Abstract
In the past several decades, the establishment of in vitro models of pluripotency has ushered in a golden era for developmental and stem cell biology. Research in this arena has led to profound insights into the regulatory features that shape early embryonic development. Nevertheless, an integrative theory of the epigenetic principles that govern the pluripotent nucleus remains elusive. Here, we summarize the epigenetic characteristics that define the pluripotent state. We cover what is currently known about the epigenome of pluripotent stem cells and reflect on the use of embryonic stem cells as an experimental system. In addition, we highlight insights from super-resolution microscopy, which have advanced our understanding of the form and function of chromatin, particularly its role in establishing the characteristically “open chromatin” of pluripotent nuclei. Further, we discuss the rapid improvements in 3C-based methods, which have given us a means to investigate the 3D spatial organization of the pluripotent genome. This has aided the adaptation of prior notions of a “pluripotent molecular circuitry” into a more holistic model, where hotspots of co-interacting domains correspond with the accumulation of pluripotency-associated factors. Finally, we relate these earlier hypotheses to an emerging model of phase separation, which posits that a biophysical mechanism may presuppose the formation of a pluripotent-state-defining transcriptional program.
More than 600 million years ago, the first multicellular metazoan gained the ability to regenerate entirely from a single cell (Müller 2001). The significance of this evolutionary feat is underscored by the unique difficulty of animal multicellularity. Crucially, all animal life is bound by the concept of “division of labor.” Neither phototrophs nor chemotrophs, cells in metazoans must sacrifice a unicellular feeding and reproductive life cycle for the combined benefit of the organism (Cavalier-Smith 2017). Thus, for animals, a profound level of cooperation is necessary to balance the selective disadvantage imposed by the need for specialized cell types. Precise details of this evolutionary process remain unclear, but phylogenomic analyses and recent transcriptomic data from sponges—our closest extant relatives to primitive metazoans—hint that we may have descended from an “ancestral pluripotent stem cell,” capable of switching between different cell types or transcriptional programs (Sebé-Pedrós et al. 2017; Sogabe et al. 2019). From this unlikely beginning, a fundamental mystery of complex life emerges: how can a single cell, with one genome, become a functional and reproductively competent multicellular organism?
Some organisms, such as the flatworm Planaria, possess adult stem cells capable of this coordinated regeneration (Morgan 1898; Rink 2013). Most animals, however, reproduce sexually and produce germ cells, which mature into gametes (oocytes or sperm) that unite to form the single-cell zygote. As the genetic content of the zygote is preserved in each cell of the adult body, it is important to study principles of epigenetic regulation if we wish to understand how developmental decision-making precedes the establishment of cellular identity. In this regard, insights from pluripotent stem cell models have been particularly revealing. A growing body of evidence suggests that a categorically distinct epigenetic program separates somatic cells from those in the earliest stages of development.
Previous work has shown that pluripotent cells possess a diffuse, flexible chromatin structure and an active epigenetic landscape, which can be dramatically influenced by environmental signals (Schlesinger and Meshorer 2019). This takes the form of “open” chromatin in the pluripotent nucleus, where a milieu of accessible chromatin permits cross talk between chromatin remodeling complexes and transcription factors (TFs), resulting in elevated transcription (Efroni et al. 2008; Gaspar-Maia et al. 2011). In the past decade, our understanding of the nature of these phenomena has been bolstered by several technological advances. For instance, using super-resolution microscopy techniques, we are now able to visualize chromatin organization at the level of nucleosomes. In addition, rapid advances in high-throughput sequencing have improved the resolution of Hi-C chromatin–chromatin interaction maps, which have been generated for many different cell types, including pluripotent stem cells. These techniques have led to insights into the 3D organization of the pluripotent genome, suggesting a role for pluripotency-factor-associating hubs that govern the transcriptional activity of particular subregions of the nucleus (Gorkin et al. 2014). However, despite the ever-improving detail from which we can now infer structure and function, a precise mechanism for how the characteristic “plastic state” of pluripotent nuclei is established and maintained remains far from understood. To this end, recent models of phase separation are providing clues toward an integrative account of pluripotency, namely, that the combinatorial binding of pluripotency factors may result in emergent properties that exist only in the pluripotent state.
In this review, we paint a picture of the pluripotent nucleus from the perspective of its unique epigenome, dynamic chromatin structure, and higher-order organizational features.
PLURIPOTENT STEM CELLS AND THEIR EXPERIMENTAL BEGINNINGS
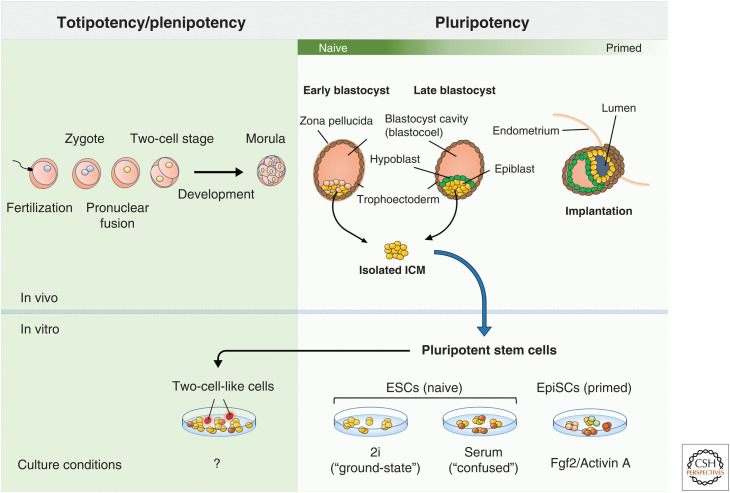
Pluripotency describes the intrinsic potential of a cell to give rise to any cell type or tissue in the developed organism. In eutherians (mammals who possess an internal placenta), the blastocyst forms shortly after fertilization of the zygote and comprises the inner cell mass (ICM) enclosed by an outer shell of cells called the trophoectoderm. Cells in the ICM are pluripotent and, as such, will ultimately give rise to the embryo proper. Before implantation of the embryo in the uterus wall, the ICM segregates into the pluripotent epiblast and the hypoblast, which partitions the epiblast and the blastocyst cavity. The pluripotent state of cells in the blastocyst is sustained until implantation around day 4.5 in mouse (day 7 in human) of embryonic development (Bergh and Navot 1992; Plusa and Hadjantonakis 2014), when the three primary germ layers, endoderm, ectoderm, and mesoderm, begin to form (Fig. 1). Here, gastrulation begins, and cells in the epiblast begin to differentiate toward somatic as well as primordial germ cell (PGC) lineages. The developmental potential of pluripotent cells is eclipsed only by the zygote itself and its initial cleavage, known as the two-cell stage. These cells are totipotent and can uniquely become the trophoblast lineages, which eventually form the bulk of the placenta (Kunath and Rossant 2009; Wu et al. 2017).
Figure 1.
Stem cells in early development and in vitro. Early development in eutherians begins following fertilization (fusion of the male and female gamete). The single-cell zygote soon forms with the potential to generate a complete organism. This ability is referred to as totipotency and is lost following development past the two-cell stage (in mouse). Formation of the blastocyst coincides with the onset of pluripotency within the inner cell mass (ICM). These cells are innately capable of differentiation to all three germ lineages. Isolation and culture of cells from the ICM produces what are known as “pluripotent stem cells” (PSCs). These in vitro models of the pluripotent ICM vary in the precise developmental stage that they resemble along the naive-primed axis, which has been shown to be highly culture-condition-dependent. Cultures of PSCs contain rare populations of two-cell-like cells that mirror the transcriptome of the in vivo two-cell-stage embryo. However, as of yet, no conditions have been defined for the maintenance of these cells in culture. (ESCs) embryonic stem cells, (EpiSCs) epiblast stem cells.
The first cultures of mouse embryonic stem cells (ESCs) were established in 1981 by isolating cells from the ICM and growing them on top of a feeder layer of mouse embryonic fibroblasts (MEFs) (Evans and Kaufman 1981; Martin 1981). These cells were shown to be capable of contributing to all adult tissues, including the germ line, upon injection into a blastocyst-stage embryo (Bradley et al. 1984; Nagy et al. 1990). This ability, known as chimera formation, remains the gold standard for developmental competency of in vitro pluripotent models. Later, culturing of pluripotent-state cells from human in vitro fertilized (IVF) blastocysts was similarly accomplished (Thomson et al. 1998). However, our knowledge of the quality of their pluripotency is limited due to ethical concerns, which prohibit the generation of fully developed human chimeras (Mascetti and Pedersen 2016).
REGULATORY NETWORKS UNDERLYING PLURIPOTENCY
Initial breakthroughs in understanding pluripotency in mouse arose from investigations into the core transcriptional circuitry, signaling pathways, and chemical requirements involved in the maintenance of pluripotent ESCs (for review, see Boiani and Schöler 2005). The presence of leukemia inhibitory factor (LIF) and bone morphogenetic proteins (BMPs) was found to sustain the pluripotent state of mouse ESCs in the absence of a feeder layer. LIF and BMPs act in concert to activate the JAK-STAT3 pathway and stimulate expression of Id genes, respectively, promoting ESC self-renewal and inhibition of differentiation (Smith et al. 1988; Ying et al. 2003). Later, a cocktail of two inhibitors (2i), of the mitogen-activated protein kinase (MEK) and glycogen synthase kinase 3 (GSK3) pathways, was shown to maintain the “ground state” (i.e., ICM-like) pluripotency of mouse ESCs (Ying et al. 2008). Key pluripotency TFs operating at the core of pluripotency were also identified, including the three master regulators OCT4, NANOG, and SOX2 (Schöler 1991; Yuan et al. 1995; Mitsui et al. 2003). These factors were shown to act cooperatively, largely co-occupying the same target genes, including each other, revealing that the pluripotent state could be perpetuated in a self-regulating manner (Boyer et al. 2005; for review, see Li and Belmonte 2018). In 2006, experiments by Yamanaka and colleagues demonstrated that somatic cells could be reprogrammed into induced pluripotent stem cells (iPSCs) by ectopic expression of only four “master” TFs, OCT4, SOX2, c-MYC, and KLF4 (Takahashi and Yamanaka 2006). This seminal report further paved the way for extensive characterization of the pluripotency gene regulatory network, and how the binding of TFs reshape the epigenetic landscape to promote pluripotency. This realization also prompted the discovery of a growing number of chromatin-related factors that regulate pluripotency and early ESC differentiation, including BAF250b, BPTF, esBAF, BAF155, CHD1, BAF60a, and the linker histone chaperone SET (Landry et al. 2008; Yan et al. 2008; Gaspar-Maia et al. 2009; Ho et al. 2009; Schaniel et al. 2009; Alajem et al. 2015; Edupuganti et al. 2017). In spite of these discoveries, a great deal of work remains in discerning the epigenetic principles and chromatin organization of the pluripotent state.
THE EPIGENETIC LANDSCAPE OF PLURIPOTENCY
Epigenetics, the nongenetic inheritable form of gene regulation, naturally underlies any functional distinction between pluripotent and differentiated cells. At the chromatin level, two types of epigenetic modifications—DNA methylation and posttranslational histone marks—define the epigenetic landscape. The pattern of epigenetic marks and their distribution across the genome dictate where—and importantly—how much, transcription can take place. This ultimately contributes to the establishment of cellular identity.
DNA Methylation
The methylome of the early embryo undergoes profound reprogramming from fertilization of the zygote through to gastrulation of the postimplantation blastocyst. In ESCs, methylation defects do not impair stem cell identity; however, upon differentiation, DNA methyltransferase (DNMT) mutants do not up-regulate lineage-specific markers and fail to silence pluripotency factors (Jackson et al. 2004; Smith and Meissner 2013). In vivo, nuclei of pluripotent cells are widely depleted of DNA methylation—a leftover from the wave of global demethylation triggered by pronuclear fusion (Smith and Meissner 2013). This process is thought to occur mainly through passive loss (i.e., dilution following DNA replication) by nuclear exclusion or inactivation of DNMTs, as opposed to active demethylation through the TET3 methylcytosine dioxygenase, which occurs in PGCs (Ko et al. 2005; Hirasawa and Sasaki 2009; von Meyenn et al. 2016; Greenberg and Bourc'his 2019). Some regions, such as transposable elements and imprinting control regions, resist this demethylation and are protected by recruitment of residual DNMT proteins, a process that is mediated by factors such as UHRF1, STELLA (DPPA3), and ZFP57/KAP1 (Nakamura et al. 2007; Rowe et al. 2010; Quenneville et al. 2011; Iurlaro et al. 2017; Li et al. 2018). This hypomethylated state is sustained until the exit from pluripotency, which coincides with the re-establishment of methylation.
Histone Modifications
In broad terms, pluripotent cells possess a relative abundance of active histone marks, including H3K4 methylation and H3/H4 acetylation, and, relative to somatic cells, are depleted in repressive marks, such as H3K27me3, H3K9me3, and H3K9me2 (Meshorer et al. 2006; Efroni et al. 2008; Fussner et al. 2010; Hawkins et al. 2010; Liu et al. 2015; Qiao et al. 2015). Posttranslational histone modifications also exhibit distinct patterns of localization in pluripotent cells. A large body of evidence suggests that epigenetic modifiers and preestablished histone modifications are required to regulate gene expression during development (Meissner et al. 2008; Mohn et al. 2008; Athanasiadou et al. 2010; Gao et al. 2018). A prominent mechanistic theory for how this can occur is “promoter bivalency.” This epigenetic feature describes the presence of opposing histone modifications, H3K4me3 and H3K27me3, which are often located at the promoters of developmentally important genes (Bernstein et al. 2006). In pluripotent ESCs, the repressive H3K27me3 mark—catalyzed by polycomb repressive complex 2 (PRC2)—keeps genes silent during an undifferentiated state, while the simultaneous presence of active H3K4me3—deposited by COMPASS family proteins—prevents these genes from becoming permanently silenced, allowing prompt activation upon differentiation (Margueron and Reinberg 2011; Shilatifard 2012). Another modification associated with pluripotency is H3K56 acetylation. H3K56ac can be found in ESCs at the binding sites of the core pluripotency factors, OCT4, SOX2, and NANOG (Xie et al. 2009), and marks pluripotency-related enhancers (Livyatan et al. 2015). Although only OCT4 has been shown to directly interact with H3K56ac, loss of SIRT6, an H3K56-targeting NAD-dependent deacetylase, results in differentiation defects and activation of OCT4, SOX2, and NANOG (Tan et al. 2013; Etchegaray et al. 2015).
It remains unclear, however, to what extent the epigenetic features observed in ESCs are characteristic of the early embryo (Harikumar and Meshorer 2015; Yagi et al. 2017). As it stands, our knowledge of epigenetic modifications in pluripotency comes predominantly from in vitro models, owing to the large number of cells required for standard ChIP-seq protocols. Moreover, a plethora of studies have noted that changing the growth conditions of ESCs has a significant effect on the epigenetic landscape (Marks et al. 2012; Ficz et al. 2013; Habibi et al. 2013; Leitch et al. 2013). Encouragingly, improvements in ChIP-seq and alternative methods such as CUT&RUN (antibody-targeted MNase digestion) have been recently developed for epigenetically profiling individual embryos or even single cells (Hainer et al. 2019). For instance, by adapting ChIP-seq for low-input samples, one study confirmed that broad H3K4me3 enrichment is a feature of the preimplantation embryo; conversely, the authors found that bivalency at promoters was relatively rare and unstable (Liu et al. 2016). Additionally, a recent report observed functional bivalency in postimplantation epiblasts but not at earlier pluripotent stages (Xiang et al. 2020). Despite being powerful models of early embryonic states, the debate continues regarding ESCs and their in vivo relevance.
STATES OF PLURIPOTENCY
The pluripotent-stage embryo can be separated into two categories: naive and primed. These terms reflect populations in the pre- and postimplantation epiblast and exist in both mouse and human (Guo et al. 2016b). Importantly, these populations can be broadly recapitulated in vitro (Nichols and Smith 2009). This has led to extensive characterization of molecular and functional features unique to each state (for review, see Weinberger et al. 2016). Culturing mouse ESCs in either 2i/LIF- or serum/LIF-containing media mimics the naive state, whereas “primed” epiblast stem cells (EpiSCs) are derived from the postimplantation epiblast and require supplementation of FGF2 and Activin A (Ying et al. 2008). Crucially, although both states are pluripotent, only the naive form is fully capable of contributing to chimera formation upon ex vivo injection into the blastocyst (Brons et al. 2007; Tesar et al. 2007).
Naive and Primed Pluripotency
Smith and colleagues proposed in 2008 that ESCs grown in 2i/LIF conditions more accurately represent the preimplantation ICM, that is, the in vivo naive state (Ying et al. 2008). Hence, this has been described as “ground-state” pluripotency, as opposed to a “confused” pluripotent state of ESCs grown in serum/LIF conditions (Schlesinger and Meshorer 2019). This notion has been explored further in studies comparing epigenomic and transcriptomic profiles of ESCs cultured in either condition. Two clear hallmarks of “ground-state” pluripotency have emerged: globally depleted DNA methylation and local reduction of H3K27me3 at developmental genes (Marks et al. 2012). In contrast, the “confused” state is typified by a more heterogeneous transcription profile, particularly of pluripotency factors and bivalent-marked genes (Kolodziejczyk et al. 2015; Guo et al. 2016a). This is perhaps a reflection of a more chaotic signaling environment caused by the undefined mixture of factors present in the serum itself (see Schlesinger and Meshorer 2019 for a systematic comparison of ESCs in 2i vs. serum conditions).
Describing a definitive epigenome for primed pluripotency has proven more difficult. Regardless of culture condition, naive and primed cells clearly possess distinct developmental capacities. ESCs and EpiSCs were recently shown to vary in their propensity to differentiate toward endoderm, ectoderm, and mesoderm lineages (Ghimire et al. 2018). However, inconsistencies between cells in the postimplantation blastocyst and EpiSCs, their in vitro counterparts, have thwarted attempts to identify precise epigenetic differences (Takahashi et al. 2018). Although EpiSCs have more promoter-associated hypermethylation and exhibit enhancer switching at pluripotency genes, the same differences are not observed in vivo (Factor et al. 2014; Veillard et al. 2014; Takahashi et al. 2018). These discrepancies are complicated further by the poorly conserved nature of “primed” features like X-chromosome inactivation. For instance, although dosage compensation of X-linked genes is a feature of the “primed” state, the initiation of this process is not generalizable among eutherians (Sado and Sakaguchi 2013). In addition, only recently have culture conditions for a “naive” pluripotent state been established for human cells, albeit with relatively limited success. The difficulty of establishing human ESCs of a comparable developmental stage to naive mouse ESCs reflects our incomplete understanding of the core principles of pluripotency (Rossant 2015; Ying and Smith 2017; Yilmaz and Benvenisty 2019).
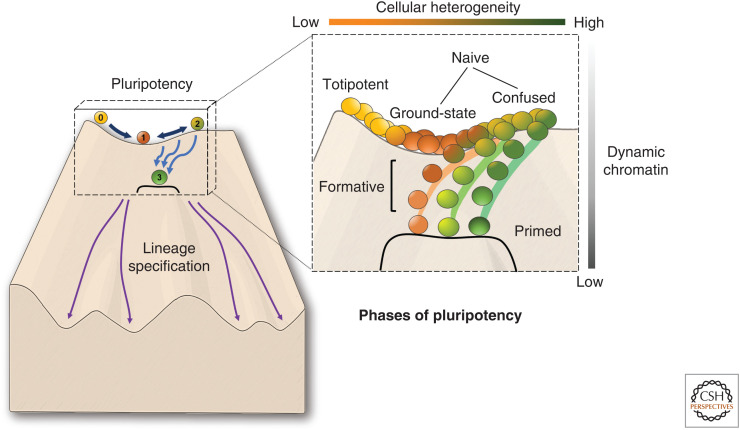
The Pluripotency Continuum
Ultimately, experimental pluripotent models appear to provide “snapshots” of pluripotency, rather than behave as mirrors of precise developmental stages. Indeed, a third, “formative” or “epiblast-like” phase, resembling embryonic day 5.5, has been proposed to exist between the naive and primed states (Kalkan and Smith 2014; Smith 2017; for review, see Morgani et al. 2017). In addition, recent single-cell analyses have found that a subpopulation of naive cells possess a transcriptome with primed-like expression, and a similar naive-primed transition is apparent across species during early embryonic development (Messmer et al. 2019). This echoes the theorizing of many who have speculated that pluripotent cells lie on a developmental continuum of successive phases (Fig. 2; Ohtsuka and Dalton 2008; Hough et al. 2009; Loh and Lim 2010; Tesar 2016). The heterogeneity of “confused” ESCs cultured in serum could represent a mixture of naive and primed cells or cells in the process of transitioning between the two states. Interestingly, many of the epigenetic differences observed between ESCs and EpiSCs reiterate the differences between “ground-state” versus “confused” ESC populations. For example, increases in global levels of DNA methylation and focal enrichments of H3K27me3 at developmental promoters appear to correlate with being further along on the ground-state/confused/primed axis; this is also true for clusters of H3K9me3 at pericentromeric repeats (Marks et al. 2012; Juan et al. 2016; Tosolini et al. 2018). The broad range of epigenetic landscapes that are permissive of a pluripotent state is indicative that a holistic characterization of pluripotency is needed—one that accounts for the spatial and chemical properties of the nucleus.
Figure 2.
Pluripotency lies on a continuum of successive phases. The figure is based on “Waddington's landscape,” and shows the journey taken by a naive pluripotent cell (ball labeled 1) toward a more differentiated (but still pluripotent) primed state (ball labeled 3). The status of cells transitioning between these two states is not well defined (balls labeled 2). Color of the balls represents the overall transcriptomic state. (Inset, right) Represents “pluripotent phases” as serial populations of cells with differing levels of heterogeneity rather than discrete developmental stages. Importantly, ground-state pluripotency seems to occupy a “lower energy” state in terms of chromatin mobility compared to both two-cell-stage embryos and serum-cultured embryonic stem cells (ESCs) (Bošković et al. 2014), which may also explain the relatively lower transcriptional promiscuity among naive pluripotent populations (Tosolini et al. 2018). Prior to commitment to a particular primed state, cells may be found in a heterogeneous mix of transcriptomic and metabolic states. This has been somewhat recapitulated in vitro with serum-grown ESCs, which have been described to possess a “confused” transcriptome. However, to what extent there exists a phenotypically distinct “formative” phase between naive and primed pluripotency, or whether this state is strictly necessary for lineage commitment, remains poorly understood.
CHROMATIN STRUCTURE IN PLURIPOTENT NUCLEI
In mammalian cells, as much as 2 m of DNA can fit comfortably within a nucleus of 10 μm in diameter. Besides the staggering degree of coordination required for this feat alone, cells must also retain their cellular identity and the appropriate transcriptomic activity. This is facilitated by the structural material of the genome, or chromatin, which is comprised of repetitive units of histone–DNA complexes called nucleosomes (Kornberg 1974; Oudet et al. 1975). Nucleosomes consist of a histone octamer wrapped by 145–147 bp of DNA, separated by linker DNA of variable length and further compacted by linker histone H1 (Korolev et al. 2018). Early images from electron microscopy (EM) revealed that nucleosome fibers are roughly 10-nm wide and resemble “beads on a string” (Oudet et al. 1975; Kornberg and Klug 1981). Later, the first X-ray diffraction images of isolated chromatin informed 30-nm fiber models of chromatin folding, and others predicted more complex patterns of folding, including the 120-nm chromonema (Belmont and Bruce 1994). It was conceivable, therefore, that a “higher-order structure” of these nucleosomes followed hierarchical principles of organization, and that this could be functionally relevant for gene regulation.
However, with the advent of advanced EM techniques, the existence of these structures in situ has been disputed. By using transmission EM (TEM) and cryo-EM, several groups were unable to detect 30-nm fibers in the chromatin of intact nuclei (Eltsov et al. 2008; Maeshima et al. 2014; Maeshima 2020). Indeed, beyond the level of the 10-nm nucleosome fiber, larger folded structures in interphase chromatin are often thought to be artefacts of fixation procedures or exist only in vitro (Kuznetsova and Sheval 2016). Currently, models depicting less-ordered chains of zigzagging nucleosomes are considered to be more accurate representations of interphase chromatin (Fussner et al. 2011; Grigoryev et al. 2016; Bascom and Schlick 2017; Krietenstein et al. 2020).
Nucleosome Organization
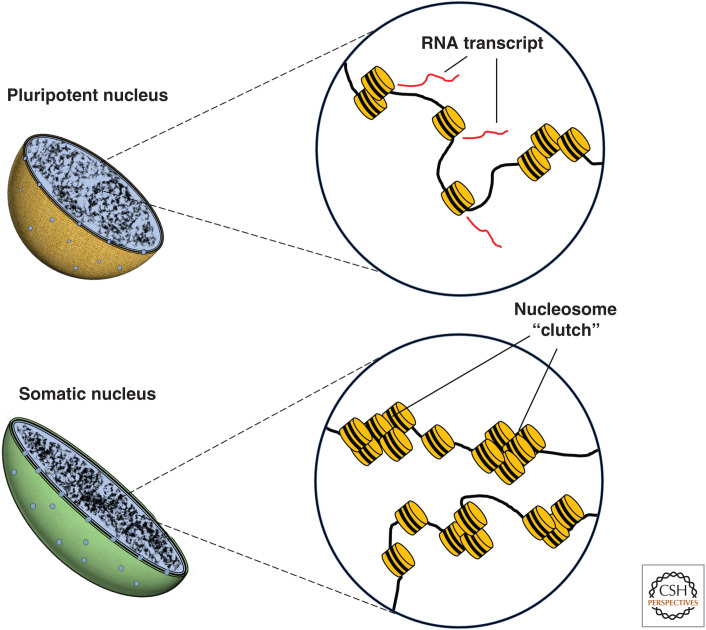
At the nanoscale level, nucleosomes have been observed to form discrete “clutches,” or clusters, that differ in size and distribution depending on developmental stage (Ricci et al. 2015). Imaging of mouse ESCs at this scale has been accomplished with various super-resolution techniques, including structured illumination microscopy (SIM) and stochastic optical reconstruction microscopy (STORM) (for review, see Ricci et al. 2017). STORM imaging of H2B in ESCs and in ESC-derived neuronal progenitor cells (NPCs) revealed that NPCs have a higher number of nucleosomes per clutch. Moreover, clutch size could predict how well human iPSCs had been reprogrammed toward pluripotency (Ricci et al. 2015). These results strengthened previous observations that used electron spectroscopic imaging (ESI) to visualize chromatin structure in pluripotent cells. These studies showed that cells in early embryonic tissue have mesh-like, more widely dispersed fibers of nucleosomes, compared with differentiated fibroblasts, which are enriched in dense regions of 10-nm nucleosomal fibers (Ahmed et al. 2010; Fussner et al. 2010). Heterochromatin, which is associated with transcriptional repression, contains a higher density of nucleosomes than euchromatin, confirming early histological observations that there are fewer clusters of heterochromatin in undifferentiated cells (Park et al. 2004; Meshorer and Misteli 2006; Imai et al. 2017).
Together, these studies demonstrate that a diffuse nucleosomal organization is a distinguishing structural feature of the pluripotent genome (Fig. 3). Intuitively, the dual requirements of stem cells to self-renew and differentiate presumes a high degree of flexibility in gene regulation, and, therefore, being able to easily switch between different arrangements of chromatin may facilitate this process. However, it is worth considering alternative morphological interpretations of chromatin organization. For example, a recent study creatively illustrated that chromatin reorganization can occur in response to mechanical stress. Nava et al. (2020) showed that reduction of H3K9me3 levels could be triggered by cell stretching, which reduces heterochromatin at the nuclear periphery and protects cells from mechanically induced DNA damage. The tightly packed chromatin of differentiated cells may reflect a physiological need for morphological compaction when compared with the characteristically large nuclei of ESCs. Indeed, it has been suggested that in quiescent cells, the main function of chromatin compaction may be to reduce cellular size by reducing nuclear volume, rather than to regulate global transcription (Pombo and Dillon 2015). The decondensed chromatin in ESCs may have as much to do with their unspecialized nature as their need for transcriptional flexibility.
Figure 3.
Nucleosome density in interphase nuclei of pluripotent and somatic cells. Pluripotent cells possess smaller and relatively sparse clusters of nucleosomes (“clutches”) compared with somatic cells. In more differentiated cells, a dense arrangement of “clutches” results in more condensed chromatin, preventing the accessibility of transcriptional machinery, such as Pol II, and reducing transcription (Ricci et al. 2015).
In any case, it is safe to conclude that imaging-based studies strongly support earlier hypotheses that an “open” chromatin configuration is a hallmark of pluripotency (this theory has been covered at length in the following reviews: Meshorer and Misteli 2006; Koh et al. 2010; Gaspar-Maia et al. 2011; Chen and Dent 2014; Kobayashi and Kikyo 2015; Percharde et al. 2017; Schlesinger and Meshorer 2019). Here, it is important to note that although it has been proposed that chromatin density and not higher-order organization separates “closed” from “accessible” chromatin (Ou et al. 2017), the density of heterochromatin has been measured to be only 1.53-fold higher than neighboring euchromatin (Imai et al. 2017). This suggests that the accumulation of non-nucleosomal proteins or RNA molecules may play a significant role in contributing to the “closed” quality of heterochromatin. Indeed, elucidating the relationship between chromatin structure and the function of chromatin-associating factors will aid our understanding of how transcriptional activity is organized in the nucleus.
OPEN CHROMATIN
Several decades prior to the discovery of the molecular structure of DNA, Emil Heitz (1928) precociously observed two types of differentially stained regions of chromosomes, which he termed “euchromatin” and “heterochromatin.” Although classically used to distinguish cytologically between light- and heavy-staining DNA, today these terms are more commonly used to describe respective regions of active or inactive transcription (Woodcock and Ghosh 2010). The transcriptionally repressive environment associated with heterochromatin is thought to reflect its condensed structure, which prevents physical access by transcriptional machinery. In contrast, euchromatin is considered to be more open and accessible to regulatory proteins and thus more transcriptionally permissive.
The concept of “open chromatin” also applies to the characteristically flexible and dynamic nature of the pluripotent nucleus. That is, in addition to having a higher euchromatin-to-heterochromatin ratio, pluripotent cells have hyperdynamic nucleosomes as evidenced by fluorescence recovery after photobleaching (FRAP) experiments, where heterochromatic regions of mouse ESC nuclei undergo rapid exchange of core histones H2B and H3, linker histone H1, and heterochromatin protein 1 (HP1) (Meshorer et al. 2006). Not limited to mammals, loosely bound chromatin has also been observed in Drosophila embryos (Bhattacharya et al. 2009); moreover, totipotent planarian neoblasts are largely depleted in heterochromatin (Morita et al. 1969). Mechanistically, several factors were suggested to contribute to chromatin plasticity in ESCs, including histone hyperacetylation and the almost complete lack of the nuclear intermediate filament protein lamin A (Melcer et al. 2012). As noted above, several chromatin remodeling proteins were also implicated in maintaining open chromatin and/or pluripotency in ESCs. CHD1, a well-studied chromatin remodeling factor, is required to prevent spurious formation of heterochromatin in ESCs (Gaspar-Maia et al. 2009). CHD1 is implicated in the maintenance of pluripotency in ESCs and transcription initiation via binding to the mediator coactivator complex at active genes (Lin et al. 2011). Unsurprisingly, serum-grown euchromatin-rich ESCs are transcriptionally promiscuous and exhibit low levels of genome-wide gene expression (Efroni et al. 2008). More recently, single-cell RNA-seq studies have shown that the transcriptomes of undifferentiated cells are also more diverse, demonstrating widespread transcriptome sampling in pluripotent stem cells, which become gradually restricted upon differentiation (Gulati et al. 2020).
Genome Accessibility
TFs exert their effect on gene expression by binding to DNA; chromatin plasticity is thought to facilitate this by allowing regulatory proteins access to nucleosome-bound DNA (Klemm et al. 2019). Therefore, genome accessibility is a central variable that links the flexibility of chromatin structure with transcriptional control by regulatory factors. Human ChIP-seq data from the Encyclopedia of DNA Elements (ENCODE) project estimates that almost 95% of TFs bind to accessible DNA, despite accessible regions comprising as little as 4% of the genome (Thurman et al. 2012; Kelley et al. 2016).
Measurement of genome accessibility can be assessed indirectly through fragmentation by nucleases, such as DNase-I, or by insertion of sequencing adaptors, as in transposase-accessible chromatin sequencing (ATAC-seq). Using these techniques, a number of groups have shown that the genome of pluripotent cells is more accessible than differentiated cells. For example, a study mapping DNase-I hypersensitive sites (DHSs) revealed that ESCs had almost double the frequency of DHSs than NPCs, and the depletion of HMGN, a nucleosome-binding protein, was shown to lead to a significant reduction in DHSs (Deng et al. 2013). ATAC-seq experiments have further supported these findings. Differentiation of ESCs toward definitive endoderm results in a 13% net loss of ATAC-seq peaks globally (Simon et al. 2017). Moreover, an interesting study used an allele-specific ATAC-seq method to show that promoter elements in ESCs that were biallelically accessible became monoallelically accessible after NPC differentiation (Xu et al. 2017). This complements a previous observation that monoallelic transcription is more than fivefold higher in NPCs than in ESCs (Eckersley-Maslin et al. 2014).
Micrococcal nuclease (MNase) digestion is also used to measure genome accessibility and is a classic method for detecting regions of nucleosome occupancy. One study found that, globally, differentiating cells are more resistant to MNase digestion than ESCs. The bromodomain factor, ATAD2, which is predominantly expressed in ESCs, was found partly responsible for increasing chromatin sensitivity to MNase digestion (Morozumi et al. 2016). Moreover, the ESC-specific chromatin remodeling BAF complex, esBAF, was found to possess an additional role in maintaining nucleosome-depleted regions in pluripotent cells (Ho et al. 2009; Hainer et al. 2015). Intriguingly, the same group demonstrated that esBAF activity at these regions was necessary for binding of the pluripotency factor, KLF4 (Hainer and Fazzio 2015). This suggests that synergy may exist between chromatin remodeling complexes and pluripotency TF activity, especially as components of the BAF complex have been previously shown to enhance iPSC reprogramming efficiency (Singhal et al. 2010).
Chromatin Remodeling and Pluripotency
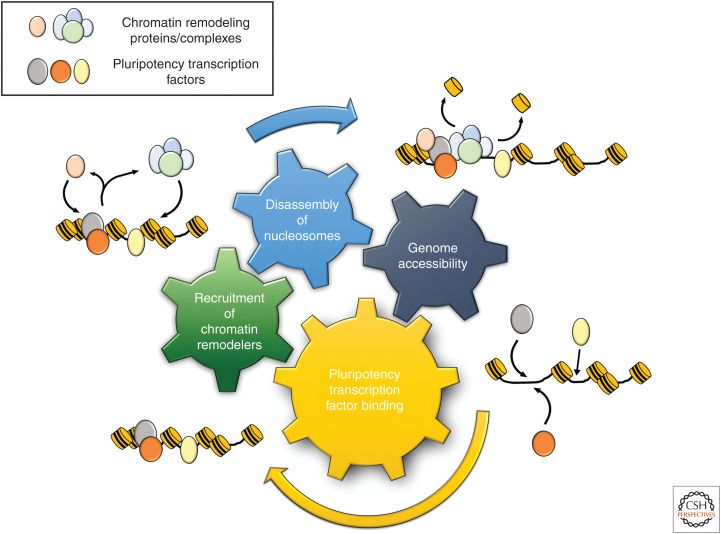
Intriguingly, members of the “pluripotent essentialome” (factors necessary for maintenance of pluripotency) are very highly enriched in the protein interactome of OCT4, SOX2, and NANOG (Wang et al. 2006; Yilmaz et al. 2018). Many of these proteins are involved in chromatin remodeling, as mentioned above, adding to mounting evidence that cross talk between chromatin-modifying complexes and core pluripotency factors is a fundamental mechanism for maintaining pluripotency (for review, see Orkin and Hochedlinger 2011). Greater chromatin accessibility in pluripotent cells, therefore, may be the consequence of positive reinforcement of nucleosome disassembly and the binding of regulatory factors, amplifying the existing feedforward loops maintained by the pluripotency network (Fig. 4). Indeed, a recent study has shown that subpopulations of human pluripotent stem cells with high capacities for self-renewal have greater chromatin accessibility at regions of pluripotency TF binding (Lau et al. 2020). Moreover, evidence from zebrafish embryos indicates that TFs are in passive competition with core histones for binding to DNA, and the nuclear ratio between histones and DNA-binding factors is important for the onset of zygotic transcription (Joseph et al. 2017).
Figure 4.
Synergy between chromatin remodeling and pluripotency factor binding. Four hallmarks of pluripotent stem cells are represented by the colored gears. The binding of pluripotency factors (e.g., SOX2, NANOG, OCT4) can bring about the recruitment of chromatin remodeling complexes (e.g., esBAF, CHD1, p300). This leads to histone exchange as nucleosomes are disassembled, which in turn promotes accessible regions of DNA and permits further binding of transcription factors.
3D ORGANIZATION OF THE PLURIPOTENT GENOME
How the genome is spatially organized is intrinsically linked to the establishment of cell-type identity. Although, as discussed earlier, the precise nature of higher-order chromatin structure is controversial, a conserved hierarchy of topological organization can be observed throughout interphase. Perhaps the most obvious example of this is the arrangement of chromosomes, which occupy distinct territories in interphase nuclei (Cremer and Cremer 2010).
Chromosome Organization
At the chromosome level, the genomes of most cell types are similarly arranged both in human and mouse, although some features of cell specificity do exist. Whereas each possess established chromosome territories, pluripotent ESCs and differentiated cell types differ in their radial positioning of individual chromosomes, which is related to gene density, as revealed by dual-color 3D fluorescence in situ hybridization (3D-FISH) experiments (Mayer et al. 2005). In addition, repositioning of chromosomal arms has been observed in the case of NANOG, which is highly expressed during pluripotency, where the short arm of chromosome 12 (12p) is found closer to the center of the nucleus in human ESCs than in differentiated cells (Wiblin et al. 2005). Heterochromatic chromosomal subregions, such as the centromere and pericentromere, which are called “chromocenters” when imaged in stained nuclei, cluster differently depending on cell type. For instance, mouse myoblasts and fibroblasts have smaller but a greater number of chromocenters than ESC nuclei, whereas fewer chromocenters can be seen in macrophages and postmitotic neurons (Mayer et al. 2005; Aoto et al. 2006; Kobayakawa et al. 2007). Intriguingly, loss of NANOG causes chromocenters in ESCs to resemble a more differentiated state (Novo et al. 2016). Notably, visible chromocenters are absent in human cells, but staining for centromere protein C (CENP-C) reveals that centromeres in human ESCs localize further away from the nuclear periphery than most other cell types (Wiblin et al. 2005).
Chromatin Loops, Domains, and Interaction Hubs
Organization at a finer scale occurs through chromatin looping and formation of topologically associating domains (TADs) or lamina-associated domains (LADs). Chromatin loops and TADs have been identified using chromosome conformation capture (3C)-based technologies, which map chromatin–chromatin interactions through formaldehyde cross-linking. LADs, which are detected with DNA adenine methyltransferase identification (DamID), are located at the nuclear periphery and correlate with gene silencing (Gonzalez-Sandoval and Gasser 2016). Chromosome regions that interact with the nuclear lamina make up around 40% of the genome and are mostly conserved across cell lineages, as evidenced by DamID experiments, where 73%–87% of LADs were found to overlap between several stages of development (Peric-Hupkes et al. 2010). Nonetheless, many LAD-associated loci reposition themselves relative to the nuclear interior following differentiation, and stem-cell-related genes, such as NANOG, KLF4, and OCT4, show more frequent interactions with the nuclear lamina in ESC-derived NPCs (Peric-Hupkes et al. 2010). As these loci generally encompass a single gene or transcriptional unit, this alludes to a functional role for regulation of nuclear architecture (Melcer and Meshorer 2010). However, the relationship between spatial relocalization of gene loci and changes in transcription is not always apparent. For instance, in the case of the Hoxd cluster, although the entire locus loops away from its chromosome territory upon differentiation in mouse ESCs, individual genes in the cluster do not become activated in a concurrent fashion (Morey et al. 2007).
A fundamental regulatory feature of metazoan nuclei is the existence of long-range enhancer–promoter interactions. CTCF-mediated chromatin looping on the kilobase scale is one mechanism by which this is thought to occur (Rao et al. 2014). Genome-wide maps of contacts that occur between any two genomic loci can be detected using Hi-C, a 3C-derived method, which incorporates proximity-based ligation with high-throughput sequencing (Dekker et al. 2002; Lieberman-Aiden et al. 2009). Overall, long-range contacts are weaker and less prevalent in mouse ESCs compared with somatic cells, and chromatin topology is progressively established during ESC lineage progression (Pękowska et al. 2018). This is in line with the more globally open chromatin configuration of ESCs, as a diffuse chromatin environment would hinder the formation of long-range interactions. In contrast, interactions between pluripotency factor binding sites are enriched in ESCs and the pluripotency factors themselves contribute to formation of these contacts (Apostolou et al. 2013; Denholtz et al. 2013; de Wit et al. 2013; Wei et al. 2013). These observations are supported by Hi-C maps generated during reprogramming of B cells into iPSCs. Strikingly, these experiments revealed that following reprogramming, ESC-specific contacts became more frequent at genes related to developmental processes, the formation of which preceded changes in transcription (Stadhouders et al. 2018). In support of this finding, another study, using a technique called promoter capture Hi-C, found that developmental pathway–related genes are enriched in ESC-specific promoter–enhancer interactions; a large amount of distinct cell-type-specific contacts were detected for both ESCs and fetal liver cells at promoters interacting with more than 10 enhancers (Schoenfelder et al. 2018).
Similarly, Hi-C studies have found that the position of TADs and their boundaries do not considerably vary across diverse mammalian cell types (Dixon et al. 2012; Rao et al. 2014). However, at a subset of TADs, changes in the frequency of intradomain interactions can occur following human ESC differentiation; this corresponds with minor changes in expression (with a bias toward repression) of genes within these domains (Dixon et al. 2015). These findings were corroborated by an ultra-deep Hi-C study, which compared two types of self-interacting compartments in serum grown ESCs with NPCs and neurons. These compartments, referred to as “A” or “B,” are distinct networks of loci whose intradomain genes fall within active or inactive chromatin, respectively. Bonev et al. found that co-interactions within compartment “A” were weaker, and compartment “B” co-interactions became stronger following neuronal differentiation of ESCs, suggesting that “switching on and off” of particular TADs occurs in a cell-type-sensitive manner. Overall, the authors found that differentiation results in stronger intra-TAD contacts and weaker inter-TAD contacts (Bonev et al. 2017). In stark contrast, a recent study showed the opposite trend. Barrington et al. revealed that ESCs cultured in 2i conditions had, on average, weaker intradomain interactions, weaker TAD borders, and stronger interdomain contacts than neural stem cells (NSCs) (Barrington et al. 2019). This discrepancy indicates that environmental signals significantly influence TAD organization in ESCs. Moreover, it suggests that 2i-cultured, “ground state” ESCs are less organized topologically and that, conversely, ESCs cultured in serum may have already acquired some lineage-specific genomic features as their transcriptional heterogeneity would imply. Nevertheless, both results suggest that during development, formation, and dissolution of TADs occur in a distinctly cell-type-specific manner. Here, it is worth mentioning that the presence and absence of contacts in Hi-C maps may not reflect the state of each cell in a population, especially as the expression of some pluripotency factors, such as Nanog and Rex1, have been shown to be highly heterogeneous in ESCs (Toyooka et al. 2008; Clancy et al. 2016). Furthermore, it is also important to note that as chromatin–chromatin interaction maps are generated from a population of cells, due to cell-to-cell heterogeneity, they are not wholly representative of “true” or stable chromatin contacts. Indeed, these may be highly transient as single-cell Hi-C and imaging experiments have demonstrated (Nagano et al. 2013; Cattoni et al. 2017; Finn et al. 2019).
Nevertheless, evidence from chromatin–chromatin interaction studies reveal that overall, topological organization does not profoundly differ between pluripotent and differentiated cells. Rather, interactions at specific loci or domains appear to be locally implicated in an ESC-specific manner. In addition, these interactions are often located at genes in the core pluripotency network. However, it remains to be seen to what extent these interactions precede function. There is a growing consensus, at least in the realm of genome topology, that function drives structure more frequently than not (Finn and Misteli 2019). In the case of pluripotent stem cells, both form and function are likely intertwined, and the transcription of pluripotency factors and the formation of hubs of interactions may be costabilizing events (Fig. 5). Indeed, further chromatin interaction studies at higher resolutions are needed to disentangle the relationship between transcription and spatial organization. To this end, the establishment of a micrococcal nuclease (micro-C) method for profiling of sub-TAD contacts in mammalian cells is an extremely promising development (Hsieh et al. 2015, 2020). Micro-C has revealed that compared with human ESCs, human fetal fibroblasts (HFFs) possess dramatically increased compartment strength, as well as an increase in the number of long-range contacts (Krietenstein et al. 2020). This ultrahigh, single-nucleosome resolution (Hsieh et al. 2020) analysis of genome organization in ESCs and HFFs provides the most comprehensive chromatin interactions catalog in both cell types, supporting the hyperplastic chromatin model of the pluripotent cell (Schlesinger and Meshorer 2019).
Figure 5.
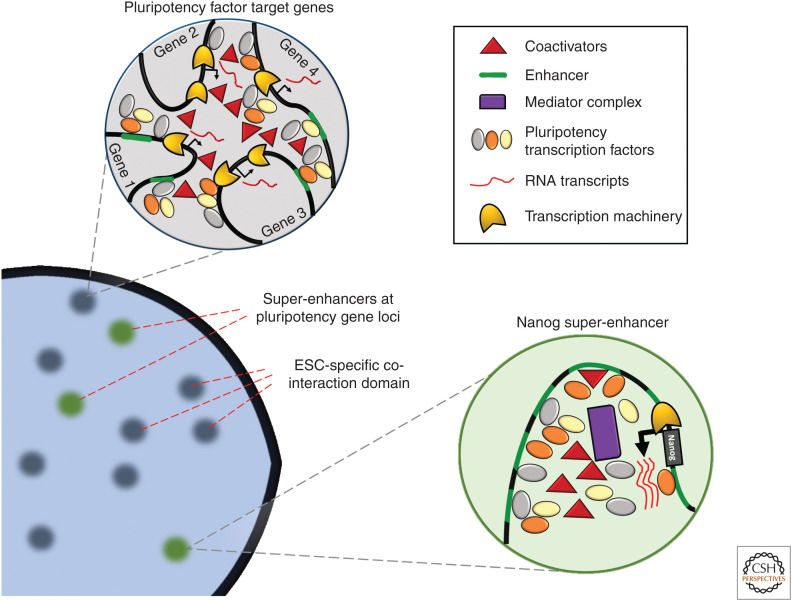
Transcriptional “hubs” of pluripotency factor accumulation. In pluripotent cells, master transcription factors are highly expressed from loci that are regulated by regions of densely packed enhancers termed “super-enhancers” (SEs). The combinatorial binding of pluripotency factors themselves at pluripotency gene loci stabilizes their high levels of expression. Large quantities of pluripotency factors in turn can accumulate at their target genes throughout the genome. As each pluripotency factor can itself form complexes with other coactivators, multiple loci may be brought together spatially through the binding of enhancers/promoters of different genes. (ESC) embryonic stem cell.
CONCLUSIONS AND FUTURE PERSPECTIVES
As we have seen, with the constant development of new methods, mostly relying on high-throughput sequencing readouts, and more recently, super-resolution imaging, the 2D and 3D genome has been studied in exceedingly finer detail in pluripotent and differentiated cells. Whereas the organization of the pluripotent genome (e.g., chromosome territories, the marking of DNA by cytosine methylation and of chromatin by histone modifications, chromatin–chromatin, and chromatin–lamina interactions) is globally not unlike its somatic counterpart, it is nonetheless unique in specific chromatin features, which, collectively, confer fluid-like properties. How then do pluripotent ESCs maintain their hyperplastic chromatin? The emerging concept of “biomolecular condensates” (Banani et al. 2017) or “transcriptional condensates” (Shrinivas et al. 2019) may provide some clues. Working with knockin fluorescent mouse ESC strains, Boija et al. (2018) showed that nuclear condensates formed by the mediator complex at ESC SEs are dependent on OCT4, which incorporates into these nuclear condensates together with MED1 and other mediator subunits. Similarly, MED1 and BRD4 form transcriptional condensates on pluripotent SEs, including the Nanog SE. Because phase-separated condensates are formed by the nonstructured activation domains of many TFs (Boija et al. 2018), it is tempting to speculate that the core transcriptional circuitry operating in pluripotent cells might be unique in its ability to form dynamic transcriptional condensates, antagonizing chromatin phase separation, particularly as there is a high level of combinatorial binding of OCT4, SOX2, and NANOG at enhancers associated with mediator colocalization (Göke et al. 2011). In contrast, somatic TFs might mediate stronger long-range interactions by fusing several distant high-density regions (Shrinivas et al. 2019), promoting heterochromatin-favoring phase separation, genome aggregation, and restricted dynamics. Supporting this model, histone acetylation, which is prevalent in ESCs (Bártová et al. 2008; Bian et al. 2009; Hezroni et al. 2011) and was shown to support chromatin plasticity and high mobility of chromatin proteins in ESCs (Melcer et al. 2012), was also shown recently to constrain phase separation (Gibson et al. 2019). These ideas, however, remain to be tested.
In closing, beyond the organizational and structural elements of the pluripotent genome described herein, a great deal more work—both theoretical and experimental—will be needed to bridge the gap between our understanding of defining features of pluripotency (e.g., open chromatin, active epigenetic marks) and the biological necessity of these traits in the early embryo. For instance, could sensitivity to developmental cues be an emergent property of a heterogeneous population capable of promiscuous transcription? Moreover, to what extent is pluripotency a feature of an individual cell versus that of the population as a whole? Indeed, future studies should attempt to contextualize features of the pluripotent genome with respect to its developmental milieu. The coming decades will no doubt offer deeper insights into the mysterious phenomenon of pluripotency.
ACKNOWLEDGMENTS
This work was supported by the Israel Science Foundation (ISF 1140/2017 to E.M.). E.M. is the incumbent of the Arthur Gutterman Professor Chair for Stem Cell Research. P.S.L.L. is supported by the H2020 Marie Skłodowska-Curie Innovative Training Network – EpiSyStem, Grant No. 765966.
Footnotes
Editors: Ana Pombo, Martin W. Hetzer, and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. 2010. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE 5: e10531 10.1371/journal.pone.0010531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alajem A, Biran A, Harikumar A, Sailaja BS, Aaronson Y, Livyatan I, Nissim-Rafinia M, Sommer AG, Mostoslavsky G, Gerbasi VR, et al. 2015. Differential association of chromatin proteins identifies BAF60a/SMARCD1 as a regulator of embryonic stem cell differentiation. Cell Rep 10: 2019–2031. 10.1016/j.celrep.2015.02.064 [DOI] [PubMed] [Google Scholar]
- Aoto T, Saitoh N, Ichimura T, Niwa H, Nakao M. 2006. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev Biol 298: 354–367. 10.1016/j.ydbio.2006.04.450 [DOI] [PubMed] [Google Scholar]
- Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, et al. 2013. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell 12: 699–712. 10.1016/j.stem.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadou R, de Sousa D, Myant K, Merusi C, Stancheva I, Bird A. 2010. Targeting of de novo DNA Methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PLoS ONE 5: e9937 10.1371/journal.pone.0009937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrington C, Georgopoulou D, Pezic D, Varsally W, Herrero J, Hadjur S. 2019. Enhancer accessibility and CTCF occupancy underlie asymmetric TAD architecture and cell type specific genome topology. Nat Commun 10: 2908 10.1038/s41467-019-10725-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bártová E, Galiová G, Krejcí J, Harničarová A, Strašák L, Kozubek S. 2008. Epigenome and chromatin structure in human embryonic stem cells undergoing differentiation. Dev Dyn 237: 3690–3702. 10.1002/dvdy.21773 [DOI] [PubMed] [Google Scholar]
- Bascom G, Schlick T. 2017. Linking chromatin fibers to gene folding by hierarchical looping. Biophys J 112: 434–445. 10.1016/j.bpj.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Bruce K. 1994. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J Cell Biol 127: 287–302. 10.1083/jcb.127.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh PA, Navot D. 1992. The impact of embryonic development and endometrial maturity on the timing of implantation. Fertil Steril 58: 537–542. 10.1016/S0015-0282(16)55259-5 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Talwar S, Mazumder A, Shivashankar GV. 2009. Spatio-temporal plasticity in chromatin organization in mouse cell differentiation and during Drosophila embryogenesis. Biophys J 96: 3832–3839. 10.1016/j.bpj.2008.11.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y, Alberio R, Allegrucci C, Campbell KH, Johnson AD. 2009. Epigenetic marks in somatic chromatin are remodelled to resemble pluripotent nuclei by amphibian oocyte extracts. Epigenetics 4: 194–202. 10.4161/epi.4.3.8787 [DOI] [PubMed] [Google Scholar]
- Boiani M, Schöler HR. 2005. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol 6: 872–881. 10.1038/nrm1744 [DOI] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. 2018. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175: 1842–1855.e16. 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171: 557–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bošković A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Ram EVSR, Meshorer E, Torres-Padilla ME. 2014. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev 28: 1042–1047. 10.1101/gad.238881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956. 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A, Evans M, Kaufman MH, Robertson E. 1984. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309: 255–256. 10.1038/309255a0 [DOI] [PubMed] [Google Scholar]
- Brons IGM, Smithers LE, Trotter MWB, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195. 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- Cattoni DI, Cardozo Gizzi AM, Georgieva M, Di Stefano M, Valeri A, Chamousset D, Houbron C, Déjardin S, Fiche JB, González I, et al. 2017. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat Commun 8: 1753 10.1038/s41467-017-01962-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2017. Origin of animal multicellularity: precursors, causes, consequences—the choanoflagellate/sponge transition, neurogenesis and the Cambrian explosion. Philos Trans R Soc Lond B Biol Sci 372: 20150476 10.1098/rstb.2015.0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Dent SYR. 2014. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 15: 93–106. 10.1038/nrg3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy CE, An G, Cannon WR, Liu Y, May EE, Ortoleva P, Popel AS, Sluka JP, Su J, Vicini P, et al. 2016. Multiscale modeling in the clinic: drug design and development. Ann Biomed Eng 44: 2591–2610. 10.1007/s10439-016-1563-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. 2010. Chromosome territories. Cold Spring Harb Perspect Biol 2: a003889 10.1101/cshperspect.a003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Deng T, Zhu ZI, Zhang S, Leng F, Cherukuri S, Hansen L, Mariño-Ramírez L, Meshorer E, Landsman D, Bustin M. 2013. HMGN1 modulates nucleosome occupancy and DNase I hypersensitivity at the CpG island promoters of embryonic stem cells. Mol Cell Biol 33: 3377–3389. 10.1128/MCB.00435-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, Ernst J, Pellegrini M, Plath K. 2013. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell 13: 602–616. 10.1016/j.stem.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Bouwman BA, Zhu Y, Klous P, Splinter E, Verstegen MJ, Krijger PH, Festuccia N, Nora EP, Welling M, et al. 2013. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature 501: 227–231. 10.1038/nature12420 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Thybert D, Bergmann JH, Marioni JC, Flicek P, Spector DL. 2014. Random monoallelic gene expression increases upon embryonic stem cell differentiation. Dev Cell 28: 351–365. 10.1016/j.devcel.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti RR, Harikumar A, Aaronson Y, Biran A, Sailaja BS, Nissim-Rafinia M, Azad GK, Cohen MA, Park JE, Shivalila CS, et al. 2017. Alternative SET/TAFI promoters regulate embryonic stem cell differentiation. Stem Cell Rep 9: 1291–1303. 10.1016/j.stemcr.2017.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RDG, Buetow KH, et al. 2008. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2: 437–447. 10.1016/j.stem.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltsov M, MacLellan KM, Maeshima K, Frangakis AS, Dubochet J. 2008. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci 105: 19732–19737. 10.1073/pnas.0810057105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, Martinez-Pastor B, Walsh RM, Sommer CA, Lienhard M, Kugel S, et al. 2015. The histone deacetylase Sirt6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol 17: 545–557. 10.1038/ncb3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156. 10.1038/292154a0 [DOI] [PubMed] [Google Scholar]
- Factor DC, Corradin O, Zentner GE, Saiakhova A, Song L, Chenoweth JG, McKay RD, Crawford GE, Scacheri PC, Tesar PJ. 2014. Epigenomic comparison reveals activation of “seed” enhancers during transition from naive to primed pluripotency. Cell Stem Cell 14: 854–863. 10.1016/j.stem.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul YL, Walter J, et al. 2013. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13: 351–359. 10.1016/j.stem.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EH, Misteli T. 2019. A genome disconnect. Nat Genet 51: 1205–1206. 10.1038/s41588-019-0476-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EH, Pegoraro G, Brandão HB, Valton AL, Oomen ME, Dekker J, Mirny L, Misteli T. 2019. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176: 1502–1515.e10. 10.1016/j.cell.2019.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussner E, Ahmed K, Dehghani H, Strauss M, Bazett-Jones DP. 2010. Changes in chromatin fiber density as a marker for pluripotency. Cold Spring Harb Symp Quant Biol 75: 245–249. 10.1101/sqb.2010.75.012 [DOI] [PubMed] [Google Scholar]
- Fussner E, Ching RW, Bazett-Jones DP. 2011. Living without 30 nm chromatin fibers. Trends Biochem Sci 36: 1–6. 10.1016/j.tibs.2010.09.002 [DOI] [PubMed] [Google Scholar]
- Gao Y, Gan H, Lou Z, Zhang Z. 2018. Asf1a resolves bivalent chromatin domains for the induction of lineage-specific genes during mouse embryonic stem cell differentiation. Proc Natl Acad Sci 115: E6162–E6171. 10.1073/pnas.1801909115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460: 863–868. 10.1038/nature08212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. 2011. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 12: 36–47. 10.1038/nrm3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghimire S, der Jeught MV, Neupane J, Roost MS, Anckaert J, Popovic M, Nieuwerburgh FV, Mestdagh P, Vandesompele J, Deforce D, et al. 2018. Comparative analysis of naive, primed and ground state pluripotency in mouse embryonic stem cells originating from the same genetic background. Sci Rep 8: 5884 10.1038/s41598-018-24051-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK. 2019. Organization of chromatin by intrinsic and regulated phase separation. Cell 179: 470–484.e21. 10.1016/j.cell.2019.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, Jung M, Behrens S, Chavez L, O'Keeffe S, Timmermann B, Lehrach H, Adjaye J, Vingron M. 2011. Combinatorial binding in human and mouse embryonic stem cells identifies conserved enhancers active in early embryonic development. PLoS Comput Biol 7: e1002304 10.1371/journal.pcbi.1002304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A, Gasser SM. 2016. On TADs and LADs: spatial control over gene expression. Trends Genet 32: 485–495. 10.1016/j.tig.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Gorkin DU, Leung D, Ren B. 2014. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 14: 762–775. 10.1016/j.stem.2014.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MVC, Bourc'his D. 2019. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20: 590–607. 10.1038/s41580-019-0159-6 [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Bascom G, Buckwalter JM, Schubert MB, Woodcock CL, Schlick T. 2016. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc Natl Acad Sci 113: 1238–1243. 10.1073/pnas.1518280113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati GS, Sikandar SS, Wesche DJ, Manjunath A, Bharadwaj A, Berger MJ, Ilagan F, Kuo AH, Hsieh RW, Cai S, et al. 2020. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367: 405–411. 10.1126/science.aax0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Pinello L, Han X, Lai S, Shen L, Lin TW, Zou K, Yuan GC, Orkin SH. 2016a. Serum-based culture conditions provoke gene expression variability in mouse embryonic stem cells as revealed by single-cell analysis. Cell Rep 14: 956–965. 10.1016/j.celrep.2015.12.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, von Meyenn F, Santos F, Chen Y, Reik W, Bertone P, Smith A, Nichols J. 2016b. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep 6: 437–446. 10.1016/j.stemcr.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HHD, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, et al. 2013. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13: 360–369. 10.1016/j.stem.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Hainer SJ, Fazzio TG. 2015. Regulation of nucleosome architecture and factor binding revealed by nuclease footprinting of the ESC genome. Cell Rep 13: 61–69. 10.1016/j.celrep.2015.08.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Gu W, Carone BR, Landry BD, Rando OJ, Mello CC, Fazzio TG. 2015. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev 29: 362–378. 10.1101/gad.253534.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Bošković A, McCannell KN, Rando OJ, Fazzio TG. 2019. Profiling of pluripotency factors in single cells and early embryos. Cell 177: 1319–1329.e11. 10.1016/j.cell.2019.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A, Meshorer E. 2015. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep 16: 1609–1619. 10.15252/embr.201541011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491. 10.1016/j.stem.2010.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E. 1928. Das heterochromatin der moose. I Jahrb Wiss Botanik 69: 762–818 (in German). [Google Scholar]
- Hezroni H, Tzchori I, Davidi A, Mattout A, Biran A, Nissim-Rafinia M, Westphal H, Meshorer E. 2011. H3K9 histone acetylation predicts pluripotency and reprogramming capacity of ES cells. Nucleus 2: 300–309. 10.4161/nucl.2.4.16767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Sasaki H. 2009. Dynamic transition of Dnmt3b expression in mouse pre- and early post-implantation embryos. Gene Expr Patterns 9: 27–30. 10.1016/j.gep.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. 2009. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci 106: 5187–5191. 10.1073/pnas.0812888106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough SR, Laslett AL, Grimmond SB, Kolle G, Pera MF. 2009. A continuum of cell states spans pluripotency and lineage commitment in human embryonic stem cells. PLoS ONE 4: e7708 10.1371/journal.pone.0007708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh THS, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. 2015. Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell 162: 108–119. 10.1016/j.cell.2015.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh THS, Cattoglio C, Slobodyanyuk E, Hansen AS, Rando OJ, Tjian R, Darzacq X. 2020. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol Cell 78: 539–553.e8. 10.1016/j.molcel.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai R, Nozaki T, Tani T, Kaizu K, Hibino K, Ide S, Tamura S, Takahashi K, Shribak M, Maeshima K. 2017. Density imaging of heterochromatin in live cells using orientation-independent-DIC microscopy. Mol Biol Cell 28: 3349–3359. 10.1091/mbc.e17-06-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iurlaro M, von Meyenn F, Reik W. 2017. DNA methylation homeostasis in human and mouse development. Curr Opin Genet Dev 43: 101–109. 10.1016/j.gde.2017.02.003 [DOI] [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. 2004. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol 24: 8862–8871. 10.1128/MCB.24.20.8862-8871.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SR, Pálfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, Shevchenko A, Vastenhouw NL. 2017. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. eLife 6: e23326 10.7554/eLife.23326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Wang S, Ko KD, Zare H, Tsai PF, Feng X, Vivanco KO, Ascoli AM, Gutierrez-Cruz G, Krebs J, et al. 2016. Roles of H3K27me2 and H3K27me3 examined during fate specification of embryonic stem cells. Cell Rep 17: 1369–1382. 10.1016/j.celrep.2016.09.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T, Smith A. 2014. Mapping the route from naive pluripotency to lineage specification. Philos Trans R Soc Lond B Biol Sci 369: 20130540 10.1098/rstb.2013.0540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Snoek J, Rinn JL. 2016. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res 26: 990–999. 10.1101/gr.200535.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, Greenleaf WJ. 2019. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20: 207–220. 10.1038/s41576-018-0089-8 [DOI] [PubMed] [Google Scholar]
- Ko YG, Nishino K, Hattori N, Arai Y, Tanaka S, Shiota K. 2005. Stage-by-stage change in DNA methylation status of Dnmt1 locus during mouse early development. J Biol Chem 280: 9627–9634. 10.1074/jbc.M413822200 [DOI] [PubMed] [Google Scholar]
- Kobayakawa S, Miike K, Nakao M, Abe K. 2007. Dynamic changes in the epigenomic state and nuclear organization of differentiating mouse embryonic stem cells. Genes Cells 12: 447–460. 10.1111/j.1365-2443.2007.01063.x [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kikyo N. 2015. Epigenetic regulation of open chromatin in pluripotent stem cells. Transl Res 165: 18–27. 10.1016/j.trsl.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh FM, Sachs M, Guzman-Ayala M, Ramalho-Santos M. 2010. Parallel gateways to pluripotency: open chromatin in stem cells and development. Curr Opin Genet Dev 20: 492–499. 10.1016/j.gde.2010.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Tsang JCH, Ilicic T, Henriksson J, Natarajan KN, Tuck AC, Gao X, Bühler M, Liu P, et al. 2015. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17: 471–485. 10.1016/j.stem.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184: 868–871. 10.1126/science.184.4139.868 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Klug A. 1981. The nucleosome. Sci Am 244: 52–64. 10.1038/scientificamerican0281-52 [DOI] [PubMed] [Google Scholar]
- Korolev N, Lyubartsev AP, Nordenskiöld L. 2018. A systematic analysis of nucleosome core particle and nucleosome-nucleosome stacking structure. Sci Rep 8: 1–14. 10.1038/s41598-018-19875-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietenstein N, Abraham S, Venev SV, Abdennur N, Gibcus J, Hsieh THS, Parsi KM, Yang L, Maehr R, Mirny LA, et al. 2020. Ultrastructural details of mammalian chromosome architecture. Mol Cell 78: 554–565.e7. 10.1016/j.molcel.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Rossant J. 2009. Stem cells in extraembryonic lineages. In Essentials of stem cell biology, 2nd ed., pp. 137–144. Academic, San Diego: 10.1016/B978-0-12-374729-7.00015-9 [DOI] [Google Scholar]
- Kuznetsova MA, Sheval EV. 2016. Chromatin fibers: from classical descriptions to modern interpretation. Cell Biol Int 40: 1140–1151. 10.1002/cbin.10672 [DOI] [PubMed] [Google Scholar]
- Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MSH, et al. 2008. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet 4: e1000241 10.1371/journal.pgen.1000241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau KX, Mason EA, Kie J, De Souza DP, Kloehn J, Tull D, McConville MJ, Keniry A, Beck T, Blewitt ME, et al. 2020. Unique properties of a subset of human pluripotent stem cells with high capacity for self-renewal. Nat Commun 11: 2420 10.1038/s41467-020-16214-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, et al. 2013. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol 20: 311–316. 10.1038/nsmb.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Izpisua Belmonte JC. 2018. Deconstructing the pluripotency gene regulatory network. Nat Cell Biol 20: 382–392. 10.1038/s41556-018-0067-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Chen J, Liu W, Lai W, Liu B, Li X, Liu L, Xu S, Dong Q, et al. 2018. Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature 564: 136–140. 10.1038/s41586-018-0751-5 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Lehmann LW, Bonora G, Sridharan R, Vashisht AA, Tran N, Plath K, Wohlschlegel JA, Carey M. 2011. Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev 25: 2198–2209. 10.1101/gad.17554711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhang Z, Wu H, Jiang Y, Meng L, Xiong J, Zhao Z, Zhou X, Li J, Li H, et al. 2015. Recognition of H3K9 methylation by GLP is required for efficient establishment of H3K9 methylation, rapid target gene repression, and mouse viability. Genes Dev 29: 379–393. 10.1101/gad.254425.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang C, Liu W, Li J, Li C, Kou X, Chen J, Zhao Y, Gao H, Wang H, et al. 2016. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537: 558–562. 10.1038/nature19362 [DOI] [PubMed] [Google Scholar]
- Livyatan I, Aaronson Y, Gokhman D, Ashkenazi R, Meshorer E. 2015. BindDB: an integrated database and webtool platform for “reverse-ChIP” epigenomic analysis. Cell Stem Cell 17: 647–648. 10.1016/j.stem.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Loh KM, Lim B. 2010. Recreating pluripotency? Cell Stem Cell 7: 137–139. 10.1016/j.stem.2010.07.005 [DOI] [PubMed] [Google Scholar]
- *.Maeshima K. 2020. The chromatin fiber. Cold Spring Harb Perspect Biol 10.1101/cshperpsect.a040675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Imai R, Tamura S, Nozaki T. 2014. Chromatin as dynamic 10-nm fibers. Chromosoma 123: 225–237. 10.1007/s00412-014-0460-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The polycomb complex PRC2 and its mark in life. Nature 469: 343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Francis Stewart A, Smith A, et al. 2012. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149: 590–604. 10.1016/j.cell.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638. 10.1073/pnas.78.12.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascetti VL, Pedersen RA. 2016. Contributions of mammalian chimeras to pluripotent stem cell research. Cell Stem Cell 19: 163–175. 10.1016/j.stem.2016.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S. 2005. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol 6: 44 10.1186/1471-2121-6-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770. 10.1038/nature07107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcer S, Meshorer E. 2010. The silence of the LADs: dynamic genome-lamina interactions during ESC differentiation. Cell Stem Cell 6: 495–497. 10.1016/j.stem.2010.05.006 [DOI] [PubMed] [Google Scholar]
- Melcer S, Hezroni H, Rand E, Nissim-Rafinia M, Skoultchi A, Stewart CL, Bustin M, Meshorer E. 2012. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nat Commun 3: 910 10.1038/ncomms1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. 2006. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 7: 540–546. 10.1038/nrm1938 [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10: 105–116. 10.1016/j.devcel.2005.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer T, von Meyenn F, Savino A, Santos F, Mohammed H, Lun ATL, Marioni JC, Reik W. 2019. Transcriptional heterogeneity in naive and primed human pluripotent stem cells at single-cell resolution. Cell Rep 26: 815–824.e4. 10.1016/j.celrep.2018.12.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642. 10.1016/S0092-8674(03)00393-3 [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D. 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766. 10.1016/j.molcel.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Morey C, Silva NRD, Perry P, Bickmore WA. 2007. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development 134: 909–919. 10.1242/dev.02779 [DOI] [PubMed] [Google Scholar]
- Morgan TH. 1898. Experimental studies of the regeneration of Planaria maculata. Archiv für Entwickelungsmechanik der Organismen 7: 364–397. 10.1007/BF02161491 [DOI] [Google Scholar]
- Morgani S, Nichols J, Hadjantonakis AK. 2017. The many faces of pluripotency: in vitro adaptations of a continuum of in vivo states. BMC Dev Biol 17: 7 10.1186/s12861-017-0150-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Best JB, Noel J. 1969. Electron microscopic studies of planarian regeneration. I: Fine structure of neoblasts in Dugesia dorotocephala. J Ultrastruct Res 27: 7–23. 10.1016/S0022-5320(69)90017-3 [DOI] [PubMed] [Google Scholar]
- Morozumi Y, Boussouar F, Tan M, Chaikuad A, Jamshidikia M, Colak G, He H, Nie L, Petosa C, de Dieuleveult M, et al. 2016. Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells. J Mol Cell Biol 8: 349–362. 10.1093/jmcb/mjv060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller WEG. 2001. Review: how was metazoan threshold crossed? The hypothetical Urmetazoa. Comp Biochem Physiol A Mol Integr Physiol 129: 433–460. 10.1016/S1095-6433(00)00360-3 [DOI] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gócza E, Diaz EM, Prideaux VR, Iványi E, Markkula M, Rossant J. 1990. Embryonic stem cells alone are able to support fetal development in the mouse. Development 110: 815–821. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64–71. 10.1038/ncb1519 [DOI] [PubMed] [Google Scholar]
- Nava MM, Miroshnikova YA, Biggs LC, Whitefield DB, Metge F, Boucas J, Vihinen H, Jokitalo E, Li X, García Arcos JM, et al. 2020. Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell 181: 800–817.e22. 10.1016/j.cell.2020.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. 2009. Naive and primed pluripotent states. Cell Stem Cell 4: 487–492. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- Novo CL, Tang C, Ahmed K, Djuric U, Fussner E, Mullin NP, Morgan NP, Hayre J, Sienerth AR, Elderkin S, et al. 2016. The pluripotency factor Nanog regulates pericentromeric heterochromatin organization in mouse embryonic stem cells. Genes Dev 30: 1101–1115. 10.1101/gad.275685.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka S, Dalton S. 2008. Molecular and biological properties of pluripotent embryonic stem cells. Gene Ther 15: 74–81. 10.1038/sj.gt.3303065 [DOI] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. 2011. Chromatin connections to pluripotency and cellular reprogramming. Cell 145: 835–850. 10.1016/j.cell.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O'Shea CC. 2017. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357: eaag0025 10.1126/science.aag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P, Gross-Bellard M, Chambon P. 1975. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell 4: 281–300. 10.1016/0092-8674(75)90149-X [DOI] [PubMed] [Google Scholar]
- Park SH, Park SH, Kook MC, Kim EY, Park S, Lim JH. 2004. Ultrastructure of human embryonic stem cells and spontaneous and retinoic acid-induced differentiating cells. Ultrastruct Pathol 28: 229–238. 10.1080/01913120490515595 [DOI] [PubMed] [Google Scholar]
- Pękowska A, Klaus B, Xiang W, Severino J, Daigle N, Klein FA, Oleś M, Casellas R, Ellenberg J, Steinmetz LM, et al. 2018. Gain of CTCF-anchored chromatin loops marks the exit from naive pluripotency. Cell Syst 7: 482–495.e10. 10.1016/j.cels.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Bulut-Karslioglu A, Ramalho-Santos M. 2017. Hypertranscription in development, stem cells, and regeneration. Dev Cell 40: 9–21. 10.1016/j.devcel.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusa B, Hadjantonakis AK. 2014. Embryonic stem cell identity grounded in the embryo. Nat Cell Biol 16: 502–504. 10.1038/ncb2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Dillon N. 2015. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 16: 245–257. 10.1038/nrm3965 [DOI] [PubMed] [Google Scholar]
- Qiao Y, Wang R, Yang X, Tang K, Jing N. 2015. Dual roles of histone H3 lysine 9 acetylation in human embryonic stem cell pluripotency and neural differentiation. J Biol Chem 290: 2508–2520. 10.1074/jbc.M114.603761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44: 361–372. 10.1016/j.molcel.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP. 2015. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160: 1145–1158. 10.1016/j.cell.2015.01.054 [DOI] [PubMed] [Google Scholar]
- Ricci MA, Cosma MP, Lakadamyali M. 2017. Super resolution imaging of chromatin in pluripotency, differentiation, and reprogramming. Curr Opin Genet Dev 46: 186–193. 10.1016/j.gde.2017.07.010 [DOI] [PubMed] [Google Scholar]
- Rink JC. 2013. Stem cell systems and regeneration in planaria. Dev Genes Evol 223: 67–84. 10.1007/s00427-012-0426-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. 2015. Mouse and human blastocyst-derived stem cells: vive les differences. Development 142: 9–12. 10.1242/dev.115451 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- Sado T, Sakaguchi T. 2013. Species-specific differences in X chromosome inactivation in mammals. Reproduction 146: R131–R139. 10.1530/REP-13-0173 [DOI] [PubMed] [Google Scholar]
- Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. 2009. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells 27: 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S, Meshorer E. 2019. Open chromatin, epigenetic plasticity, and nuclear organization in pluripotency. Dev Cell 48: 135–150. 10.1016/j.devcel.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Mifsud B, Senner CE, Todd CD, Chrysanthou S, Darbo E, Hemberger M, Branco MR. 2018. Divergent wiring of repressive and active chromatin interactions between mouse embryonic and trophoblast lineages. Nat Commun 9: 4189 10.1038/s41467-018-06666-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler HR. 1991. Octamania: the POU factors in murine development. Trends Genet 7: 323–329. 10.1016/0168-9525(91)90422-M [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Degnan B, Ruiz-Trillo I. 2017. The origin of Metazoa: a unicellular perspective. Nat Rev Genet 18: 498–512. 10.1038/nrg.2017.21 [DOI] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Cell Dev Biol 81: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrinivas K, Sabari BR, Coffey EL, Klein IA, Boija A, Zamudio AV, Schuijers J, Hannett NM, Sharp PA, Young RA, et al. 2019. Enhancer features that drive formation of transcriptional condensates. Mol Cell 75: 549–561.e7. 10.1016/j.molcel.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon CS, Downes DJ, Gosden ME, Telenius J, Higgs DR, Hughes JR, Costello I, Bikoff EK, Robertson EJ. 2017. Functional characterisation of cis-regulatory elements governing dynamic Eomes expression in the early mouse embryo. Development 144: 1249–1260. 10.1242/dev.147322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Schöler HR. 2010. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141: 943–955. 10.1016/j.cell.2010.04.037 [DOI] [PubMed] [Google Scholar]
- Smith A. 2017. Formative pluripotency: the executive phase in a developmental continuum. Development 144: 365–373. 10.1242/dev.142679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. 2013. DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220. 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. 1988. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336: 688–690. 10.1038/336688a0 [DOI] [PubMed] [Google Scholar]
- Sogabe S, Hatleberg WL, Kocot KM, Say TE, Stoupin D, Roper KE, Fernandez-Valverde SL, Degnan SM, Degnan BM. 2019. Pluripotency and the origin of animal multicellularity. Nature 570: 519–522. 10.1038/s41586-019-1290-4 [DOI] [PubMed] [Google Scholar]
- Stadhouders R, Vidal E, Serra F, Stefano BD, Dily FL, Quilez J, Gomez A, Collombet S, Berenguer C, Cuartero Y, et al. 2018. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet 50: 238–249. 10.1038/s41588-017-0030-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Kobayashi S, Hiratani I. 2018. Epigenetic differences between naïve and primed pluripotent stem cells. Cell Mol Life Sci 75: 1191–1203. 10.1007/s00018-017-2703-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Xue Y, Song C, Grunstein M. 2013. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc Natl Acad Sci 110: 11493–11498. 10.1073/pnas.1309914110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ. 2016. Snapshots of pluripotency. Stem Cell Rep 6: 163–167. 10.1016/j.stemcr.2015.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199. 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosolini M, Brochard V, Adenot P, Chebrout M, Grillo G, Navia V, Beaujean N, Francastel C, Bonnet-Garnier A, Jouneau A. 2018. Contrasting epigenetic states of heterochromatin in the different types of mouse pluripotent stem cells. Sci Rep 8: 1–14. 10.1038/s41598-018-23822-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. 2008. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135: 909–918. 10.1242/dev.017400 [DOI] [PubMed] [Google Scholar]
- Veillard AC, Marks H, Bernardo AS, Jouneau L, Laloë D, Boulanger L, Kaan A, Brochard V, Tosolini M, Pedersen R, et al. 2014. Stable methylation at promoters distinguishes epiblast stem cells from embryonic stem cells and the in vivo epiblasts. Stem Cells Dev 23: 2014–2029. 10.1089/scd.2013.0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Meyenn F, Iurlaro M, Habibi E, Liu NQ, Salehzadeh-Yazdi A, Santos F, Petrini E, Milagre I, Yu M, Xie Z, et al. 2016. Impairment of DNA methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol Cell 62: 848–861. 10.1016/j.molcel.2016.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444: 364–368. 10.1038/nature05284 [DOI] [PubMed] [Google Scholar]
- Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. 2013. Klf4 organizes long-range chromosomal interactions with the Oct4 locus in reprogramming and pluripotency. Cell Stem Cell 13: 36–47. 10.1016/j.stem.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Weinberger L, Ayyash M, Novershtern N, Hanna JH. 2016. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol 17: 155–169. 10.1038/nrm.2015.28 [DOI] [PubMed] [Google Scholar]
- Wiblin AE, Cui W, Clark AJ, Bickmore WA. 2005. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci 118: 3861–3868. 10.1242/jcs.02500 [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Ghosh RP. 2010. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol 2: a000596 10.1101/cshperspect.a000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lei L, Schöler HR. 2017. Totipotency in the mouse. J Mol Med 95: 687–694. 10.1007/s00109-017-1509-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang Y, Xu Q, Zhou C, Liu B, Du Z, Zhang K, Zhang B, Wang X, Gayen S, et al. 2020. Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency. Nat Genet 52: 95–105. 10.1038/s41588-019-0545-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, et al. 2009. Histone H3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 33: 417–427. 10.1016/j.molcel.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Carter AC, Gendrel AV, Attia M, Loftus J, Greenleaf WJ, Tibshirani R, Heard E, Chang HY. 2017. Landscape of monoallelic DNA accessibility in mouse embryonic stem cells and neural progenitor cells. Nat Genet 49: 377–386. 10.1038/ng.3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M, Yamanaka S, Yamada Y. 2017. Epigenetic foundations of pluripotent stem cells that recapitulate in vivo pluripotency. Lab Invest 97: 1133–1141. 10.1038/labinvest.2017.87 [DOI] [PubMed] [Google Scholar]
- Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, Piao Y, Aiba K, Matoba R, Wang W, Ko MSH. 2008. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells 26: 1155–1165. 10.1634/stemcells.2007-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Benvenisty N. 2019. Defining human pluripotency. Cell Stem Cell 25: 9–22. 10.1016/j.stem.2019.06.010 [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Peretz M, Aharony A, Sagi I, Benvenisty N. 2018. Defining essential genes for human pluripotent stem cells by CRISPR-Cas9 screening in haploid cells. Nat Cell Biol 20: 610–619. 10.1038/s41556-018-0088-1 [DOI] [PubMed] [Google Scholar]
- Ying QL, Smith A. 2017. The art of capturing pluripotency: creating the right culture. Stem Cell Rep 8: 1457–1464. 10.1016/j.stemcr.2017.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115: 281–292. 10.1016/S0092-8674(03)00847-X [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev 9: 2635–2645. 10.1101/gad.9.21.2635 [DOI] [PubMed] [Google Scholar]