Abstract
In eukaryotes, most secretory and membrane proteins are targeted by an N‐terminal signal sequence to the endoplasmic reticulum, where the trimeric Sec61 complex serves as protein‐conducting channel (PCC). In the post‐translational mode, fully synthesized proteins are recognized by a specialized channel additionally containing the Sec62, Sec63, Sec71, and Sec72 subunits. Recent structures of this Sec complex in the idle state revealed the overall architecture in a pre‐opened state. Here, we present a cryo‐EM structure of the yeast Sec complex bound to a substrate, and a crystal structure of the Sec62 cytosolic domain. The signal sequence is inserted into the lateral gate of Sec61α similar to previous structures, yet, with the gate adopting an even more open conformation. The signal sequence is flanked by two Sec62 transmembrane helices, the cytoplasmic N‐terminal domain of Sec62 is more rigidly positioned, and the plug domain is relocated. We crystallized the Sec62 domain and mapped its interaction with the C‐terminus of Sec63. Together, we obtained a near‐complete and integrated model of the active Sec complex.
Keywords: post‐translational translocation, protein translocation, sec complex, signal sequence
Subject Categories: Membranes & Trafficking, Structural Biology
Cryo‐electron microscopy and X‐ray crystallography studies reveal the molecular architecture of the engaged eukaryotic heptameric Sec complex, binding the protein signal sequence in an open conformation.

Introduction
In eukaryotes, the majority of secretory and membrane proteins is targeted by an N‐terminal signal sequence to the Sec61 complex, the protein‐conducting channel (PCC) at the endoplasmic reticulum (ER) membrane (Blobel & Dobberstein, 1975; Rapoport et al, 2017; Gemmer & Förster, 2020). Here, the Sec61 complex is gated open by the signal sequence and can facilitate either translocation of proteins into the ER lumen or insertion of membrane proteins by laterally releasing them into the lipid bilayer. The highly conserved heterotrimeric Sec61 complex (SecYEG complex in prokaryotes) consists of the large Sec61α subunit (SecY in E. coli; Sec61p in Saccharomyces cerevisiae) with 10 transmembrane helices (TMs) and the two small single‐spanning Sec61β (SecG in E.c.; Sbh1 in S.c.) and Sec61γ (SecE in E.c.; Sss1 in S.c.; Görlich & Rapoport, 1993; Hanada et al, 1994). It can operate in two modes, either co‐translationally bound to a translating ribosome or post‐translationally. For the co‐translational mode, the Sec61α subunit serves as a high‐affinity ribosome receptor, whereas in both modes, Sec61α recognizes and is gated by an N‐terminal signal sequence. The signal sequence typically consists of a 15–20 amino acid long hydrophobic stretch and is often flanked by positively charged or hydrophilic residues. Most membrane proteins employ the co‐translational mode and—at least in S. cerevisiae—carry a more hydrophobic signal sequence or a signal anchor sequence, which later serves as TM. Both are recognized by the signal recognition particle (SRP) as soon as they emerge from the ribosomal exit tunnel (Wild et al, 2004; Nyathi et al, 2013). At the membrane, the SRP‐bound ribosome‐nascent chain complex (RNC) docks to the SRP receptor (SR) and is subsequently handed over to the Sec61 complex. As a result, the nascent peptide can be threaded from the ribosomal tunnel exit directly into the protein‐conducting channel.
In contrast, the post‐translational translocation mode is mostly employed by soluble and secretory proteins, which in yeast carry less hydrophobic signal sequences that are not recognized by SRP (Ng et al, 1996; Ast et al, 2013). As a consequence, these proteins are fully synthesized, released from the ribosome, and kept in an unfolded, translocation competent state by chaperones (Ngosuwan et al, 2003). Without engaging SRP, the signal sequences of these proteins are directly recognized by a specialized (heptameric) channel complex, commonly termed the Sec complex or post‐translocon (the Sec complex from here on). It consists of the trimeric Sec61 complex, the additional Sec62 and Sec63 subunits as well as the additional subunits Sec71 and Sec72 in yeast (Deshaies et al, 1991; Panzner et al, 1995; Plath et al, 1998). Sec63 is an ER‐resident membrane protein with three TM helices at its N‐terminus. On the luminal side of the ER membrane, Sec63 carries between TM2 and TM3 a J‐domain that is able to recruit the Hsp70‐type chaperone BiP (Kar2p in yeast) to the complex. BiP/Kar2p has been shown to act like a molecular ratchet by iterative binding to the translocating peptide, thereby providing the driving force for unidirectional translocation (Matlack et al, 1999). On the cytoplasmic side, the C‐terminus of Sec63 forms a large domain harboring an acidic stretch at its ultimate C‐terminus that interacts with its partner, Sec62. Similar to Sec63, also Sec62 is an essential protein, but its exact function during protein translocation is less well defined. Topologically, Sec62 features two TM helices flanked by two cytoplasmic domains (Deshaies & Schekman, 1989, 1990). While acting most likely in concert with Sec63 during translocation, Sec62 was shown by chemical cross‐linking to be in direct proximity to inserting signal sequences (Lyman & Schekman, 1997; Matlack et al, 1999; Dünnwald et al, 1999),. Moreover, it was suggested to stabilize binding of the signal sequence to Sec61α and thereby promoting channel gating (Wu et al, 2018). In contrast to Sec62 and Sec63, the fungus‐specific subunits Sec71 and Sec72 are not essential. Only Sec71 carries a TM helix, but its cytoplasmic domain binds tightly to the Sec72 subunit, thereby integrating it into the Sec complex. Together, both proteins were shown to aid in protein translocation by interacting with cytosolic chaperones, thereby facilitating the handover of the secretory peptide from the chaperones to the Sec complex (Feldheim et al, 1993; Feldheim & Schekman, 1994; Tripathi et al, 2017).
The structure of the heterotrimeric Sec61 PCC is well‐established in several functional states (Rapoport et al, 2017; van den Berg et al, 2004; Park et al, 2014; Voorhees et al, 2014; Voorhees & Hegde, 2016; Kater et al, 2019). It forms an hourglass‐shaped aqueous channel across the membrane with a central constriction dividing cytosolic and luminal sides, and a short helical plug domain in the luminal region of the channel (van den Berg et al, 2004). The central constriction, the so‐called pore ring, was shown to act as the vertical gate for translocating peptides (Cannon et al, 2005), whereas for channel opening the plug domain has to relocate to a more peripheral conformation to allow transit of the peptide. Signal sequences as well as hydrophobic TM segments were found to bind at a distinct side of Sec61α, the so‐called lateral gate, which is located at the interface between the N‐ and the C‐terminal halves of the protein (Plath et al, 1998, 2003; van den Berg et al, 2004; Gogala et al, 2014; Voorhees & Hegde, 2016; Kater et al, 2019; Li et al, 2016; Ma et al, 2019). This lateral gate—formed by the Sec61α TMs 2 and 3 of the N‐terminal and TMs 7 and 8 of the C‐terminal half—was observed in a variety of conformations ranging from fully closed in the idle state, to partially open when primed with the ribosome or SecA (in prokaryotes; Zimmer et al, 2008; Park et al, 2014; Voorhees et al, 2014; Braunger et al, 2018), and fully open when engaged with a signal sequence or TM domain (Frauenfeld et al, 2011; Park et al, 2014; Voorhees & Hegde, 2016; Li et al, 2016; Ma et al, 2019). Notably, it has been observed that the signal sequence driven opening of the lateral gate can destabilize the plug domain, thereby opening the aqueous conduit for the peptide to the luminal side (Tam et al, 2005; Voorhees & Hegde, 2016; Li et al, 2016; Ma et al, 2019).
Two recent cryo‐EM studies provided the overall architecture of the heptameric post‐translocon in the idle state, i.e., lacking a signal sequence or translocating peptide (Wu et al, 2018; Itskanov & Park, 2018). In these structures, the Sec71‐Sec72 dimer and the Sec63 C‐terminal domain are packed upon the cytosolic face of the Sec61 complex. As a result, the C‐terminal cytoplasmic loops of Sec61α are masked in a way that all ribosome interactions are sterically prohibited, explaining why the Sec complex can exclusively exert the post‐translational mode. Moreover, compared to ribosome‐primed (Voorhees et al, 2014; Braunger et al, 2018) or signal sequence‐engaged Sec61α (Voorhees & Hegde, 2016), the lateral gate of Sec61α in both structures is stabilized in a more open conformation. This is mainly due to Sec63, which interacts with Sec61α via its three TM domains that bind to both, the N‐ and C‐terminal half (Wu et al, 2018; Itskanov & Park, 2018). Despite some differences between the studies regarding the plug conformation, the highly similar open conformation of the lateral gate was interpreted to facilitate insertion of and gating by the less hydrophobic signal sequences employed in the post‐translational mode. Yet, it is not clear how exactly these signal sequences engage the heptameric complex and how the structurally unknown Sec62 subunit may contribute.
Here we present a cryo‐EM structure of the heptameric Sec complex bound to a substrate carrying the signal sequence of prepro‐α factor (ppαF) at an average resolution of 4.4 Å. Compared to the idle structures (Wu et al, 2018; Itskanov & Park, 2018), the signal sequence is inserted into the groove of the lateral gate of Sec61α and the channel adopts an even more open conformation with the plug domain relocated. The signal sequence adopts a helical conformation and appears at a position reminiscent to those previously seen in co‐translational Sec61 and post‐translational‐acting bacterial SecYEG complexes (Voorhees et al, 2014; Li et al, 2016; Ma et al, 2019). We can localize the two TMs of Sec62 in the vicinity of the signal sequence, whereas the cytosolic domains of Sec62, a short C‐terminal and a globular N‐terminal domain, appeared rather flexible. Therefore, we crystallized the cytoplasmic domain at the N‐terminus of Sec62 and mapped its interaction site with the C‐terminus of Sec63. Thus, our study provides an integrated near‐complete model of the active Sec complex.
Results
Structural analysis of the apo and signal sequence‐engaged Sec complexes
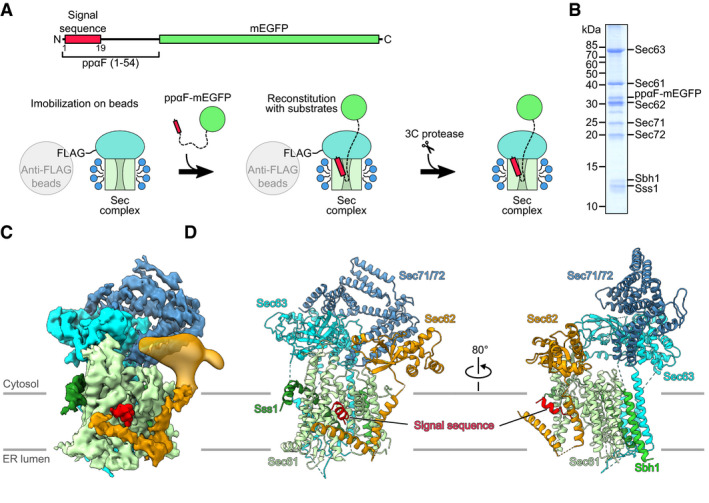
The heptameric Sec complex from the baker’s yeast Saccharomyces cerevisiae was previously shown to be fully active in detergent solution (Matlack et al, 1997). Employing very similar conditions, we reconstituted the signal sequence‐bound post‐translocon using solubilized and purified yeast heptameric Sec complex and the well‐characterized N‐terminal signal sequence of prepro‐α factor (ppαF; Panzner et al, 1995; Matlack et al, 1997). In order to prevent full translocation of ppαF, we generated a construct containing the first 54 amino acids of ppαF followed by mEGFP (ppαF‐mEGFP; Fig 1A, upper panel). After verifying that the ppαF‐mEGFP construct was indeed bound to the heptameric complex in a pull‐down assay (Fig EV1), we reconstituted and purified the complex by an on‐bead scheme (Fig 1A, lower panel): Firstly, the Sec complex was immobilized on anti‐FLAG M2 agarose beads and then incubated with a molar excess of ppαF‐mEGFP. Subsequently, the reconstituted complex was cleaved off from the beads and subjected to anion exchange chromatography for further purification and concentration. The concentrated sample contained all seven components of the heptameric Sec complex and the ppαF‐mEGFP (Figs 1B and EV1E) and was subjected to cryo‐EM and single particle analysis (Fig EV1F and G). After routine processing (Appendix Fig S1) and classification, promising 3‐dimensional (3D) classes with clear density for the heptameric complex were further classified using a mask for the TM region of the complex. This yielded two exclusive 3D classes differing in overall conformation and the presence of an additional distinct density in the open lateral gate, which we assigned to the ppαF signal sequence. In agreement with the roughly estimated stoichiometry between ppαF‐mEGFP and the Sec complex on the SDS–PAGE gel (Fig 1B), the complexes in the signal sequence‐engaged state represented about half of the particles, while the other half represented empty Sec complexes, from here on called the apo state (Appendix Fig S1A).
Figure 1. Preparation and cryo‐EM structure of signal sequence‐bound Sec complex.
-
ASchematic diagram depicting the recombinant ppαF‐mEGFP substrate (upper panel) and purification strategy for the signal sequence‐bound Sec complex (lower panel): Detergent‐solubilized heptameric Sec complex (TM domains: green; cytosolic domains: cyan; detergent micelles: blue) FLAG‐tagged on Sec62 was immobilized on anti‐FLAG beads and incubated with molar excess of purified recombinant ppαF‐mEGFP substrate. Substrate‐bound Sec complex was eluted with 3C protease.
-
BSDS–PAGE of the reconstituted signal sequence‐bound Sec complex stained with Coomassie Blue. The full gel of the reconstitution is shown in Fig EV1E.
-
CCryo‐EM reconstruction as obtained after focused refinement and post‐processing. The map was segmented and color‐coded by the individual Sec complex components. For clarity, density for the detergent micelle was masked and the contour levels for the for Sec62 TMs and Sec71/72 densities are slightly lower. For the Sec62 cytosolic domain, the map is low‐pass filtered to 15 Å.
-
DMolecular model of the signal sequence‐bound Sec complex. The model for the Sec62 N‐terminal cytoplasmic domain (Sec62 domain) is derived from the crystal structure as determined in this work.
Figure EV1. Purification and cryo‐EM of the Sec complex.

-
ASDS–PAGE analysis of the purification of the heptameric Sec complex containing FLAG‐tagged Sec62. After solubilization, the tagged complex was applied to anti‐FLAG‐M2 resin and the eluate was subjected to ion‐exchange chromatography using Q sepharose (S: solubilized microsomes; FT: flow‐through from the anti‐FLAG resin; 3C: eluate after 3C cleavage; E: eluate from Q sepharose; C: concentrated heptameric Sec complex sample).
-
BCryo‐EM map of the (heptameric) Sec complex (apo state) shown after focused refinement and post‐processing. The map is shown at contour level 0.02 except for the Sec71/72 part, which is at 0.018. Individual subunits of the Sec complex are color‐coded as in Fig 1C.
-
CMolecular model of the Sec complex in apo state.
-
DSDS–PAGE analysis of pull‐down assays of the purified Sec complex with the signal sequence‐containing mEGFP‐tagged ppαF or its translocation defect mutant ppαFm3. Pull downs were performed at varying molar ratios (B: bead‐bound fraction; FT, unbound flow‐through). Semi‐quantitative assessment of the Sec63 bands on the gel using ImageJ shows that the mutant signal sequence binding to the Sec complex appears weaker than the wildtype, as expected by weaker activity of the mutant in translocation (Allison & Young, 1989).
-
ESDS–PAGE analysis of the purification of the signal sequence‐bound Sec complex. Lanes are labeled as in (A) except: FT1: flow‐through after binding of the Sec complex on beads; FT2: flow‐through after binding of ppαF‐mEGFP to the Sec complex immobilized on beads; W1‐3: washes after ppαF‐mEGFP binding.
-
FA representative cryo‐EM micrograph as obtained from TITAN KRIOS equipped with a K2 direct electron detector. The scale bar represents 20 nm.
-
GSelected 2D class averages. The box size was chosen to be 200 pixels resulting in a box width of 212 Å.
The maps of the apo and the signal sequence‐engaged Sec complexes were refined to an overall resolution of 4.3 and 4.4 Å, respectively (Appendix Fig S1). All TM helices of the Sec complex were clearly resolved, which allowed for the unambiguous rigid body fitting of available structures into both maps with only minor adjustments (see Materials and Methods and Fig EV2). The overall architecture of our Sec complex that lacked additional density was very similar to the two previous structures (Fig EV1B and C). The cytosolic domains of Sec63 and Sec71/72 were located on top of Sec61α and the three TM helices of Sec63 transversed the membrane at the backside of the Sec61 complex as observed before (Wu et al, 2018; Itskanov & Park, 2018). Also, as seen in previous reconstructions, less well‐resolved density was present for the cytosolic N‐terminal domain of Sec62 that we identified based on our crystal structure (see below). In the apo structure, the lateral gate is open and the plug is clearly present below the pore ring, thus, our apo map is closest to the structure described by the Rapoport lab (PDB 6ND1; Wu et al, 2018; Fig EV3A). The overall structure of the engaged state was similar to the apo state, yet, we observed a more pronounced density for the cytosolic domain of Sec62 and additional α‐helical densities for the ppαF signal sequence in the lateral gate of Sec61α as well as the two TM helices of Sec62 (Figs 1C and D, and EV2A and B).
Figure EV2. Fitting of molecular models into the cryo‐EM maps of ss‐bound and apo Sec complex.

-
AView focusing on the lateral gate of Sec61α. In the ss‐bound state (left), the signal sequence of ppαF (red) is inserted into the lateral gate (TM2 and TM7) and the two Sec62 TM helices are stabilized. In the apo state (right), density for the plug (P) is present below the central pore.
-
BView focusing on the N‐terminal half of Sec61α (TMs 2, 3 and 4). In the ss‐bound state (left), Sec62 TM1 is bound near Sec61α TM2.
-
CView focusing on the C‐terminal half of Sec61α (TMs 6‐9).
-
DView focusing on the cytosolic domains of the Sec complex.
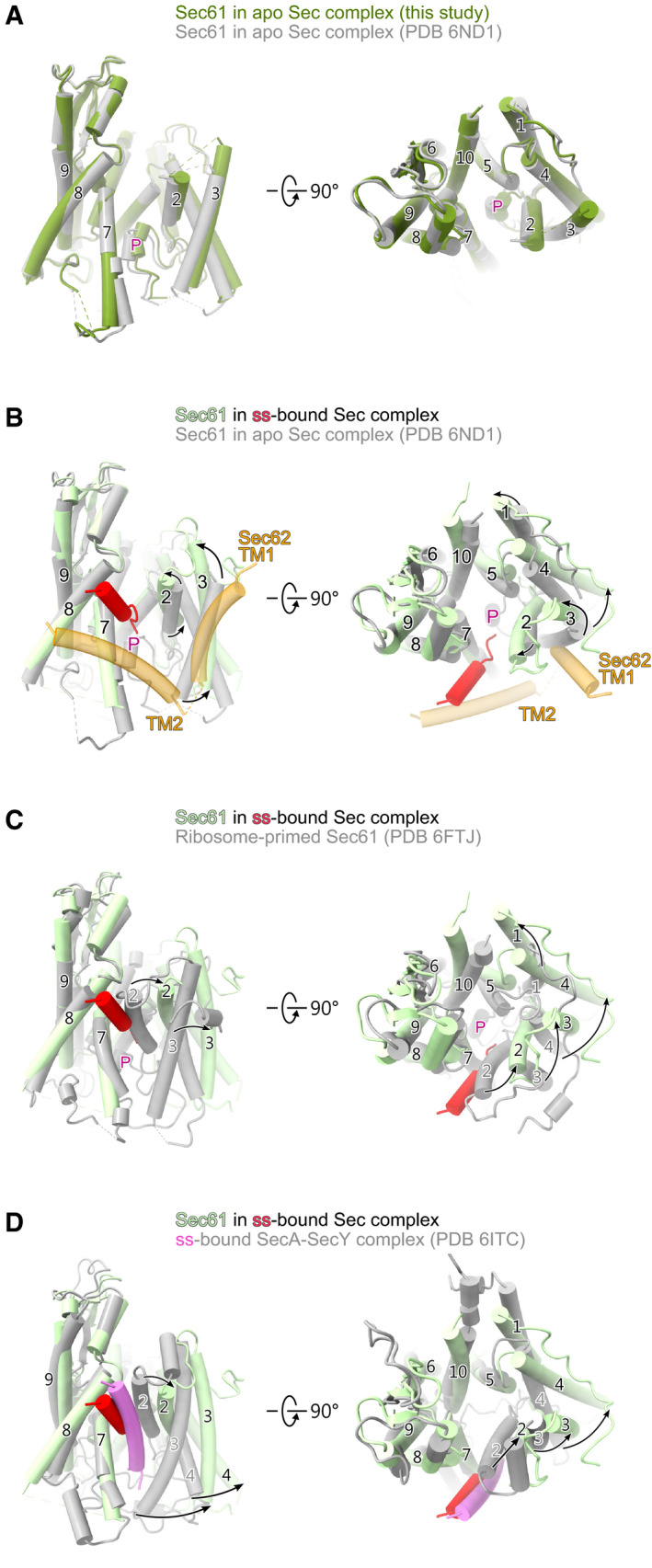
Figure EV3. Comparison of lateral gate conformation.
-
AComparison of Sec61α in the apo state (this study) with Sec61α in the apo Sec complex as shown in the Wu study (PDB 6ND1).
-
BComparison of Sec61α in the ss‐bound Sec complex with Sec61α in the apo Sec complex as shown in the Wu study (PDB 6ND1). The ppαF‐ss and Sec62 TM helices are shown in addition.
-
CComparison of Sec61α in the ss‐bound Sec complex with ribosome‐primed apo Sec61 complex as shown in the Braunger study (PDB 6FTJ)
-
DComparison of Sec61α in the ss‐bound heptameric Sec complex with SecY as found in the study of Ma et al showing the ss‐bound SecA‐SecY complex (PDB 6ITC).
Data information: All structures are shown from a view focusing on the lateral gate (left) and from the top (cytosolic side; right). Structure alignments are based on TMs 7–9 of Sec61α. Black arrows indicate the movement of the helices of the Sec61 complex. The plug is indicated by “P”.
Conformation of ppαF signal sequence, the Sec61 complex and Sec62
When we compared the reconstruction of the apo state with that of the engaged one, we observed extra density within the lateral gate in close contact with TM7 and parallel to TM2 of Sec61α (Fig 1C, red). Due to its unique appearance and position in the lateral gate, this density was assigned as the α‐helical signal sequence of ppαF (Figs 1D and 2A and B). Its position is very similar to those signal sequences observed in the structures of co‐translationally acting Sec61 or the bacterial SecYEG complex (Voorhees & Hegde, 2016; Li et al, 2016; Ma et al, 2019). Its position is also fully consistent with previous cross‐linking data suggesting that the ppαF signal sequence localizes in close proximity to TMs 2 and 7 of the Sec complex (Plath et al, 1998, 2003). Moreover, we observed two additional α‐helical densities close to the lateral gate extending toward the Sec62 density at the cytosolic side (Wu et al, 2018; Itskanov & Park, 2018) (Fig 1B, orange). We noticed that these densities were more pronounced in the engaged state and, since all other TMs of the complex had already been identified, we assigned them to the remaining TM1 and TM2 of Sec62.
Figure 2. Conformation of the signal sequence‐bound heptameric Sec complex.

-
ACut side view of the cryo‐EM density of the signal sequence‐bound (up) and the apo (bottom) Sec complex highlighting the ppαF signal sequence (ss; red) and Sec62 cytosolic and transmembrane domains (orange). Both maps were low‐pass filtered to 8 Å. The density of the Sec62 cytosolic C‐terminal domain connected to the Sec62 TM2 is marked by an arrowhead.
-
BTwo views showing the conformations of the Sec61 complex (green) with the signal sequence (ss; red) bound to the lateral gate (upper panels) and in the apo state (lower panels). The two Sec62 TMs (orange) are stabilized in ss‐bound state. The plug is indicated by “P” in the apo state.
-
C–EComparison of Sec61 complex conformations in the ss‐bound (pale green) with the apo (dark green) Sec complex from this study (C), with the apo Sec complex from Itskanov (PDB 6N3Q, (D)) and with ss‐opened ribosome‐bound Sec61 complex (PDB 3JC2, (E)). Alignments of structures were based on TMs 7‐9. The black arrows indicate the movement of Sec61α TM helices of the Sec61 complex. The plug is indicated by “P”.
For the conformation of the channel entity itself, the Sec61 complex, we observed a conformational shift of TMs 2 and 3 away from TMs 7 and 8 when compared to the apo state (Figs 2C and D and EV3B, and Movie EV1) and the ribosome‐primed Sec61 (PDB 6FTJ; Braunger et al, 2018; Fig EV3C), resulting in an even more open lateral gate. This conformation was most similar to the already more open structure described by the Park lab (PDB 6N3Q; Itskanov & Park, 2018; Fig 2D), but with an even wider lateral gate (~5.5° relative to TM5). Furthermore, and contrary to the structure by the Park lab, the plug density was not present anymore in its pore closing position and was likely delocalized by the signal sequence guided inserting peptide. Again, the overall position of our ppαF signal sequence is largely consistent with the position of a more hydrophobic co‐translational signal sequence of preprolactin (PDB 3JC2; Voorhees & Hegde, 2016; Fig 2E) and with bacterial post‐translational signal sequences observed in SecA‐SecY complexes (PDB 6ITC; Li et al, 2016; Ma et al, 2019; Fig EV3D). However, in our structure that lateral gate is significantly wider than in any other known structure of signal sequence‐bound Sec61 channels (Figs 2E and EV3D).
Interestingly, the observation that the TM helices of Sec62 were positioned close to the inserted ppαF signal sequence suggests that Sec62 may act by being in direct contact with signal sequences, which would also be in agreement with previous chemical cross‐linking data (Plath et al, 1998, 2003). In fact, in our reconstruction the still somewhat more flexible TM2 of Sec62 could directly contact the signal sequence whereas TM1 was in proximity close enough to possibly contact the lateral gate helices TMs 2 and 3 of Sec61α (Fig 2B, upper panel). Overall, the more rigid conformation of Sec62 in the engaged state hints at a stabilizing function for the active open channel conformation. This notion is supported by the absence of the Sec62 TM helices in the apo state and a less pronounced appearance of the N‐terminal cytosolic domain of Sec62 (Fig 2A, lower panel) indicating an overall flexibility of Sec62 in the inactive apo state.
Structural assignment of Sec62‐N to the architecture of the Sec complex
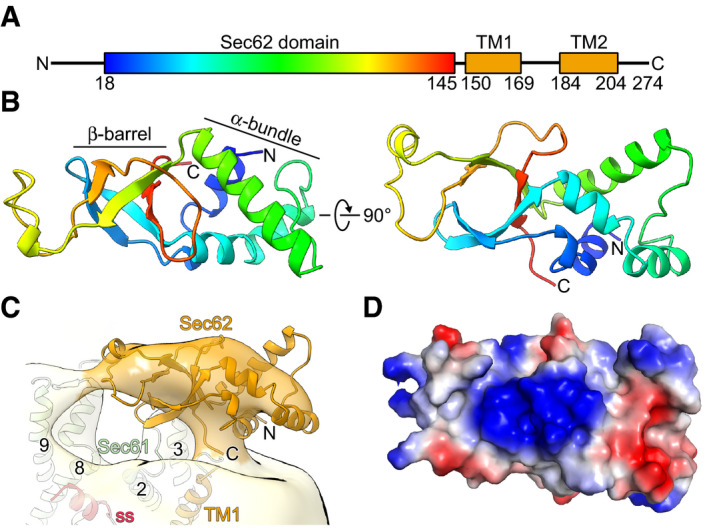
Although the density for the Sec62 cytosolic domain is more rigid in the engaged state, a molecular interpretation was not possible due to limited local resolution in our cryo‐EM map. Thus, we determined the structure of the N‐terminal cytoplasmic domain of Sec62 (Sec62‐N; residues 18–145; Fig 3A) by X‐ray crystallography to a resolution of 2.5 Å (Table 1). The structure of Sec62‐N displays an elongated shape and can be divided into two tightly interacting lobes: a four‐helix bundle (lobe 1) and a β‐barrel (lobe 2) (Fig 3B). Surprisingly, no structural homologue was found, neither by sequence similarity nor by structure comparison using the DALI server (Holm, 2019). Thus, we named this novel domain the “Sec62 domain”.
Figure 3. Crystal structure of Sec62 domain.
-
ADomain organization of Sec62. The N‐terminal cytoplasmic domain (Sec62 domain) is colored in rainbow.
-
BTwo views on the crystal structure of the Sec62 domain consisting of an α‐helical bundle and a β‐barrel domain.
-
CFit of the Sec62 domain crystal structure into the respective cryo‐EM density of the ss‐bound heptameric Sec complex as in Fig 1C. The map is low‐pass filtered to 15 Å.
-
DSurface representation of the Sec62 domain colored according to the electrostatic potential. The viewing angle is the same as in the right panel in (B).
Table 1.
Data collection and refinement statistics for the X‐ray structure of the Sec62 domain
| Sec62 (19‐145) | |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.97958 |
| Space group | P6522 |
| Cell dimensions | |
| a, b, c (Å) | 99.206, 99.206, 155.657 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 44.41‐2.54 (2.63‐2.54) a |
| Total reflections | 592,956 (56,970) a |
| Unique reflections | 15,552 (1,495) a |
| R merge | 0.1075 (1.886) a |
| I/σI | 26.53 (1.99) a |
| Completeness (%) | 99.66 (97.57) a |
| Redundancy | 38.1 (37.6) a |
| CC1/2 | 1 (0.949) a |
| Refinement | |
| R work/ R free | 0.2123/0.2439 |
| No. atoms | 2,283 |
| Protein | 2,108 |
| Ligand/ion | 99 |
| Water | 76 |
| B‐factors | 75.34 |
| Protein | 74.04 |
| Ligand/ion | 104.16 |
| Water | 74.06 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.73 |
| Ramachandran plot | |
| Ramachandran favored (%) | 98.40 |
| Ramachandran outliers (%) | 0.00 |
Values in parentheses are for highest‐resolution shell.
The dimension of Sec62‐N agreed overall with the EM density at the given local resolution of about 15 Å, and we positioned it accordingly into the engaged state adjacent to the front side of Sec61α in order to allow for connectivity with the putative TMs of Sec62 (Fig 3C). The apparent flexibility of this domain as indicated by the limited resolution did not allow for secondary structure‐based fitting and more exact positioning. Notably, in our map no large additional or connecting density was present to explain the previously described interaction between the Sec62 and Sec63 cytosolic domains, despite the essential nature of this interaction for translocation activity (Wittke et al, 2000). However, the interaction between Sec63 and Sec62‐N was previously shown to critically rely on the acidic C‐terminus of Sec63, in particular the small region comprising the amino acids 650–663 (Wittke et al, 2000). Moreover, the protein kinase CK2‐dependent phosphorylation of the threonines 652 and 654 of Sec63 is a requirement for the binding of Sec62‐N (Wang & Johnsson, 2005). This feature is conserved because also mammalian Sec62 interacts with the phosphorylated acidic C‐terminus of Sec63 in vitro (Ampofo et al, 2013). Therefore, it is likely that the interaction site of Sec62 for the Sec63 C‐terminus harbors a positively charged patch. Indeed, the electrostatic potential surface map of the Sec62 domain revealed a pronounced positively charged surface in the β‐barrel lobe (Fig 3D) as a candidate region. A sequence alignment of the Sec62 domain showed several positively charged residues which are conserved in fission yeast and three of them (R27, R51, and R104) are even conserved from yeast to humans (Appendix Fig S2).
To better characterize the Sec62/Sec63 interaction by biochemical and biophysical means, we employed two synthetic biotin‐labeled peptides representing residues 621–647 (C1) and 648‐663 (C2) of Sec63 (Fig 4A) for in vitro pull‐down assays with the purified Sec62‐N. In agreement with the previous studies (Wittke et al, 2000; Wang & Johnsson, 2005), the pull‐down assay showed that the Sec62 domain could be co‐purified using both C1 and phosphorylated C2 (C2P), however not with unphosphorylated C2 (Fig 4B). Isothermal titration calorimetry (ITC) experiments further confirmed that Sec63‐C2P and Sec62‐N interact with nanomolar affinity (Fig 4C), whereas no binding was observed for unmodified Sec63‐C2 (Appendix Fig S3).
Figure 4. Interaction between Sec62 and Sec63.

-
ADomain organization of Sec63. The sequence of the acidic patch on the C‐terminus—divided into C1 and C2 regions—is shown in the lower panel. The two phosphorylated threonines (T652 and T654) are marked in the C2 region. The J‐domain of Sec63 is indicated by “J”.
-
BPull‐down assay of Sec62 domain with different biotin‐labeled Sec63 C‐terminal peptides. Sec63‐C2P denotes di‐phosphorylated Sec63‐C2.
-
CITC‐thermogram obtained from ITC after titration of Sec62 domain with increasing amounts of Sec63‐C2P.
-
DDifference in HDX between the Sec62 domain in complex with Sec63‐C2P and the Sec62 domain alone. The graph depicts the summed means of differences ± 3 SD (n = 3 technical replicates) for each peptide plotted according to the midpoint of each peptide.
-
ECrystal structure of Sec62 domain color‐coded according the difference in HDX as shown in (D). The critical residues constituting the interaction interface between Sec63‐C2P and Sec62 are labeled.
-
FPull‐down assay of the Sec62 domain and a mutant in the interaction interface (Sec62R51E) with biotin‐labeled Sec63‐C2P.
Phosphorylated C‐terminus of Sec63 interacts with a basic patch at Sec62‐N
To delineate the interaction site of Sec63‐C2P at Sec62‐N, we employed hydrogen–deuterium exchange mass spectrometry (HDX‐MS; Kochert et al, 2018). To that end, the Sec62 domain alone or in presence of Sec63‐C2P was incubated in deuterated buffer and allowed to exchange hydrogen through deuterium. After stopping the exchange reaction with low pH and proteolytic digestion of the proteins, the deuterium incorporation into the Sec62 domain was determined by mass spectrometry. In the Sec62/Sec63‐C2P complex, large parts of the Sec62 domain incorporated less deuterium (Fig 4D, Appendix Fig S4) than Sec62 alone evidencing extensive Sec63‐C2P‐dependent conformational changes. Regions of most severe HDX reduction map to the β‐barrel lobe 2 of Sec62‐N domain essentially representing the highly positively charged candidate patch identified in our crystal structure while the α‐helical bundle centering around the N‐terminus of Sec62 domain was less affected (Fig 4E; compare also to: Fig 3D). Thus, considering the highly negatively charged C‐terminus of the Sec63, we concluded that this patch of Sec62 N‐terminal domain with its multitude of arginine and lysine residues would represent the interaction platform for Sec63‐C2P (Fig 4E). To test this notion, we mutated the central and conserved Arg51 of Sec62 to Glu, which resulted in complete inhibition of binding to the Sec63‐C2P in our pull‐down assay (Fig 4F). Taken together, the crystal structure of the N‐terminal domain of Sec62 revealed a novel fold, which can be positioned in the cryo‐EM density near the signal sequence‐binding site at the lateral gate. Moreover, a highly basic patch present in the β‐barrel lobe of Sec62‐N serves as highly specific binding site for the phosphorylated and acidic C‐terminus of the Sec63.
Discussion
By using the recombinant ppαF‐mEGFP as a substrate and on‐bead in vitro reconstitution, we were able to obtain cryo‐EM structures of the heptameric Sec complex in both substrate‐engaged and apo states. Despite the comparably low resolution—likely due to intrinsic flexibility of these highly dynamic complexes—we could unambiguously assign all α‐helices in the transmembrane moiety of the complex allowing a detailed description of the overall architecture and conformation of the signal sequences‐engaged state, as well as comparisons with previously described structures.
When comparing all three available Sec complex structures in the apo state, certain dynamics in Sec61α lateral gate opening and plug position become evident. Notably, the conformation of Sec61α in our apo state resembles the one described in Wu et al (2018) (Fig EV3A) whereas our structure of the signal sequence‐engaged state with the more open lateral gate is more similar to the apo‐state Sec complex described in Itskanov & Park (2018) (Fig 2D). Interestingly, in all three maps of apo states (Wu et al, 2018; Itskanov & Park, 2018; this study; Fig EV4) extra densities were present at lower contour levels within the lateral gate, that may stem from a bound lipid or detergent molecule. In fact, in the study of Itskanov & Park (2018) detergents LMNG and CHS were used for solubilization of the Sec complex, whereas digitonin or its analog glyco‐diosgenin were used in Wu et al (2018) and in our study, respectively. This indicates that the lateral gate opening of the Sec complex may be affected by size, bulkiness, and chemical properties of different detergents or lipids, that may partly mimic the signal sequence or act as place holders for the signal sequence. However, upon engagement with the ppαF signal sequence the lateral gate opened even wider than in other signal sequence‐engaged structures. This widened structure may be further stabilized by the Sec62 TMs. Notably, so far, the essential function for Sec62 during translocation remained largely elusive and no densities for the Sec62 TMs could be observed in previous structures due to flexibility (Wu et al, 2018; Itskanov & Park, 2018). In agreement with a previous chemical cross‐linking study (Plath et al, 1998, 2003), we observe a relative rigidification of the Sec62 TMs in the signal sequence‐bound state, and particularly its TM2 may help stabilize the signal sequence surrounded by lipids at the lateral gate. This stabilization could be necessary for recognition of the less hydrophobic post‐translational signal sequences, thereby explaining Sec62’s crucial role for post‐translational translocation. On the other hand, it should be noted that in our map the density for the Sec62 TM2 does not span completely across the membrane, which might be caused by the presence of detergent. Notably, lateral gate widening upon signal sequence recognition is known to result in a destabilization and relocation of the plug helix, thereby removing the obstruction from the aqueous peptide channel. Also in our more open signal sequence‐bound state, the plug helix became flexible and relocated in order to open the central pore. Although we cannot visualize the translocating peptide in the central pore of Sec61α, this plug relocation was expected because it is required to open the channel and allow translocation of the secretory protein to the ER lumen.
Figure EV4. Detergents or lipids bound to the lateral gate of Sec complex.

-
ACryo‐EM map and fitted model of the ss‐bound Sec complex from this study.
-
BCryo‐EM map and fitted model of the apo Sec complex from this study.
-
C, DCryo‐EM maps and fitted models of the apo Sec complexes from the Itskanov study (C; PDB 6N3Q and EMD‐0336) and Wu study (D; PDB 6ND1 and EMD‐0440).
Data information: All maps are low‐pass filtered to 8 Å. The black triangle marks extra density likely attributing for a bound detergent or lipid molecule and “P” marks the plug.
Despite a near‐complete structure of the heptameric Sec complex bound to a signal sequence, density for the cytosolic N‐ and C‐termini of Sec62 was weak or less resolved than the rest of the complex. Yet, both termini were shown to be binding sites for the Sec complex (Wittke et al, 2000). While the interaction partner for the minor binding site in the Sec62 C‐terminus is still unknown, Sec62 mainly uses its positively charged N‐terminal domain to interact with the acidic C‐terminal 14 residues of Sec63, which is regulated by phosphorylation (Wittke et al, 2000; Wang & Johnsson, 2005). Mutations to these basic residues also led to deficiency in translocation of less hydrophobic transmembrane segments (Jung et al, 2014). Our crystal structure of the Sec62 domain together with the HDX analysis and binding assays show that Sec62 utilizes the basic patch on its N‐terminal domain to interact with the ultimate C‐terminus (Sec63‐C2 comprising the last 16 residues 648–663) of Sec63 and that binding is dependent on di‐phosphorylation of Sec63‐C2 (Figs 3D and 4). Besides, the more N‐terminal fragment of the Sec63 C‐terminus (Sec63‐C1; residues 621–647) also shows interaction with the Sec62 domain. However, given that the last Sec63 residue in our model (D612) is more than 40 Å away from the Sec62 basic patch and that the linker between Sec63‐C1 and the Sec63 globular domain has only 8 residues, we assume that Sec63‐C2P and not Sec63‐C1 can span the distance and therefore acts as the primary binding partner for the Sec62 domain (Fig EV5). Alternatively, Sec63‐C1 may also interact with the C‐terminus of Sec62 which is also positively charged (Appendix Fig S2). We observe a low‐resolved unassigned cytosolic density connecting to the Sec62 TM2 which may attribute for the C‐terminus of Sec62. Accordingly, this density is close to the Sec63 C‐terminus (Figs 2A and EV5), further supporting the idea that Sec63‐C1 may interact with the C‐terminus of Sec62. Along those lines, not only deletion of the N‐terminus but also of the C‐terminal 35 residues of Sec62 weakens the interaction with the Sec complex and causes defects in protein translocation (Wittke et al, 2000). Considering that the TM2 of Sec62 stabilizes the signal sequence, the N‐ and C‐termini of Sec62 likely serve as anchors on the Sec complex, bringing the flexible TMs of Sec62 in close proximity to the lateral gate in order to prime it for signal sequence engagement.
Figure EV5. Localization of Sec62 in the Sec complex.

Cryo‐EM reconstruction of the ss‐bound Sec complex low‐pass filtered to 8 Å to visualize the rather flexible C‐terminus of Sec63 (highlighted by a cyan dashed line).
Taken together, our structures of the signal sequence‐engaged Sec complex and the Sec62 domain provide a refined model for post‐translational protein translocation across the ER membrane (Fig 5): To enable translocation of post‐translational clients in the ER, the Sec61 complex assembles with Sec62, Sec63 and in yeast with accessory factors Sec71 and Sec72 into the Sec complex (post‐translocon). Due to its interaction with Sec63, Sec61α is already in a conformation with a pre‐opened lateral gate as observed in all three available structures of the apo state (our structure and Wu et al, 2018; Itskanov & Park, 2018). In this conformation, it is already primed for signal sequence engagement while the plug still blocks the Sec61α channel for the translocating peptide. Within this assembly, Sec62 is very flexible and contacts the Sec complex primarily via the acidic C‐terminal tail of Sec63 with the basic patch of the N‐terminal domain of Sec62. Upon substrate engagement, the signal sequence binds to the groove at the lateral gate and is further stabilized by the TM2 of Sec62. Consequently, also TM1 of Sec62 is brought closer to the lateral gate and displaces TM3 of Sec61α. This movement causes the lateral gate to widen even more, followed by plug displacement and translocation of the polypeptide chain.
Figure 5. Model for substrate engagement of the Sec complex.
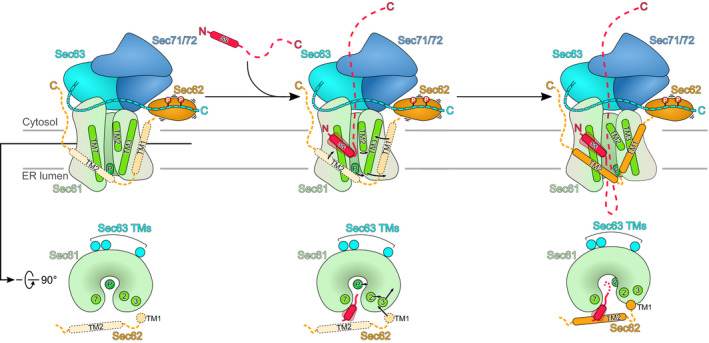
In the apo state of Sec complex, the Sec61 channel is closed by the plug while the lateral gate is already open. Sec62 flexibly associates with the Sec complex mainly through a basic surface on its Sec62 domain interacting with the acidic C‐terminus of Sec63. During the insertion of a post‐translational client, its signal sequence (ss) binds to the groove at lateral gate supported by the Sec62 TM2. At the same time, the Sec62 TM1 pushes the Sec61α TM3 outward leading to an even more open lateral gate. This also leads to removal of the plug away from the pore ring, allowing the translocating peptide to be gated through the Sec61α central pore.
Materials and Methods
Yeast strains and plasmids
For purification of the heptameric Sec complex, sequences coding for a 3C cleavage site followed by eight histidines and a triple FLAG sequence (3C‐His8‐3X‐FLAG) tag was inserted downstream of the SEC62 gene of wild type Saccharomyces cerevisiae (S.c.) strain W303. For the ppαF‐mEGFP construct, the sequence coding for the first 54 amino acids of ppαF followed by mEGFP was cloned into a modified pET28a vector that adds an N‐terminal SUMO tag to the translated insert. The translocation defect mutant ppαFm3 (Allison & Young, 1989) was constructed by introducing an A13E mutation into the ppαF‐mEGFP construct using site‐directed mutagenesis. The N‐terminal cytosolic domain of Sec62 from S.c (Sec62 domain; residues 18–145) and a R51E mutant of this domain were also cloned into the same modified pET28a or into a modified pGEX‐6P‐1 plasmid.
Purification of ppαF‐mEGFP
The ppαF‐mEGFP constructs were overexpressed in the E. coli strain BL21 (DE3) and affinity‐purified using Ni‐NTA agarose, followed by on‐column cleavage of the SUMO tag by the Ulp1 protease at 4°C overnight. The eluted proteins were further purified using size‐exclusion chromatography with a Superdex 200 column (GE Healthcare) in buffer containing 20 mM HEPES pH 7.4, 150 mM NaCl, and 1 mM dithiothreitol (DTT).
Purification of apo and ppαF‐mEGFP‐bound heptameric Sec complex
The yeast strain expressing C‐terminally tagged endogenous Sec62 protein was grown in YPD at 30°C overnight and then diluted into a large culture and incubated again at 30°C until reaching an optical density of OD600 ~ 5. The cells were pelleted and resuspended in lysis buffer (20 mM HEPES pH 7.4, 100 mM KOAc, 2 mM Mg(OAc)2, 1 mM DTT, 0.5 mM PMSF) supplemented with EDTA‐free protease inhibitor cocktail (Roche) and lysed using a microfluidizer (Microfluidics). The lysate was centrifuged in an SLA‐1500 rotor (Thermo) at 29,800 g at 4°C for 20 min. The supernatant was centrifuged in a Ti45 rotor (Beckman Coulter) at 185,500 g at 4°C for 1 h. The pelleted rough microsomes were flash‐frozen in liquid nitrogen and stored at −80°C until use.
For purification of the apo heptameric Sec complex, the frozen microsomes were thawed and resuspended with a Dounce homogenizer in solubilization buffer (20 mM HEPES pH 7.4, 0.75 M KOAc, 2.5 mM Mg(OAc)2, 0.4 M sucrose, 0.5 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 3% glyco‐diosgenin (GDN, Anatrace)) and protease inhibitor cocktail. After incubation with stirring at 4°C for 1 h, the solubilized microsomes were centrifuged in a Ti45 rotor at 126,000 g at 4°C for 1 h. The supernatant was incubated with anti‐FLAG M2 agarose beads (Sigma) at 4°C for 1 h. The beads were washed three times in wash buffer (20 mM HEPES pH 7.4, 100 mM KOAc, 2.5 mM Mg(OAc)2, 10% glycerol, 1 mM DTT, 0.02% GDN) and the complex was eluted in wash buffer containing home‐made 3C protease at 20°C for 1 h. The eluted sample was diluted two times with Q buffer (20 mM HEPES pH 7.4, 2.5 mM Mg(OAc)2, 1 mM DTT, 0.02% GDN), subjected to Q Sepharose Fast Flow (GE Healthcare), and then eluted in Q buffer with additional 1 M KOAc. The buffer of the eluted complex was exchanged to Sec buffer (20 mM HEPES pH 7.4, 100 mM KOAc, 2.5 mM Mg(OAc)2, 1 mM DTT, 0.02% GDN) and concentrated to ~5 mg/ml using a 100‐kDa‐cutoff Amicon membrane (GE Healthcare).
For on‐bead reconstitution of ppαF‐bound heptameric Sec complex, the same protocol was applied except that, after washing the anti‐FLAG M2 beads, the ppαF‐mEGFP was added to the complex in varying molar ratios. For the cryo‐EM sample, a 10:1 ratio of substrate to Sec complex was used. Here, the sample was incubated at 30°C for 20 min followed by three washing steps. The rest of the purification remained the same as described for the apo heptameric complex. The final purified reconstituted ppαF‐Sec complex was concentrated to ~5 mg/ml.
Purification of the Sec62 domain
Constructs for the N‐terminal cytosolic Sec62 domain (including selenomethionine sample for crystallization) were overexpressed in the E. coli BL21(DE3) strain. The proteins were initially purified using Ni‐NTA agarose, followed by 3C or Ulp1 protease on‐column cleavage at 4°C overnight. The proteins were further purified using anion exchange and size‐exclusion chromatography. The final buffer condition was 20 mM Tris–HCl, pH 8.0, 300 mM NaCl, and 5 mM DTT. For crystallization, the final buffer was 20 mM Tris–HCl, pH 8.0, 100 mM NaCl, and 5 mM DTT.
Sec63 C‐terminal peptides
Biotin‐labeled Sec63 C‐terminal peptides used in pull‐down, ITC, and HDX‐MS experiments were synthesized by GL Biochem (Shanghai).
In vitro pull‐down assays
For binding assays with the heptameric Sec complex and ppαF‐mEGFP, purified Sec complex (0.8 μM) and purified ppαF‐mEGFP or ppαFm3‐mEGFP (0.8, 1.6 or 4 μM) were incubated in Sec buffer at 30°C for 20 min. The protein samples were then immobilized on GFP‐Trap magnetic agarose beads (Chromotek) at 4°C for 30 min. The flow‐through was collected and the beads were washed three times with Sec buffer. The bound proteins and the flow‐through were subjected to SDS–PAGE analysis followed by Coomassie Brilliant Blue staining.
For binding assays with Sec63 C‐terminal peptides and the Sec62 domain, Sec63‐C1 and Sec63‐C2P peptides with a biotin tag were incubated with the purified Sec62 domain (residues 18‐145) or the R51E mutant Sec62 domain in binding buffer (20 mM HEPES pH 7.4, 100 mM NaCl, 5% glycerol, 0.05% Triton X‐100, and 1 mM DTT) for 2 h at 4°C. The protein samples were then immobilized on 25 μl of Streptavidin resin (GE Healthcare) for 20 min at 4°C. The resin was washed three times with binding buffer, and bound proteins were eluted using sample buffer. The final sample was subjected to SDS–PAGE and stained with Coomassie Brilliant Blue.
Cryo‐EM sample preparation and data acquisition
The reconstituted ppαF‐Sec complex (3.5 μl) was applied on glow‐discharged Quntifoil R2/2 UltrAuFoil grids, blotted for 2 s at 4°C and 100% humidity, and immediately plunge‐frozen in liquid ethane using a Vitrobot Mark IV (FEI). Two sets of Cryo‐EM data were acquired on a Titan Krios electron microscope (FEI) using a K2 detector (Gatan) and GIF energy filter. For dataset 1, a total of 6,243 dose‐fractionated movies were collected with 40 frames, an exposure of 0.9 e−/frame/Å2, and a magnification resulting in an image pixel size of 1.059 Å per pixel. For dataset 2, a total of 8,802 dose‐fractionated movies were collected with 40 frames, an exposure of 1.15 e−/frame/Å2, and a magnification resulting in an image pixel size of 1.059 Å per pixel.
Image processing
The original movies were first subjected to motion correction and dose weighting using MotionCor2 (Zheng et al, 2017), and the CTF parameters were estimated using CTFFIND4 (Rohou & Grigorieff, 2015). The does‐weighted micrograph sums were visually inspected to remove bad micrographs. A total of 5,112 micrographs of dataset 1 and 5,118 micrographs of dataset 2 were selected for further processing in RELION‐3 (Zivanov et al, 2018) as shown in Fig EV1F and G and Appendix Fig S2A. For dataset 1, after auto‐picking and several rounds of 2D classifications, 453,116 particles were selected for 3D refinement and then classified into six classes. One class (C6) showed clear secondary structure of the Sec complex. The other classes were subjected to another round of refinement and classification with a mask around Sec61 complex/Sec63/Sec71/Sec72. The class with clear secondary structure density in transmembrane region was merged with C6 and refined with the same mask. This refined map was used as a template for picking particles in dataset 2 using RELION. A total of 117,117 particles were selected after several rounds of 2D classifications. These particles were then merged with the particles from dataset 1, and the same 3D classification process was performed again to obtain classes with clear secondary structure density in transmembrane region. The resulted particles were refined with the same Sec61 complex/Sec63/Sec71/Sec72 mask, and classification was done with a mask around the transmembrane domain of the Sec61 complex without alignment. One class showing extra density of signal sequence and Sec62 TMs was refined with a mask around Sec61 complex/Sec63/Sec71/Sec72 plus signal sequence and Sec62 TMs, yielding a 4.5 Å resolution map after post‐processing. The other class showing the heptameric Sec complex in the apo state was refined with the Sec61 complex/Sec63/Sec71/Sec72 mask and post‐processed, yielding a map with 4.4 Å resolution. Two rounds of random‐phase 3D classification (Gong et al, 2016) were performed in cryoSPARC (Punjani et al, 2017) to further remove bad particles and particles at the edge of micrographs were also removed. The resulting particle stacks were subjected to 3D refinement in RELION, yielding a 4.4 Å resolution map for the signal sequence‐engaged state and 4.3 Å for the apo state. Neither CTF refinement nor Bayesian polishing could further improve the resolution. Local resolution filtered maps were calculated using RELION.
Model building
All models were built in COOT (Emsley et al, 2010). For the signal sequence‐bound Sec complex, the structure of yeast heptameric Sec complex (PDB 6N3Q) was used to rigid body fit into the map. For the apo Sec complex, the structure of another yeast Sec complex (PDB 6ND1) was used to rigid body fit into the map. Figures of models and maps were generated using UCSF ChimeraX (Goddard et al, 2018) and PyMOL (Schrödinger).
Crystallization, data collection, and structure determination
Crystals of the cytosolic N‐terminal domain of Sec62 (res. 18–145) were grown at 4°C using the hanging drop vapor diffusion method by mixing equal volumes of the purified protein complex (20 mg/ml) and crystallization buffer (100 mM MES pH 5.7–5.9, 2 M (NH4)2SO4). Crystals were transiently transferred into a cryoprotectant buffer containing reservoir buffer and additional 20% glycerol (v/v) before they were flash‐frozen in a cold nitrogen stream at −173°C. All data were collected in 0.97958 Å wavelength at ESRF (The European Synchrotron Radiation Facility, France). The data were processed using the program XDS package (Kabsch, 2010).
Phases were initially determined by the single‐wavelength anomalous dispersion (SAD) using the phasing module Autosol; density modification and automatic model building were performed using the AutoBuild of program package PHENIX. The final model was manually built using Coot. All refinements were performed using the refinement module phenix.refine of the PHENIX package(Adams et al, 2010). The model quality was validated using the MolProbity of the PHENIX package, which indicated good stereochemistry according to the Ramachandran plot for the structure (favored: 98.4%, outliers: 0.0%).
Hydrogen–deuterium exchange (HDX) coupled to mass spectrometry (MS)
Samples for HDX‐MS of Sec62 were prepared with the aid of a two‐arm robotic autosampler (LEAP Technologies). 7.5 µl (50 µM) of Sec62 or Sec62/Sec63‐C2P peptide (50 µM each) were mixed with 67.5 µl of D2O‐containing buffer (10 mM HEPES‐Na, 100 mM NaCl, final pH 7.4) to start H/D exchange. After labeling for 0.25/0.5/1/10/100 min at 25°C, 55 µl of the reaction was added to 55 µl quench buffer (400 mM KH2PO4/H3PO4, 2 M guanidine‐HCl, pH 2.2) kept at 1°C and 95 µl of the resulting mixture immediately injected into an ACQUITY UPLC M‐class system with HDX technology (Waters; Wales et al, 2008). Non‐deuterated samples of Sec62 were prepared similarly except that non‐deuterated buffer was employed. Sec62 was digested either with immobilized porcine pepsin or protease type XIII from Aspergillus saitoi (Cravello et al, 2003) at 12°C and 100 µl/min flow rate of water + 0.1% (v/v) formic acid, and the resulting peptides were collected on a trap column (2 mm × 2 cm) filled with POROS 20 R2 material (Thermo Scientific) kept at 0.5°C. The trap column was after 3 min placed in line with an ACQUITY UPLC BEH C18 1.7 µm 1.0 × 100 mm column (Waters) and the peptides eluted at 0.5°C using a gradient of water + 0.1% formic acid (A) and acetonitrile + 0.1% formic acid (B) at 30 µl/min flow rate: linear increase from 5 to 35% B within 7 min followed by a ramp to 85% B within 1 min and hold at 85% B for 2 min. The peptides were ionized by electrospray ionization at 250°C source capillary temperature and a spray voltage of 3.0 kV and mass spectra acquired on a G2‐Si HDMS mass spectrometer with ion mobility separation (Waters) over a range of 50 to 2000 m/z in HDMSE or HDMS mode for undeuterated and deuterated samples, respectively (Geromanos et al, 2009; Li et al, 2009). [Glu1]‐Fibrinopeptide B standard (Waters) was employed for lock mass correction. All measurements were performed in triplicates (individual labeling reactions). Between samples, the protease column was washed three times with 80 µl of 4% (v/v) acetonitrile and 0.5 M guanidine hydrochloride and blank runs performed between samples. Peptides were identified and their deuterium incorporation determined with the PLGS and DynamX 3.0 softwares (both Waters) as described previously (Osorio‐Valeriano et al, 2019). Hereby, the data originating from digestion with porcine pepsine or protease type XIII from A. saitoi were merged. In cases where peptides were found in both datasets, the ones with the higher quality (as judged by intensity, mass error or the presence of overlapping ions) were used for analysis.
Author contributions
JC and RB designed the study. T‐HW with help from TB and BB prepared the signal sequence‐bound heptameric Sec complex. T‐HW processed the cryo‐EM data and together with JC built the molecular models. T‐HW, JC, TB, GB, and RB interpreted the structure. OB collected cryo‐EM data. JC purified and crystallized the Sec62 domain and performed binding assays. WS and GB performed HDX experiments. T‐HW, TB, GB, JC, and RB wrote the manuscript with contributions from all authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Source Data for Expanded View
Acknowledgements
The authors thank H. Sieber, J. Musial, C. Ungewickell, and S. Rieder for technical assistance and L. Kater and K. Best for support with the pre‐processing pipeline of cryo‐EM data. We thank K. Lammens and the staff members in ESRF for the help of X‐ray data collection. We thank the Deutsche Forschungsgemeinschaft for support through the Collaborative Research Council (CRC) 174 (to R.B. and G.B.) and the DFG‐core facility for interactions, dynamics, and macromolecular assembly structure (to G.B.). Open Access funding enabled and organized by ProjektDEAL.
The EMBO Journal (2021) 40: e105643.
[Correction added on 5 February 2021 after first online publication: Publishing license has been changed]
Contributor Information
Jingdong Cheng, Email: jcheng@genzentrum.lmu.de.
Roland Beckmann, Email: beckmann@genzentrum.lmu.de.
Data availability
The X‐ray structure of the Sec62 domain is available in the Protein Data Bank under accession code 6ZZZ.
The cryo‐EM structures of the Sec complex have been deposited in the Protein Data Bank under accession code 7AFT (ss‐engaged state), and in the Electron Microscopy Data Bank under accession codes EMD‐11774 (ss‐engaged state) and EMD‐11775 (apo state).
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L‐W, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DS, Young ET (1989) Mutations in the signal sequence of prepro‐alpha‐factor inhibit both translocation into the endoplasmic reticulum and processing by signal peptidase in yeast cells. Mol Cell Biol 9: 4977–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampofo E, Welker S, Jung M, Müller L, Greiner M, Zimmermann R, Montenarh M (2013) CK2 phosphorylation of human Sec63 regulates its interaction with Sec62. Biochim Biophys Acta 1830: 2938–2945 [DOI] [PubMed] [Google Scholar]
- Ast T, Cohen G, Schuldiner M (2013) A network of cytosolic factors targets SRP‐independent proteins to the endoplasmic reticulum. Cell 152: 1134–1145 [DOI] [PubMed] [Google Scholar]
- van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA (2004) X‐ray structure of a protein‐conducting channel. Nature 427: 36–44 [DOI] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B (1975) Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane‐bound ribosomes of murine myeloma. J Cell Biol 67: 835–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunger K, Pfeffer S, Shrimal S, Gilmore R, Berninghausen O, Mandon EC, Becker T, Förster F, Beckmann R (2018) Structural basis for coupling protein transport and N‐glycosylation at the mammalian endoplasmic reticulum. Science 360: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon KS, Or E, Clemons WM, Shibata Y, Rapoport TA (2005) Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol 169: 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravello L, Lascoux D, Forest E (2003) Use of different proteases working in acidic conditions to improve sequence coverage and resolution in hydrogen/deuterium exchange of large proteins. Rapid Commun Mass Spectrom 17: 2387–2393 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R (1991) Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane‐bound multisubunit complex. Nature 349: 806–808 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R (1989) SEC62 encodes a putative membrane protein required for protein translocation into the yeast endoplasmic reticulum. J Cell Biol 109: 2653–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R (1990) Structural and functional dissection of Sec62p, a membrane‐bound component of the yeast endoplasmic reticulum protein import machinery. Mol Cell Biol 10: 6024–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünnwald M, Varshavsky A, Johnsson N (1999) Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. MBoC 10: 329–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D, Schekman R (1994) Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J Cell Biol 126: 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D, Yoshimura K, Admon A, Schekman R (1993) Structural and functional characterization of Sec66p, a new subunit of the polypeptide translocation apparatus in the yeast endoplasmic reticulum. Mol Biol Cell 4: 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfeld J, Gumbart J, van der Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K et al (2011) Cryo–EM structure of the ribosome–SecYE complex in the membrane environment. Nat Struct Mol Biol 18: 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmer M, Förster F (2020) A clearer picture of the ER translocon complex. J Cell Sci 133: jcs231340 [DOI] [PubMed] [Google Scholar]
- Geromanos SJ, Vissers JPC, Silva JC, Dorschel CA, Li G‐Z, Gorenstein MV, Bateman RH, Langridge JI (2009) The detection, correlation, and comparison of peptide precursor and product ions from data independent LC‐MS with data dependant LC‐MS/MS. Proteomics 9: 1683–1695 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE (2018) UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogala M, Becker T, Beatrix B, Armache J‐P, Barrio‐Garcia C, Berninghausen O, Beckmann R (2014) Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506: 107–110 [DOI] [PubMed] [Google Scholar]
- Gong X, Qian H, Zhou X, Wu J, Wan T, Cao P, Huang W, Zhao X, Wang X, Wang P et al (2016) Structural insights into the niemann‐pick C1 (NPC1)‐Mediated cholesterol transfer and ebola infection. Cell 165: 1467–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA (1993) Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75: 615–630 [DOI] [PubMed] [Google Scholar]
- Hanada M, Nishiyama KI, Mizushima S, Tokuda H (1994) Reconstitution of an efficient protein translocation machinery comprising SecA and the three membrane proteins, SecY, SecE, and SecG (p12). J Biol Chem 269: 23625–23631 [PubMed] [Google Scholar]
- Holm L (2019) Benchmarking fold detection by DaliLite vol 5. Bioinformatics 35: 5326–5327 [DOI] [PubMed] [Google Scholar]
- Itskanov S, Park E (2018) Structure of the posttranslational Sec protein‐translocation channel complex from yeast. Science 363: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Kim JEH, Reithinger JH, Kim H (2014) The Sec62–Sec63 translocon facilitates translocation of the C‐terminus of membrane proteins. J Cell Sci 127: 4270–4278 [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010) XDS. Acta Cryst D 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater L, Frieg B, Berninghausen O, Gohlke H, Beckmann R, Kedrov A (2019) Partially inserted nascent chain unzips the lateral gate of the Sec translocon. EMBO Rep 20: e48191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochert BA, Iacob RE, Wales TE, Makriyannis A, Engen JR (2018) Hydrogen‐deuterium exchange mass spectrometry to study protein complexes. Methods Mol Biol 1764: 153–171 [DOI] [PubMed] [Google Scholar]
- Li G‐Z, Vissers JPC, Silva JC, Golick D, Gorenstein MV, Geromanos SJ (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9: 1696–1719 [DOI] [PubMed] [Google Scholar]
- Li L, Park E, Ling J, Ingram J, Ploegh H, Rapoport TA (2016) Crystal structure of a substrate‐engaged SecY protein‐translocation channel. Nature 531: 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman SK, Schekman R (1997) Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell 88: 85–96 [DOI] [PubMed] [Google Scholar]
- Ma C, Wu X, Sun D, Park E, Catipovic MA, Rapoport TA, Gao N, Li L (2019) Structure of the substrate‐engaged SecA‐SecY protein translocation machine. Nat Commun 10: 2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack KE, Misselwitz B, Plath K, Rapoport TA (1999) BiP acts as a molecular ratchet during posttranslational transport of prepro‐alpha factor across the ER membrane. Cell 97: 553–564 [DOI] [PubMed] [Google Scholar]
- Matlack KES, Plath K, Misselwitz B, Rapoport TA (1997) Protein transport by purified yeast sec complex and Kar2p without membranes. Science 277: 938–941 [DOI] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P (1996) Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol 134: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngosuwan J, Wang NM, Fung KL, Chirico WJ (2003) Roles of cytosolic Hsp70 and Hsp40 molecular chaperones in post‐translational translocation of presecretory proteins into the endoplasmic reticulum. J Biol Chem 278: 7034–7042 [DOI] [PubMed] [Google Scholar]
- Nyathi Y, Wilkinson BM, Pool MR (2013) Co‐translational targeting and translocation of proteins to the endoplasmic reticulum. Biochimica et Biophysica Acta (BBA) ‐ Molecular . Cell Res 1833: 2392–2402 [DOI] [PubMed] [Google Scholar]
- Osorio‐Valeriano M, Altegoer F, Steinchen W, Urban S, Liu Y, Bange G, Thanbichler M (2019) ParB‐type DNA segregation proteins are CTP‐dependent molecular switches. Cell 179: 1512–1524 [DOI] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA (1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81: 561–570 [DOI] [PubMed] [Google Scholar]
- Park E, Ménétret J‐F, Gumbart JC, Ludtke SJ, Li W, Whynot A, Rapoport TA, Akey CW (2014) Structure of the SecY channel during initiation of protein translocation. Nature 506: 102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA (1998) Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94: 795–807 [DOI] [PubMed] [Google Scholar]
- Plath K, Wilkinson BM, Stirling CJ, Rapoport TA (2003) Interactions between sec complex and prepro‐α‐factor during posttranslational protein transport into the endoplasmic reticulum. MBoC 15: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA (2017) cryoSPARC: algorithms for rapid unsupervised cryo‐EM structure determination. Nat Methods 14: 290–296 [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Li L, Park E (2017) Structural and mechanistic insights into protein translocation. Annu Rev Cell Dev Biol 33: 369–390 [DOI] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N (2015) CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192: 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PCK, Maillard AP, Chan KKY, Duong F (2005) Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J 24: 3380–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Mandon EC, Gilmore R, Rapoport TA (2017) Two alternative binding mechanisms connect the protein translocation Sec71‐Sec72 complex with heat shock proteins. J Biol Chem 292: 8007–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Fernández IS, Scheres SHW, Hegde RS (2014) Structure of the mammalian ribosome‐Sec61 complex to 3.4 Å resolution. Cell 157: 1632–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Hegde RS (2016) Structure of the Sec61 channel opened by a signal sequence. Science 351: 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales TE, Fadgen KE, Gerhardt GC, Engen JR (2008) High‐speed and high‐resolution UPLC separation at zero degrees Celsius. Anal Chem 80: 6815–6820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Johnsson N (2005) Protein kinase CK2 phosphorylates Sec63p to stimulate the assembly of the endoplasmic reticulum protein translocation apparatus. J Cell Sci 118: 723–732 [DOI] [PubMed] [Google Scholar]
- Wild K, Halic M, Sinning I, Beckmann R (2004) SRP meets the ribosome. Nat Struct Mol Biol 11: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Wittke S, Dünnwald M, Johnsson N (2000) Sec62p, A component of the endoplasmic reticulum protein translocation machinery, contains multiple binding sites for the sec‐complex. Mol Biol Cell 11: 3859–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Cabanos C, Rapoport TA (2018) Structure of the post‐translational protein translocation machinery of the ER membrane. Nature 566: 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J‐P, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat Methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA (2008) Structure of a complex of the ATPase SecA and the protein‐translocation channel. Nature 455: 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH (2018) New tools for automated high‐resolution cryo‐EM structure determination in RELION‐3. Elife 7: e42166 [DOI] [PMC free article] [PubMed] [Google Scholar]


