Abstract
Background
This study developed a diagnostic tool to automatically detect normal, unclear and tumor images from colonoscopy videos using artificial intelligence.
Methods
For the creation of training and validation sets, 47,555 images in the jpg format were extracted from colonoscopy videos for 24 patients in Korea University Anam Hospital. A gastroenterologist with the clinical experience of 15 years divided the 47,555 images into three classes of Normal (25,895), Unclear (2038) and Tumor (19,622). A single shot detector, a deep learning framework designed for object detection, was trained using the 47,255 images and validated with two sets of 300 images—each validation set included 150 images (50 normal, 50 unclear and 50 tumor cases). Half of the 47,255 images were used for building the model and the other half were used for testing the model. The learning rate of the model was 0.0001 during 250 epochs (training cycles).
Results
The average accuracy, precision, recall, and F1 score over the category were 0.9067, 0.9744, 0.9067 and 0.9393, respectively. These performance measures had no change with respect to the intersection-over-union threshold (0.45, 0.50, and 0.55). This finding suggests the stability of the model.
Conclusion
Automated detection of normal, unclear and tumor images from colonoscopy videos is possible by using a deep learning framework. This is expected to provide an invaluable decision supporting system for clinical experts.
Keywords: Colon, Neoplasm, Artificial intelligence
Background
Colorectal cancer is a leading cause of disease burden in the world. It was the third and second greatest sources of cancer incidence and mortality in the world for year 2018, respectively—it accounted for 10.2% (1,849,518) of new cancer cases (18,078,957) and 9.2% (880,792) of total cancer deaths (9,555,027) [1, 2]. This global pattern is consistent with its local counterpart in Korea. Colorectal cancer ranked second and third in terms of cancer incidence and mortality in the country for year 2016, respectively—it was responsible for 12.3% (28,127) of new cancer cases (229,180) and 10.7% (8,358) of total cancer deaths (78,194) [3]. Indeed, its economic burden became more significant in the country during 2000–2010. Its ranking and amount registered a rapid rise from the 5th/837 in 2000 to the 3rd/2,210 in 2010 (million US$ for the total population) [4]. Colonoscopy is an effective way to screen colorectal tumors and prevent colorectal cancer [5, 6]. However, its performance depends on various factors including tumor size and screening conditions. Its sensitivity can be as low as 0.75 depending on tumor size [5, 6]. This situation gets even worse with image blurring from screen shaking or fluid injection. However, the recent development of artificial intelligence (AI) is expected to provide an invaluable decision supporting system for endoscopists to overcome this challenge.
The artificial neural network is a popular AI model including one input layer, one, two or more hidden layers and one output layer. Neurons in a previous layer unite with the weights in the next layer. This process can be denoted as the feedforward algorithm. Then, these weights are refined by the amounts of their contributions for a difference between the actual and predicted final outputs. This process can be denoted as the backpropagation algorithm. These processes are iterated until a certain criterion is met for the accurate prediction of the dependent variable [7, 8]. The convolutional neural network (CNN) is an artificial neural network including convolutional layers. In the convolutional layer, a feature detector slides across input data and the dot product of its elements and their input data counterparts is computed. This process leads to effective identification of the CNN for specific features of the input data [8, 9]. Based on a recent review, the CNN is expected to aid in endoscopists’ accurate diagnosis of gastrointestinal regions [10]. Especially, some studies report that the CNN outperformed endoscopists for the classification of colorectal tumors (86% vs. 74%) [11, 12]. However, little study has been done and more effort is needed on this topic. In this context, this study developed a diagnostic tool to automatically detect normal, unclear and tumor images from colonoscopy videos using the CNN.
Methods
Study participants
This study was approved by the Institutional Review Board (IRB) of Korea University Anam Hospital on October, 17, 2019 (IRB No. 2019AN0424). Informed consent was waived by the IRB. For the creation of training and validation sets, 47,555 images in the jpg format were extracted from colonoscopy videos for 24 patients in Korea University Anam Hospital (Additional file 1: Table S1). The resolution of a video was 720 × 480 i60. A gastroenterologist with the clinical experience of 15 years divided the 47,555 images into three classes of Normal (25,895), Unclear (2038) and Tumor (19,622). The class of Unclear included blurred images with screen shaking, fluid injection and other causes.
Model development
A single shot detector (SSD) [9, 13], a deep learning framework designed for object detection, was trained using the 42,555 images and validated with two sets of 300 images—each validation set included 150 images (50 normal, 50 unclear and 50 tumor cases). Half of the 47,255 images were used for building the model and the other half were used for testing the model. This process of model building and testing (“training process”) was repeated 250 times (250 epochs) to improve the model. The two validation sets with 300 images total, were completely separate from the training process and were used for validating the model. Here, the training set of 47,255 images and one validation set of 150 images (validation set 1) came from 5 patients while the other validation set of 150 images (validation set 2) came from other 19 patients (Table S1, supplementary information). The learning rate of the model was 0.0001 during the 250 epochs. SSD, which does not require the stages of proposal generation and feature resampling, is faster than another detection model, Faster R-CNN [14]. Indeed, SSD has an important advantage as compared to CNNs for disease classification. These models only classify a single disease, i.e., testing whether it belongs to a certain category (e.g., normal vs. tumor). They do not provide additional information on the regions of interest (i.e., the locations of the lesions). On the contrary, SSD output covers not only the types of various diseases but also the locations of their lesions, which helps clinicians improve their diagnostic criteria.
Performance measures
Accuracy, precision, recall, and F1 score are the performance measures of the model [9]. These measures were calculated for three thresholds of the intersection over union (IOU), i.e., 0.45, 0.50, and 0.55 [9, 15]. The model can be considered stable when its performance measures show no or little changes with respect to the three IOU thresholds. The Python programming language (v.3.52) and a graphics card (GeForce GTX 1080 Ti D5X 11 GB) were used for the analysis.
Results
After 250 training epochs, the test loss of the model decreased from 11.66 to 1.79 (Fig. 1). Table 1 shows the confusion matrix of the model, which compares the predicted classes against the true classes for 150 images in each of the validation sets 1 and 2. The prediction was repeated 10 times, and the average over 10 runs is presented in Table 1. The confusion matrix had no change with respect to the IOU threshold (0.45, 0.50, and 0.55). This finding suggests the stability of the model. The accuracy, precision, recall, and F1 score of the model are shown in Table 2. These values were derived from Table 1, which represents the confusion matrix of the model with the average over 10 runs of the prediction. These performance measures also had no change with respect to the IOU threshold (0.45, 0.50, and 0.55). The respective accuracy measures of the validation sets 1 and 2 were 0.9733 and 9067. The respective precision results of the validation sets 1 and 2 were: (1) 1.0000 and 1.0000 for Normal, (2) 1.0000 and 1.0000 for Unclear, (3) 1.0000 and 0.9231 for Tumor and (4) 1.0000 and 0.9744 for the average of the three groups. The respective recall measures of the validation sets 1 and 2 were: (1) 1.0000 and 0.9800 for Normal, (2) 1.0000 and 0.7800 for Unclear, (3) 0.9200 and 0.9600 for Tumor and (4) 0.9733 and 0.9067 for the average of the three groups. Similarly, the respective F1 scores of the validation sets 1 and 2 were: (1) 1.0000 and 0.9899 for Normal, (2) 1.0000 and 0.8764 for Unclear, (3) 0.9583 and 0.9412 for Tumor and (4) 0.9865 and 0.9393 for the average of the three groups. Both the validation sets 1 and 2 were completely separate from the training process and the unit of analysis is a normal, unclear or tumor image (not a patient). For this reason, both the validation sets 1 and 2 were used for validating the model in this study. However, the validation set 1 came from 5 patients (the sources of the training set of 47,255 images as well), while the validation set 2 came from other 19 patients (Table S1, supplementary information). The validation set 2 is expected to be more reliable than the validation set 1. Examples of correctly classified cases are presented in Fig. 2.
Fig. 1.
Test loss during training epochs
Table 1.
Confusion matrix
| Predicted | True | |||
|---|---|---|---|---|
| Background | Normal | Unclear | Tumor | |
| Validation set 1 (5 patients) | ||||
| Backgrounda | 0 | 0 | 0 | 4 |
| Normal | 0 | 50 | 0 | 0 |
| Unclear | 0 | 0 | 50 | 0 |
| Tumor | 0 | 0 | 0 | 46 |
| Predicted | True | |||
|---|---|---|---|---|
| Background | Normal | Unclear | Tumor | |
| Validation set 2 (19 patients) | ||||
| Background | 0 | 1 | 7 | 2 |
| Normal | 0 | 49 | 0 | 0 |
| Unclear | 0 | 0 | 39 | 0 |
| Tumor | 0 | 0 | 4 | 48 |
aBackground indicates that the model does not bring any detection result
Table 2.
Model performance
| Validation set 1 | Validation set 2 | |
|---|---|---|
| Accuracy | 0.9733 | 0.9067 |
| Normal | ||
| Precision | 1.0000 | 1.0000 |
| Recall | 1.0000 | 0.9800 |
| F1 score | 1.0000 | 0.9899 |
| Unclear | ||
| Precision | 1.0000 | 1.0000 |
| Recall | 1.0000 | 0.7800 |
| F1 score | 1.0000 | 0.8764 |
| Tumor | ||
| Precision | 1.0000 | 0.9231 |
| Recall | 0.9200 | 0.9600 |
| F1 score | 0.9583 | 0.9412 |
| Class average | ||
| Precision | 1.0000 | 0.9744 |
| Recall | 0.9733 | 0.9067 |
| F1 score | 0.9865 | 0.9393 |
Fig. 2.
Examples of predicted classes and boxes. a Tumor, b unclear and c normal
Discussion
In this study, 47,555 images were extracted from colonoscopy videos for 24 patients in a general hospital, and a deep learning framework (SSD) was developed to automatically detect normal, unclear and tumor images. The performance of the model was excellent in standard measures. The average accuracy, precision, recall, and F1 score over the category were 0.9067, 0.9744, 0.9067 and 0.9393, respectively. These performance measures had no change with respect to the IOU threshold (0.45, 0.50, and 0.55). This finding suggests the stability of the model. A recent review shows that the development and application of the CNN has been popular and successful in gastrointestinal endoscopy with the range of its accuracy from 75.1 to 94.0% [10]. Specifically, the CNN was reported to be better than endoscopists for the classification of colorectal tumors, that is, 86% versus 74% in terms of accuracy [11, 12]. But it has been very rare in this area to develop and apply SSD with multiple classes such as normal, unclear and tumor images. In this vein, this study developed a diagnostic tool to automatically detect normal, unclear and tumor images from colonoscopy videos using SSD. The performance of SSD with three classes in this study was comparable to the very best of the existing literature with binary classes.
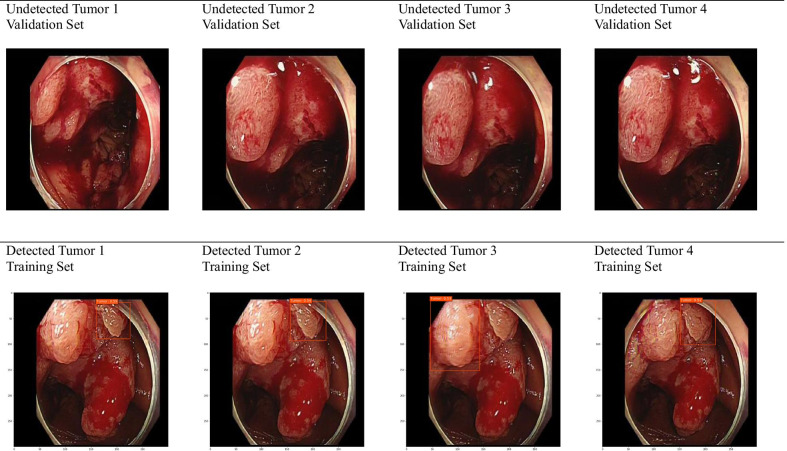
However, this study had some limitations. Four undetected tumors in the validation set 1 are displayed in the first row of Fig. 3 (These tumors were classified as background with no detection results). For comparison, four detected tumors from the training set are presented in the second row. The undetected tumors in the validation set look bigger and more evenly spread than do their detected counterparts from the training set. The former look more homogenous than do the latter in terms of color as well. This would explain why the model predicted the undetected tumors to be background with no detection result. One effective solution would be to expand the training set with this type of tumors and to perform additional training of the model. Indeed, it would be a good topic for future research to diversify the classes of colonoscopy images in terms of tumor’s shape, color and severity.
Fig. 3.
Undetected and detected tumors
Conclusion
Automated detection of normal, unclear and tumor images from colonoscopy videos is possible by using a deep learning framework. This is expected to provide an invaluable decision supporting system for clinical experts.
Supplementary information
Additional file 1: Table S1. Descriptive statistics of study participants.
Acknowledgements
Not applicable.
Abbreviations
- AI
Artificial Intelligence
- CNN
Convolutional Neural Network
- SSD
Single Shot Detector
Authors’ contributions
KSL, SHS, SHP and ESK (corresponding author) have directly participated in the planning, execution and analysis of the study. All authors read and approved the final manuscript.
Funding
This work was supported by the Technology Development Program funded by the Ministry of SMEs and Startups of South Korea (Program No. S2680996), the Korea University Grant, the Technology Innovation Program funded by the Ministry of Trade, Industry and Energy of South Korea (Program No. 20003767), and the Ministry of Science and ICT of South Korea under the Information Technology Research Center support program supervised by the IITP (Institute for Information & Communications Technology Planning & Evaluation) (Program No. IITP-2018-0-01405). This fund provided resources for the design of the study, the collection, analysis and interpretation of the data and the writing of the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board (IRB) of Korea University Anam Hospital on October, 17, 2019 (IRB No. 2019AN0424). The IRB granted permissions to access the clinical/personal patient data used in this study. Informed consent was waived by the IRB.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12911-020-01314-8.
References
- 1.1International Agency for Research on Cancer. Globocan 2018. https://gco.iarc.fr/today/data/factsheets/cancers/10_8_9-Colorectum-fact-sheet.pdf. Accessed 17 April 17 2020
- 2.GBD 2017 Colorectal Cancer Collaborators The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2019;4:913–933. doi: 10.1016/S2468-1253(19)30345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung KW, Won YJ, Kong HJ, Lee ES. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2016. Cancer Res Treat. 2019;51:417–430. doi: 10.4143/crt.2019.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee KS, Chang HS, Lee SM, Park EC. Economic burden of cancer in Korea during 2000–2010. Cancer Res Treat. 2015;47:387–398. doi: 10.4143/crt.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitlock EP, Lin JS, Liles E, Beil TL, Fu R. Screening for colorectal cancer: a targeted, updated systematic review for the U.S. Preventive Services Task Force. Ann Intern Med. 2008;149:638–658. doi: 10.7326/0003-4819-149-9-200811040-00245. [DOI] [PubMed] [Google Scholar]
- 6.Lin JS, Piper MA, Perdue LA, Rutter CM, Webber EM, O'Connor E, et al. Screening for colorectal cancer: updated evidence report and systematic review for the US Preventive Services Task Force. JAMA. 2016;315:2576–2594. doi: 10.1001/jama.2016.3332. [DOI] [PubMed] [Google Scholar]
- 7.Han J, Micheline K. Data mining: concepts and techniques. 2. San Francisco: Elsevier; 2006. [Google Scholar]
- 8.Lee KS, Ahn KH. Application of artificial intelligence in early diagnosis of spontaneous preterm labor and birth. Diagnostics (Basel) 2020;10:733. doi: 10.3390/diagnostics10090733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee KS, Kwak HJ, Oh JM, Jha N, Kim YJ, Kim W, et al. Automated detection of TMJ osteoarthritis based on artificial intelligence. J Dent Res. 2020;99:1363–1367. doi: 10.1177/0022034520936950. [DOI] [PubMed] [Google Scholar]
- 10.Min JK, Kwak MS, Cha JM. Overview of deep learning in gastrointestinal endoscopy. Gut Liver. 2019;13:388–393. doi: 10.5009/gnl18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.11Ribeiro E, Uhl A, Häfner M. Colonic polyp classification with convolutional neural networks. In: Proceedings of IEEE 29th international symposium on computer-based medical systems, pp 253–58. 2016.
- 12.Zhang R, Zheng Y, Mak TWC, Yu R, Wong SH, Lau JY, et al. Automatic detection and classification of colorectal polyps by transferring low-level CNN features from nonmedical domain. IEEE J Biomed Health Inform. 2017;21:41–47. doi: 10.1109/JBHI.2016.2635662. [DOI] [PubMed] [Google Scholar]
- 13.13Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu C, et al. SSD: Single Shot MultiBox Detector. arXiv: 1512.02325 [cs.CV].
- 14.14Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. arXiv: 1506.01497 [cs.CV]. [DOI] [PubMed]
- 15.Everingham M, Gool LV, Williams CKI, Winn J, Zisserman A. The PASCAL visual object classes (VOC) challenge. Int J Comput Vis. 2010;88:303–338. doi: 10.1007/s11263-009-0275-4. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Descriptive statistics of study participants.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.