
We determined structures for three new crystal forms of E. coli HslU or HslUV by molecular replacement (Table 1).
Table 1.
Crystallographic statistics
| PDB entry | 6PXI | 6PXL | 6PXK |
|---|---|---|---|
| Protein | HslUV | HslU | HslU |
| Resolution (Å) | 3.45 | 3.74 | 3.65 |
| Space group | P321 | C2 | P2 |
| Unit cell | |||
| A (Å) | 168.60 | 414.59 | 200.03 |
| B(Å) | 168.60 | 92.33 | 91.20 |
| C(Å) | 162.89 | 200.85 | 201.80 |
| α | 90° | 90° | 90° |
| β | 90° | 99.43° | 99.43° |
| γ | 120° | 90° | 90° |
| No. of unique reflections | 29,212 | 68,314 | 15,427 |
| Redundancy | 2.2 | 3.8 | 5.0 |
| Completeness (%) | 81.7 (59.3) | 95.3 (77.1) | 98.4 (92.5) |
| CC-1/2 | 0.985 (0.593) | 0.981 (0.669) | 0.998 (0.609) |
| Rsym | 0.114 | 0.121 | 0.105 |
| Rpim | 0.081 | 0.068 | not calculated |
| Rwork/Rfree | 0.257/0.285 | 0.220/0.265 | 0.223/0.271 |
| MolProbity score (percentile) | 1.04 (100) | 1.31 (100) | 1.30 (100) |
| Clash score (percentile) | 2.54 (100) | 5.71 (100) | 5.9 (100) |
| Ramachandran outliers (%) | 0 | 0 | 0 |
| Ramachandran favored (%) | 98.58 | 98.15 | 99.26 |
| Bad bonds/angles | 0/0 | 0/0 | 0/0 |
| Cβ deviations | 0 | 0 | 0 |
Completeness and CC-1/2 values in parentheses are for the highest resolution shell.
Our new structures were similar to previous ones with the notable exception that a trimeric plug, formed by residues 183–206 of the I-domain, completely blocked the axial channel of the HslU ring (Figures 1C–1E). Because protein substrates must access the axial channel of HslU to be engaged, unfolded, and translocated into the peptidase chamber of HslV, this plug would inhibit substrate degradation. The plug consisted of amino acids from three of the six HslU subunits (Figures 1C–1E), each comprising an extended region (residues 183–188), a sharp turn (residues 189–191), and an α helix (residues 192–206). We could not reliably model connections between plug elements and other regions of the I-domain in specific subunits. Importantly, however, a crystallographic 3-fold axis related plug elements in the 6PXI structure, indicating that I-domain elements from three alternating subunits of the HslU hexamer must form the plug.
The three α helices that comprise the plug center packed together via hydrophobic contacts involving the side chains of Met192, Met195, Leu199, and Phe203. Hence, replacing the nonpolar Leu199 with a polar Gln199 in the L199QHslU variant should destabilize helix-helix packing and plug stability, thereby reducing autoinhibition and providing a structural rationale for the ability of L199QHslUV to degrade some protein substrates more rapidly than wild-type HslUV (WTHslUV; Baytshtok et al., 2016). In prior biochemical experiments, the subtilisin and chymotrypsin endoproteases were found to cleave sites within the plug region of L199QHslU faster than in WTHslU (Baytshtok et al., 2016), supporting a model in which destabilization of the mutant plug results in partial melting and increased susceptibility to proteolytic cleavage. Additional hydrophobic packing contacts within the plug included Val184 and Ile186 from the extended region, which occupy peripheral grooves between plug helices, and Met202 from the helix. Notably, residues that form hydrophobic interactions within the plug are highly conserved in HslU orthologs (Figure 1F).
A Pro189-Pro190-Gly191 tripeptide formed a sharp turn between the extended region and α helix of the plug. The negatively charged side chains of Glu193 and Glu194 following this turn were also close to the positively charged side chains of Arg101 (94% conserved) and Lys293 (92% conserved) in the HslU axial channel, suggesting that favorable electrostatic interactions also stabilize the plugged conformation. Conservation of the plug residues that form the turn and electrostatic interactions within the HslU channel (Figure 1F) provide additional support for a model in which the autoinhibited conformation is functionally important.
Electron microscopy of closed- and open-channel HslU structures
To investigate channel accessibility in WTHslU and L199QHslU in a non-crystalline environment, we used negative-stain electron microscopy. Both enzyme samples were equilibrated at room temperature prior to application to grids and staining. Class-average images of WTHslU, viewed approximately down the axis of the AAA+ ring, showed a shallow axial depression or blocked channel (Figure 2A, top row), similar to projections of plugged crystallographic HslU hexamers filtered to 15 Å (Figure 2A, bottom row). Most class averages for L199QHslU hexamers (Figure 2B, top row), by contrast, resembled open-channel crystal structures filtered to 15 Å (Figure 2B, bottom row). These results provide further evidence that the axial channel of WTHslU at room temperature is plugged in most enzymes and that the L199Q mutation shifts the equilibrium to favor the open-channel conformation.
Figure 2. Effects of L199Q mutation on HslU channel size.
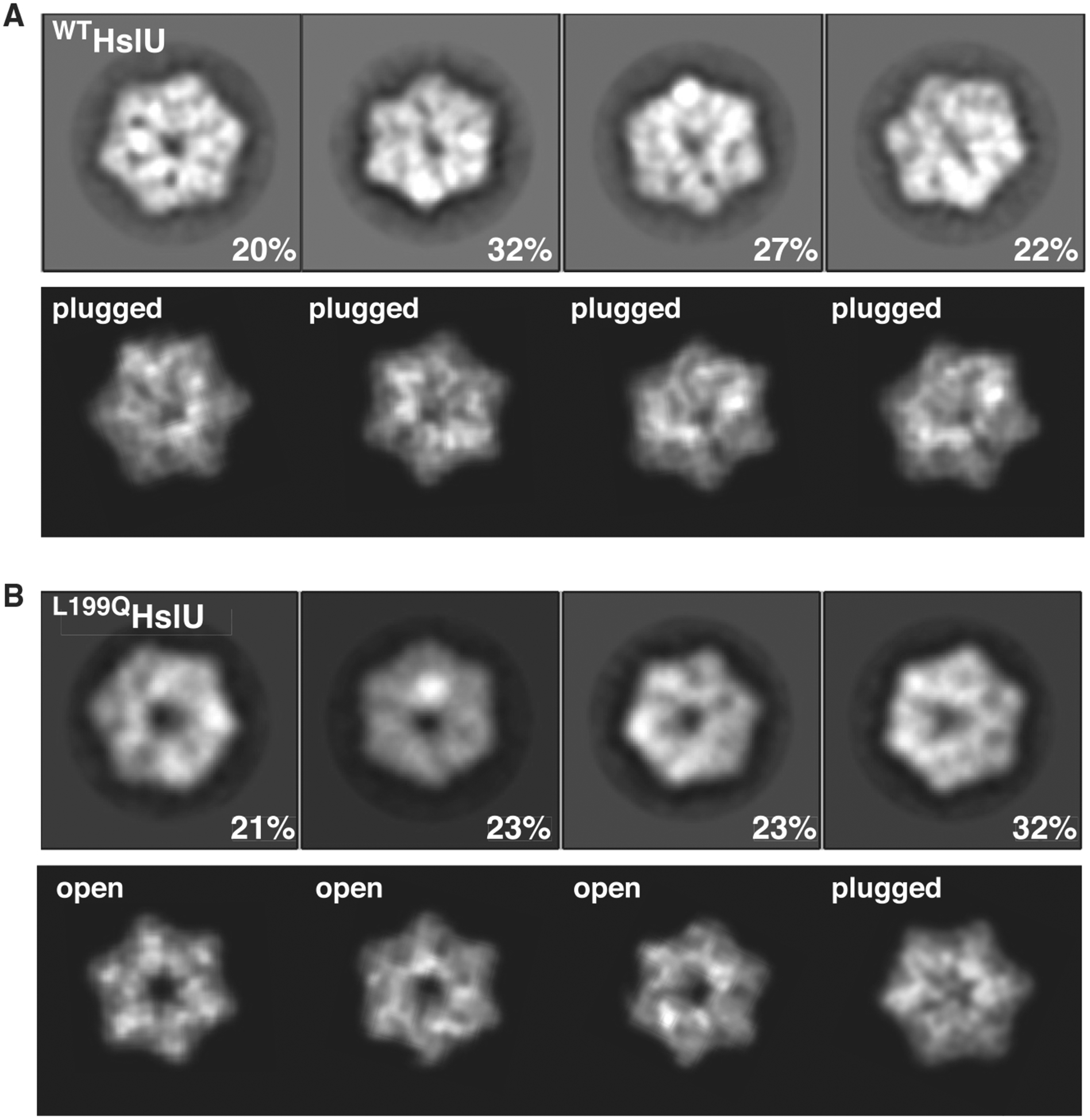
(A) Class-average negative-stain electron microscopy (EM) images of WTHslU (top row) compared with projections of a plugged hexamer (PDB: 6PXK) filtered to 15-Å resolution (bottom row). Samples were prepared at room temperature.
(B) Class-average images of L199QHslU (top row) compared with filtered images of an open-channel hexamer (PDB: 5JI3) or the plugged hexamer (PDB: 6PXK).
Plug stability affects temperature-dependent activity
We propose that high temperature destabilizes the plug and favors the active open-channel conformation of HslUV, whereas low temperature stabilizes the inactive plugged-channel conformation (Figure 3A). As a first test of this model, we measured the rates of ATP hydrolysis by WTHslUV and L199QHslUV at 25°C, 35°C, 45°C, and 55°C without or with the I37AArc-cp6GFP-st11-ssrA protein substrate (Figures 3B and 3C). These data fit well to melting curves calculated assuming that the high-temperature conformations of WTHslUV and L199QHslUV had the same ATPase activity, whereas the low-temperature conformations were inactive in ATP hydrolysis. Without protein substrate, the fitted temperatures of half-maximal activity (TM) were 42°C for L199QHslUV and 52°C for WTHslUV (Figure 3B). Protein substrate reduced the TM values by 10°C–12°C for both enzymes (Figure 3C), indicating that binding of the I37AArc-cp6GFP-st11-ssrA substrate preferentially stabilizes the active enzyme conformation. The lower TM values for L199QHslUV than for WTHslUV, both with and without protein substrate, strongly support our model that plug melting results in temperature-dependent relief of autoinhibition.
Figure 3. Activities of WTHslUV and mutants with destabilized plugs.
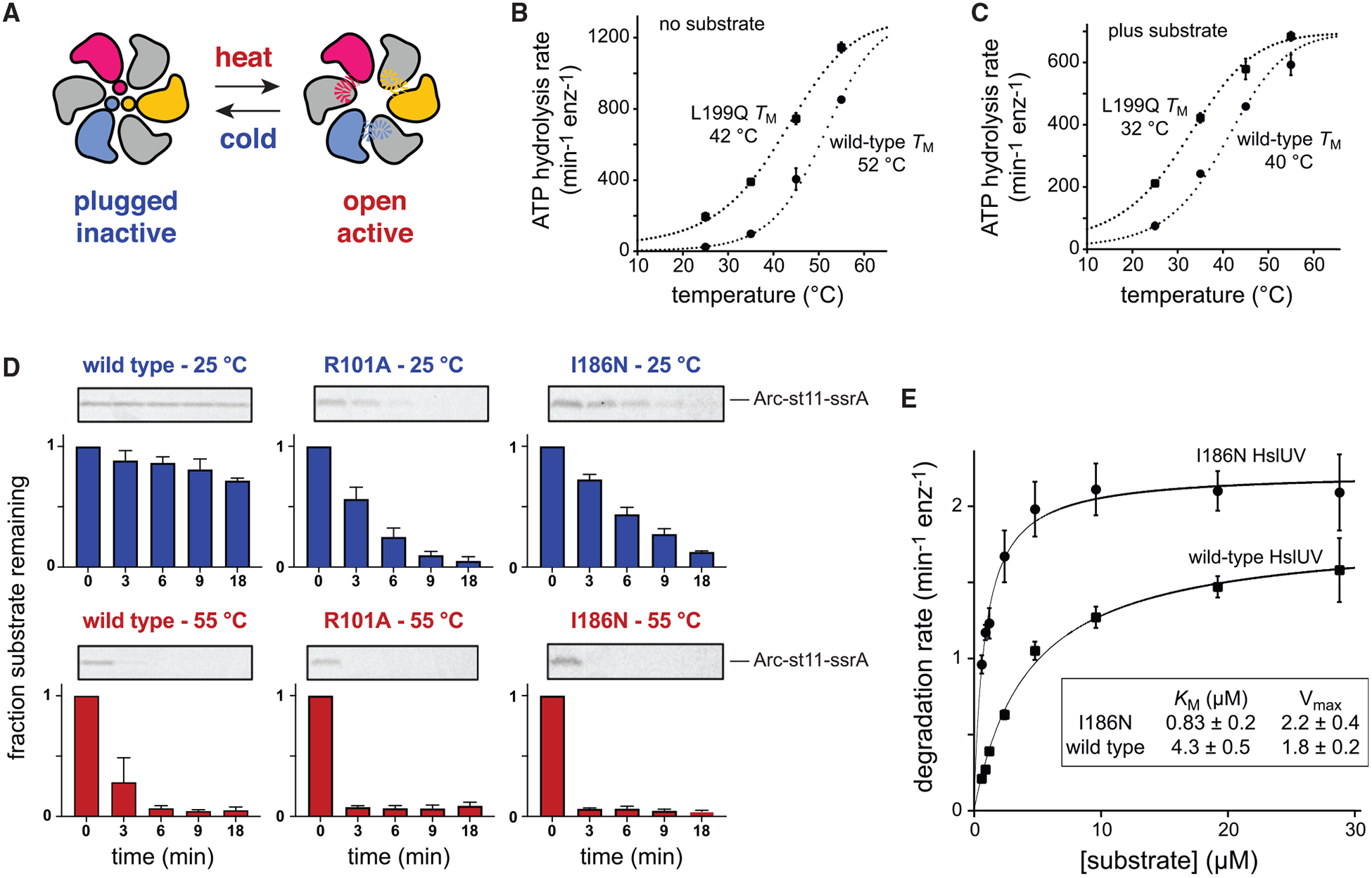
(A) Model for activation of HslU by thermal melting of the autoinhibitory plug.
(B) Temperature dependence of ATP hydrolysis by WTHslUV or L199QHslUV (0.3 μM HslU6, 0.9 μM HslV12). Values are averages (±SD) of three independent experiments. The dotted lines are fits to melting curves.
(C) Same as in (B) but in the presence of the I37AArc-cp6GFP-st11-ssrA substrate (50 μM).
(D) SDS-PAGE was used to assay the kinetics of degradation of the Arc-st11-ssrA substrate (5 μM) at 25°C or 55°C by WTHslUV, R101AHslUV, or I186NHslUV (0.3 μM HslU6, 0.9 μM HslV12). In each subpanel, a representative gel is shown at the top, and the mean (±SD) of substrate remaining in three independent replicates is shown in the graph below.
(E) Michaelis-Menten plots and steady-state kinetic parameters for I37AArc-cp6GFP-st11-ssrA degradation at 37°C by WTHslUV or I186NHslUV (0.3 μM HslU6, 0.9 μM HslV12). Values plotted are averages (±SD) of three independent replicates.
As a second test of our temperature-dependent activation model, we used SDS-PAGE to measure HslUV degradation of an Arc-st11-ssrA substrate at 25°C or 55°C (Figure 3D). As expected from findings of a previous study (Burton et al., 2005), degradation by the WT enzyme was very slow at 25°C and much faster at 55°C. We also assayed degradation of the same substrate by R101AHslUV, constructed to remove favorable electrostatic interactions between Arg101 in the axial channel and glutamates in the autoinhibitory plug, and by I186NHslUV, which replaces a hydrophobic residue that normally stabilizes plug packing with a polar residue (Figures 1D and 1E). At 25°C, both the R101AHslUV and I186NHslUV enzymes degraded Arc-st11-ssrA substantially faster than did WTHslUV (Figure 3D), as expected by our model. At 55°C, all three enzymes degraded Arc-st11-ssrA more rapidly than at the lower temperature, with WTHslUV degradation being slower than degradation by the mutant enzymes. In each case, faster substrate degradation at the higher temperature probably results from a combination of reduced autoinhibition and reduced stability of the native portion of the Arc-st11-ssrA substrate. Importantly, these results support a model in which the R101A and I186N mutations destabilize the autoinhibited conformation at low temperatures and thereby increase protease activity relative to the WT enzyme.
As a third test of our model, we determined steady-state degradation rates of different concentrations of the I37AArc-cp6GFP-st11-ssrA substrate at a temperature (37°C) at which both WTHslUV and I186NHslUV have substantial activity and fit the data to the Michaelis-Menten equation to determine KM and Vmax values (Figure 3E). At low substrate concentrations, degradation by I186NHslUV was ~6-fold faster than by WTHslUV, largely as a consequence of a tighter KM for the mutant (Figure 3E). By contrast, degradation of high concentrations of this substrate by I186NHslUV was only ~1.2-fold faster than by WTHslUV. These results support a model in which plug destabilization, as a consequence of reduced hydrophobic packing, makes the axial channel accessible in a larger fraction of the population of I186N mutant enzymes at low substrate concentrations, thereby resulting in tighter substrate binding and faster degradation. Very high substrate concentrations, in turn, appear to shift the equilibrium toward the open-channel conformation of WTHslUV, thereby reducing the activity difference between the mutant and WT enzymes.
Conclusions and biological inferences
We have identified a new conformation of the HslUV protease in which part of the I-domain forms a plug that blocks the axial channel of the AAA+ HslU ring and prevents productive substrate engagement and thus degradation. This plugged-channel inactive conformation is in dynamic equilibrium with an open-channel active conformation, with higher temperature and/or higher protein-substrate concentrations favoring the active conformation.
In cells, direct thermal activation of HslUV by melting of the autoinhibitory plug at heat shock temperatures (Figure 3A) would increase protease activity almost immediately. Temperature-induced expression increases of HslU and HslV would then further amplify HslUV proteolytic capacity. Following a return to lower temperatures, levels of the HslUV enzyme would be expected to remain high for several generations before reduced expression and cell division allowed a return to the low-temperature steady state. During this transition from high to low temperatures, increased plug-mediated autoinhibition could contribute to cell fitness by minimizing rogue HslUV proteolysis and/or reducing excessive ATP hydrolysis.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests for information and resources should be directed to and will be fulfilled by the Lead Contact, Robert T. Sauer (bobsauer@mit.edu).
Materials Availability
This study generated new His6-tagged mutants of E. coli HslU containing the R101A and I186N mutations. Plasmids expressing these mutant enzymes are available from the lead contact upon request without restriction.
Data and Code Availability
Coordinates, structure factors and electron-density maps for the 6PXI, 6PXL, and 6PXK crystal structures are available from the RCSB Protein Data Bank (https://www.rcsb.org/). The biochemical data and electron micrographs supporting the current study are available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Proteins were expressed in E. coli strain X90 (λDE3) slyD::kan hslUV::tet). No additional biological strains were used in this work.
METHOD DETAILS
Genes encoding H6-tagged variants of E. coli HslU, E257QHslU, L199QHslU, I186NHslU, R101AHslU, HslV, I37AArc-cp6GFP-st11-ssrA, and Arc-st11-ssrA were expressed from pET12b or pET21b vectors (Novagen) in E. coli strain X90 (λDE3) slyD::kan hslUV::tet and proteins were purified as described (Baytshtok et al., 2016).
Crystals of HslU were grown at 4°C by the hanging-drop method after mixing 0.5 μL of selenomethionine-labeled H6-tagged HslU (15 mg/mL) in 15 mM Tris (pH 7.5), 100 mM NaCl, 20 mM MgCl2, 0.2 mM EDTA, and 5 mM ATPγS with an equal volume of well solution containing 100 mM Tris (pH 7.5), 18% PEG-3350, and either 0.1 M (PDB code 6PXK) or 0.2 M (PDB code 6PXL) ammonium sulfate. For cryo-protection, PEG-3350 was increased to 20% and 10% MPD was added to the base-well solution. HslUV crystals were grown in the same way except the protein solution contained E257QHslU (10 mg/mL) plus HslV (9.6 mg/mL) and the well solution contained 0.1 M Bis-Tris (initial pH 5.5), 1.85 M ammonium sulfate, 5% glycerol, and 5 mM ATPγS. For cryo-protection, glycerol was increased to 25%. Diffraction data for the HslUV crystals were collected on our home source (Rigaku MicroMax-007HF with a Saturn 944 detector) and for the HslU crystals at the Advanced Photon Source (APS) beamline 24-ID-E and processed using HKL2000 (Otwinowski and Minor, 1997) or XDS (Kabsch, 2010). Structures were solved by molecular replacement with PHASER (McCoy et al., 2007) using search models consisting of HslU hexamers alone or HslUV (PDB code 5JI3; Baytshtok et al., 2016). Anomalous difference maps – showing the selenomethionine side chains of residues 187, 192, 195, and 202 – were used to assist model building into plug density. Phenix was used to refine structures (Adams et al., 2010), Coot was used for model building (Emsley et al., 2010), and MolProbity was used to assess model geometry (Chen et al., 2010).
Negative-stain EM experiments were performed as described (Baytshtok et al., 2016), using WT WTHslU or L199QHslU (1.5 μM) and ATPγS (5 mM) in 20 mM HEPES (pH 7.5), 5 mM MgCl2, 500 mM NaCl, 10% glycerol (v/v), and 0.032% Igepal CA-630. Particles with tilted six-fold symmetry axis were removed by two rounds of 2D classification. For WTHslU, 10927 particles from 23 micrographs were used to generate representative 2D class averages; for L199QHslU, 5567 particles from 20 micrographs were used. The e2pdb2mrc and e2project3d utilities in EMAN2 (Tang et al., 2007) were used to generate 2D projections from PDB files and Relion 3.0.8 (Scheres, 2012) was used for 2D classification.
Degradation and ATPase assays were performed in 25 mM HEPES (pH 7.5), 5 mM KCl, 20 mM MgCl2, 10% glycerol, and 0.032% Igepal CA-630 as described (Baytshtok et al., 2016). ATPase rates at different temperatures were measured using a Spectramax M5 plate reader (Molecular Devices) by using an NADH-coupled assay (Nørby, 1988) with or without 50 μM I37AArc-cp6GFP-st11-ssrA. Using the Solver tool of Microsoft Excel, the temperature dependencies of ATPase rates were globally fitted to the equation max/(1+exp(ΔH/R·(1/TM-1/T))), where max is the maximum ATPase rate, T is the temperature in Kelvin, TM is the temperature at 50% activity, ΔH is the enthalpy at TM, and R is the universal gas constant. Degradation of I37AArc-cp6GFP-st11-ssrA by WTHslUV or I186NHslUV was monitored by loss of fluorescence (excitation 467 nm; emission 511 nm). For degradation of Arc-st11-ssrA (5 μM) by wild-type or mutant enzymes (0.3 μM HslU6, 0.9 μM HslV12), proteins were preincubated for 1 min at 25°C or 55°C before adding an ATP regeneration system (2.5 mM ATP, 7.5 mM phosphoenolpyruvate, 1 mM NADH, 18.8 U/ml pyruvate kinase, 21.5 U/ml lactate dehydrogenase) to initiate degradation. Aliquots were removed at different times, quenched by addition of Tricine sample buffer (Bio-Rad) with β-mercaptoethanol, and flash frozen in liquid nitrogen. Samples were then boiled for 5 min and electrophoresed on 15% Tris-Tricine SDS-PAGE gels. After electrophoresis, gels were stained for 10 min using a Coomassie-blue solution, destained overnight in water, scanned using a Typhoon FLA 9500 Imager (GE Healthcare), and the amount of substrate remaining at each time point was quantified using ImageQuant TL (GE Healthcare).
QUANTIFICATION AND STATISTICAL ANALYSIS
Crystallographic unit-cell parameters, data completeness and redundancy, CC1/2 values, and merging R-factors (Table 1) were obtained using HKL2000 (Otwinowski and Minor, 1997), which was used to index, integrate, and scale the diffraction data. Model-refinement statistics (Rwork, Rfree; Table 1) were obtained from the phenix.refine program in Phenix (Adams et al., 2010) and are shown in Table 1. Statistics for the quality of the final crystallographic models (Table 1) were determined using MolProbity (Chen et al., 2010). Biochemical experiments were performed using a minimum of three independent replicates. Biochemical data were plotted as averages ± one standard deviation (SD), as described in the Figure 3 legend, using Prism (GraphPad). The errors for KM and Vmax (Figure 3E) were calculated by non-linear least-squares fitting to the Michealis-Menten equation using Prism (GraphPad).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| X90 (λDE3) slyD::kan hslUV::tet | Baytshtok et al., 2016 | N/A |
| Proteins and Enzymes | ||
| HslU (His6 tagged) | Bochtler et al., 2000 | N/A |
| L199QHslU (His6 tagged) | Baytshtok et al., 2016 | N/A |
| R101AHslU (His6 tagged) | This study | N/A |
| i186NHsiu (His6 tagged) | This study | N/A |
| e257QHsiu (His6 tagged) | Yakamavich et al., 2008 | N/A |
| HslV (His6 tagged) | Bochtler et al., 2000 | N/A |
| I37AArc-cp6GFP-st11-ssrA | Baytshtok et al., 2016 | N/A |
| Arc-st11-ssrA | Milla et al., 1993 | N/A |
| pyruvate kinase | Sigma-Aldrich | N/A |
| lactate dehydrogenase | Sigma-Aldrich | N/A |
| Deposited Data | ||
| E257QHsU-HslV-ADP, plugged | This study | PDB: 6PXI |
| WTHslU- ADP, plugged | This study | PDB: 6PXL |
| WTHslU- ADP, plugged | This study | PDB: 6PXK |
| Software and Algorithms | ||
| Coot | Emsley et al., 2010 | N/A |
| ChimeraX | Goddard et al., 2018 | N/A |
| Ctffind3 | Mindell and Grigorieff, 2003 | N/A |
| EMAN2 | Tang et al., 2007 | N/A |
| HKL2000 | Otwinowski and Minor, 1997 | N/A |
| Phenix | Adams et al., 2010 | N/A |
| PyMOL | Schrodinger, LLC | N/A |
| Relion | Scheres, 2012 | N/A |
| XDS | Kabsch, 2010 | N/A |
Highlights.
A trimeric structure can plug the axial channel of the AAA+ HslU ring hexamer
This autoinhibitory plug is observed in crystal structures and by electron microscopy
Temperature-induced melting of the plug activates ATP hydrolysis and proteolysis
Low-temperature autoinhibition limits rogue degradation after recovery from heat shock
ACKNOWLEDGMENTS
This work was supported by NIH grant AI-15706. NECAT beamline studies at APS were supported by NIGMS (P41 GM103403) and DOE (DE-AC02–06CH11357) grants.
Footnotes
DECLARATION OF INTERESTS
We, the authors and our immediate family members, have no competing financial interests to declare.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baytshtok V, Fei X, Grant RA, Baker TA, and Sauer RT (2016). A structurally dynamic region of the HslU intermediate domain controls protein degradation and ATP hydrolysis. Structure 24, 1766–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, and Huber R (2000). The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403, 800–805. [DOI] [PubMed] [Google Scholar]
- Burton RE, Baker TA, and Sauer RT (2005). Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat. Struct. Mol. Biol 12, 245–251. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Cryst 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla ME, Brown BM, and Sauer RT (1993). P22 Arc repressor: enhanced expression of unstable mutants by addition of polar C-terminal sequences. Protein Sci. 2, 2198–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, and Grigorieff N (2003). Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol 142, 334–347. [DOI] [PubMed] [Google Scholar]
- Nørby JG (1988). Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 156, 116–119. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–325. [DOI] [PubMed] [Google Scholar]
- Rohrwild M, Coux O, Huang HC, Moerschell RP, Yoo SJ, Seol JH, Chung CH, and Goldberg AL (1996). HslV-HslU: A novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc. Natl. Acad. Sci. USA 93, 5808–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrwild M, Pfeifer G, Santarius U, Müller SA, Huang HC, Engel A, Baumeister W, and Goldberg AL (1997). The ATP-dependent HslVU protease from Escherichia coli is a four-ring structure resembling the proteasome. Nat. Struct. Biol 4, 133–139. [DOI] [PubMed] [Google Scholar]
- Sauer RT, and Baker TA (2011). AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem 80, 587–612. [DOI] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, and McKay DB (2000). Crystal and solution structures of an HslUV protease-chaperone complex. Cell 103, 633–643. [DOI] [PubMed] [Google Scholar]
- Sundar S, Baker TA, and Sauer RT (2012). The I domain of the AAA+ HslUV protease coordinates substrate binding, ATP hydrolysis, and protein degradation. Protein Sci. 21, 188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, and Chung CH (2001). Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure 9, 1107–1116. [DOI] [PubMed] [Google Scholar]
- Yakamavich JA, Baker TA, and Sauer RT (2008). Asymmetric nucleotide transactions of the HslUV protease. J. Mol. Biol 380, 946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Coordinates, structure factors and electron-density maps for the 6PXI, 6PXL, and 6PXK crystal structures are available from the RCSB Protein Data Bank (https://www.rcsb.org/). The biochemical data and electron micrographs supporting the current study are available from the Lead Contact upon request.


