Abstract
Background
The Barotse floodplains of the upper Zambezi River and its tributaries are a highly dynamic environment, with seasonal flooding and transhumance presenting a shifting mosaic of potential larval habitat and human and livestock blood meals for malaria vector mosquitoes. However, limited entomological surveillance has been undertaken to characterize the vector community in these floodplains and their environs. Such information is necessary as, despite substantial deployment of insecticide-treated nets (ITNs) and indoor residual spraying (IRS) against Anopheles vectors, malaria transmission persists across Barotseland in Zambia’s Western Province.
Methods
Geographically extensive larval surveys were undertaken in two health districts along 102 km of transects, at fine spatial resolution, during a dry season and following the peak of the successive wet season. Larvae were sampled within typical Anopheles flight range of human settlements and identified through genetic sequencing of cytochrome c oxidase I and internal transcribed spacer two regions of mitochondrial and nuclear DNA. This facilitated detailed comparison of taxon-specific abundance patterns between ecological zones differentiated by hydrological controls.
Results
An unexpected paucity of primary vectors was revealed, with An. gambiae s.l. and An. funestus representing < 2% of 995 sequenced anophelines. Potential secondary vectors predominated in the vector community, primarily An. coustani group species and An. squamosus. While the distribution of An. gambiae s.l. in the study area was highly clustered, secondary vector species were ubiquitous across the landscape in both dry and wet seasons, with some taxon-specific relationships between abundance and ecological zones by season.
Conclusions
The diversity of candidate vector species and their high relative abundance observed across diverse hydro-ecosystems indicate a highly adaptable transmission system, resilient to environmental variation and, potentially, interventions that target only part of the vector community. Larval survey results imply that residual transmission of malaria in Barotseland is being mediated predominantly by secondary vector species, whose known tendencies for crepuscular and outdoor biting renders them largely insensitive to prevalent vector control methods.
Keywords: Primary vector, Secondary vector, Anopheles, Exophagy, Malaria, Residual transmission, Larvae, COI, ITS2
Background
Without the ambitious global control efforts of the last 20 years, it is estimated that malaria would have killed 995,000 people in 2018; instead, there were 405,000 deaths [1]. Ninety-three percent of malaria cases occur in sub-Saharan Africa [1], where six species of Anopheles mosquito are thought to be responsible for 95% of transmission [2]. These primary vector species dominate transmission because of their propensity to obtain blood meals from humans, but the associated endophagic and endophilic behaviors (preference for feeding and resting indoors) render them vulnerable to interventions using insecticide-treated nets (ITNs) and indoor residual spraying (IRS) of insecticides. Together, these vector control methods accounted for 78% of the dramatic global reduction in malaria transmission achieved between 2000 and 2015, during which time an estimated 663 million clinical cases were averted [3].
Traditional indoor-focused interventions may fail to control transmission by exophagic and exophilic mosquitoes, however. This was strikingly demonstrated in the Kilombero Valley in Tanzania, where the introduction of ITNs was less than half as effective at suppressing outdoor-biting An. arabiensis mosquitoes than endophagic An. gambiae sensu stricto (s.s.) [4]. Many other exophagic and exophilic anopheline mosquito species are typically zoophagic, but those that also feed on humans can consequently play a role as secondary vectors of malaria. Although less efficient than primary vectors [5], secondary vectors can augment or sustain malaria transmission alongside primary vectors and where they are sufficiently abundant may be locally important as main vectors in their own right [2, 5, 6]. Indeed, secondary vectors may be assuming an increasingly significant role in transmission as primary vector populations are suppressed. However, the same outdoor behaviors that make secondary vectors less susceptible to indoor interventions may also permit them to remain largely undetected by surveillance methods dominated by indoor sampling [7, 8]. Lack of detection can be compounded by errors in morphological identification and also by molecular tests seeking to identify primary vectors designated a priori from well-established studies in geographically disparate areas [7]. Outdoor collections, although undertaken comparatively rarely, frequently reveal substantially greater vector diversity than could be inferred from prevailing indoor trapping [7].
Successful vector control interventions must be tailored according to vector community composition [9], and therefore accurate species identification is fundamental. DNA-based methods are becoming invaluable in anopheline identification; they have facilitated differentiation between morphologically indistinguishable sibling species (e.g. [10]), highlighted inaccuracies in morphological identification (e.g. [11, 12]) and revealed new or cryptic species (e.g. [13]), even among heavily studied complexes such as An. gambiae sensu lato (s.l.) [14]. Sequencing both the mitochondrial DNA (mtDNA) cytochrome c oxidase I (COI) gene and nuclear DNA (nDNA) second internal transcribed spacer (ITS2) region has been advocated [70, 71], in recognition of the challenges posed by issues of database coverage [15] and species labelling [7] to universal identification from a single region. Accurate assignment of species—and thus bionomic traits—to specimens allows intervention strategies to target vulnerable behaviors. Subsequent characterization of the spatio-temporal distribution of vector communities can not only facilitate the targeting of interventions, but also monitoring of species’ responses [7]. It is crucial that survey campaigns aiming to describe the entire vector community recognize inherent biases in surveillance approaches so that both endophagic and exophagic populations are sampled. Surveys of larval habitats localized around human habitation avoid such biases by sampling both populations before they become stratified by adult behaviors. Combining DNA-based species identification alongside extensive sampling of larval habitats thus offers considerable, and to date largely unfulfilled, potential to gain a holistic overview of malaria vector communities.
Concerted national malaria control efforts in Zambia have led to a nationwide decline in parasite prevalence among children under 5 from 22 to 9% between 2006 and 2018 [16]. However, rural areas such as much of Western Province continue to suffer disproportionately. Ecologically, the region is dominated by the Barotse floodplain, comprising a vast network of wetlands flooded seasonally by the Zambezi River and its tributaries [17–19], which makes up the area traditionally known as Barotseland. Inundation of the floodplain in the wet season drives an eight-fold increase in the extent of potential mosquito larval habitat [20], and there is evidence of an upward trend in inundation extent over the last decade [17, 21]. Many of the floodplain’s 300,000 human inhabitants [22, 23] practice transhumance in response to the hydrologically dynamic landscape, grazing their cattle in the floodplain in the dry season and shifting them to the uplands when the floodplain becomes inundated [19, 23]. The resultant shifting mosaic of both larval habitat and blood sources for malaria vectors sustains endemic year-round malaria (Fig. 1). The Western Province Health Office has primary-care health facilities distributed throughout the province, including several in the floodplain (Fig. 1). Despite annual application of IRS (30% of households in 2018; [16]) and distribution campaigns tripling the proportion of households with an ITN per sleeping space between 2010 and 2018, malaria parasite prevalence among under-5s in Western Province is currently 10%, twice as high as 2010 levels [16].
Fig. 1.
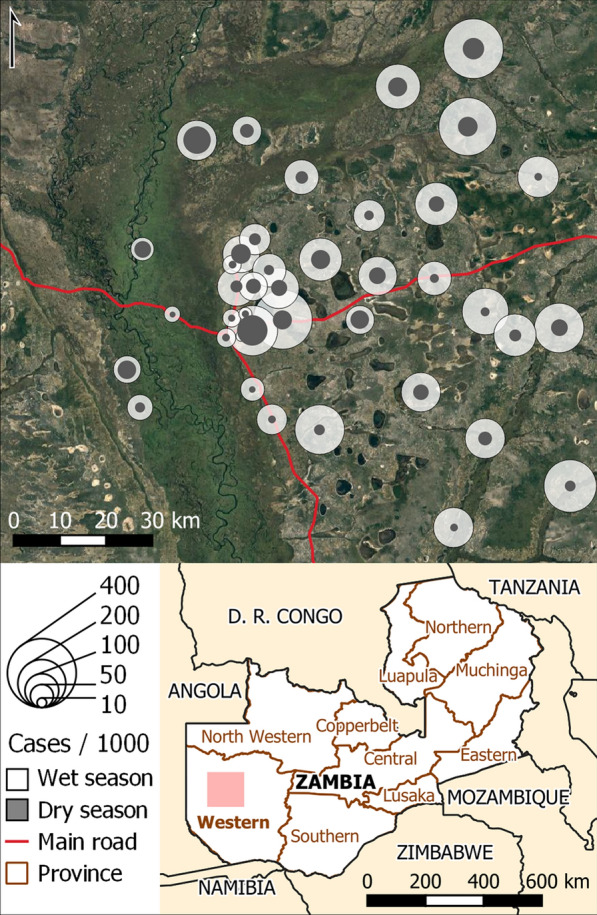
Average seasonal malaria incidence rates in health facilities in Limulunga and Mongu districts, Western Province, Zambia. Rates reported as cases per 1000 population; malaria data provided by Zambian government. Wet season: March–May, averaged 2014–2018; dry season Sept–Nov, 2014–2017. Pink rectangle in inset indicates study extent. Basemap: Google satellite imagery; TerraMetrics
Both secondary vectors and exophagic primary vectors can evade indoor interventions and may be implicated in the residual malaria transmission that persists in Barotseland, and so a detailed understanding of the complete vector community of this area is needed to inform future malaria control interventions. A recent molecular-based study of adult specimens collected in eastern Zambia revealed an unexpectedly high diversity of anophelines including primary and secondary vector species, as well as previously undescribed species [12]. The aim of the present study was therefore to employ the same DNA-based species identification methods, in conjunction with a geographically extensive survey of larval habitats, to characterize distribution and abundance of Anopheles species assemblages across Barotseland in Zambia’s Western Province. Specific objectives were to identify: candidate primary and secondary vector species present, variation in distribution and abundance of species in the wet and dry seasons and any distinct spatial pattern in species distribution, for example in association with human communities or habitat types.
Methods
Sampling strategy
The study area in Limulunga and Mongu districts of Western Province of Zambia was partitioned into five broad ecological zones (Fig. 2), defined to represent the different hydrological drivers and vector habitat provision in a typical year, and described below. Larval sampling was undertaken in each zone during the dry season (September–October 2017) and after peak flooding (May–June 2018); Zambezi River discharge in 2018 was the second highest recorded since 1990 (Chalo, C. & Willis, T., 2020, personal communication). The main Zambezi floodplain zone is affected predominantly by overbank flow from the Zambezi channel in the wet season, and consists of a mosaic of seasonally flooded grassland, channels and water bodies, interspersed with seasonally occupied human settlements located on mounds known as mazulu (see [23]) and one large settlement in the middle of the floodplain. Relatively few water bodies persist in the dry season, being confined to main channels and disconnected features formerly part of active channels [24]. Water bodies in the floodplain edge zone, however, persist for much of the year as upland dambos maintain a high water table, which forms seepage zones at the foot of the escarpment (see [23]). This zone, along the eastern edge of the Zambezi floodplain, supports high and year-round human populations because of the agricultural opportunity provided by fertile soils and nutrient-enriched springs [23, 25, 26]. Water bodies in the Luena flats zone also persist for much of the year [20], driven largely by the flood regime of the Luena River, resulting in extensive areas of grassland [23] among a highly diffuse anabranching river system [20], which contrasts with the Zambezi; human settlements are restricted to the floodplain edge. East of the Barotse floodplain, aquatic habitats in the narrower Lui valley zone are also formed by local, not Zambezi, flooding [26, 27], with human settlements concentrated along the valley edge. The dambo zone too is found largely to the east of the Barotse floodplain: dambos are shallow depressions, lined with organic sediments (often peat), which are seasonally or permanently waterlogged, forming an important dry season water source for closely associated human settlements [28, 29].
Fig. 2.
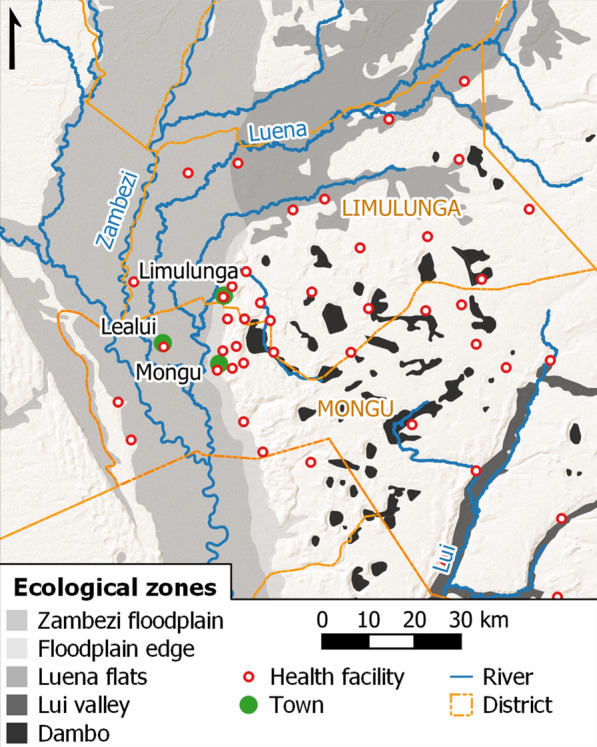
Ecological zones with different hydrological regimes identified in Western Province, Zambia. Health facilities in Limulunga and Mongu districts are shown. Basemap: ESRI Shaded Relief (2020)
Entomological field surveys
We selected two or more health facilities located in representative habitat in each of the ecological zones, with the exception of the Lui valley where only one health facility was chosen because of access constraints. Using knowledge from local health facility staff of current hydrological conditions, two nearby villages in the facility’s catchment area were selected for survey on each sample day. The survey team deployed considerable expertise in sampling Anopheles larval habitats gained on previous projects in An. gambiae s.s., An. arabiensis and An. funestus dominated vector systems [30, 31]. Entomological surveys were undertaken along two line transects radiating from each village periphery to characterize the diversity and abundance of anopheline larvae at increasing distances from human habitation [32]. Each transect was up to 1.5 km long, access permitting; approximately 90% of Anopheles gambiae s.l. were found to remain within 1.5 km of their larval habitat in similar rural floodplain savannah in The Gambia [32], so a transect of this length was expected to encounter the larval habitats of the majority of anophelines feeding on people in each village. Regular sampling points were located at 100 m intervals along each transect, with additional opportunistic sampling points between the regular ones if water bodies were encountered within 5 m of the transect line.
At each sampling point, an area within a 5 m radius was searched for potential larval habitat, and each water body was geolocated with a GPS handset (Garmin eTrex). A purposive dipping strategy was employed [33–35] to search for mosquito larvae in likely microhabitats, particularly along the water body periphery, among clumps of emergent vegetation and under floating vegetation or debris. Up to 40 dips were taken from each water body with standard 350 ml dippers (BioQuip, USA). The contents of each dip were examined in a white plastic tray, and mosquito larvae were differentiated into anophelines and culicines morphologically and by body position on the water surface. A random sample of up to ten Anopheles larvae was collected from each sampling point and stored individually in 95% ethanol for genetic identification.
Ethical considerations
An ethical approval waiver was provided by the University of Zambia’s Biomedical Research Ethics Committee (Ref 018-08-17). The Barotseland Royal Establishment granted their approval for entomological surveys to be conducted around villages in the study area. District Health Office staff accompanied the field survey team; at the beginning of each day’s fieldwork, the survey team checked in with the nearest health facility and sought permission from village chiefs to undertake fieldwork following introductory discussions.
Specimen identification
DNA was obtained from larval specimens (crushed with a mounted needle) using a standard CTAB-phenol/chloroform/isoamyl alcohol extraction protocol [36]. Larvae were identified as far as possible to species by sequencing of the mtDNA COI gene and nDNA ITS2 region following protocols outlined in Lobo et al. [12]. ITS2 genotyping was restricted to a subset of individuals from each species or group delineated by COI analyses [12]. Sequence chromatograms were manually trimmed in Chromas (Technelysium Pty Ltd, Australia) and aligned using the Clustal W multiple alignment program [37] implemented in BioEdit [38]. Haplotypes were generated using DNA SP6 [39].
Specimen identities were inferred by a multi-layered consensus of three approaches. First, following Lobo et al. [12], COI sequences were analyzed using the Basic Local Alignment Search Tool nucleotide (BLASTn) to query the National Center for Biotechnology Information (NCBI) nucleotide database (nt) employing the megablast algorithm and applying a standardized cut-off of ≥ 95% similarity. Second, results from BLASTn searches were compared with maximum likelihood phylogenetic trees constructed in MEGA X [40]. Third, ITS2 sequences for the subset of samples based on COI BLASTn results and phylogeny were also analyzed using BLASTn, with the same threshold of ≥ 95% similarity to database accessions.
Statistical analyses
Fieldwork and laboratory records were compiled in Access (Microsoft) databases and joined to location record shapefiles produced from gpx files from GPS handsets in QGIS 3.10-A Coruña [41]. To enable comparison of the abundance of a species between transect points, the sum total of anophelines recorded in all dips at a transect point was multiplied by the proportion of larvae sampled from that transect point that was molecularly identified as this species. This estimate of the total number of larvae of this species encountered at the transect point was then standardized for the sampling effort by reporting as an encounter rate per ten dips.
Statistical comparisons were undertaken in SPSS [42] on untransformed data [43]; comparisons between dry and wet seasons, or between ecological zones, were made using appropriate parametric (Student’s t test; Odds Ratio; ANOVA) or non-parametric tests (Kolmogorov-Zmirnov Z; Kruskal-Wallis H with stepwise step-down post-hoc comparisons and adjusted p value for multiple comparisons).
Results
Larval habitat sampling
The distribution of water bodies encountered in sampling in the 2017 dry season and following the peak of the 2018 wet season conformed to expectations for the ecological zones outlined above; see [20] for a detailed spatio-temporal characterization of water bodies across this region of Barotseland during this period. Sampling was undertaken along 70 km of transects in the dry season and 32 km of transects in the wet season (Fig. 3). A similar number of transects was completed in both seasons, but difficult access to field study sites and the frequent presence of impassably deep water resulted in a lower average transect length in the wet season (mean ± standard deviation (SD): dry season 1316 ± 309 m, n = 53; wet season 666 ± 317 m, n = 48; Student’s t test 10.321, df 99, p < 0.001). Consequently, there were 18 transect points on average along dry season transects (SD 3.2), while wet season transects averaged nine transect points (SD 3.3). Water was encountered at a significantly higher proportion of transect points in the wet season than in the dry season (80.1 and 44.6%, respectively; odds ratio [OR] 5.01, 95% CI 3.823–6.566; p < 0.001 [44]). The presence of larger water bodies in the wet season necessitated a larger number of dips per transect point to ensure representative sampling of the higher level of variance expected across such water bodies (mean dips ± SD: wet season 23 ± 5.4; dry season 13 ± 4.7).
Fig. 3.
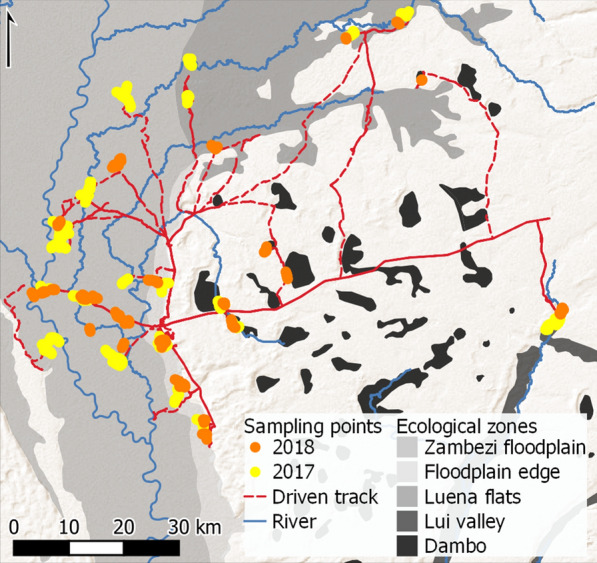
Distribution of sampling points across ecological zones in dry season and wet season entomological surveys. Dry season: Sept–Oct 2017; wet season: May–June 2018, following peak inundation. Sampling undertaken in Limulunga and Mongu districts in Zambia’s Western Province. Basemap: ESRI Shaded Relief (2020)
A significantly higher proportion of transect points where water was found (‘wet transect points’) contained mosquito larvae (including anophelines) in the wet season (95%) than in the dry season (62.3%; OR 11.49, CI 6.781–19.47, p < 0.001). A similar pattern was observed in anopheline distribution, whereby 85% of wet transect points in the wet season were positive for anophelines compared to 43.3% in the dry season (OR 7.405, CI 5.184–10.578, p < 0.001). Approximately one third of mosquito larvae encountered were field-identified as anophelines (3045 of 11504 larvae; n = 13,335 dips), and the average number per ten dips in wet season surveys was double that of the dry season average (2.8 ± 9.13 and 1.4 ± 6.47, respectively; Kolmogorov-Smirnov Z 4.974, p < 0.001).
Genetic identification of larvae
A total of 1034 specimens were collected for individual DNA analysis (of the 3045 encountered). A 318 bp fragment of the COI region was aligned across 903 specimens, with sequences resolving 413 haplotypes. BLASTn searches of the NCBI nt database revealed matches at ≥ 95% identity for 83% (n = 340) of these haplotypes, corresponding to 81% of specimens. Thirty-nine specimens were identified as culicines (non-anophelines). The above-threshold BLASTn hits to anophelines could be partitioned into five groups (Table 1). The majority of haplotypes (n = 203) and individuals (n = 475) were assigned to the An. coustani group (> 95% identity with at least one of An. coustani, An. conferre (cf.) coustani 1 [12], An. cf. coustani 2 [12], An. tenebrosus or An. ziemanni); 157 specimens were identified as An. squamosus and 42 as An. species O/15 [11]. Only 14 were identified as An. gambiae s.l. (> 95% identity with at least one of An. arabiensis, An. coluzzii, An. fontenillei or An. gambiae s.s.) and 1 as An. funestus s.s. A group of 175 specimens returned NCBI nt matches below the 95% sequence similarity threshold, but where the closest matches on the database were all to anopheline species. These individuals do not match with sequence similarity of > 95% to any of the taxa identified by Lobo et al. [12] or St Laurent et al. [11], despite 100% query coverage with longer sequences deposited by these studies in NCBI nt. Individuals in this group were therefore categorized as “Unknown Anopheles species” (Table 1 and Fig. 4; further details in Additional file 1). A maximum parsimony phylogenetic tree of haplotypes from this study and reference sequences from recent studies in Zambia and Kenya [11, 12] supported the COI-assigned identities (Additional file 2).
Table 1.
Species and species-group identities assigned to Anopheles larvae based on mitochondrial and nuclear DNA sequences
| Final ID | Potential member species of group/complex (if applicable) | COI-assigned specimens (haplotypes) | Additional ITS2-assigned specimens | Dry season | Wet season | Total |
|---|---|---|---|---|---|---|
| An. coustani group | An. coustani s.s., An. cf. coustani 1, An. cf. coustani 2, An. tenebrosus, An. ziemanni | 475 (203) | 88 | 153 | 410 | 563 |
| An. funestus | – | 1 (1) | 0 | 1 | 0 | 1 |
| An. gambiae s.l. | An. arabiensis, An. gambiae s.s. | 14 (6) | 3 | 10 | 7 | 17 |
| An. species O/15 | – | 42 (22) | 0 | 1 | 41 | 42 |
| An. squamosus | – | 157 (79) | 0 | 19 | 138 | 157 |
| Unknown An. species | – | 175 (71) | 52 | 63 | 152 | 215 |
| Total | 864 (382) | 143 | 247 | 748 | 995 |
Identities inferred from above-threshold matches to cytochrome c oxidase I and/or internal transcribed spacer region 2 sequences on the National Center for Biotechnology Information nucleotide database (NCBI nt). Sampling undertaken in dry season (Sept–Oct 2017) and wet season (May–June 2018) in Limulunga and Mongu Districts, Western Province, Zambia. Of 3045 field-identified anophelines encountered, 1034 specimens were collected for genetic identification; 39 were subsequently identified as culicines and not reported in this table. “Unknown An. species” indicates below-threshold (<95%) matches to Anopheles sequences in NCBI nt; 12 specimens assigned to this group from COI sequences were subsequently re-assigned to other taxa based on ITS2 sequences
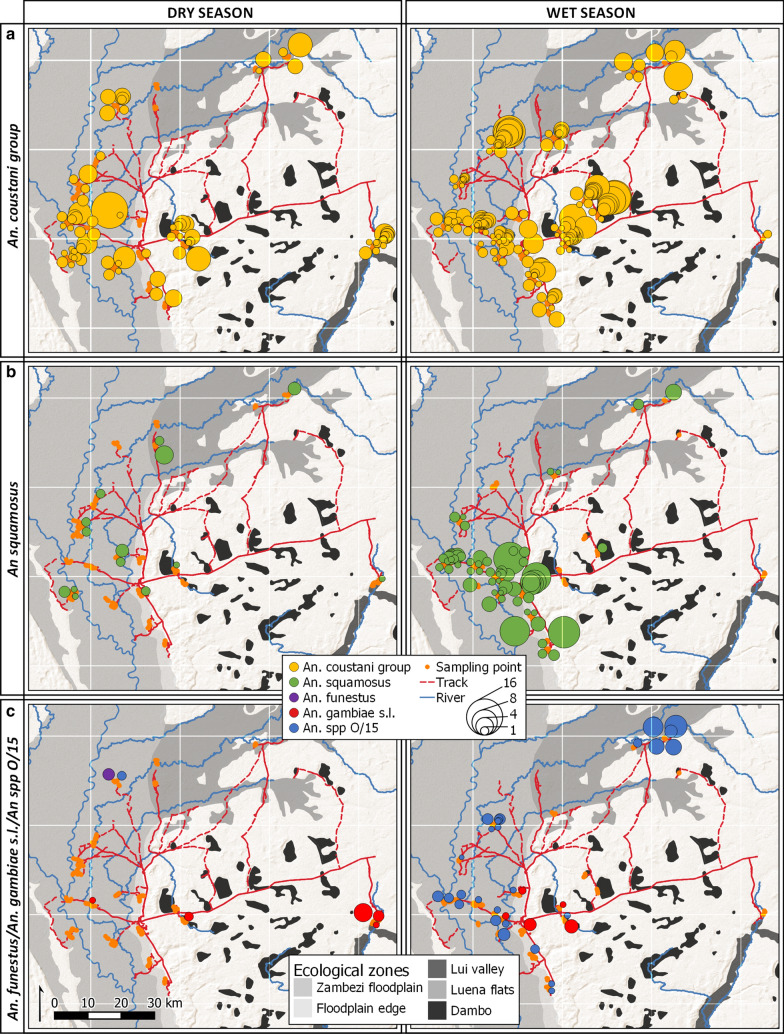
Fig. 4.
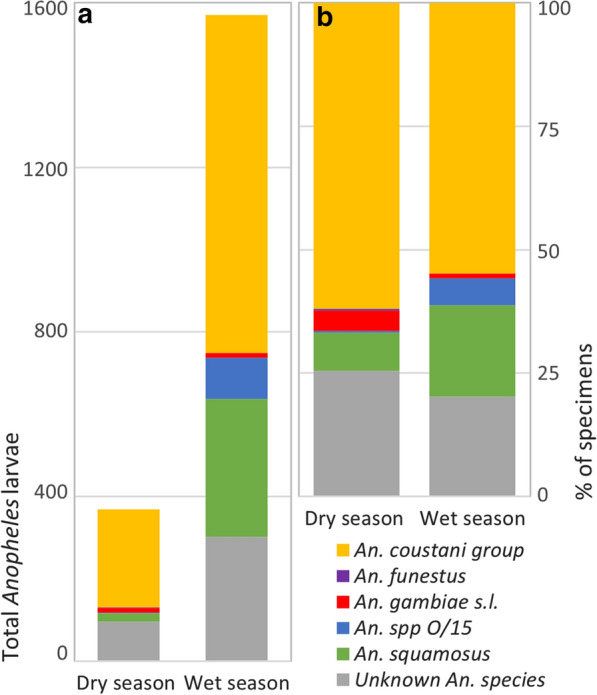
Abundance and species composition of larval Anopheles communities in dry and wet seasons, western Zambia. a Estimated total abundance of Anopheles species (/groups), calculated by applying species proportions from a subset of sampled larvae (b) to the total number of surveyed larvae per transect point and summing for all transect points. b Composition of specimens identified from cytochrome c oxidase I and/or internal transcribed spacer region 2 DNA sequences. “Unknown An. species” assigned to specimens whose alignment with NCBI nt accessions fell below the 95% identity threshold, but most closely matched to anopheline sequences. Sampling undertaken in Limulunga and Mongu districts, Western Province; dry season: Sept–Oct 2017; wet season: May–June 2018. Basemap: ESRI Shaded Relief (2020)
COI identities were supported by representative ITS2 sequences for the An. coustani group (matches with An. cf. coustani 1, An. coustani) and An. funestus. ITS2 sequences supporting COI-assigned An. gambiae s.l. specimens indicated the presence of An. arabiensis (60%) and An. gambiae s.s. (40%); none of these specimensʼ ITS2 sequences demonstrated a match with An. coluzzii or An. fontenillei, and neither have been found in Zambia, although the former is known to occur in neighboring countries [45]. ITS2 sequences from specimens identified from COI sequences as An. species O/15 failed to return a match with ≥ 95% identity to any accession. Among 27 ITS2 sequences from COI-assigned An. squamosus specimens, 56% matched with An. cf. coustani 1, 11% equally with An. species O/15 and An. cf. coustani 1, 7% matched with An. pharoensis, while the remainder were unmatched at ≥ 95% identity, likely reflecting the paucity of ITS2 accessions for An. squamosus on NCBI nt (n = 3). In cases where individuals exhibited above-threshold COI assignments with apparently discordant ITS2 scores, we presently assign identification based on their COI identity and discuss the potential reasons for these discordances and implication for final species proportions.
The ITS2 sequencing also included some individuals without COI sequences (n = 131) and some assigned to the unknown anopheline category based on COI sequences (n = 12). ITS2 sequences for these specimens permitted additional assignments to the An. coustani group (88 individuals), An gambiae s.l. (3 individuals; An. arabiensis (n = 1) and An. gambiae s.s. (n = 2)) and the unknown anopheline category (52 individuals which aligned at < 95% similarity with NCBI nt Anopheles sequences; details in Additional file 1). Final species totals are reported in Table 1 and proportions observed in the dry and wet seasons shown in Fig. 4.
The majority of Anopheles larvae identified by sequencing in both the dry season (n = 247) and wet season (n = 748) were identified as An. coustani group (62 and 55%, respectively; Fig. 4). Species considered primary malaria vectors were rare in the sample; a single An. funestus s.s. specimen was found in the wet season, while very small numbers of An. gambiae s.l. were found in both the dry (n = 10) and wet (n = 7) seasons. An. squamosus made up a substantially greater proportion of sequence-identified samples in the wet (18%) than in the dry season (8%). An. species O/15 was represented by a single individual in the dry season, but made up 5% of sequence-identified specimens in the wet season. Between a fifth and a quarter of sequences did not align with > 95% identity to any sequence on the NCBI nt database, and were designated unknown An. species (dry season: 26%; wet season: 20%).
Spatial distribution of species
Due to significant differences between ecological zones in the number of dips taken per transect point, comparisons between ecological zones are made on a standardized metric, whereby average total abundance values per transect point are presented per ten dips, i.e. with constant sampling effort.
There was no difference in the standardized mean total Anopheles larvae per transect point between ecological zones in the dry season (F = 0.984, df 4, p = 0.416). In the wet season, Anopheles larvae distribution differed significantly between ecological zones (Kruskal-Wallis H = 24.938, df 4, p < 0.001); fewer Anopheles larvae were found per transect point in Zambezi floodplain and floodplain edge habitats than in the Luena and Lui catchments and in dambos (stepwise step-down comparisons; adjusted (adj) p < 0.05; Fig. 6a).
Fig. 6.
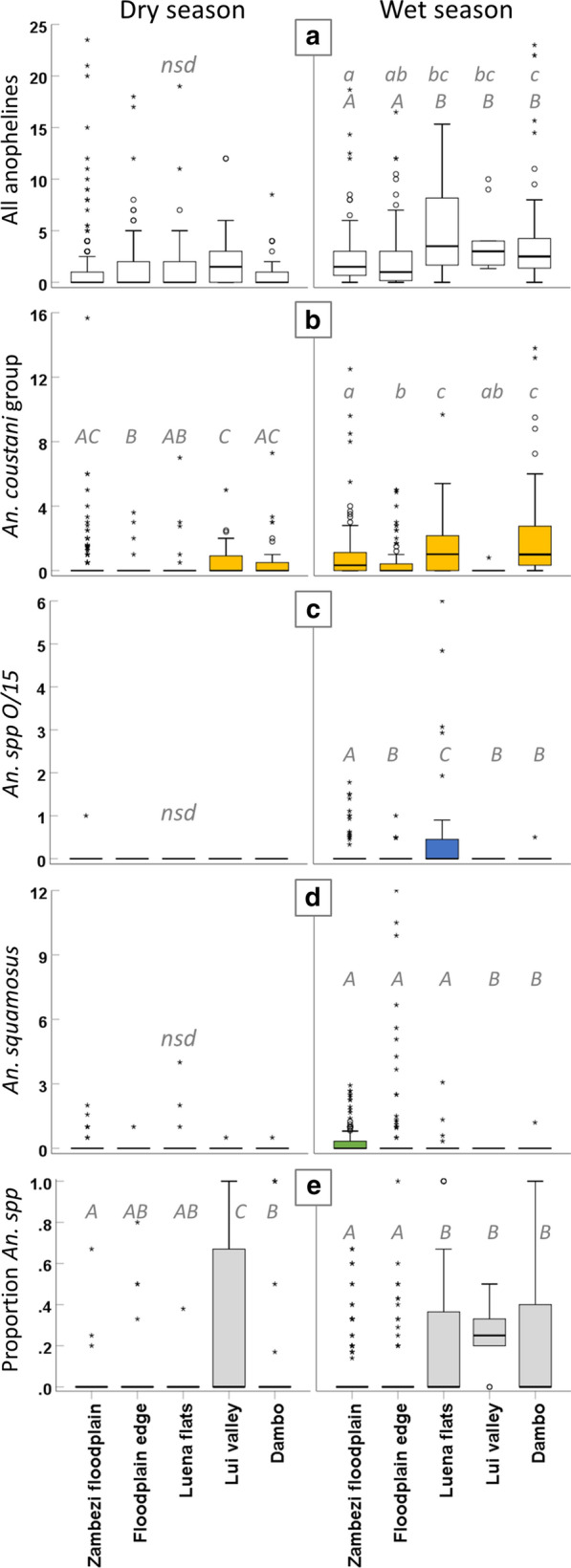
Average abundance of Anopheles larvae across ecological zones in dry and wet seasons, western Zambia. Boxes indicate the interquartile range (IQ) of total values per transect point, with median plotted as bold line; whiskers extend to minimum and maximum values within 1.5 times the IQ; outliers (values within 1.5 to 3 times the IQ) are indicated by circles, and extreme values (> 3 times the IQ) by asterisks. a total anopheline larvae per ten dips; b total An. coustani group larvae; c total An. spp O/15; d total An. squamosus; e proportion of unknown anophelines per transect point. Taxon-specific totals (b–d) are the product of total anopheline count and species proportion in the subsample from each transect point, standardized per ten dips. Within panels, letters in italics denote statistical comparisons. Ecological zones that do not share a letter within a panel are significantly different. Lower case letters refer to median values (independent-samples median test); upper case letters refer to distribution (Kruskal-Wallis test); stepwise step-down comparisons with adjusted p-value for multiple comparisons. Nsd indicates no significant differences within panel. Dry season sampling undertaken Sept–Oct 2017 (left column); wet season sampling May–June 2018 after peak inundation (right column)
In the dry season, An. coustani group larvae were distributed ubiquitously across all ecological zones (Fig. 5a), although significantly fewer were found in floodplain edge habitats than in all others except the Luena flats; abundance in the Lui valley was higher than in the Luena flats (Kruskal-Wallis H = 29.435, df 4, p < 0.001; stepwise step-down comparisons with adj p < 0.05 Fig. 6b). An. squamosus larvae were present in all ecological zones (Fig. 5b), and there was no significant difference between zones in the average standardized total per transect point (H = 3.216, df 4, p = 0.522; Fig. 6d). Only ten An. gambiae s.l. larvae were encountered in the dry season, in a range of ecological zones, although eight of these were found in three adjacent transect points within 140 m in the Lui valley. Another An. gambiae s.l. was found in a pool adjacent to a tributary of the Zambezi in the floodplain, and another in a dambo immediately east of Mongu (Fig. 5c). One An. funestus larva was found in a pool on a transect in the Luena flats, and a single An. species O/15 larva was found on the neighboring transect from the same village. The proportion of unknown anophelines was lowest in the floodplain and significantly higher in the Lui valley ecological zone than in all other zones (H = 86.214, df 4, p < 0.001; stepwise step-down comparisons, adj p < 0.05; Fig. 6e).
Fig. 5.
Distribution of Anopheles larvae across ecological zones in dry and wet seasons, western Zambia. a An. coustani group; b An. squamosus; c An. funestus, An. gambiae s.l. and An. spp O/15. Dry season sampling undertaken Sept–Oct 2017 (left column); wet season sampling May–June 2018 after peak inundation (right column). Symbol area proportional to total larvae per transect point (product of total anopheline count and species proportion in subsample), standardized per ten dips. Basemap: ESRI Shaded Relief (2020)
After the peak of the 2018 wet season, An. coustani group mosquitoes were again ubiquitous across the sampled area (Fig. 5a), but median abundance was significantly higher in the dambo and Luena floodplain ecological zones than in the floodplain edge and Lui valley zones (H = 48.563, df 4, p < 0.001; stepwise step-down comparisons, adj p < 0.05; Fig. 6b). In contrast to the dry season, distributions of An. squamosus abundance in the Luena and Zambezi floodplains and along the floodplain edge ranked significantly higher than those of dambo habitats (H = 19.227, df 4, p = 0.001; stepwise step-down comparisons, adj p < 0.05; Fig. 6d), and the species was absent from the Lui valley (Fig. 5b). Known primary vector species were limited to An. gambiae s.l. in the wet season (Fig. 5c): two individuals at one dambo transect point within 600 m of the dry season transect point where An. gambiae s.l. occurred, with another individual in the adjacent dambo to the north; three in the saturated floodplain edge immediately around Mongu and one in Zambezi floodplain habitat analogous to the Zambezi floodplain location of the dry season specimen. An. species O/15 occurred predominantly in floodplain habitats, with the highest abundance in the Luena flats followed by the main Zambezi floodplain and significantly lower abundance in floodplain edge and dambo habitats; it was absent from the Lui valley (H = 19.169, df 4, p = 0.001; stepwise step-down comparisons, adj p < 0.05; Figs. 5c, 6c). The proportion of unknown anophelines also varied between ecological zones in the wet season, being significantly higher in the Lui valley, Luena flats and dambo habitats than habitats in the Zambezi floodplain and along its edge (H = 38.834, df 4, p < 0.001; stepwise step-down comparisons, adj p < 0.05; Fig. 6e).
Discussion
The combination of a geographically extensive field survey of anopheline larvae, spatially stratified by hydro-ecology, and a genetic identification approach has revealed a complex and dynamic assemblage of potential malaria vectors across Barotseland in western Zambia. Anopheline larvae were found throughout this hydrodynamically complex area and in all ecological zones, but as expected were more widespread and abundant in the wet season (higher proportion of transect points). They were significantly more abundant in the Luena, Lui and dambo ecological zones than in the Zambezi floodplain and floodplain edge in the wet season. Within this widespread anopheline distribution there were substantial differences in abundance and distribution of different species, and differences between wet and dry seasons. A key finding was that in terms of both distribution and abundance, the anopheline population was dominated by secondary vector species. The An. coustani group dominated in both wet and dry seasons (55 and 62%, respectively), followed by An. squamosus, which was more prevalent in wet than dry season (18 and 8%, respectively), and An. spp O/15 in the wet season only (5%). These secondary vector species/species groups were widespread across the region, in both wet and dry seasons, although several species (An. coustani, An. spp O/15) were more abundant in Zambezi and Luena floodplain habitats. Primary vector species (An. arabiensis, An. gambiae s.s. and An. funestus s.s.) were relatively rare in both seasons and had very localized distributions consistent between seasons. A group of anopheline individuals that could not be identified to known species comprised 20 and 26% of the dry and wet season population, respectively.
Application of molecular techniques consistent with previous studies ensured that species assignments are comparable with those from eastern Zambia [12] and western Kenya [11]. As in these study areas, we uncovered an unexpected diversity of potential vector species, but with a surprising scarcity of primary vectors (< 2%) among 995 sequenced anopheline larvae. Bias for specific feeding or resting behaviors was avoided by larval sampling and so potentially better represents the whole anopheline community within 1.5 km of villages than adult trapping; although this sampling does not directly demonstrate exposure of the human population to the potential vector species encountered [7], persistent malaria prevalence in the region [16] indicates the presence of substantial numbers of vectors. Given the known diversity of the Anopheles genus, with over 140 species in sub-Saharan Africa [5], and the abundance of non-human blood sources across Barotseland (especially livestock [18]), it is expected that non-vector anophelines will be represented in larval sampling. Nonetheless, village-centered surveys encompassing all water body types encountered within known flight range [32] of human blood meals failed to reveal substantial numbers of primary vector species either in the dry season or following the peak of the wet season. Reduced average transect length in the wet season effectively further concentrated sampling on the peridomestic environment, which might have been expected to bias the sample towards more anthropophilic species, yet a smaller proportion of the sample in this season consisted of primary vector taxa.
The primary malaria vector species in Zambia are thought to be An. arabiensis, An. funestus s.s. and An. gambiae s.s., although detailed entomological studies have been undertaken only comparatively recently [46, 47]. Despite the continued high prevalence of malaria in Barotseland, An. gambiae complex and An. funestus s.s. represented only 1.7 and 0.1% of anophelines identified by genetic methods in this study. Our result is in contrast to the study by Lobo et al. (2015; [12]), which sampled adult anophelines in villages in eastern Zambia and described a predominance of An. funestus s.s. (55% of specimens) followed by An. arabiensis and implicated both species in malaria transmission. An. funestus s.s. is also thought to dominate transmission in northern Zambia, followed by An. gambiae s.s. [48], while in southern Zambia An. arabiensis is thought to be the primary vector [47]. However, although An. funestus and An. gambiae s.l. accounted for 29% and 9%, respectively, of indoor and outdoor collections of adult anophelines in Western Province villages in 2013 [49], they were not the most abundant species in that collection (some of which originated in our study area). All three of these species exhibit large human blood indices in Zambia [50], and although An. arabiensis is generally considered to be less anthropophilic than An. gambiae s.s. and An. funestus, it is more anthropophilic in Zambia than elsewhere in Africa [47].
It is unknown whether the larval community we recorded is a product of the suppression of populations of endophilic and endophagic primary vectors by increased interventions [16], as has been documented elsewhere (e.g. [4, 51, 52]), or is representative of a natural species assemblage of anopheline vectors in Barotseland that has not been historically dominated by primary vector species. The distribution of An. gambiae s.l. found in the present study was highly clustered, with eight of ten specimens found within 140 m of each other in the dry season. Sampling after the peak of the subsequent wet season found three of seven An. gambiae s.l. associated with the same village, and all specimens across both seasons were closely associated with people (within 600 m of a village). This conforms to the established tendency of An. gambiae s.l. to breed in close proximity to human settlements [53, 54] and limit dispersal from these blood sources [32]. The range of aquatic habitats surveyed in our study encompassed streams and large, permanent and heavily vegetated larval habitats typically associated with An. funestus and smaller, more ephemeral ones often favored by An. gambiae s.l. [48, 55–57], so we do not think biased habitat sampling can explain the low abundance of primary vector species detected. Combined sampling of larval habitat and adults will be needed to further resolve the dynamic relationships among primary and secondary vector populations around villages and across the wider landscape.
An. coustani group species comprise up to 65% of surveyed larvae, which supports the limited sampling of adult mosquitoes in Western Province in 2013 that found 60% to be An. coustani [49]. The molecularly identified An. coustani group in our study contains several closely related species that cannot be further resolved from COI sequences; although some may be separable based on morphology, a number of species remain virtually indistinguishable [58]. In addition, molecular identification of anophelines in eastern Zambia by Lobo et al. has suggested the presence of further morphologically cryptic sibling species, denoted An. cf. coustani 1 and 2 [12] (one of which may represent An. crypticus, the most recently identified cryptic species in the group [59]).
An. coustani group was distributed across all ecological zones in this study, although consistently less abundant in floodplain edge habitats. Significantly higher abundance in permanently waterlogged dambo habitats across both seasons reflects the recognized preference of An. coustani s.s. for more established water bodies [60]. The species displays a preference for natural vegetated water bodies and an aversion to temporary, non-vegetated pools elsewhere [61], and this is supported by its ubiquitous presence in vegetated habitats [20] across Barotseland in this study. Until recently, An. coustani has been considered a secondary vector as it was seen as largely zoophilic [61], but it was recently demonstrated to be the main vector in a village in Madagascar [62] and its secondary vector status is increasingly being reconsidered. An. coustani exhibits high levels of anthropophily in some settings [15, 62, 63], practices endophagy as well as (predominantly) exophagy [64–66] and shows a markedly high rate of early biting [62, 63, 67]. It has tested positive for Plasmodium infection in Ethiopia [65], Kenya [66] and Madagascar [67]; in Zambia, An. coustani, An. cf. coustani 1 and An. cf. coustani 2 have all tested positive [12]. Even low infection rates, in combination with these behavioral traits and locally high abundance [62, 67], may allow An. coustani to play a substantive role in malaria transmission.
Among other potential members of the taxon designated as An. coustani group in this study, An. tenebrosus is assumed not to be a competent vector because of its low parity and long gonotrophic cycle; it has not been detected with malaria parasites [7, 61], and records of presence in Zambia are historical [45]. These traits are also associated with An. ziemanni, and although it has occasionally been found to harbor Plasmodium, it has historically not been considered a significant vector [7, 61]. Nonetheless, in some areas it demonstrates anthropophily [68, 69], and it may be locally important as a vector in an area of low transmission in Cameroon [70]. Habitat preferences for both these species are also poorly characterized and thought to conform broadly with those of An. coustani; both are typically associated with permanent water [61].
An. squamosus was also abundant in our larval collections during the wet season. This species is more zoophilic and exophagic than An. coustani, although an unexpectedly high degree of anthropophily has been revealed in southern Zambia [63] and Madagascar [71], and Plasmodium infection has incriminated the species as a vector in both countries [63, 71]. In Barotseland An. squamosus displayed a habitat-specific distribution, predominantly in floodplains and floodplain edge zones. Larvae have been recorded previously from a wide range of habitats, provided they are at least partially vegetated [61], but relatively little is known about this species’ habitat associations.
The third abundant non-primary vector anopheline identified in Barotseland was An. species O/15, previously identified in Kenya as a potential sibling species to An. coustani [11]. An. species O/15 is not considered a malarial vector in Kenya (negative for Plasmodium falciparum; [11]), and there is no further information on bionomics or larval habitat preferences. In the present study An. species O/15 constituted 5% of the wet season sample and was significantly more prevalent in floodplain habitats than in other ecological zones.
In eastern Zambia, 39% of 18 delineated anopheline taxa [12], and 53% of 17 taxa in Kenya [11], were designated as ‘unknown’ because of the lack of conclusive similarity to database sequences. While some taxa may represent novel or cryptic species, the lack of an identity may also result from the absence of DNA sequences from known species. In the present study, identifications were based on matches at ≥ 95% similarity to published COI sequences, supported by ITS2 matches using the same threshold, and specimens that failed to yield an above-threshold match (but had consistent below-threshold matches to anopheline sequences, see Additional file 1) were designated as “unknown Anopheles species.” Some of these “unknown” individuals may represent further examples of the cryptic species diversity uncovered by recent studies [11, 12], as we know that these individuals do not match to the taxa identified in these studies even though we had high sequence coverage of the same DNA regions; others are likely to be known species that are poorly represented in the NCBI nt database. The occurrence of unidentified anophelines does not affect the key conclusion of this study, because where these individuals are matched (at < 95% similarity) to a sympatrically occurring species, the majority cluster with secondary vector taxa, and we can be confident that they are not primary vectors well represented in the published database. The occurrence of potential further cryptic anopheline species in this and other areas should be taken account of in future studies.
A key remaining question is how representative our comprehensive larval survey is of the distribution of adult Anopheles vectors and hence malaria transmission hazard. In some settings, larval densities have been found to be a poor predictor of adult abundance [72], while in others there is very close correlation [73]. If the larval composition is representative, there are significant implications for malaria vector control efforts in the region.
Conclusions
The diversity of candidate vector species and their high relative abundance observed across diverse hydro-ecosystems indicate a highly adaptable transmission system, resilient to environmental variation and, potentially, interventions that target only part of the vector community. Larval survey results imply that residual transmission of malaria in Barotseland is being mediated predominantly by secondary vector species, whose known tendencies for crepuscular and outdoor biting renders them largely insensitive to prevalent vector control methods.
Supplementary Information
Additional file 1: Table S1. Cytochrome c oxidase 1 (COI) sequences for unknown Anopheles species. Table S2: second internal transcribed spacer region (ITS2) sequences for unknown Anopheles species.
Additional file 2: Figure S1. Maximum likelihood phylogenetic tree of cytochrome c oxidase I (COI) sequences from Anopheles larvae sampled in western Zambia.
Acknowledgements
The authors would like to thank our collaborators in the University of Barotseland (UBL) for their support, particularly Mukelebai Ndiyoi and Dr Muyoba Macwani. We are very grateful to the communities across Barotseland for their warm welcome, engagement, and guidance. We thank the UBL field team for their dedication to fieldwork under sometimes challenging conditions: Favourite Imasiku, Chikenge Henry, Kundananji Silwenga, Matthewz Kamangala, Munukayumbwa Sitwala and Sekeli Lyayo. Thanks too to James Thomas for his contribution to fieldwork, and to Nicolae Adrian Macarie for his commitment to sample processing in the lab. We are very grateful to Alexandra Malloy for her enthusiastic help with project logistics. The comments of two anonymous reviewers improved this manuscript.
Abbreviations
- BLASTn
Basic Local Alignment Search Tool (nucleotide)
- NCBI
National Center for Biotechnology Information
Authors contributions
DC, CT, DS, FL, and AS conceptualized the study. DC, CT, MS, MM and AJH acquired funding. DC, CT, VS, DS, FL and JS contributed to the study design. DC, CT, VS and TW undertook collection of field data and samples; DC, NMcK and AH performed laboratory analyses. DC, NMcK, AH and PS interpreted results of genetic analyses. DC drafted the manuscript; CT, NMcK, AH, DS, MS, AJH and PS critically reviewed and edited subsequent drafts. All authors read and approved the final manuscript.
Funding
This research was undertaken as part of the FLOODMAL project, funded by the UK Natural Environment Research Council (Grant Ref: NE/P013481/1). The funding body played no role in the design of the study, the collection, analysis and interpretation of data, or in writing the manuscript.
Availability of data and materials
The COI and ITS2 sequence datasets generated by and analyzed in the current study are available in the NCBI GenBank nucleotide archive with accession numbers MW167786–MW168163 for COI sequences and MW166490–MW166863 for ITS2 sequences. Additional data on specimens assigned to “unknown Anopheles species” category are provided within Additional file 1. A maximum likelihood phylogenetic tree of COI haplotypes is provided in Additional file 2.
Ethics approval and consent to participate
An ethical approval waiver was provided by the University of Zambia’s Biomedical Research Ethics Committee (Ref 018-08-17). The Barotseland Royal Establishment granted their approval for entomological surveys to be conducted around villages in the study area. The field survey team was accompanied by District Health Office staff; at the beginning of each day’s fieldwork, the survey team checked in with the nearest health facility, and sought permission from village chiefs to undertake fieldwork following introductory discussions.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-020-04540-1.
References
- 1.WHO . World Malaria Report. Geneva: World Health Organisation; 2019. [Google Scholar]
- 2.Mouchet J, Carnevale P, Coosemans M, Julvez J, Manguin S, Richard-Lenoble D, et al. Biodiversité du paludisme dans le monde. Montrouge, France: Editions John Libbey Eurotext; 2004. [Google Scholar]
- 3.Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell TL, Lwetoijera DW, Maliti D, Chipwaza B, Kihonda J, Charlwood JD, et al. Impact of promoting longer-lasting insecticide treatment of bed nets upon malaria transmission in a rural Tanzanian setting with pre-existing high coverage of untreated nets. Malar J. 2010;9:187. doi: 10.1186/1475-2875-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afrane YA, Bonizzoni M, Yan G. Secondary malaria vectors of sub-Saharan Africa: threat to malaria elimination on the continent? Curr Top Malar. 2016;20:473. [Google Scholar]
- 6.Antonio-Nkondjio C, Kerah CH, Simard F, Awono-Ambene P, Chouaibou M, Tchuinkam T, et al. Complexity of the malaria vectorial system in Cameroon: contribution of secondary vectors to malaria transmission. J Med Entomol. 2006;43:1215–1221. doi: 10.1093/jmedent/43.6.1215. [DOI] [PubMed] [Google Scholar]
- 7.Stevenson JC, Norris DE. Implicating cryptic and novel anophelines as malaria vectors in Africa. Insects. 2017;8:1. doi: 10.3390/insects8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sougoufara S, Ottih EC, Tripet F. The need for new vector control approaches targeting outdoor biting anopheline malaria vector communities. Parasit Vectors. 2020;13:295. doi: 10.1186/s13071-020-04170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson HM, Dornhaus A, Beeche A, Borgemeister C, Gottlieb M, Mulla MS, et al. Ecology: a prerequisite for malaria elimination and eradication. PLoS Med. 2010;7:e1000303. doi: 10.1371/journal.pmed.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 11.St Laurent B, Cooke M, Krishnankutty SM, Asih P, Mueller JD, Kahindi S, et al. Molecular characterization reveals diverse and unknown malaria vectors in the western Kenyan highlands. Am J Trop Med Hyg. 2016;94:327–335. doi: 10.4269/ajtmh.15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobo NF, St Laurent B, Sikaala CH, Hamainza B, Chanda J, Chinula D, et al. Unexpected diversity of Anopheles species in eastern Zambia: implications for evaluating vector behavior and interventions using molecular tools. Sci Rep. 2015;5:17952. doi: 10.1038/srep17952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spillings BL, Brooke BD, Koekemoer LL, Chiphwanya J, Coetzee M, Hunt RH. A new species concealed by Anopheles funestus Giles, a major malaria vector in Africa. Am J Trop Med Hyg. 2009;81:510–515. doi: 10.4269/ajtmh.2009.81.510. [DOI] [PubMed] [Google Scholar]
- 14.Barron MG, Paupy C, Rahola N, Akone-Ella O, Ngangue MF, Wilson-Bahun TA, et al. A new species in the major malaria vector complex sheds light on reticulated species evolution. Sci Rep. 2019;9:14753. doi: 10.1038/s41598-019-49065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciubotariu II, Jones CM, Kobayashi T, Bobanga T, Muleba M, Pringle JC, et al. Genetic diversity of Anopheles coustani in high malaria transmission foci in southern and central Africa. J Med Entomol. 2020;57:1782–1792. doi: 10.1093/jme/tjaa132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ministry of Health . Zambia malaria indicator survey. Zambia: Lusaka; 2018. [Google Scholar]
- 17.Zimba H, Kawawa B, Chabala A, Phiri W, Selsam P, Meinhardt M, et al. Assessment of trends in inundation extent in the Barotse floodplain, upper Zambezi river basin: a remote sensing-based approach. J Hydrol Reg Stud. 2018;15:149–170. doi: 10.1016/j.ejrh.2018.01.002. [DOI] [Google Scholar]
- 18.IUCN: Barotse floodplain, Zambia: Local economic dependence on wetland resources. In: Integrating wetland economic values into river basin management. IUCN, Harare, Zimbabwe: The World Conservation Union; 2003
- 19.Moore AE, Cotterill FPD, Main MPL, Williams HB. The Zambezi river. In: Gupta A, editor. Large rivers: geomorphology and management. Chichester: John Wiley; 2007. [Google Scholar]
- 20.Hardy A, Ettritch G, Cross DE, Bunting P, Liywalii F, Sakala J, et al. Automatic detection of open and vegetated water bodies using Sentinel 1 to map African malaria vector mosquito breeding habitats. Remote Sensing. 2019;11:593. doi: 10.3390/rs11050593. [DOI] [Google Scholar]
- 21.Cai XL, Haile AT, Magidi J, Mapedza E, Nhamo L. Living with floods—household perception and satellite observations in the Barotse floodplain. Zambia Phys Chem Earth. 2017;100:278–286. doi: 10.1016/j.pce.2016.10.011. [DOI] [Google Scholar]
- 22.Central Statistical Office . Population and demographic projections 2011–2035. Zambia: Lusaka; 2013. [Google Scholar]
- 23.Turpie J, Smith B, Emerton L, Barnes J. Economic valuation of the Zambezi basin wetlands. IUCN, The World Conservation Union Regional Office for Southern Africa: Harare, Zimbabwe; 1999. [Google Scholar]
- 24.Lewin J, Ashworth PJ. The negative relief of large river floodplains. Earth Sci Rev. 2014;129:1–23. doi: 10.1016/j.earscirev.2013.10.014. [DOI] [Google Scholar]
- 25.Flint LS. Socio-ecological vulnerability and resilience in an arena of rapid environmental change: community adaptation to climate variability in the Upper Zambezi floodplain. In: Working paper on social-ecological resilience. Kyoto: Enda Tiers Monde-Programme Energie; 2008
- 26.Timberlake J. Biodiversity of the Zambezi Basin wetlands: Review and preliminary assessment of available information. Zimbabwe: Harare; 1997. [Google Scholar]
- 27.McCartney M, Nyambe IA. Ecosystem services. In: Lautze J, Phiri Z, Smakhtin V, Saruchera D, editors. The Zambezi river basin: water and sustainable development. London, UK: Routledge, Taylor & Francis Group; 2017
- 28.Burrough SL, Thomas DSG, Orijemie EA, Willis KJ. Landscape sensitivity and ecological change in western Zambia: the long-term perspective from dambo cut-and-fill sediments. J Quaternary Sci. 2015;30:44–58. doi: 10.1002/jqs.2757. [DOI] [Google Scholar]
- 29.Heyden CJ. The hydrology and hydrogeology of dambos: a review. Prog Phys Geogr. 2004;28:544–564. doi: 10.1191/0309133304pp424oa. [DOI] [Google Scholar]
- 30.Bøgh C, Clarke SE, Jawara M, Thomas CJ, Lindsay SW. Localized breeding of the Anopheles gambiae complex (Diptera: Culicidae) along the River Gambia, west Africa. Bull Entomol Res. 2003;93:279–287. doi: 10.1079/BER2003239. [DOI] [PubMed] [Google Scholar]
- 31.Hardy AJ, Gamarra JG, Cross DE, Macklin MG, Smith MW, Kihonda J, et al. Habitat hydrology and geomorphology control the distribution of malaria vector larvae in rural Africa. PLoS ONE. 2013;8:e81931. doi: 10.1371/journal.pone.0081931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas CJ, Cross DE, Bøgh C. Landscape movements of Anopheles gambiae malaria vector mosquitoes in rural Gambia. PLoS ONE. 2013;8:e68679. doi: 10.1371/journal.pone.0068679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derua YA, Alifrangis M, Hosea KM, Meyrowitsch DW, Magesa SM, Pedersen EM, et al. Change in composition of the Anopheles gambiae complex and its possible implications for the transmission of malaria and lymphatic filariasis in north-eastern Tanzania. Malaria J. 2012;11:188. doi: 10.1186/1475-2875-11-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fillinger U, Lindsay SW. Suppression of exposure to malaria vectors by an order of magnitude using microbial larvicides in rural Kenya. Tropical Med Int Health. 2006;11:1629–1642. doi: 10.1111/j.1365-3156.2006.01733.x. [DOI] [PubMed] [Google Scholar]
- 35.Majambere S, Lindsay SW, Green C, Kandeh B, Fillinger U. Microbial larvicides for malaria control in the Gambia. Malaria J. 2007;6:76. doi: 10.1186/1475-2875-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winnepenninckx B, Backeljau T, De Wachter R. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993;9:407. doi: 10.1016/0168-9525(93)90102-N. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. Clustal-W - Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall TA. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/nt. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 39.Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DNASp 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.QGIS Development Team: QGIS geographic information system. Open Source Geospatial Foundation Project; 2020
- 42.IBM Corp.: IBM SPSS statistics for windows, version 25.0. Armonk, NY; 2017
- 43.O’Hara RB, Kotze DJ. Do not log-transform count data. Methods Ecol Evol. 2010;1:118–122. doi: 10.1111/j.2041-210X.2010.00021.x. [DOI] [Google Scholar]
- 44.Rita H, Komonen A. Odds ratio: an ecologically sound tool to compare proportions. Ann Zool Fenn. 2008;45:66–72. doi: 10.5735/086.045.0106. [DOI] [Google Scholar]
- 45.Irish SR, Kyalo D, Snow RW, Coetzee M. Updated list of Anopheles species (Diptera: Culicidae) by country in the Afrotropical region and associated islands. Zootaxa. 2020;4747:401–449. doi: 10.11646/zootaxa.4747.3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chanda E, Hemingway J, Kleinschmidt I, Rehman AM, Ramdeen V, Phiri FN, et al. Insecticide resistance and the future of malaria control in Zambia. PLoS ONE. 2011;6:e24336. doi: 10.1371/journal.pone.0024336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kent RJ, Thuma PE, Mharakurwa S, Norris DE. Seasonality, blood feeding behavior, and transmission of Plasmodium falciparum by Anopheles arabiensis after an extended drought in southern Zambia. Am J Trop Med Hyg. 2007;76:267–274. doi: 10.4269/ajtmh.2007.76.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson JC, Pinchoff J, Muleba M, Lupiya J, Chilusu H, Mwelwa I, et al. Spatio-temporal heterogeneity of malaria vectors in northern Zambia: implications for vector control. Parasit Vectors. 2016;9:510. doi: 10.1186/s13071-016-1786-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masaninga F, Muleba M, Masendu H, Songolo P, MweeneNdumba I, MazabaLiwewe ML, et al. Distribution of yellow fever vectors in Northwestern and Western Provinces, Zambia. Asian Pac J Trop Med. 2014;7(S1):88–S92. doi: 10.1016/S1995-7645(14)60210-8. [DOI] [PubMed] [Google Scholar]
- 50.Das S, Henning TC, Simubali L, Hamapumbu H, Nzira L, Mamini E, et al. Underestimation of foraging behaviour by standard field methods in malaria vector mosquitoes in southern Africa. Malar J. 2015;14:12. doi: 10.1186/s12936-014-0527-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bayoh MN, Mathias DK, Odiere MR, Mutuku FM, Kamau L, Gimnig JE, et al. Anopheles gambiae: Historical population decline associated with regional distribution of insecticide-treated bed nets in western Nyanza Province. Kenya Malar J. 2010;9:62. doi: 10.1186/1475-2875-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sougoufara S, Harry M, Doucoure S, Sembene PM, Sokhna C. Shift in species composition in the Anopheles gambiae complex after implementation of long-lasting insecticidal nets in Dielmo. Senegal Med Vet Entomol. 2016;30:365–368. doi: 10.1111/mve.12171. [DOI] [PubMed] [Google Scholar]
- 53.Getachew D, Balkew M, Tekie H. Anopheles larval species composition and characterization of breeding habitats in two localities in the Ghibe river basin, southwestern Ethiopia. Malar J. 2020;19:65. doi: 10.1186/s12936-020-3145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kweka EJ, Zhou G, Munga S, Lee MC, Atieli HE, Nyindo M, et al. Anopheline larval habitats seasonality and species distribution: A prerequisite for effective targeted larval habitats control programmes. PLoS ONE. 2012;7:e52084. doi: 10.1371/journal.pone.0052084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das S, Muleba M, Stevenson JC, Norris DE. Habitat partitioning of malaria vectors in Nchelenge District. Zambia Am J Trop Med Hyg. 2016;94:1234–1244. doi: 10.4269/ajtmh.15-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J, et al. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic precis. Parasit Vectors. 2010;3:117. doi: 10.1186/1756-3305-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nambunga IH, Ngowo HS, Mapua SA, Hape EE, Msugupakulya BJ, Msaky DS, et al. Aquatic habitats of the malaria vector Anopheles funestus in rural south-eastern Tanzania. Malar J. 2020;19:219. doi: 10.1186/s12936-020-03295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coetzee M. Key to the females of Afrotropical Anopheles mosquitoes (Diptera: Culicidae) Malar J. 2020;19:70. doi: 10.1186/s12936-020-3144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coetzee M. Anopheles crypticus, new species from South Africa is distinguished from Anopheles coustani (Diptera: Culicidae) Mosq Syst. 1994;26:125–131. [Google Scholar]
- 60.Kiszewski AE, Teffera Z, Wondafrash M, Ravesi M, Pollack RJ. Ecological succession and its impact on malaria vectors and their predators in borrow pits in western Ethiopia. J Vector Ecol. 2014;39:414–423. doi: 10.1111/jvec.12117. [DOI] [PubMed] [Google Scholar]
- 61.Gillies MT, De Meillon B. The Anophelinae of Africa south of the Sahara (Ethiopian zoogeographical region) Publ South Afr Inst Med Res. 1968;54:1–343. [Google Scholar]
- 62.Goupeyou-Youmsi J, Rakotondranaivo T, Puchot N, Peterson I, Girod R, Vigan-Womas I, et al. Differential contribution of Anopheles coustani and Anopheles arabiensis to the transmission of Plasmodium falciparum and Plasmodium vivax in two neighboring villages of Madagascar. bioRxiv. 2019;13:430. doi: 10.1186/s13071-020-04282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fornadel CM, Norris LC, Franco V, Norris DE. Unexpected anthropophily in the potential secondary malaria vectors Anopheles coustani s.l. and Anopheles squamosus in Macha, Zambia. Vector Borne Zoonotic Dis. 2011;11:1173–1179. doi: 10.1089/vbz.2010.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Degefa T, Yewhalaw D, Zhou G, Lee MC, Atieli H, Githeko AK, et al. Indoor and outdoor malaria vector surveillance in western Kenya: implications for better understanding of residual transmission. Malar J. 2017;16:443. doi: 10.1186/s12936-017-2098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Degefa T, Zeynudin A, Godesso A, Michael YH, Eba K, Zemene E, et al. Malaria incidence and assessment of entomological indices among resettled communities in Ethiopia: a longitudinal study. Malar J. 2015;14:24. doi: 10.1186/s12936-014-0532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mwangangi JM, Muturi EJ, Muriu SM, Nzovu J, Midega JT, Mbogo C. The role of Anopheles arabiensis and Anopheles coustani in indoor and outdoor malaria transmission in Taveta District, Kenya. Parasit Vectors. 2013;6:114. doi: 10.1186/1756-3305-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nepomichene TNJJ, Tata E, Boyer S. Malaria case in Madagascar, probable implication of a new vector Anopheles coustani. Malaria J. 2015;14:475. doi: 10.1186/s12936-015-1004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamau L, Mulaya N, Vulule JM. Evaluation of potential role of Anopheles ziemanni in malaria transmission in western Kenya. J Med Entomol. 2006;43:774–776. doi: 10.1603/0022-2585(2006)43[774:eoproa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 69.Nyirakanani C, Chibvongodze R, Kariuki L, Habtu M, Masika M, Mukoko D, et al. Characterization of malaria vectors in Huye District, southern Rwanda. Tanzania J Health Res. 2017;19:1–10. [Google Scholar]
- 70.Tabue RN, Nem T, Atangana J, Bigoga JD, Patchoke S, Tchouine F, et al. Anopheles ziemanni a locally important malaria vector in Ndop Health District, North West Region of Cameroon. Parasit Vectors. 2014;7:262. doi: 10.1186/1756-3305-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tedrow RE, Rakotomanga T, Nepomichene T, Howes RE, Ratovonjato J, Ratsimbasoa AC, et al. Anopheles mosquito surveillance in Madagascar reveals multiple blood feeding behavior and Plasmodium infection. PLoS Negl Trop Dis. 2019;13:e0007176. doi: 10.1371/journal.pntd.0007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dambach P, Machault V, Lacaux JP, Vignolles C, Sie A, Sauerborn R. Utilization of combined remote sensing techniques to detect environmental variables influencing malaria vector densities in rural west Africa. Int J Health Geogr. 2012;11:8. doi: 10.1186/1476-072X-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Machault V, Gadiaga L, Vignolles C, Jarjaval F, Bouzid S, Sokhna C, et al. Highly focused anopheline breeding sites and malaria transmission in Dakar. Malar J. 2009;8:138. doi: 10.1186/1475-2875-8-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Cytochrome c oxidase 1 (COI) sequences for unknown Anopheles species. Table S2: second internal transcribed spacer region (ITS2) sequences for unknown Anopheles species.
Additional file 2: Figure S1. Maximum likelihood phylogenetic tree of cytochrome c oxidase I (COI) sequences from Anopheles larvae sampled in western Zambia.
Data Availability Statement
The COI and ITS2 sequence datasets generated by and analyzed in the current study are available in the NCBI GenBank nucleotide archive with accession numbers MW167786–MW168163 for COI sequences and MW166490–MW166863 for ITS2 sequences. Additional data on specimens assigned to “unknown Anopheles species” category are provided within Additional file 1. A maximum likelihood phylogenetic tree of COI haplotypes is provided in Additional file 2.