Bacteria have evolved a variety of signal transduction mechanisms that generate different outputs in response to external stimuli. Chemosensory pathways are widespread in bacteria and are among the most complex signaling mechanisms, requiring the participation of at least six proteins.
KEYWORDS: signaling, Pseudomonas aeruginosa, chemosensory pathway, chemotaxis
SUMMARY
Bacteria have evolved a variety of signal transduction mechanisms that generate different outputs in response to external stimuli. Chemosensory pathways are widespread in bacteria and are among the most complex signaling mechanisms, requiring the participation of at least six proteins. These pathways mediate flagellar chemotaxis, in addition to controlling alternative functions such as second messenger levels or twitching motility. The human pathogen Pseudomonas aeruginosa has four different chemosensory pathways that carry out different functions and are stimulated by signal binding to 26 chemoreceptors. Recent research employing a diverse range of experimental approaches has advanced enormously our knowledge on these four pathways, establishing P. aeruginosa as a primary model organism in this field. In the first part of this article, we review data on the function and physiological relevance of chemosensory pathways as well as their involvement in virulence, whereas the different transcriptional and posttranscriptional regulatory mechanisms that govern pathway function are summarized in the second part. The information presented will be of help to advance the understanding of pathway function in other organisms.
INTRODUCTION
Chemosensory pathways represent a major mechanism in bacterial signal transduction and the corresponding genes have been identified in more than half of the sequenced bacterial genomes (1). The key element of a chemosensory pathway is the ternary complex between chemoreceptors, the CheA autokinase and the CheW coupling protein. Stimulus detection by the chemoreceptor initiates conformational changes that modulate CheA activity, in turn regulating the flux of phosphoryl groups to the CheY response regulator. The intracellular level of CheY-P controls the response output. In addition, the control of the chemoreceptor methylation state by the opposing activities of the CheR methyltransferase and the CheB methylesterase was identified as an essential mechanism to adapt pathway sensitivity to temporal changes in chemoeffector concentration (2, 3).
Much of what we know in this field is due to the study of Escherichia coli, a bacterium that has a single pathway, stimulated by five chemoreceptors that mediate chemotaxis (2–5). However, genome analyses of other bacteria revealed that chemoreceptor-based signaling is often more complex. First, the number of chemoreceptors is frequently higher than that in E. coli, reaching in some cases up to 80 (6). Second, apart from the core pathway signaling proteins mentioned above, there is a significant mechanistic diversity, as shown by additional signaling proteins present in only some pathways (1). Third, many bacteria possess more than one copy of chemosensory signaling genes, which is indicative for multiple pathways (1, 6, 7). Importantly, not all pathways mediate chemotaxis but can carry out other functions (8, 9). In fact, chemosensory pathways have been classified based on their evolutionary history into 19 different classes, 17 of which (classes F1 to F17) are associated with flagellar motility, whereas the remaining two classes are either associated with type four pilus-based motility (class TFP) or comprise systems with alternative cellular functions such as the control of second messenger levels (class ACF) (1).
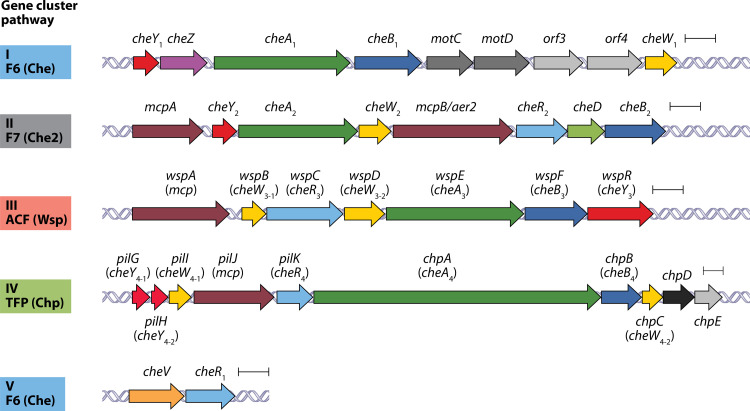
Pseudomonas aeruginosa PAO1 is the main reference strain for research on this opportunistic human pathogen. This strain possesses five gene clusters encoding proteins that assemble into four chemosensory pathways that each belong to a different class and carry out a distinct function (Fig. 1). Proteins encoded by gene clusters I and V form an F6 pathway (Che) for chemotaxis, cluster II encodes an F7-type pathway (Che2) of unknown function, cluster III encodes an ACF-type pathway (Wsp [wrinkly spreader phenotype]) that modulates bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) levels, whereas cluster IV (Chp [chemosensory pili]) is a member of the TFP class and is associated with twitching motility (Fig. 2).
FIG 1.
Chemosensory pathways of P. aeruginosa PAO1. A schematic representation shows the five gene clusters that encode chemosensory signaling proteins. Genes are annotated according to UniProt. Indicated are the gene clusters and the classification of chemosensory pathways as previously described (1). Che, chemotaxis; Wsp, wrinkly spreader phenotype; Chp, chemosensory pili. The F7 pathway is of unknown function. Scale bars, 0.5 kbp.
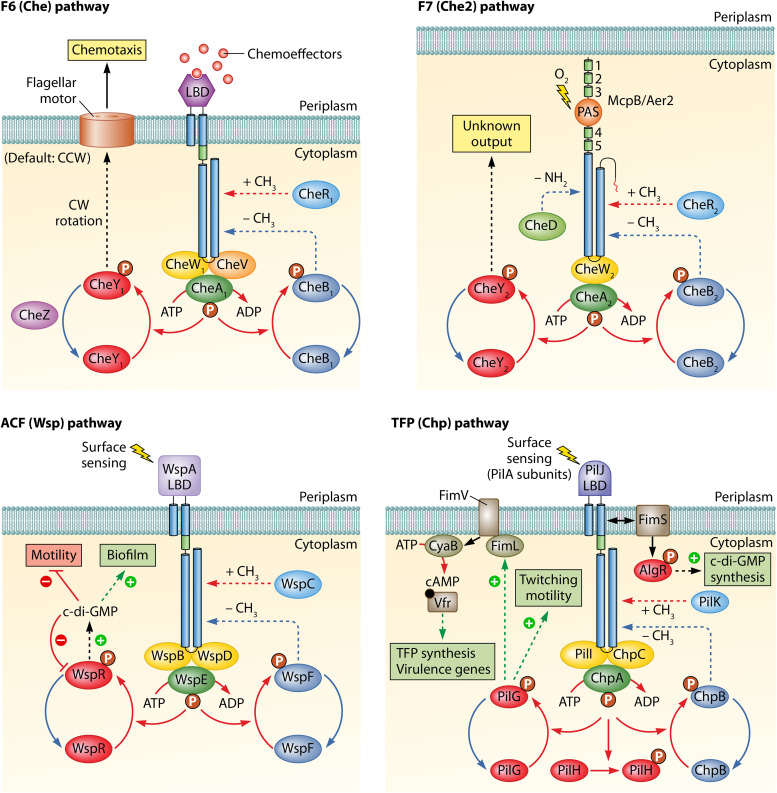
FIG 2.
The protein interaction network of the four chemosensory pathways of P. aeruginosa. The pathway output is highlighted by boxes. The color code of signaling proteins corresponds to that of Fig. 1. CheA/WspE/ChpA, histidine kinase; CheR/WspC/PilK, methyltransferase; CheB/WspF/ChpB, methylesterase; CheW/WspB/WspD/PilI/ChpC, CheW-type coupling protein; CheV, CheV-type coupling protein; CheY/WspR/PilG/PilH, CheY-type response regulator.
The input into these four pathways is mediated by the recognition of different stimuli at 26 chemoreceptors (Fig. 3 and Table 1). However, and as detailed in this review, P. aeruginosa also employs additional proteins that participate and modulate pathway function. Research conducted mainly over the last 10 years combining microbiological work with approaches in the fields of biophysics, bioinformatics, structural biology, cryo-electron tomography, omics, or single cell studies, among others, have enormously advanced our knowledge of these four pathways. Due to these advances, P. aeruginosa is now a central model organism to study the complexity of chemosensory pathway signaling. In this article, we review the current knowledge on the molecular mechanisms that control pathway function and their physiological relevance. We also review data on the regulatory mechanisms that govern pathway expression and function.
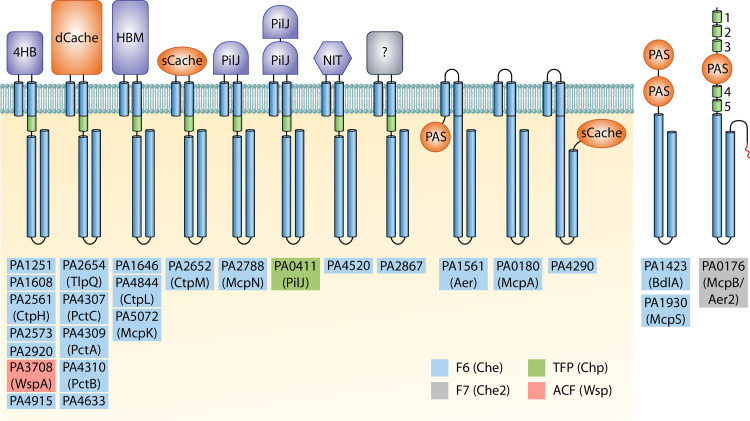
FIG 3.
Chemoreceptor repertoire of P. aeruginosa PAO1. Ligand-binding domains with parallel helix or α/β folds are shown in blue and orange, respectively. HAMP (histidine kinases, adenyl cyclases, methyl-accepting proteins, and phosphatases) and signaling domains are represented as green cylinders, whereas transmembrane regions and signaling domains are shown in blue. No HAMP domains were identified in Aer, BdlA, McpA, McpS, and PA4290. McpB/Aer2 is the only chemoreceptor that carries a C-terminal pentapeptide (in red) that acts as an additional CheR binding site (11). 4HB, four helix bundle; Cache, calcium channels and chemotaxis receptors; HBM, helical bimodular; PilJ, N-terminal domain of type IV pilus chemoreceptor; NIT, nitrate and nitrite sensing; PAS, Per-Arnt-Sim. Chemoreceptors are present in higher oligomeric states in vivo but are shown as monomers for simplicity. The assignment of P. aeruginosa PAO1 chemoreceptors to their respective chemosensory pathways was reported by Ortega et al. (10).
TABLE 1.
Chemoreceptors for which information is available on function and/or signal sensed
| Locus tag | Name | LBD type | Effector(s) (binding mode) | Function/comment | Reference(s) |
|---|---|---|---|---|---|
| PA0176 | Aer2/McpB | PAS | O2, NO, CO, cyanide (direct) | Stimulates the Che2 pathway; involved in virulence | 66, 67, 82, 88, 89 |
| PA0180 | CttP/McpA | none | Chloroethylenes (unknown) | Chemotaxis | 76 |
| PA0411 | PilJ | 2×PilJ | Phosphatidylethanolamine?, PilA? | Stimulates the Chp pathway | 129 |
| PA1423 | BdlA | 2×PAS | Unknown | Involved in biofilm dispersion | 70–72 |
| PA1561 | Aer/TlpC | PAS | O2 (unknown) | Aerotaxis | 66, 67 |
| PA1930 | McpS | 2×PAS | Unknown | Modulates chemotaxis and chemoreceptor clustering | 69 |
| PA2561 | CtpH | 4HB | Inorganic phosphate (direct) | Chemotaxis | 45 |
| PA2573 | - | 4HB | Unknown | Involved in virulence | 77 |
| PA2652 | CtpM | sCache | Malate, citramalate, methylsuccinate, bromosuccinate, citraconate (direct) | Chemotaxis | 63, 64 |
| PA2654 | TlpQ | dCache | Histamine, putrescine, cadaverine, spermidine, agmatine, ethylenediamine (direct), ethylene (unknown) | Chemotaxis | 30, 39 |
| PA2788 | McpN | PilJ | Nitrate (direct) | Chemotaxis | 46 |
| PA3708 | WspA | 4HB | Growth on solid surfaces (unknown), ethanol (unknown) | Stimulates the Wsp pathway | 15, 101 |
| PA4307 | PctC | dCache | γ-Aminobutyrate, histidine, proline (direct), histamine (unknown) | Chemotaxis | 33–36, 38, 39, 178 |
| PA4309 | PctA | dCache | Seventeen amino acids (direct), histamine (unknown), chloroethylenes, chloroform (unknown) | Chemotaxis | 33–36, 38, 39 |
| PA4310 | PctB | dCache | Five amino acids (direct) | Chemotaxis | 33–36, 38 |
| PA4844 | CtpL | HBM | Inorganic phosphate (indirect, via PstS), chloroaniline, catechol (unknown) | Chemotaxis | 45, 52 |
| PA5072 | McpK | HBM | α-Ketoglutarate (direct) | Chemotaxis | 60 |
ASSIGNING CHEMORECEPTORS TO CHEMOSENSORY PATHWAYS
In strain PAO1, only four of the chemoreceptor genes (mcpA, mcpB/aer2, wspA, and pilJ) are clustered together with signaling genes (Fig. 1), whereas the remaining chemoreceptor genes are scattered over the genome. A central question in the study of complex chemosensory systems is to identify which chemoreceptor feeds into which pathway. This aspect involves analyzing whether one particular receptor is specific for a given pathway or whether it can stimulate various pathways. A bioinformatic study has advanced our understanding of this issue (10). In this work, clusters of orthologous groups (COGs) of chemoreceptors were generated from completed Pseudomonadales genomes and sequence conservation of the CheA/CheW binding sites in chemoreceptor COGs was investigated. This analysis resulted in a single pattern for 23 chemoreceptors, whereas a different profile was obtained for each of McpB/Aer2, WspA, and PilJ—three chemoreceptors encoded within gene clusters II, III, and IV, respectively (Fig. 1). Based on the observation that CheR methyltransferases appear to be pathway specific (11, 12), the authors investigated the conservation of chemoreceptor methylation sites, and the results agreed with the sequence analysis of the CheA/CheW binding sites (10). This study resulted in a model in which the F7, Wsp, and Chp pathways are each stimulated by a single chemoreceptor, McpB/Aer2, WspA, and PilJ, respectively, which correspond to the chemoreceptors encoded in the gene cluster of their cognate pathway (Fig. 1 and 3). The remaining 23 chemoreceptors (22 encoded by genes scattered over the genome and mcpA as part of cluster II) are predicted to feed into the F6 pathway that mediates chemotaxis. This model agrees with a large amount of experimental evidence (11, 13–19) and suggests that the primary physiological role of chemosensory signaling in P. aeruginosa is chemotaxis.
DIVERSITY OF P. AERUGINOSA CHEMORECEPTORS
The prototypal chemoreceptor is composed of a conserved cytosolic signaling domain and an extracytoplasmic ligand-binding domain (LBD) that is flanked by two transmembrane regions (4). Typically, chemoreceptors are activated by the direct binding of signal molecules or signal-loaded ligand-binding proteins to the LBD. Genome analyses revealed that bacterial chemoreceptors employ more than 80 different types of LBDs and the topological diversity includes membrane-bound receptors with cytosolic LBDs, receptors with a C-terminal LBDs, or cytosolic receptors (20). As illustrated in Fig. 3, this diversity is well reflected in the 26 P. aeruginosa PAO1 chemoreceptors. The majority of chemoreceptors are transmembrane receptors with a periplasmic LBD, whereas others possess a cytosolic sensor domain. Conversely, the transmembrane receptor McpA does not have an LBD, and three other receptors (BdlA, McpS, and McpB/Aer2) lack transmembrane regions and are likely to be soluble receptors present in the cytosol (Fig. 3). From the structural point of view, LBDs can be classified into domains composed of parallel helices (e.g., 4HB [four-helix bundle], HBM [helical bimodular], PilJ [N-terminal domain of type IV pilus chemoreceptor], and NIT [nitrate and nitrite sensing]) or domains that show an α/β-fold (e.g., sCache, dCache, and PAS) (Fig. 4). Almost half of the PAO1 chemoreceptors possess 4HB and dCache LBDs that are the most abundant sensor domains in bacterial chemoreceptors (20, 21). Knowledge of the function and signal(s) recognized by P. aeruginosa chemoreceptors is summarized in Table 1.
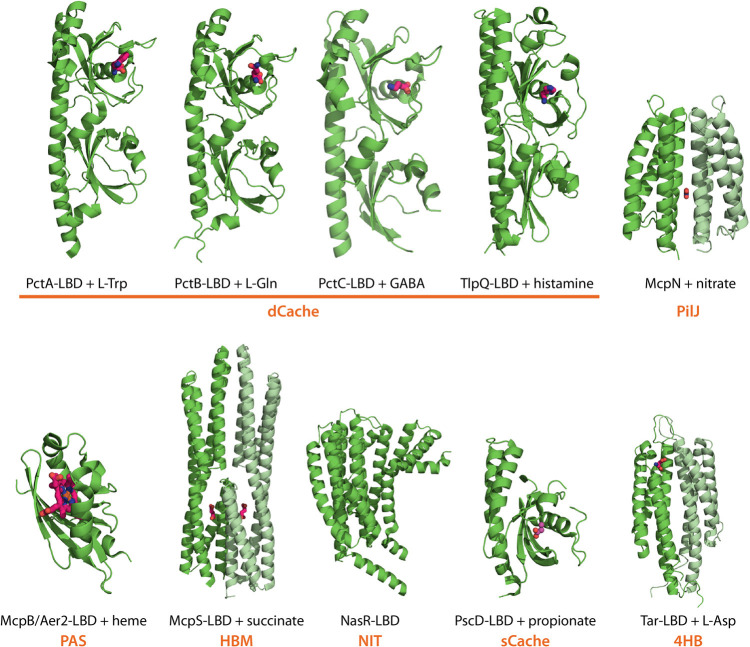
FIG 4.
Diversity of P. aeruginosa chemoreceptor LBDs. 3D structures of LBDs from PctA (PDB ID 5T7M), PctB (5LTO), PctC (5LTV) (38), TlpQ (6FU4) (39), McpN (6GCV) (46), and McpB/Aer2 (4HI4) (88) are shown (all P. aeruginosa). For the remaining protein families (Fig. 3), the structures of homologous domains from other species are shown, namely, P. putida McpS (HBM domain, 2YFB) (59), Klebsiella oxytoca NasR (NIT domain, 4AKK) (196), P. syringae PscD (sCache domain, 5G4Z) (197), and Salmonella enterica serovar Typhimurium Tar (4HB domain, 2LIG) (198). Bound ligands are shown in stick mode, and the LBD type is shown in orange. The monomers of LBD dimers are shown in different shades of green.
SIGNALING THROUGH THE F6 (Che) PATHWAY
Mode of P. aeruginosa Chemotaxis
Chemotaxis in P. aeruginosa is mediated by an F6 pathway, which contrasts with the chemosensory pathway of E. coli that belongs to class F7 (1). The origin of this divergence is discussed further below. In addition, the mechanism of chemotaxis is also different in both organisms. E. coli has several flagella, and chemotaxis is based on the ligand-mediated alteration of the ratio between clockwise (CW) and counterclockwise (CCW) flagellar rotation, causing cell tumbling and running, respectively, which enable bacterial reorientation in chemical gradients (3). In contrast, P. aeruginosa has a single flagellum, and CW rotation does not induce tumbling but straight backward movements, corresponding to a change in swimming direction by 180° (22). This “run-reverse-turn” mechanism of P. aeruginosa is thus different to the “run-and-tumble” mode of E. coli (23). In addition, a pause phase has been observed in the P. aeruginosa motor. Since pause duration was shown to correlate with turn angle sizes, it was concluded that a central element in mediating a chemotactic response in P. aeruginosa is the modulation of pause frequency and duration. In contrast to E. coli, the CW/CCW bias in the absence and presence of a chemoeffector stays constant in P. aeruginosa. As a result, chemotaxis is achieved by increasing the durations of both CW and CCW rotations when swimming up a chemoattractant gradient and decreasing rotation durations when swimming down the gradient, regardless of whether such movement is a forward or backward run (22).
Molecular Mechanisms and Chemoreceptors for Chemotaxis
Early studies reported that P. aeruginosa is attracted by different amino acids, organic acids, aromatic compounds, sugars, oligopeptides, phytohormones, or inorganic compounds (24–31) and repelled by other compounds, including chloroform and thiocyanic esters (32, 33). More recent work has brought insight into the molecular mechanisms involved and has led to the identification of many of the corresponding chemoreceptors (Table 1).
Amino acids.
P. aeruginosa is attracted to all proteinogenic amino acids by the concerted action of three paralogous chemoreceptors, PctA, PctB, and PctC (34, 35), and chemoreceptor activation occurs by direct ligand binding to the dCache-type LBDs of these three receptors (36) (Fig. 4). Whereas PctA is a broad ligand range receptor that responds to the majority of proteinogenic amino acids, PctB and PctC have a ligand preference for l-Gln and γ-aminobutyrate (GABA), respectively (35, 36). Chimeric receptors generated by replacing the E. coli Tar-LBD with either the PctA- or PctB-LBD were introduced into E. coli, and fluorescence resonance energy transfer (FRET) experiments were conducted to determine the 50% effective concentration (EC50; i.e., chemoeffector concentration at which signal response is half-maximal) for each of the ligands recognized. Interestingly, for both proteins the signal input, represented by the dissociation constant (KD) of the ligands for the purified PctA/PctB-LBDs, was found to correlate with the EC50 values representing the signaling output (37), indicating that ligand affinity determines the onset of chemotaxis. In a more recent study, the evolutionary history of the three Pct receptors was established. A sequence alignment of Pct homologs showed a significant sequence diversion between the LBDs, whereas the signaling domains were almost identical, suggesting a particularly rapid evolution of the LBDs (38). Using phylogenetic profiling and protein sequence analyses, it was shown that pctC and pctB originated through two independent pctA gene duplications from a common ancestor of P. aeruginosa (38). Many bacteria possess paralogous chemoreceptors (38) that may have similar and overlapping ligand profiles, but further studies are necessary to verify whether the evolution of narrow ligand range receptors from broad range chemoreceptors is a more general mechanism. The three-dimensional (3D) structures of the three Pct LBDs in complex with amino acids (Fig. 4) revealed changes at multiple positions in the ligand binding sites, suggesting that the evolution of chemoreceptors with novel ligand profiles is highly complicated (38).
PctA and PctC also appear to mediate chemotaxis to compounds other than amino acids. The complementation of a P. aeruginosa mutant deficient in histamine chemotaxis with plasmids harboring either pctA or pctC conferred histamine chemotaxis (see below) (39). However, binding studies did not provide any evidence for direct histamine binding, suggesting that these receptors may be stimulated instead by binding a histamine-loaded binding protein (39). The notion that PctA, PctB, and PctC mediate responses to multiple chemoeffectors is also consistent with the observation that mutation of their corresponding genes led to abolished repellent responses to trichloroethylenes and chloroform (33).
Several studies suggest that the Pct receptors are related to virulence. Whereas PctA and PctC show a wide phylogenetic distribution among pseudomonads, PctB is exclusively found in P. aeruginosa (38). It was hypothesized that PctB-mediated glutamine chemotaxis, the most abundant amino acid in human plasma (40), may be related to virulence and the birth of this chemoreceptor might have been especially beneficial for efficient host colonization by P. aeruginosa (38). Another study reported that a triple mutant ΔpctABC was less efficient in immobilizing along wounds of human cystic fibrosis (CF) airway epithelial cells (41), thus revealing the potential importance of amino acid sensing chemoreceptors for host infection. PctA and PctB protein levels were downregulated in P. aeruginosa isolated from the sputum of CF patients, suggesting that these receptors are important during the initial stages of infection (42).
Inorganic phosphate.
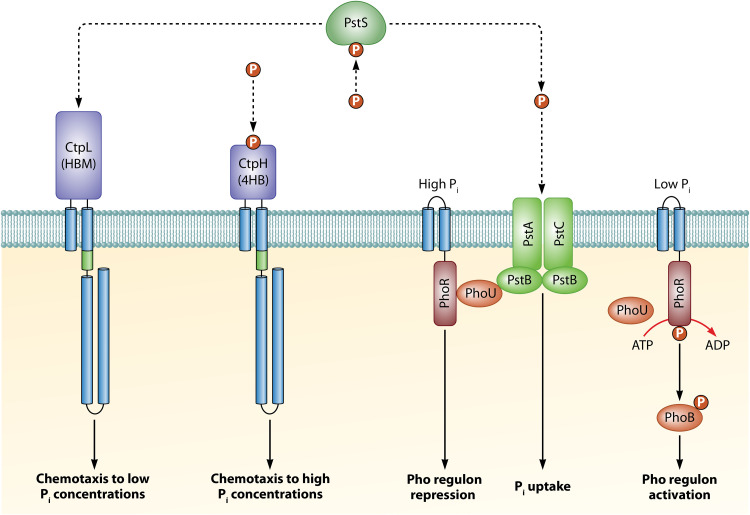
Inorganic phosphate (Pi) is a central signal molecule controlling P. aeruginosa virulence, and the reduction in the Pi concentration increased the transcript levels of many virulence factor genes (43). In addition, Pi starvation was found to shift P. aeruginosa toward the expression of a lethal phenotype against Caenorhabditis elegans (44). Remarkably, P. aeruginosa exhibits strong chemoattraction to Pi and two receptors, CtpL and CtpH, mediate this response (45). Whereas CtpL mediated responses to low Pi concentrations, CtpH responded to high Pi concentrations (45). Interestingly, Pi chemotaxis was only observed under Pi-limiting conditions, which is due to the fact that Pi downregulates the expression of both ctpL and ctpH (43, 45). In this respect, close parallels exist to the mechanism of nitrate chemotaxis (see below), a behavior that was only observed under nitrate limiting conditions since nitrate was shown to reduce transcript levels of mcpN encoding the nitrate chemoreceptor (46).
CtpH and CtpL differ in structure and function. Whereas CtpH has a 4HB-type LBD, CtpL has an HBM LBD, a domain that is composed of two structural modules and significantly larger than the 4HB domain (Fig. 4) (47). Significantly, only CtpH recognizes Pi directly, whereas CtpL is stimulated by the binding of the Pi-loaded ligand binding protein PstS (48). This periplasmic protein forms part of the Pi uptake system and binds to the PstABC transporter providing the substrate to be transported (49). Further research indicated that PstS binding to the PstABC transporter generates a molecular stimulus that is transmitted to the PhoR/PhoB two-component system causing transcriptional control in response to Pi (49). This pivotal role of PstS is further illustrated by the fact that it is the most abundant protein in P. aeruginosa when grown under Pi-limiting conditions (48, 50) and that pstS transcript levels showed very large increases when exposed to Pi starvation (43). PstS thus serves as a model illustrating the coordination of chemotaxis, transport, and transcriptional regulation by a periplasmic ligand-binding protein (Fig. 5).
FIG 5.
Model of the interwoven signaling processes that mediate phosphate chemotaxis, transport, and transcriptional regulation. Chemoreceptors and PhoR form oligomeric assemblies in vivo but are shown as monomers for simplicity. HAMP domains are shown as green cylinders. (Based on data from references 48 and 49.)
For the large majority of so far characterized chemoreceptors, a direct signal binding mechanism has been established (20). CtpL is currently the only P. aeruginosa chemoreceptor that has been shown to be stimulated by a ligand-binding protein. However, indirect stimulation mechanisms may be much more abundant as the scarceness of experimental data may rather be due to the technical complexity of identifying indirect binding. An import issue to take into account for experiments to identify indirect binding is the frequently strict control of ligand-binding protein expression. For example, the pull-down experiments leading to the identification of PstS as a CtpL ligand were only successful with protein extracts of P. aeruginosa grown under severe Pi limitation (48). The notion that indirect chemoreceptor stimulation is more frequent than what current experimental data would indicate is also consistent with the high abundance of periplasmic ligand binding proteins, namely, 98 in the case of P. aeruginosa (51). It is thus likely that the model established for Pi responses also applies to other signals such as histamine, as discussed below. On the other hand, by analogy to the PctABC chemoreceptors, CtpL also carries out an alternative function since a ctpL mutant failed to respond chemotactically to 4-chloroaniline and catechol (52).
Nitrate.
P. aeruginosa, when grown under nitrate-limiting conditions, shows strong chemotaxis to nitrate, a molecule that serves as a nitrogen source for growth and supports anaerobic respiration. P. aeruginosa has a single chemoreceptor, PA4520, with an NIT-type LBD (Fig. 3 and 4) (20), a domain family that was predicted to bind nitrate (53). However, PA4520 does not bind nitrate, nor does the deletion of its gene alter nitrate chemotaxis (46). Instead, nitrate chemotaxis is mediated by the McpN chemoreceptor that binds specifically nitrate at its periplasmic PilJ-type LBD. The 3D structure of the McpN-LBD (Fig. 4) can be closely superimposed onto that of Tar-LBD that belongs to a different protein family (4HB). Although the structure of both domains is conserved, they employ a different ligand-binding mode: whereas Tar-LBD binds two molecules of aspartate at two binding sites in the dimer interface (54), McpN-LBD recognizes a single nitrate molecule at a single site at the dimer symmetry axis (Fig. 4) (46). The LBD of the McpN chemoreceptor is very similar to that of the nitrate responsive sensor kinases NarX and NarQ (55), indicating that the mechanism of nitrate sensing in chemoreceptors and sensor kinases is similar.
Histamine and polyamines.
P. aeruginosa is attracted by the plant hormone ethylene. This response was abolished in a mutant of the TlpQ chemoreceptor (30), but binding studies with the purified TlpQ-LBD did not provide any evidence for direct ethylene recognition (39). Instead, high-throughput ligand screening approaches revealed that the TlpQ-LBD recognizes specifically five polyamines and histamine (39). Remarkably, TlpQ shows very high ligand-binding affinity with a KD value of 56 nM for spermidine (39) corresponding to the highest affinity ever measured for a chemoreceptor ligand. Three other ligands—putrescine, cadaverine, and agmatine—showed affinities between 134 and 150 nM, whereas histamine and ethylenediamine had KD values of 640 and 1,700 nM, respectively. Due to these high binding affinities, significant chemotactic responses were noted at low ligand concentrations such as 500 nM histamine (39). Dissociation constants in the nanomolar range are frequently observed for dCache domains (56), and it appears plausible that the capacity to sense multiple ligands with high affinity may be a cause for the high abundance of dCache domains in bacterial signal transduction systems (21). In contrast, responses to high histamine concentrations, such as 50 mM, were mediated by the dCache-containing receptors PctA and PctC. However, histamine did not bind to the LBDs of either receptor, indicating that binding may occur in an indirect manner involving histamine-binding proteins (39). Histamine is a central signal molecule in animals that coordinates local immune responses and controls the activity of immune cells. P. aeruginosa PAO1 infection was found to greatly increase neutrophil histamine content and secretion (57), and bacterial migration to infection sites increases cell density that, in turn, may modulate quorum- sensing-mediated gene expression (39).
Organic acids.
Organic acids are preferred C sources for P. aeruginosa (58) and, so far, two receptors have been identified that mediate taxis to these compounds. McpS of P. putida KT2440 was the first receptor identified with an HBM-type LBD and responded to a number of different Krebs cycle intermediates (59). Ligand screening of its homolog in P. aeruginosa, McpK, revealed that it binds specifically α-ketoglutarate (60). Interestingly, McpK-LBD bound its ligand with positive cooperativity, in contrast to the well-established sensing mechanism of Tar that binds aspartate with very strong negative cooperativity (61, 62).
Initial studies at the Harwood laboratory showed that deletion of the gene encoding the chemoreceptor CtpM resulted in a loss of malate chemotaxis (63). Subsequent biochemical studies showed that malate is recognized directly by the sCache-type LBD of CtpM (64). High-throughput screening experiments led to the identification of four additional ligands that are similar to malate, namely, citramalic, bromosuccinic, methylsuccinic, and citraconic acids (64). Surprisingly, whereas the first two compounds triggered a chemotactic response, the latter two compounds failed to do so. Chemotaxis competition assays revealed that methylsuccinic and citraconic acids reduced malate chemotaxis in a dose-dependent manner (64). These compounds were thus termed antagonists since, although they bind to the same site at the CtpM-LBD (64), they fail to trigger downstream signaling. Similar observations have been made for the Tar chemoreceptor (65), and the existence of signal antagonists offers the possibility to specifically interfere with chemotactic signaling.
Other chemoreceptors predicted to stimulate the F6 pathway.
(i) Aer (TlpC).
This receptor shows the same topology and domain arrangement as E. coli Aer (Fig. 3) and was found to mediate aerotaxis (66, 67). P. aeruginosa strains adapt their gene expression to hypoxia conditions during pulmonary infections (68), and Aer protein levels were downregulated in strains isolated from CF patients compared to the laboratory strain PAO1 (42).
(ii) McpS and BdlA.
McpS is a cytosolic receptor containing two tandem PAS-type LBDs (Fig. 3). Its overexpression resulted in a dominant-negative effect on chemotaxis and caused a loss of polar chemoreceptor clustering (69). The chemoreceptor BdlA (biofilm dispersion locus A) has the same domain arrangement as McpS, and initial experiments showed that a bdlA mutant was deficient in biofilm dispersion, a phenotype that is most likely caused by the increased c-di-GMP levels of the mutant (70). The N-terminal PAS domain of BdlA was found to contain bound heme, suggesting gas sensing, and removal of this domain abolished the capacity to disperse biofilm (71). Data were presented suggesting that BdlA may employ an unorthodox signaling mechanism. The full-length protein is inactive, but phosphorylation of a tyrosine residue at the segment which links both PAS domains with the signaling domain was identified as a signal required for proteolytic cleavage of the N-terminal PAS domain, leading to protein activation (72). Furthermore, the cleaved N-terminal PAS domain was found to be required for correct signaling (72). Cytosolic receptors with double PAS domains are relatively frequent (73), but no evidence has been obtained so far to suggest that other receptors may employ the BdlA mechanism. The inactivation of bdlA, resulting in the inability to disperse biofilms, correlated with a reduced pathogenicity in acute virulence models but, on the other hand, rendered P. aeruginosa more resistant upon chronic infection of the murine lung (74). BdlA has been predicted to feed into the F6 pathway (chemotaxis) (10) but was shown to modulate c-di-GMP levels (70). A study of Comamonas testosteroni revealed cross talk between chemosensory pathways mediating chemotaxis and biofilm formation (75), which may also occur in this case. The signals recognized by BdlA and McpS are unknown, but BdlA-dependent biofilm dispersion was induced by specific nutrients in the medium (70, 71).
(iii) McpA.
This atypical receptor lacks an LBD (Fig. 3) and was found to mediate chemoattraction to trichloroethylene (76). Although the mcpA gene forms part of the gene cluster encoding the F7 pathway (Fig. 1), the receptor was proposed to feed into the F6 pathway (10), a notion that is supported by McpA colocalization with F6 pathway signaling complexes (14). Furthermore, transcriptomic studies revealed that the expression patterns of mcpB/aer2 and cluster II genes (Fig. 1) were generally similar but different than those of mcpA (Tables 2 to 4), indicating that the latter gene is not subject to the regulatory processes controlling the F7 pathway.
TABLE 2.
Transcriptional regulation of chemoreceptor and pathways genes in P. aeruginosa by different regulatory systems
| Gene cluster/ORF | Fold change(s)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Transcriptional regulators (reference) |
Two-component systems (reference) |
Sigma factors (reference[s]) |
|||||||
| WT/ΔampRb (182) | ΔbswR/WTc (183) | ΔfleQ/WTd (156) | ΔpilR/WTe (181) | ΔfleR/WTf (156) | ΔfliA/WT (156, 184) | ΔrpoS/WT (184, 185) | ΔrpoN/WT (156, 184) | ΔsigX/WT (184) | |
| Gene cluster I (F6)g | –2.3/–3.0* | 3.0/4.2* | –2.3/–2.9* | ||||||
| Gene cluster II (F7)g | –5.2/–73.9* | –5.5/–28.4*† | –2.5/–2.8* | ||||||
| Gene cluster III (Wsp)g | –1.8/–3.3*† | –2.4/–2.7* | |||||||
| Gene cluster IV (Chp)g | –1.9/–2.4* | –2.7/–2.8* | |||||||
| Gene cluster V (F6) | –2.7 | –2.3/–3.3* | –3.1 | –2.5 | |||||
| PA0176 (McpB) | –5.2 | –8.8/–15.6† | |||||||
| PA0180 (McpA) | –5.5/–8.6† | ||||||||
| PA0411 (PilJ) | –2.7 | ||||||||
| PA1251 | 2.3 | 28.7 | |||||||
| PA1423 (BdlA) | –6.5 | –55.1 | |||||||
| PA1561 (Aer) | –2.8 | –2.9 | –8.8 | –4.4 | |||||
| PA1608 | –2.3 | –14.2 | –3.3 | ||||||
| PA1646 | 2.0 | –4.2 | 5.4 | ||||||
| PA1930 (McpS) | 3.9 | –16.3/–20.5† | –8.6 | ||||||
| PA2561 (CtpH) | –2.0 | –4.2 | |||||||
| PA2573 | –8.4/–12.1† | ||||||||
| PA2652 (CtpM) | –2.7 | –2.3 | –10.7 | ||||||
| PA2654 (TlpQ) | –4.3 | –3.7 | –57.9 | ||||||
| PA2788 (McpN) | –12.4 | –6.8 | –21.3 | ||||||
| PA2867 | –3.4 | –4.1 | –3.4 | –5.7 | –14.4 | –2.1 | |||
| PA2920 | –2.6 | ||||||||
| PA3708 (WspA) | –2.3 | –2.3 | |||||||
| PA4290 | 3.0 | ||||||||
| PA4307 (PctC) | –12.0 | –4.2 | –6.8/–15.0† | –6.5 | –4.5 | ||||
| PA4309 (PctA) | –2.1 | –2.6 | –8.4 | –20.5 | |||||
| PA4310 (PctB) | –80.6 | –3.2 | –32.5 | –21.1/–83.7† | –45.7 | –3.9 | |||
| PA4520 | –2.8 | –4.0 | |||||||
| PA4633 | –2.9 | –4.5/–6.0† | –11.8 | ||||||
| PA4844 (CtpL) | |||||||||
| PA4915 | –12.7 | –6.6/–11.0† | |||||||
| PA5072 (McpK) | –5.9 | ||||||||
*, Both the lowest and the highest fold changes among the differentially expressed genes per cluster are listed; †, values for two different studies are listed. WT, wild type.
Signal: cell wall muropeptides (179).
Signal: unknown.
Signal: PilA (181). Current data support that PilA acts as an intramembrane inhibitory signal through its interactions with the sensor kinase PilS.
Signal: unknown.
At least two genes per gene cluster have to be altered for listing.
TABLE 3.
Transcriptional regulation of chemoreceptor and pathways genes in P. aeruginosa by different signal molecules
| Gene cluster/ORF | Fold change(s) |
|||||
|---|---|---|---|---|---|---|
| Signal molecules and second messengers (reference[s]) |
Quorum-sensing signaling (reference) |
|||||
| Pi |
c-di-GMP |
cAMP |
ppGpp |
Acyl-homoserine lactones |
||
| WT (0.2 mM Pi)/ WT (1 mM Pi) (43) |
ΔwspF pelA pslBCDa/ ΔpelA pslBCD ΔwspFa/WT (8, 111) |
ΔcyaABb/WT (186) | ΔrelA ΔspoTc/WT (158) | WT/ΔlasR ΔrhlR (187) | ΔlasI ΔrhlI + AHLd/ ΔlasI ΔrhlI (188) |
|
| Gene cluster I (F6)e | 1.6/2.2f | |||||
| Gene cluster II (F7)e | 4.0/15.4f | 1.5/2.7f | 3.7/4.6f | 1.9g | ||
| Gene cluster III (Wsp)e | ||||||
| Gene cluster IV (Chp)e | –1.6/–1.9f | –2.6/–4.6f | 1.6/3.3f | 1.5/1.7f | ||
| Gene cluster V (F6) | 2.4/2.8f | |||||
| PA0176 (McpB) | 15.4 | 3.9 | ||||
| PA0180 (McpA) | ||||||
| PA0411 (PilJ) | 3.1 | 3.3 | 1.5 | |||
| PA1251 | ||||||
| PA1423 (BdlA) | 2.7 | 2.1 | 5.2 | |||
| PA1561 (Aer) | 2.2 | 2.7 | ||||
| PA1608 | 2.0 | 4.2 | 2.2 | |||
| PA1646 | 4.7 | |||||
| PA1930 (McpS) | 3.0 | 3.9 | 3.8 | |||
| PA2561 (CtpH) | 2.8 | |||||
| PA2573 | 5.0 | 3.9 | 2.7 | |||
| PA2652 (CtpM) | 2.1 | |||||
| PA2654 (TlpQ) | 2.4 | 2.8 | ||||
| PA2788 (McpN) | 2.2 | |||||
| PA2867 | 2.5 | 2.3 | 1.6 | |||
| PA2920 | 2.2 | 3.7 | ||||
| PA3708 (WspA) | ||||||
| PA4290 | 3.0 | |||||
| PA4307 (PctC) | 6.2/7.2h | 3.5 | 2.4 | |||
| PA4309 (PctA) | 2.1 | 1.6 | ||||
| PA4310 (PctB) | 1.7 | |||||
| PA4520 | 2.6 | 2.1 | ||||
| PA4633 | 2.3 | 3.5 | ||||
| PA4844 (CtpL) | 7.1 | |||||
| PA4915 | 2.1 | 2.2 | ||||
| PA5072 (McpK) | 2.2 | |||||
Increased c-di-GMP levels.
Reduced cAMP levels.
ppGpp-deficient mutant.
1 μM 3OC12-HSL and 2 μM C4-HSL were added.
At least two genes per gene cluster have to be altered for listing.
The lowest and highest fold changes among differentially expressed genes per cluster are listed.
A single fold change value was given for the entire F7 gene cluster.
Values for two different studies are listed.
TABLE 4.
Transcriptional regulation of chemoreceptor and pathways genes in P. aeruginosa during biofilm formation and infection
| Gene cluster/ORF | Fold change(s)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| Biofilm formation on abiotic surfaces (reference[s]) |
Virulence/in vivo infection (reference) |
|||||||
| Biofilm/planktonic (159, 189) | Planktonic/dispersed cells (159) | Anaerobic biofilm/planktonic (190) | Human sputum/in vitrob (191) | Human sputum/in vitrob (161) | Human burn wound infection/in vitro (191) | Mice lung infection/in vitro (192) | Human urine/in vitro (193) | |
| Gene cluster I (F6)c | –3.3/–3.7* | –3.0/–6.3* | –2.5/–4.1* | –2.2/–6.5* | ||||
| Gene cluster II (F7)c | –3.5/–12.5* | 4.3/13.1* | –3.9/–42.2* | –3.0/–25.0* | –4.6/–32.2* | –4.4/–21.4* | –5.7/–15.9* | –2.7/–4.5* |
| Gene cluster III (Wsp)c | 3.0/4.3* | –3.0/–5.4* | –2.3/–2.6* | |||||
| Gene cluster IV (Chp)c | –2.9/–3.0* | –2.8/–4.4* | –3.13/–12.5* | –2.5/–4.9* | ||||
| Gene cluster V (F6) | –2.8 | –5.4 | –3.65/–16.0* | –1.9 | ||||
| PA0176 (McpB) | –5.8 | 4.9/7.3† | –31.2 | –25.1 | –13.5 | –9.0 | –12.6 | –4.5 |
| PA0180 (McpA) | –3.9 | –5.1 | –4.6 | –4.3 | –5.7 | –2.7 | ||
| PA0411 (PilJ) | –3.2/–5.1† | –12.5 | 3.9 | |||||
| PA1251 | –5.7 | 8.9/9.3† | –2.9 | –2.6 | –20.8 | |||
| PA1423 (BdlA) | –3.6 | |||||||
| PA1561 (Aer) | –2.1/–7.9† | –1.5/–2.8† | –7.96 | |||||
| PA1608 | ||||||||
| PA1646 | 2.0 | 2.6 | ||||||
| PA1930 (McpS) | 20.8/31.2† | –9.3 | –33.2 | –7.9 | ||||
| PA2561 (CtpH) | 2.6 | 4.28 | 3.5 | |||||
| PA2573 | –3.1 | 12.7/22.1† | –28.8 | –6.4 | –12.2 | –2.8 | ||
| PA2652 (CtpM) | –2.5 | –4.7 | ||||||
| PA2654 (TlpQ) | –2.1 | |||||||
| PA2788 (McpN) | 3.0/5.6† | –7.4 | –8.2 | –12.4 | –4.1 | –10.9 | ||
| PA2867 | –2.1/–3.3† | –5.3 | –10.4 | –7.6 | –8.8 | |||
| PA2920 | –4.2 | 11.1/12.3† | –3.6 | –5.1 | –6.9 | –7.6 | ||
| PA3708 (WspA) | 4.3 | |||||||
| PA4290 | 4.6/13.4† | 4.2 | –2.0 | –31.8 | –21.8 | |||
| PA4307 (PctC) | –3.3/–7.4† | –3.2 | –2.9 | –2.0 | 9.61 | 2.2 | ||
| PA4309 (PctA) | –2.5 | –3.8 | –2.3 | –2.9 | –2.0 | |||
| PA4310 (PctB) | –4.3 | |||||||
| PA4520 | –3.1 | –2.6 | ||||||
| PA4633 | –2.6 | –2.9 | –2.8 | –3.6 | –2.8 | |||
| PA4844 (CtpL) | 3.7 | |||||||
| PA4915 | 3.6/7.9† | –24.1 | –3.8 | –5.7 | –2.2 | –6.0 | –2.8 | |
| PA5072 (McpK) | 3.0 | –2.5 | 5.2 | |||||
*, The lowest and highest fold change values among the differentially expressed genes per cluster are listed; †, values for two different studies are listed.
Human sputum refers to cystic fibrosis expectorated sputum.
At least two genes per gene cluster have to be altered for listing.
(iv) PA2573.
A mutant in this chemoreceptor showed highly reduced virulence in a greater wax moth (Galleria mellonella) larva model, where a significant reduction in swimming and swarming behaviors as well as impaired pyocyanin production was observed (77). Importantly, the transcript levels of many genes, including virulence and antibiotic resistance genes, were significantly altered in a pa2573-deficient mutant (77), suggesting a possible cross talk between chemotaxis receptors and transcriptional regulation. The signal(s) recognized by PA2573 are unknown, but proteomic analysis revealed that this protein was not present in anaerobic cultures (78).
SIGNALING THROUGH THE F7 (Che2) PATHWAY
Initial information on the F7 pathway which is encoded by genes of cluster II (Fig. 1) was reported by Ferrández et al. (79). It was shown that cheB2 could complement a mutant in cheB1, which is part of the F6 pathway. Overexpression of CheA2, CheB2, and CheW2 in E. coli disrupted chemotaxis indicating that these proteins interact with components of the E. coli system (79). Cluster II encodes two chemoreceptors, McpA and McpB/Aer2, but initial findings showing that cheB2 and mcpB/aer2 mutants were deficient in chemotaxis could not be reproduced in a later study which employed in-frame deletion mutants (14). Since Guvener et al. were unable to identify a general role for F7 pathway proteins in chemotaxis, these authors proposed that these proteins either respond to an attractant that has not been tested yet, that the F7 system somehow talks to the F6 system, or that the system mediates an output unrelated to chemotaxis (14). Remarkably, it was demonstrated that proteins of the F6 and F7 pathways form distinct clusters in which McpB/Aer2 is associated with the F7 protein complex and McpA is associated with the F6 complex (14). Subsequent studies using cryo-microscopy revealed that the F6 and F7 pathways form two spatially separated arrays, supporting the notion that McpA and McpB/Aer2 signal through different pathways (73). Thus, whereas deletion of mcpA had no effect on the F7 array, the mutation of mcpB/aer2 abolished F7 array formation (73). It can therefore be concluded that the function of the F7 pathway depends on just one chemoreceptor, McpB/Aer2; this finding is consistent with the bioinformatic prediction (10). However, since mcpA and mcpB/aer2 are frequently vicinal in genomes, there is evidence that these genes may have coevolved, suggesting that some functional link may exist between them (73, 80). The phylogenetic trees of McpA and McpB/Aer2 from different gammaproteobacteria were similar and congruent with the phylogeny based on CheA, CheB, and CheR (73). Initial evidence for potential mutual interactions has been obtained in P. pseudoalcaligenes that contains McpA and McpB/Aer2 homologs. Whereas the mcpA, mcpB, and aer single mutants, as well as the mcpA mcpB double mutant, had no effect on energy taxis, a significant reduction was observed in both mcpA aer and mcpB aer double mutants (80).
Work conducted with clinical isolates of P. aeruginosa revealed that the F7 pathway plays an important role in virulence (81). Thus, a high-throughput mutant screening for virulence defects in C. elegans demonstrated that the cheB2 mutant showed highly reduced virulence, a finding that was confirmed by experimentation with mice. The authors of that study also found that a cheB1 mutant, severely impaired in chemotaxis, had no virulence defect in C. elegans, suggesting that the F6 and F7 pathways carry out different functions (81). The notion that the F7 pathway is involved in virulence is also supported by the fact that its expression is regulated by different virulence-related mechanisms such as those mediated by inorganic phosphate, quorum-sensing molecules, or RpoS, as well as being strongly downregulated during biofilm formation and infection (Tables 2 to 5). Of note is that the deletion of the mcpB/aer2 gene reduced virulence in a number of animal models (82).
TABLE 5.
Posttranscriptional regulation of P. aeruginosa chemoreceptors and pathways
| Gene cluster/ORF | Fold change(s) (reference[s]) |
|||
|---|---|---|---|---|
| ΔrsmA/WT (169) | Δhfq/WT (anaerobic biofilm) (194) | Δhfq/WT (liquid cultures)(166, 195) | Δcrc/WT (166, 170) | |
| Gene cluster II (F7)a | 8.1/16.7b | |||
| PA0176 (McpB) | 13.3 | |||
| PA1251 | –8.3 | |||
| PA1561 (Aer) | –9.5 | |||
| PA1608 | –2.7 | |||
| PA2654 (TlpQ) | –2.1 | |||
| PA2788 (McpN) | –2.2 | |||
| PA2867 | –6.4 | |||
| PA4290 | –6.0/–23.9c | |||
| PA4307 (PctC) | –3.5 | 5.93 | ||
| PA4310 (PctB) | –3.3 | |||
| PA4520 | 7.3 | |||
At least two genes per gene cluster have to be altered for listing.
The lowest and highest fold change values among the differentially expressed genes per cluster are listed.
The values of two different studies are listed.
McpB/Aer2 Chemoreceptor
The chemoreceptor McpB/Aer2 lacks transmembrane regions and the full-length protein is soluble in the absence of detergent (11). The unusual architecture of McpB/Aer2 consists of three consecutive HAMP (histidine kinases, adenylyl cyclases, methyl-binding proteins, and phosphatases) domains (HAMPs 1 to 3 in Fig. 3) that are followed by a heme-containing PAS domain and two additional HAMP domains (HAMPs 4 and 5 in Fig. 3). McpB/Aer2 is the only P. aeruginosa chemoreceptor that contains a C-terminal pentapeptide (GWEEF) tethered through a flexible linker to its C-terminal end (Fig. 3) (11). In the Tar receptor, C-terminal pentapeptides form additional CheR and CheB binding sites (83), and their removal prevented efficient methylation and demethylation in vitro and abolished chemotaxis in vivo (84).
It was shown that only CheR2 but none of the remaining 3 CheR paralogs bound to the GWEEF pentapeptide of McpB/Aer2 (11), and its removal abolished CheR2 binding and methylation of McpB/Aer2. Sequence alignments of pentapeptide dependent and independent CheR proteins identified a small, 3-amino-acid insertion that is only present in pentapeptide-dependent CheRs, and removal of this sequence in CheR2 prevented pentapeptide binding (11). McpB/Aer2 and CheR2 are encoded in cluster II and form part of the same pathway (Fig. 1). The specific recognition of the McpB/Aer2 pentapeptide by CheR2 is thus a mechanism that enables the targeting of a particular receptor by a specific CheR paralog (11).
As indicated above, the initial observation that McpB/Aer2 is involved in aerotaxis (66) could not be confirmed in subsequent studies (14, 85). However, when McpB/Aer2 is expressed in a chemoreceptor-free E. coli, it mediates a repellent response to different gases (85), and subsequent studies indicated that oxygen is likely to be the physiologically relevant ligand (86). These data, combined with 3D structures of the HAMPs 1 to 3 (87) and PAS domains (88, 89), have permitted a model to be proposed. In this model, signaling is initiated by oxygen recognition at the heme-containing PAS domain. This binding causes a conformational state that is stabilized by the HAMP 2 and 3 domains (HAMP 1 and HAMPs 2 and 3 are separated by a helical extension, suggesting that HAMP 1 is not required for McpB/Aer2 function). This conformational change regulates the ability of HAMPs 4 and 5 to inhibit the kinase control module (85). Cryo-microscopy data show that the F7 array is attached to the membrane via the N-terminal part of McpB/Aer2 (73). This study also determined a distance of ∼40 nm from the CheA/CheW-containing baseplate to the inner membrane, which confirms the linear topology of McpB/Aer2, as proposed in an earlier study (90).
Evolution of F7 Pathways
Chemotaxis in E. coli is mediated by an F7 pathway, whereas in P. aeruginosa the chemotaxis pathway belongs to class F6. In contrast, the biological function of the F7 pathway in P. aeruginosa remains unknown. A recent study of the evolutionary history of F7 pathways has identified five different stages during their evolution (73). Gene clusters of stages 1 and 2 contain genes encoding McpA and McpB/Aer2 homologs and the chemoreceptor deamidase CheD. The P. aeruginosa F7 pathway corresponds to stage 2. Interestingly, all species that contain F7 pathways of stages 1 and 2, also harbor an F6 pathway. Gene clusters of stage 3 are characterized by the loss of the McpA and McpB/Aer2 chemoreceptor encoding genes and the incorporation of a Tar-like chemoreceptor gene. Frequently, species with an F7 system at stage 3 also encode F6 systems that lack CheA, CheR, and CheB, indicative of nonfunctional pathways. In stage 4, the cheY and cheZ genes derived from the F6 pathway are incorporated into F7 systems, giving rise to gene clusters with two cheY genes. Lastly, in stage 5, as observed in enteric Proteobacteria such as E. coli, cheD and the F7 cheY were lost, whereas the tar-like gene was duplicated. None of the stage 5 genomes retained genes of the F6 system. Taken together, current data indicate that F6 and F7 systems existed contemporaneously, with the F7 pathways taking over genes of the F6 system, a pathway that was subsequently lost.
McpB/Aer2 and Pathway Homologs in Other Species
McpB/Aer2 homologs have been identified in a number of species such as Vibrio cholerae, Shewanella oneidensis, or Methylomicrobium alcaliphilum (73, 91). Notably, like the P. aeruginosa receptor, the homolog in V. cholerae was found to bind oxygen with similar affinity (91). Although the output of any pathway that contains McpB/Aer2 homologs remains unknown, it is likely to be mediated by the cognate CheY response regulator. For example, V. cholerae encodes five CheY homologs and contains three chemosensory pathways that belong to the F6, F7 (stage 1), and F9 classes (73, 92, 93). The stage 1 F7 pathway is of unknown function and is encoded by gene cluster III harboring cheY4 and the mcpB/aer2-like gene (73, 94). Studies in which cheY homologs were overexpressed revealed that only F6 CheY3 appeared to be involved in modulating swimming behavior (94), which was subsequently verified by pulldown experiments showing that activated CheY3 but not CheY4 bound to immobilized FliM, the CheY target at the flagellar motor (95). Modeling and molecular dynamics simulation studies of the CheY homologs identified the structural reasons that may be responsible for the specific capacity of CheY3 to bind to FliM (96). It was thus concluded that only one of the five CheYs, CheY3, directly switches flagellar rotation (94). Altogether, current data suggest that CheYs of stages 1 and 2 F7 chemosensory pathways do not bind to the flagellar motor but may interact with an as-yet-unidentified target protein to generate the pathway output.
SIGNALING THROUGH THE ACF (Wsp) PATHWAY
Typically, the 17 classes of chemosensory pathways involved in flagellar motility contain a response regulator composed solely of a receiver (REC) domain that binds to the flagellar motor (1). In contrast, the response regulator of ACF pathways is frequently a fusion of a REC domain and other domains through which the pathway output is realized (1). Such additional domains can be classified into those related to two-component signaling (e.g., histidine kinase, histidine phosphotransfer, HTH DNA binding, AAA-ATPase and PP2C phosphatase, and additional REC domains), stimulus sensing (e.g., PAS and GAF domains), or control of c-di-GMP levels (e.g., GGDEF and EAL domains) (1). Frequently, ACF response regulators possess combinations of these domains, suggesting sophisticated regulatory mechanisms (1). In addition, the CheR of ACF pathways is frequently fused to a tetratricopeptide repeat (TPR) domain (97). The primary function of TPR domains resides in binding other proteins (98), but potential binding partners for TPR domains of CheRs remain to be identified.
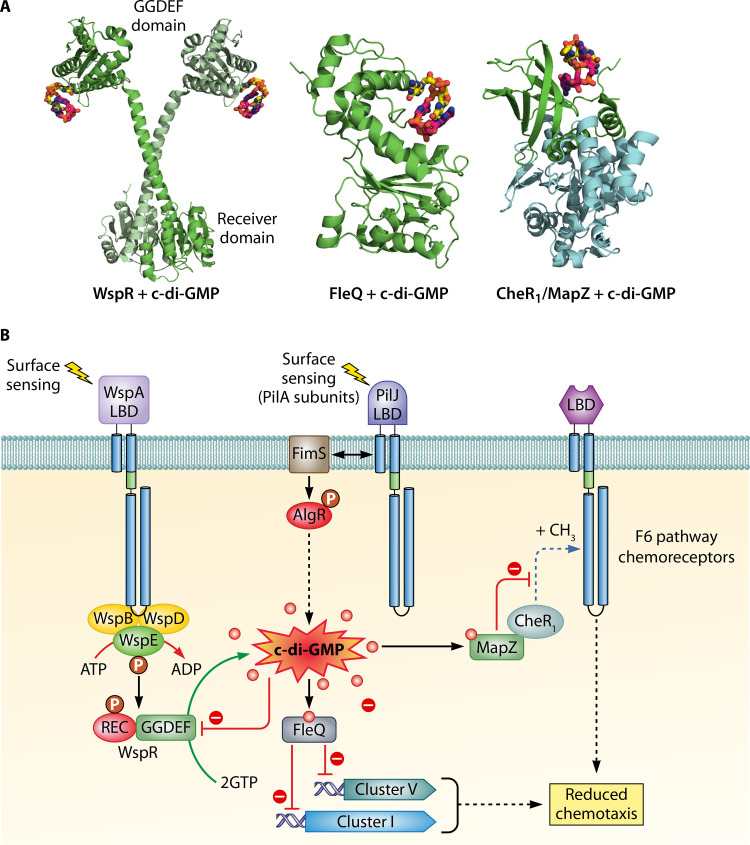
The Wsp (wrinkly spreader phenotype) pathway is the best characterized ACF pathway (8). The corresponding gene cluster III encodes a chemoreceptor, WspA, as well as all the remaining core proteins, namely, two CheW-type adaptor proteins (WspB and WspD), a REC domain-containing histidine kinase (WspE), a methyltransferase (WspC), a methylesterase (WspF), and a response regulator (WspR) (Fig. 2). The regulator WspR is a REC domain fusion with a GGDEF diguanylate cyclase domain (Fig. 6), and phosphorylation of this REC domain enhances diguanylate cyclase activity (8). Initial studies showed that a mutant defective in wspF caused elevated c-di-GMP levels leading to a wrinkly colony phenotype and enhanced biofilm formation (8). It was suggested that in the absence of the methylesterase WspF, the pathway locks into an active state, where it constantly phosphorylates WspR, causing enhanced c-di-GMP synthesis (8). Remarkably, overexpression of WspR was shown to increase c-di-GMP levels, indicative of basal protein activity (99).
FIG 6.
Regulation of chemosensory signaling by c-di-GMP. (A) 3D structures of CheR1/MapZ complex (PDB ID 5Y4R) (174), the WspR response regulator (3BRE) (105), and the transcriptional regulator FleQ (5EXX) (180) in complex with c-di-GMP, representing three different mechanisms by which c-di-GMP modulates pathway signaling. (B) Schematic view of chemosensory pathway-associated signaling by c-di-GMP. The Wsp and Chp pathways are involved in the synthesis of c-di-GMP that, in turn, reduces chemotaxis through the action of the FleQ and MapZ regulatory proteins.
Chemoreceptor WspA
The mutation of wspA in a wspF-deficient strain reversed the colony morphology from wrinkly to smooth, indicating that the WspA chemoreceptor stimulates this pathway (100); a finding that is consistent with bioinformatic predictions (Fig. 3) (10). A fluorescence-tagged WspA was found to form dynamic clusters at both polar and lateral subcellular locations. This localization differs from the mainly polar localization of chemoreceptors involved in chemotaxis (15, 100), indicating that WspA incorporates into separate signaling complexes. The WspA signaling domain was found to determine the subcellular localization, since its replacement with that of the PctA chemoreceptor caused polar localization, whereas, vice versa, the replacement of the PctA signaling domain with that of WspA caused a more disperse localization (15). Wsp pathway activation occurs by growth on surfaces (15), but the nature of the stimulus recognized by WspA is unknown. The involvement of the Wsp pathway in surface sensing is discussed further below. WspA is a transmembrane receptor containing a 4HB LBD that in other chemoreceptors binds chemoeffectors directly or via ligand binding proteins (4). Like the chemotaxis receptors PctA, PctB, PctC, or CtpM, WspA has a signaling domain composed of 40 heptad repeats. Replacement of the WspA 4HB domain with the LBDs of the former four receptors maintained the capacity of the pathway to respond to surface growth, suggesting that WspA stimulation may occur through an alternative mechanism that does not involve the 4HB domain (15).
A subsequent study has shown that ethanol increases c-di-GMP levels but not in a wspR mutant, indicating that the Wsp pathway is primarily responsible for this phenotype (101). In addition, this phenotype depended on WspA, suggesting that this chemoreceptor may respond to ethanol (101). Insight into the sensing of ethanol and other aliphatic alcohols by chemoreceptors has been gained using Ralstonia pseudosolanacearum, a plant-pathogenic bacterium that shows negative chemotaxis to these compounds (102). Surprisingly, since many chemoreceptors with different LBD types were found to participate in R. pseudosolanacearum ethanol taxis, this behavior is likely to be mediated by the modulation of common parts of these receptors, namely, the transmembrane regions and the signaling domain (102). The hypothesis that alterations in WspA transmembrane regions may cause receptor stimulation is also supported by a study showing that mutations in the pathway that determines the length of fatty acids, altering, in turn, membrane characteristics, causes constitutive c-di-GMP production by the Wsp pathway (103).
The methyltransferase WspC is subject to product feedback inhibition since it recognizes the product of the methylation reaction, S-adenosylhomocysteine, with 13-fold-higher affinity than the methylation substrate S-adenosylmethionine (SAM) (11). Consequently, an increase in SAM concentration would result in a stimulation of the methylation reaction. Data have been presented showing that an increase in the cellular SAM concentration caused c-di-GMP-mediated phenotypes and that these phenotypes depended on the Wsp signaling system (104). This research suggests that the increase in SAM concentration enhances WspA methylation, thus driving WspR activity to raise c-di-GMP levels. However, it remains to be established whether the ethanol-mediated modulation of WspA activity (101) may potentially be related to ethanol-mediated changes in cellular SAM levels.
Response Regulator WspR
The response regulator WspR consists of a phosphorylatable REC domain that is fused via a long stalk helix to the diguanylate cyclase GGDEF output domain (Fig. 6A) (105). The typical functional paradigm of response regulators is that REC domain phosphorylation by either a sensor kinase or by small molecule phosphodonors alters the activity/property of the output domain (106). This mechanism has also been observed for WspR since REC domain phosphorylation was shown to increase diguanylate cyclase activity of the regulator (8, 16). Furthermore, REC domain phosphorylation was found to induce WspR subcellular clustering, which in turn potentiated diguanylate cyclase activity (16). In addition, there is evidence for an alternative mechanism that modulates WspR activity. In the 3D structure of WspR, c-di-GMP binds to the inhibitory site of WspR (Fig. 6A), and c-di-GMP-mediated inhibition represents a product feedback regulatory mechanism (105). As a result, mutation of a key amino acid in this inhibitory site rendered the protein highly active (105). Whereas c-di-GMP free protein is present in a monomer-compact dimer-tetramer equilibrium, c-di-GMP binding to the inhibitory site of WspR shifted the protein oligomeric state to the tetramer (105, 107). This tetrameric state is active but forms the platform for the formation of elongated dimers that, in contrast to the compact dimers, are inactive (105). In addition, WspR activity was found to depend on the protein concentration (16, 105). Another study suggested that WspR activation is due to the disruption of the interface between the REC and GGDEF domains, leading to the release and activation of the GGDEF domain (108).
Interestingly, mutants defective in wspR and in the autokinase gene, wspE, had very different phenotypes, which is unexpected since the corresponding gene products form part of the same pathway. Whereas the wspE mutant showed significant increased cytotoxicity in human bronchial epithelial cells, the cytotoxicity of the wspR mutant was only about 15% of that of the parental strain, which led Gellatly et al. (109) to question whether both proteins form a two-component system. However, these data showing that WspR activity can be modulated through alternative mechanisms that do not involve phosphorylation, such as c-di-GMP binding or the protein concentration dependence of WspR activity, may explain the differences in the wspE and wspR mutant phenotypes.
Physiological Relevance of Wsp Pathway
Several studies demonstrate that the Wsp pathway modulates virulence in P. aeruginosa. Rugose small-colony variants (RSCVs) are characterized by their high fitness in biofilms and for their persistence and are therefore of interest to study chronic infections (110). As described earlier, the mutation of wspF caused the constitutive activation of WspR resulting in elevated c-di-GMP levels and the RSCV phenotype. Interestingly, CF airway infections frequently produce RSCV variants and this phenotype could be reverted to the smooth colony morphology in most of the rugose variants by the in trans expression of wspF, indicating that spontaneous mutations of wspF are a very frequent mechanism for the generation of RSCV morphotypes in vivo (111). Nevertheless, mutations in other wsp genes also occurred during P. aeruginosa evolution in CF patients (112, 113). In addition, RSCVs with a hyperbiofilm phenotype were isolated from burn wounds infected with P. aeruginosa PA14 and found to have mutations exclusively in wspF and wspA (114). Taken together, these data suggest that the Wsp pathway may have been positively selected to confer the hyperbiofilm phenotype during different types of infection.
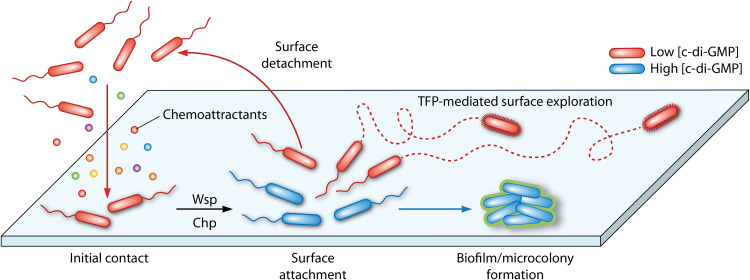
Another example that documents the physiological relevance of the Wsp system comes from single cell studies. Armbruster et al. (115) observed that the Wsp pathway generates heterogeneity in surface-attached P. aeruginosa cells at early stages of biofilm formation. Thus, two physiologically different subpopulations that differ in their c-di-GMP content were identified, namely, one with high c-di-GMP levels that produce the biofilm matrix, and a second subpopulation that has low c-di-GMP levels that permit bacteria to keep exploring the surface (Fig. 7) (115). These authors argue that both subpopulations carry out complementary functions that, when conducted in parallel, are beneficial for successful surface colonization. In another series of experiments, P. aeruginosa was coevolved with Staphylococcus aureus and, surprisingly, a large number of wsp mutants were identified, primarily in wspA and wspF genes. These mutants were characterized by a RSCV morphology and showed increased killing toward S. aureus (116), a strategy that may allow P. aeruginosa to efficiently outcompete bacterial competitors during host infection. Subsequent metabolomic approaches allowed the correlation of the enhanced antibacterial activity of the wspF mutant with an increased production of alkyl quinoline N-oxides, rhamnolipids, and hydrogen cyanide, as well as of the siderophores pyoverdin and pyochelin (117).
FIG 7.
Model of Wsp and Chp pathway-mediated surface sensing modulating surface colonization and biofilm formation in P. aeruginosa. Planktonic cells can actively (via chemotaxis) or passively (environmental changes that propel bacterial cells) interact with a surface. Initial surface contact activates the Wsp and Chp signaling pathways, leading to changes in c-di-GMP and cAMP levels. Subsequently, two physiologically different subpopulations of cells arise, which differ in their c-di-GMP content. Whereas cells with elevated c-di-GMP levels increase exopolysaccharide production to initiate biofilm formation, the subpopulation with low c-di-GMP levels either detaches or explores the surface using type IV pilus-mediated motility. Detached progeny cells retain cAMP-dependent memory of the surface, and the corresponding planktonic population has an increased ability to attach to the surface.
The discovery of the Wsp pathway in P. aeruginosa (8) motivated a number of studies that demonstrate or suggest the existence of functional Wsp pathways in other species such as P. putida (12, 118–120), P. fluorescens (121–123), Halomonas spp. (124), or Burkholderia cenocepacia (125). In addition, the analysis of the response regulator architecture of ACF-type pathways revealed REC-GGDEF fusions in a diverse range of bacterial genera such as Ralstonia, Clostridium, Mesorhizobium, Bordetella, or Janthinobacterium, suggesting a broad phylogenetic distribution of Wsp-like pathways and thus an important physiological role (1).
SIGNALING THROUGH THE TFP (Chp) PATHWAY
Type IV pili (TFP) are among the most common bacterial surface structures and are involved in adherence, motility, competence for DNA uptake, and pathogenesis (126). Motility based on TFP involves cycles of pilus extension, surface adhesion, and retraction (127). The chemosensory pilus (Chp) pathway has been associated with TFP-based motility (17, 128) and directed twitching in gradients of the phospholipid phosphatidylethanolamine (PEA) (129). Directed twitching required the extracellular phospholipase PlcB (130) and pathways that degrade the resulting long-chain fatty acids (131). Since complete metabolism of PEA-derived fatty acids was required for directed twitching, this type of motility was classified as energy taxis (131). Subsequent studies revealed that the Chp pathway also controls 3′,5′-cyclic adenosine monophosphate (cAMP) levels by modulating the activity of the primary adenylate cyclase, CyaB (18). In fact, the Chp pathway was shown to play a central role in regulating cAMP synthesis since screening of a mutant library for reduced cAMP levels resulted primarily in the identification of mutants in the chp gene cluster (18). Remarkably, cAMP signaling is closely interwoven with twitching motility since this second messenger controls TFP synthesis via its interaction with Vfr, a transcription factor that also regulates the expression of different virulence genes (Fig. 2) (18, 132, 133). In contrast to the gene cluster encoding the F6 pathway, the Chp system lacks the CheZ phosphatase and encodes a chemoreceptor (PilJ), two CheYs (PilG and PilH), and two CheWs (PilI and ChpC), as well as ChpD and ChpE, two proteins annotated in the Pfam database as an AraC-type transcriptional regulator and a LysE-type translocator, respectively (Fig. 1 and 2).
PilJ Chemoreceptor
For the PilJ chemoreceptor, a pilJ-deficient mutant was devoid of twitching motility, showed reduced cAMP levels (18, 129) and impedes the assembly and extension of TFP (18, 19). Interestingly, the deletion of the periplasmic LBD of PilJ reduced cAMP levels to a lower degree than observed for a pilJ deletion mutant (134), suggesting that the cytosolic part of PilJ is important (probably by modulating the activity of the CyaB adenylate cyclase) and that this role may by potentiated by the binding of ligands to its LBD. The PilJ-LBD has ∼200 amino acids and comprises two PilJ domains (Fig. 3 and 4).
A number of sensor kinases and chemoreceptors that contain a single periplasmic PilJ domain have been characterized and were shown to bind nitrate (46, 55). However, the amino acids involved in nitrate binding are not conserved in any of the PilJ domains of the PilJ receptor, and a microcalorimetric titration of the periplasmic part of PilJ with nitrate did not show binding (46). Interestingly, surface contact induces expression from the cAMP-dependent promoter that controls the expression of PaQa, an operon that encodes homologous components of the type II secretion system in P. aeruginosa (135). Since this induction was not observed in mutants defective in the genes encoding the PilJ receptor or the ChpA autokinase, it was hypothesized that a mechanical signal, i.e., surface contact, stimulates PilJ and induces downstream signaling (135). This hypothesis was supported by studies of the major component of TFP, the PilA protein. TFP extension and retraction is primarily due to the polymerization and depolymerization of PilA, and bacterial two-hybrid system studies suggest that PilA and PilJ interact (135). Since PilJ has a periplasmic LBD, Persat et al. propose that the PilA and PilJ interaction occurs in the periplasm. TFPs are present in either a relaxed or tense form, and current data indicate that tension generated in the TFP is the mechanical signal that is conveyed via PilA to PilJ, inducing downstream signaling. The outputs of this signaling cascade are increases in TFP extension and retraction frequencies, as well as the induction of cAMP production (Fig. 2) (135).
Additional research has shown that PilJ interacts with FimS, a transmembrane sensor kinase that forms a two-component system with AlgR (136), which ultimately controls the expression of genes involved in c-di-GMP synthesis (Fig. 2) (137). An earlier study already demonstrated that the AlgR/FimS regulatory system is required for twitching (138) and a physical interaction between PilJ and FimS may enable cross talk so that signals recognized by PilJ modulate FimS activity and vice versa. Altogether, currently available data indicate that Chp pathway-mediated signaling causes alteration in cAMP and c-di-GMP levels that, in turn, control motility, virulence, and biofilm formation (Fig. 2, 6B, and 7).
Two Response Regulators: PilG and PilH
The two CheY proteins encoded in the Chp cluster, PilG and PilH (Fig. 1 and 2), share only 29% sequence identity and mutants defective in pilG and pilH showed reduced twitching motility and piliation (18, 139). However, the molecular mechanisms giving rise to these phenotypes are different since increased cAMP and pilin levels were observed in the pilH mutant, whereas both levels were highly reduced in the pilG mutant (18, 139). PilG was found to interact via the FimL protein with FimV, the activator protein of the CyaB adenylate cyclase, thereby causing changes in cAMP levels (Fig. 2) (18, 139). In contrast, PilH was proposed to act as a phosphate sink, similar to the CheY1 of Sinorhizobium meliloti (140), a hypothesis that is based on the preferential phosphorylation of PilH by the ChpA autokinase (141).
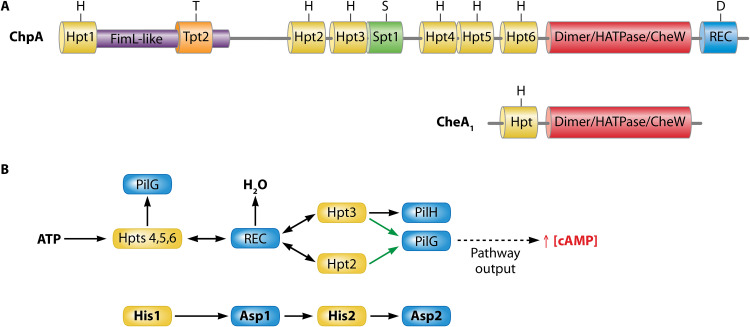
ChpA: a Very Complex Autokinase
The complexity of Chp pathway signaling may be related to the complexity of its ChpA autokinase. The ChpA protein is three times larger than the CheA1 of the F6 pathway (Fig. 1 and 8). Insertional mutants of chpA showed highly impaired twitching motility phenotypes (17, 142). The N-terminal segment of ChpA shares significant sequence similarities with the accessory protein FimL (143) (Fig. 8A), and it was proposed that FimL acts as a scaffolding protein that permits the colocalization of the Chp pathway with the TFP apparatus to coordinate cAMP-dependent signaling upon surface contact stimulation (144, 145). However, the functional relevance of the part of ChpA that is homologous to FimL remains to be explored.
FIG 8.
Complex domain arrangement and mechanism of the ChpA autokinase. (A) Domain arrangements of ChpA and CheA1. Hpt, Histidine-containing phosphotransfer domain; Tpt, Threonine-containing phosphotransfer domain; Spt, Serine-containing phosphotransfer domain; REC, response regulator receiver domain. (Based on data from references 17 and 141.) (B) The proposed mechanism of phosphoryl group flow in the Chp pathway. Black arrows represent findings based on biochemical and genetic data; green arrows indicate findings based on genetics only. (Based on data from reference 141.)
In addition to the FimL-like domain, ChpA contains nine different domains that were predicted to contain phosphorylatable amino acids, namely, six histidine (Hpt)-, one serine (Spt)-, and one threonine (Tpt)-containing phosphotransfer domains, as well as a C-terminal REC domain containing an aspartate as a phosphoryl group accepting residue (Fig. 8A). Twitching motility was either abolished or highly reduced in mutants in which the phosphoryl group accepting residues in the REC and Hpt2/Hpt3 domains, respectively, were mutated. In contrast, replacement of the phosphorylatable amino acids in the remaining domains caused only a minor reduction in twitching (142). The capacity of all eight Hpt, Spt, and Tpt domains to receive a phosphoryl group has been assessed (141). It was shown that the ChpA HATPase domain was able to phosphorylate Hpt 4 to Hpt 6, but none of the remaining domains. Transphosphorylation assays then showed that Hpt 5 and 6 can transfer the phosphoryl group to all three receiver domains of the TFP (Chp) pathway, namely, the PilG and PilH response regulators, as well as to the individual ChpA REC domain (ChpArec). However, significant differences were observed in the transphosphorylation kinetics. Whereas phosphorylation to the ChpArec domain occurred within seconds, typical for autokinase-response regulator pairs (141), transphosphorylation to PilH and particularly to PilG was significantly slower. The order of transphosphorylation rates also correlated with the rates of autodephosphorylation of the three REC domains. In subsequent studies the authors showed rapid transphosphorylation from ChpArec to the Hpt domains 2 and 3, which in turn were found to transphosphorylate primarily PilG and PilH, respectively (141). Thus, in this phosphorelay, the ChpA HATPase domain phosphorylates Hpt domains 4 to 6, followed by a rapid phosphotransfer to ChpArec, which subsequently transphosphorylates Hpt 2 and Hpt 3, domains that ultimately phosphorylate the PilH and PilG response regulators (Fig. 8B).
Phosphorelays have been observed for a number of different unorthodox two-component systems such as ArcBA (146) or TodST (147). These phosphorelays are characterized by a consecutive phosphotransfer between His1, Asp1, His2, and Asp2. Although the molecular architecture of ChpA is much more complex than that of the sensor kinases ArcB and TodS, the mechanism reported for ChpA obeys the general His1-Asp1-His2-Asp2 phosphorylation sequence observed for characterized phosphorelay systems (Fig. 8B). However, the physiological relevance of the complexity of ChpA remains to be established.
INVOLVEMENT OF Wsp AND Chp PATHWAYS IN SURFACE SENSING AND BIOFILM FORMATION
During the initiation of biofilm formation, surface sensing by P. aeruginosa leads to the production of exopolysaccharides and the suppression of surface motility, processes that ultimately promote an irreversible attachment and biofilm formation (115, 148–150). As detailed above, both the Wsp and the Chp pathways are able to respond to surfaces and are important during early stages of biofilm formation through the alteration of second messenger levels. Thus, a surface signal is recognized by the WspA chemoreceptor leading to increases in the c-di-GMP concentration that triggers biofilm formation (Fig. 2 and 7) (15, 100). In addition, the PilJ receptor and TFP also sense surfaces that, in turn, stimulate a signaling cascade that first results in cAMP synthesis and subsequently in increased c-di-GMP levels (Fig. 2 and 7), promoting, as in the case of Wsp-mediated sensing, cell attachment and biofilm formation (135, 136). However, the contribution of Wsp- and Chp-mediated sensing to the initial stages of biofilm varies between P. aeruginosa strains, as described below. TFP-mediated surface sensing and signaling via c-di-GMP has also been observed in other bacteria and may represent a more general feature (151).
A single cell study using P. aeruginosa PA14 as model revealed that the surface population behavior of planktonic cells that have previously been exposed to surfaces is different from that of surface nonsensitized planktonic cells. Data show that previously surface-exposed bacteria possess a memory that propagated across multiple generations and which manifests itself in correlated oscillations of cAMP levels and TFP activity. This behavior was observed to a lesser degree in mutants defective in pilA and pilJ, indicative of the involvement of the Chp pathway (152).
A comparative study revealed important differences in the reversible attachment of P. aeruginosa strains PAO1 and PA14 (148). PAO1 cells committed quickly to surfaces compared to PA14, resulting in a steadily progressive increase of irreversibly attached PAO1 cells. This behavior is likely due to Wsp-based surface sensing that causes c-di-GMP-mediated increases in exopolysaccharides, facilitating thus the attachment of neighboring cells. In contrast, PA14 lineages exhibit high rates of cell detachment from surfaces during the reversible attachment stage. In this strain, surface sensing is primarily mediated by the Chp pathway, causing alteration in cAMP levels that, as mentioned above, allows progeny cells to retain a memory of the surface. As a result, PA14 lineages form planktonic cells that are primed for improved surface attachment that then leads to irreversible attachment and biofilm formation (Fig. 7). Whereas the surface colonization strategy of PAO1 is aimed at recruiting neighbor cells, PA14 primes progenitor cells to optimize attachment in future generations (148).
Another layer of complexity was added by a study showing that surface attachment of P. aeruginosa PA14 increases sensitivity to quorum sensing, a process that was due to the upregulation of lasR which encodes the quorum-sensing master regulator. The corresponding mechanism appeared to be independent of PilA but dependent on the TFP retraction motors and minor pilins. Data thus suggest that there are multiple mechanisms by which TFP sense surfaces (153). In addition, a wspF-deficient mutant of P. aeruginosa PA14 showed decreased and increased levels of Las and Rhl quorum-sensing molecules, respectively, but the molecular mechanism behind these alterations remains to be elucidated (117). As discussed below, quorum-sensing mechanisms regulate the transcript levels of a number of chemoreceptor and signaling genes, including those of gene cluster IV and pilJ.
REGULATION OF CHEMOSENSORY SIGNALING PATHWAYS
Bacteria invest a considerable amount of genetic and energetic resources to synthesize and assemble the flagellar apparatus and to perform chemotaxis. Thus, some bacteria devote up to 6% of their genomes to chemotaxis and motility (154), and E. coli requires almost 3% of the total cellular protein to efficiently perform these processes (155). In order to optimize resources, the expression of chemoreceptor and signaling genes needs to be strictly regulated, and there is a wealth of transcriptomic and proteomic data available for P. aeruginosa. To get a clear picture of the transcriptional and posttranscriptional mechanisms that regulate chemosensory pathways, this information has been compiled in Tables 2 to 5.
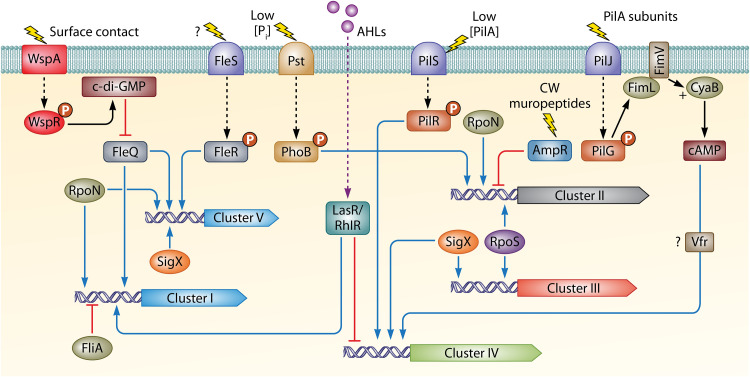
Transcriptional Regulators and Signal Molecules
Table 2 summarizes the major regulatory systems that participate in the transcriptional regulation of chemosensory genes. Data show that transcriptional regulators such as AmpR, BswR, and FleQ and two-component systems, including PilSR and FleSR, as well as different sigma factors, act either as activators or as repressors of the transcription of chemoreceptor and chemosensory pathway genes. Table 3 illustrates that this strict regulation also involves central signal molecules such as Pi, c-di-GMP, cAMP, ppGpp, or acyl-homoserine lactones.
As shown in Tables 2 and 3, there is a considerable overlap between the receptor genes differentially expressed under high c-di-GMP levels (8, 111) and those of the FleQ regulon (156), suggesting that the observed differences may be mediated by the c-di-GMP-binding regulator FleQ. It is also noteworthy that the transcription initiation of chemosensory genes is tightly controlled by different sets of sigma factors. Thus, in this regulatory process, RpoN, SigX, FliA, and RpoS play major roles by controlling the expression of 14, 11, 8, and 7 chemoreceptor genes, respectively (Table 2). As mentioned above, Pi starvation is a major signal that induces P. aeruginosa virulence (44). Studies showed that Pi starvation specifically induces the expression of cluster II (43), a pathway shown to be involved in virulence, as well as the expression of mcpB/aer2, which encodes the only receptor proposed to stimulate the F7 pathway, as discussed above (Table 3) (43). In addition, the stringent stress response signaling pathway plays an important role in the virulence capabilities of P. aeruginosa (157). This pathway is controlled by the second messenger ppGpp and transcriptional profiling of a stringent response-deficient mutant of a P. aeruginosa hypervirulent CF isolate revealed that ppGpp is involved in the regulation of 10 chemoreceptor genes and most of the chemosensory pathways (Table 3) (158). This complexity in regulation is also reflected, for example, in the fact that multiple regulators modulate the expression of the genes encoding the Aer, PctA, PctB, PctC, and PA2867 chemoreceptors. Of note is the magnitude and diversity of regulatory proteins, namely, FleQ, two-component systems and sigma factors that control transcript levels of the pctB chemoreceptor gene (Table 2). In contrast to the paralogous pctA and pctC genes that show a wider phylogenetic distribution, pctB is exclusively found in strains of P. aeruginosa (38). As mentioned above, PctB binds preferentially l-Gln, an amino acid that is very abundant in humans, and in this context it was proposed that PctB may contribute to virulence (38). A summary of the major regulatory events is shown in Fig. 9.
FIG 9.
Overview of genetic regulation of chemosensory gene clusters in P. aeruginosa. Activation and repression are represented by triangular and flat arrowheads, respectively. CW, cell wall; Pi, inorganic phosphate; AHL, N-acyl homoserine lactone.
Transcriptional Regulation during Biofilm Formation and Virulence
Transcriptional analyses conducted during biofilm formation on abiotic surfaces also highlight the complex regulation exerted on the expression of chemoreceptor genes and chemosensory pathways (Table 4). In general, a preferential downregulation in the expression of chemosensory genes was observed during biofilm formation with respect to planktonic cells. Furthermore, increased expression of chemoreceptor genes was observed in bacteria dispersed from biofilms with respect to biofilm-forming cells (Table 4), indicating that these cells are in a transition stage toward a planktonic lifestyle (159). The genes encoding the Aer aerotaxis receptor, McpB/Aer2 and PA2573, are heavily regulated during biofilm formation and dispersion. The latter receptor is of unknown function but was found to be involved in virulence (77). Of all five gene clusters, cluster II was the most heavily regulated during biofilm formation and dispersal, and biofilm formation caused a significant downregulation in the expression of this cluster. Gene cluster II harbors the mcpB/aer2 and mcpA genes, with the latter being the first gene of the cluster (Fig. 1). Transcriptomic data (Tables 2 to 4) revealed coregulation between cluster II and mcpB, but not with mcpA, indicating that mcpA is regulated differently. This finding is consistent with the notion that McpA feeds into the F6 but not the F7 pathway (10, 14).
Using different human and animal infection models, transcriptomic approaches revealed that P. aeruginosa adaptation to its hosts causes a global response that ultimately results in the repression of chemosensory genes (Table 4). Remarkably, among the most heavily regulated genes during infection are also those encoding the McpB/Aer2 receptor and the F7 pathway. A strong repression of cluster II genes was observed in all models cited in Table 4, including human or animal lung, as well as burn wound infections. Other chemoreceptors that are generally downregulated during virulence include the nitrate-responsive McpN and the amino acid chemoreceptors PctA and PctB (Table 4). It has thus been suggested that chemotaxis is important for the early stages of biofilm formation and infection (160), whereas host adaptation in P. aeruginosa induces a nonmotile and nonchemotactic state (161, 162). The importance of motility and chemotaxis in virulence is also associated with the fact that the loss of flagellar motility, in a process independent of flagellum expression, confers phagocytosis resistance due to a reduced activation of the inflammasome (163).
Posttranscriptional Regulation of Chemosensory Systems
The global regulators Hfq, Crc, and RsmA are RNA binding proteins that are central to controlling gene expression at the posttranscriptional level. Whereas RsmA modulates translation and RNA stability by binding to the 5′-untranslated regions of target mRNAs (164), the chaperone Hfq regulates mRNA stability and translation by facilitating pairing between sRNAs and their target mRNAs (165, 166). Interestingly, binding of the catabolite repression control protein Crc to its target mRNA requires Hfq in an sRNA-independent manner. Remarkably, hfq, crc, and rsmA mutants were shown to be deficient in swimming (167–169), suggesting that these regulators play a role in modulating chemotaxis. However, their implication in the posttranscriptional control of chemosensory genes in P. aeruginosa has not been a focus of attention, mainly due to the diversity of metabolic and physiological processes that are regulated by these regulatory proteins (164–166, 170). In addition, there are a number of studies of other model bacteria that correlate Hfq with the expression of chemotaxis genes (171, 172). To analyze the role of Hfq, Crc, and RsmA in posttranscriptional regulation, we have compiled transcriptomic analyses in Table 5. These studies revealed that RsmA and Crc act as positive regulators of the expression of five and two chemoreceptor genes, respectively, including mcpN, tlpQ, pctB, and pctC (Table 5). Alternatively, the RNA chaperone Hfq acts either as a positive or as a negative regulator of the expression of five chemoreceptors and gene cluster II. Remarkably, Hfq-mediated regulatory activities on chemosensory gene expression differed between biofilm and planktonic cells (Table 5). Taken together, these data suggest an involvement of the posttranscriptional regulators RsmA, Crc, and Hfq in the control of chemotaxis toward compounds such as nitrate, histamine, and amino acids.
c-di-GMP-Dependent Regulation of F6 Pathway Signaling
c-di-GMP plays a key role in regulating chemosensory pathways not only at the transcriptional level (Tables 1 and 2) but also by exerting a posttranslational control (Fig. 6). As discussed above, binding of c-di-GMP to the inhibitory site of WspR modulates its diguanylate cyclase activity, representing a product-feedback inhibition mechanism (105). However, c-di-GMP also regulates the activity of the F6 pathway by modulating chemoreceptor methylation (173). This mechanism is based on the interaction of the c-di-GMP-loaded methyltransferase-associated PilZ domain of MapZ (PA4608) with CheR1, leading to a reduction in chemoreceptor methylation and chemotaxis. The 3D structure of the CheR1/MapZ/c-di-GMP ternary complex showed that bound MapZ occupies the binding site for SAM in CheR1, impeding the binding of the methylation substrate (174, 175) (Fig. 6). Interestingly, the c-di-GMP phosphodiesterases DipA, NbdA, and RbdA were found to participate in this regulatory system by modulating c-di-GMP levels and therefore inhibiting flagellar motor switching (176). MapZ only interacts with CheR1 but none of the remaining three CheR paralogues (174). It remains to be established whether the activity of these paralogues is also regulated by similar mechanisms involving potential MapZ homologs. CheR1 was found to methylate eight chemoreceptors, namely, Aer, CtpH, CtpM, PctA, PctB, PA1251, PA1608, and PA2867, and this modification was efficiently inhibited by c-di-GMP-bound MapZ, whereas MapZ itself was unable to inhibit CheR1 activity (177). All CheR1-dependent chemoreceptors were predicted to feed into the F6 pathway (Fig. 3), whereas receptors predicted to feed into the remaining three pathways were not affected. Chemoeffectors are known for some of the methylated chemoreceptors (e.g., PctA, PctB, CtpH, and CtpM), and Sheng et al. showed that MapZ overexpression suppressed chemotaxis to their cognate ligands, as well as virulence in mouse models (177).
CONCLUSIONS AND OUTLOOK
P. aeruginosa senses different physical and chemical stimuli through four chemosensory pathways. These pathways carry out a different function, which makes P. aeruginosa an ideal model to investigate a variety of aspects of chemosensory signaling. However, further research needs to be performed in order to (i) identify the nature of the signal(s) and molecular mechanism(s) by which the WspA and PilJ chemoreceptors are activated, (ii) determine the output of the F7 pathway, (iii) identify the signals recognized by uncharacterized chemoreceptors, (iv) establish the role of periplasmic ligand binding proteins in receptor activation, (v) assess potential cross talk between different chemosensory pathways following up on evidence showing that chemotactic signaling alters transcription, (vi) unravel the molecular complexity of the Chp pathway and decipher its physiological relevance, and (vii) delve into the transcriptional, posttranscriptional, and posttranslational regulatory mechanisms that control chemosensory pathways. Furthermore, insight needs to be gained into the evolutionary reasons which underlie the existence of these sophisticated chemosensory pathways. For example, the output of the Wsp pathway is to alter c-di-GMP levels, but a number of simpler regulatory mechanisms based on single proteins or two-component systems exist that generate the same output. In this context, the reasons for the evolution of sophisticated chemosensory pathways remain to be established. There is no doubt that the extensive knowledge available on the P. aeruginosa pathways will facilitate the understanding of similar signaling and regulatory mechanisms in other bacteria.
ACKNOWLEDGMENTS
This study was supported by FEDER funds and the Fondo Social Europeo through grants from the Spanish Ministry for Science and Innovation to M.A.M. (PID2019-103972GA-I00) and the Spanish Ministry of Economy and Competitiveness to J.A.G. (BIO2016-74875-P) and T.K. (BIO2016-76779-P).
The authors declare that there are no conflicts of interest.
Biographies

Miguel A. Matilla graduated in Biological Sciences at Santiago de Compostela University. He earned his Ph.D. in Biochemistry and Molecular Biology at Granada University with the Extraordinary Doctorate prize, where he investigated the molecular mechanisms used by bacteria to adapt to life in the rhizosphere. His postdoctoral training was in the Biochemistry Department at Cambridge University (UK), where he specialized in the biosynthesis and regulation of novel antibiotics that are produced by plant-associated bacteria. Thereafter, he joined the Department of Environmental Microbiology at Estación Experimental del Zaidín (Spanish National Research Council), where he currently holds a permanent position as Research Scientist. His research combines multidisciplinary approaches from molecular microbiology, transcriptomics, proteomics, microcalorimetry, structural biology, and bacterial physiology to investigate the molecular mechanisms that modulate antibiotic production and host colonization, including chemotaxis and biofilm formation, by biocontrol bacteria and plant and human bacterial pathogens.

David Martín-Mora studied Biology at the University of Granada (Spain) and was granted with the ICARO initiation-to-research scholarship to work in the Fundación Medina (Granada). He obtained a master’s degree in Biotechnology in 2014 and his Ph.D. in 2019. Masters and Ph.D. studies were carried out in the Krell laboratory at the Estación Experimental del Zaidín in Granada. He investigated chemotaxis in Pseudomonas and in particular identified a series of novel chemoreceptors using combined in vitro and in vivo approaches. Part of his Ph.D. work was realized in the Sourjik laboratory at the Max Planck Institute for Terrestrial Microbiology (Germany). He carried out postdoctoral work at the same laboratory, developing novel biosensors.

Jose A. Gavira graduated in Chemistry (Biochemistry) from the University of Granada (UGR) and obtained his Ph.D. in protein crystallization using diffusion mass transport media working at the Spanish National Research Council (CSIC). His Ph.D. thesis was awarded with the First Prize within the Ph.D. Chemistry program of the UGR. He worked as a postdoctoral fellow at the University of Huntsville in Alabama (USA) and the Marshal Space Flight Centre (NASA) on protein crystallization methodologies and in situ resources utilization projects. Since 2003 he has worked at the Laboratory of Crystallographic Studies in Granada (Spain), where he currently holds a permanent position as a CSIC Research Scientist, leading the bio-crystallography research line. His main research interests are focused on biotechnological applications of protein/enzyme crystals for drug formulation, as self-supported catalysts and as biosensors. He collaborates with several groups to study the structural biology of chemosensory pathway proteins and enzyme fitness.

Tino Krell has studied Biochemistry at the University of Leipzig (Germany). He completed his Ph.D. and postdoctoral work at the University of Glasgow (UK), investigating the enzymes of the shikimate pathway. After two years as a Marie Curie Research fellow at the IBCP in Lyon (France) he was appointed head of a research unit at the R&D Department of Sanofi Pasteur (France), working on vaccines. In 2004, he moved to Granada (Spain) to work in the Estación Experimental del Zaidín (EEZ) institute that belongs to Spanish National Research Council CSIC. His group studies different molecular processes related to bacterial sensing and signal transduction. Initially, he studied different two-component systems, but over the last decade his research interest has turned to chemotaxis and, in particular, chemoreceptor function. Pseudomonas aeruginosa is among the model strains he uses in his research.
REFERENCES
- 1.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi S, Sourjik V. 2018. Stimulus sensing and signal processing in bacterial chemotaxis. Curr Opin Microbiol 45:22–29. doi: 10.1016/j.mib.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson JS, Hazelbauer GL, Falke JJ. 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hazelbauer GL. 2012. Bacterial chemotaxis: the early years of molecular studies. Annu Rev Microbiol 66:285–303. doi: 10.1146/annurev-micro-092611-150120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gumerov VM, Ortega DR, Adebali O, Ulrich LE, Zhulin IB. 2020. MiST 3.0: an updated microbial signal transduction database with an emphasis on chemosensory systems. Nucleic Acids Res 48:D459–D464. doi: 10.1093/nar/gkz988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamer R, Chen PY, Armitage JP, Reinert G, Deane CM. 2010. Deciphering chemotaxis pathways using cross species comparisons. BMC Syst Biol 4:3. doi: 10.1186/1752-0509-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zusman DR, Scott AE, Yang Z, Kirby JR. 2007. Chemosensory pathways, motility, and development in Myxococcus xanthus. Nat Rev Microbiol 5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 10.Ortega DR, Fleetwood AD, Krell T, Harwood CS, Jensen GJ, Zhulin IB. 2017. Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114:12809–12814. doi: 10.1073/pnas.1708842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Fontana C, Corral Lugo A, Krell T. 2014. Specificity of the CheR2 methyltransferase in Pseudomonas aeruginosa is directed by a C-terminal pentapeptide in the McpB chemoreceptor. Sci Signal 7:ra34. doi: 10.1126/scisignal.2004849. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Fontana C, Reyes-Darias JA, Munoz-Martinez F, Alfonso C, Morel B, Ramos JL, Krell T. 2013. High specificity in CheR methyltransferase function: CheR2 of Pseudomonas putida is essential for chemotaxis, whereas CheR1 is involved in biofilm formation. J Biol Chem 288:18987–18999. doi: 10.1074/jbc.M113.472605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt J, Musken M, Becker T, Magnowska Z, Bertinetti D, Moller S, Zimmermann B, Herberg FW, Jansch L, Haussler S. 2011. The Pseudomonas aeruginosa chemotaxis methyltransferase CheR1 impacts on bacterial surface sampling. PLoS One 6:e18184. doi: 10.1371/journal.pone.0018184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guvener ZT, Tifrea DF, Harwood CS. 2006. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol 61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 15.O’Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. 2012. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol 86:720–729. doi: 10.1111/mmi.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huangyutitham V, Guvener ZT, Harwood CS. 2013. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. mBio 4:e00242-13. doi: 10.1128/mBio.00242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, Semmler AB, Mellick AS, Martin PR, Alm RA, Hobbs M, Beatson SA, Huang B, Nguyen L, Commolli JC, Engel JN, Darzins A, Mattick JS. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 18.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. 2010. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLange PA, Collins TL, Pierce GE, Robinson JB. 2007. PilJ localizes to cell poles and is required for type IV pilus extension in Pseudomonas aeruginosa. Curr Microbiol 55:389–395. doi: 10.1007/s00284-007-9008-5. [DOI] [PubMed] [Google Scholar]
- 20.Ortega A, Zhulin IB, Krell T. 2017. Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 81:e00033-17. doi: 10.1128/MMBR.00033-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. 2016. Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12:e1004862. doi: 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Q, Li Z, Ouyang Q, Luo C, Gordon VD. 2016. Singly flagellated Pseudomonas aeruginosa chemotaxes efficiently by unbiased motor regulation. mBio 7:e00013. doi: 10.1128/mBio.00013-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian C, Wong CC, Swarup S, Chiam KH. 2013. Bacterial tethering analysis reveals a “run-reverse-turn” mechanism for Pseudomonas species motility. Appl Environ Microbiol 79:4734–4743. doi: 10.1128/AEM.01027-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moench TT, Konetzka WA. 1978. Chemotaxis in Pseudomonas aeruginosa. J Bacteriol 133:427–429. doi: 10.1128/JB.133.1.427-429.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moulton RC, Montie TC. 1979. Chemotaxis by Pseudomonas aeruginosa. J Bacteriol 137:274–280. doi: 10.1128/JB.137.1.274-280.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly-Wintenberg K, Montie TC. 1994. Chemotaxis to oligopeptides by Pseudomonas aeruginosa. Appl Environ Microbiol 60:363–367. doi: 10.1128/AEM.60.1.363-367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato J, Ito A, Nikata T, Ohtake H. 1992. Phosphate taxis in Pseudomonas aeruginosa. J Bacteriol 174:5149–5151. doi: 10.1128/jb.174.15.5149-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson JW, Tredgett MW, Sheehan JK, Thornton DJ, Notman D, Govan JR. 1990. Mucinophilic and chemotactic properties of Pseudomonas aeruginosa in relation to pulmonary colonization in cystic fibrosis. Infect Immun 58:1489–1495. doi: 10.1128/IAI.58.6.1489-1495.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy MJ, Lawless JG. 1985. Role of chemotaxis in the ecology of denitrifiers. Appl Environ Microbiol 49:109–114. doi: 10.1128/AEM.49.1.109-114.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HE, Shitashiro M, Kuroda A, Takiguchi N, Kato J. 2007. Ethylene chemotaxis in Pseudomonas aeruginosa and other Pseudomonas species. Microb Environ 22:186–189. doi: 10.1264/jsme2.22.186. [DOI] [Google Scholar]
- 31.Fischer RS, Song J, Gu W, Jensen RA. 1997. l-Arogenate is a chemoattractant which can be utilized as the sole source of carbon and nitrogen by Pseudomonas aeruginosa. Appl Environ Microbiol 63:567–573. doi: 10.1128/AEM.63.2.567-573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohga T, Masduki A, Kato J, Ohtake H. 1993. Chemotaxis away from thiocyanic and isothiocyanic esters in Pseudomonas aeruginosa. FEMS Microbiol Lett 113:63–66. doi: 10.1111/j.1574-6968.1993.tb06488.x. [DOI] [PubMed] [Google Scholar]
- 33.Shitashiro M, Tanaka H, Hong CS, Kuroda A, Takiguchi N, Ohtake H, Kato J. 2005. Identification of chemosensory proteins for trichloroethylene in Pseudomonas aeruginosa. J Biosci Bioeng 99:396–402. doi: 10.1263/jbb.99.396. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda A, Kumano T, Taguchi K, Nikata T, Kato J, Ohtake H. 1995. Molecular cloning and characterization of a chemotactic transducer gene in Pseudomonas aeruginosa. J Bacteriol 177:7019–7025. doi: 10.1128/jb.177.24.7019-7025.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. 1997. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 143:3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- 36.Rico-Jimenez M, Munoz-Martinez F, Garcia-Fontana C, Fernandez M, Morel B, Ortega A, Ramos JL, Krell T. 2013. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol Microbiol 88:1230–1243. doi: 10.1111/mmi.12255. [DOI] [PubMed] [Google Scholar]
- 37.Reyes-Darias JA, Yang Y, Sourjik V, Krell T. 2015. Correlation between signal input and output in PctA and PctB amino acid chemoreceptor of Pseudomonas aeruginosa. Mol Microbiol 96:513–525. doi: 10.1111/mmi.12953. [DOI] [PubMed] [Google Scholar]
- 38.Gavira JA, Gumerov VM, Rico-Jiménez M, Petukh M, Upadhyay AA, Ortega A, Matilla MA, Zhulin IB, Krell T. 2020. How bacterial chemoreceptors evolve novel ligand specificities. mBio 11:e03066-19. doi: 10.1128/mBio.03066-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corral-Lugo A, Matilla MA, Martin-Mora D, Silva Jimenez H, Mesa Torres N, Kato J, Hida A, Oku S, Conejero-Muriel M, Gavira JA, Krell T. 2018. High-Affinity chemotaxis to histamine mediated by the TlpQ chemoreceptor of the human pathogen Pseudomonas aeruginosa. mBio 9:e01894-18. doi: 10.1128/mBio.01894-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brosnan JT. 2003. Interorgan amino acid transport and its regulation. J Nutr 133:2068S–2072S. doi: 10.1093/jn/133.6.2068S. [DOI] [PubMed] [Google Scholar]
- 41.Schwarzer C, Fischer H, Machen TE. 2016. Chemotaxis and Binding of Pseudomonas aeruginosa to scratch-wounded human cystic fibrosis airway epithelial cells. PLoS One 11:e0150109. doi: 10.1371/journal.pone.0150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamath KS, Pascovici D, Penesyan A, Goel A, Venkatakrishnan V, Paulsen IT, Packer NH, Molloy MP. 2016. Pseudomonas aeruginosa cell membrane protein expression from phenotypically diverse cystic fibrosis isolates demonstrates host-specific adaptations. J Proteome Res 15:2152–2163. doi: 10.1021/acs.jproteome.6b00058. [DOI] [PubMed] [Google Scholar]
- 43.Bains M, Fernandez L, Hancock RE. 2012. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl Environ Microbiol 78:6762–6768. doi: 10.1128/AEM.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaborin A, Romanowski K, Gerdes S, Holbrook C, Lepine F, Long J, Poroyko V, Diggle SP, Wilke A, Righetti K, Morozova I, Babrowski T, Liu DC, Zaborina O, Alverdy JC. 2009. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci U S A 106:6327–6332. doi: 10.1073/pnas.0813199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu H, Kato J, Kuroda A, Ikeda T, Takiguchi N, Ohtake H. 2000. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J Bacteriol 182:3400–3404. doi: 10.1128/jb.182.12.3400-3404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin-Mora D, Ortega A, Matilla MA, Martinez-Rodriguez S, Gavira JA, Krell T. 2019. The molecular mechanism of nitrate chemotaxis via direct ligand binding to the PilJ Domain of McpN mBio 10:e02334-18. doi: 10.1128/mBio.02334-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ortega A, Krell T. 2014. The HBM domain: introducing bimodularity to bacterial sensing. Protein Sci 23:332–336. doi: 10.1002/pro.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rico-Jimenez M, Reyes-Darias JA, Ortega A, Diez Pena AI, Morel B, Krell T. 2016. Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Sci Rep 6:28967. doi: 10.1038/srep28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng YC, Lu C, Li G, Eichenbaum Z, Lu CD. 2017. Induction of the pho regulon and polyphosphate synthesis against spermine stress in Pseudomonas aeruginosa. Mol Microbiol 104:1037–1051. doi: 10.1111/mmi.13678. [DOI] [PubMed] [Google Scholar]
- 50.Zaborina O, Holbrook C, Chen Y, Long J, Zaborin A, Morozova I, Fernandez H, Wang Y, Turner JR, Alverdy JC. 2008. Structure-function aspects of PstS in multidrug-resistant Pseudomonas aeruginosa. PLoS Pathog 4:e43. doi: 10.1371/journal.ppat.0040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez M, Rico-Jimenez M, Ortega A, Daddaoua A, Garcia Garcia AI, Martin-Mora D, Torres NM, Tajuelo A, Matilla MA, Krell T. 2019. Determination of ligand profiles for Pseudomonas aeruginosa solute binding proteins. Int J Mol Sci 20:E5156. doi: 10.3390/ijms20205156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vangnai AS, Takeuchi K, Oku S, Kataoka N, Nitisakulkan T, Tajima T, Kato J. 2013. Identification of CtpL as a chromosomally encoded chemoreceptor for 4-chloroaniline and catechol in Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 79:7241–7248. doi: 10.1128/AEM.02428-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shu CJ, Ulrich LE, Zhulin IB. 2003. The NIT domain: a predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem Sci 28:121–124. doi: 10.1016/S0968-0004(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 54.Yeh JI, Biemann HP, Prive GG, Pandit J, Koshland DE Jr, Kim SH. 1996. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J Mol Biol 262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 55.Gushchin I, Melnikov I, Polovinkin V, Ishchenko A, Yuzhakova A, Buslaev P, Bourenkov G, Grudinin S, Round E, Balandin T, Borshchevskiy V, Willbold D, Leonard G, Buldt G, Popov A, Gordeliy V. 2017. Mechanism of transmembrane signaling by sensor histidine kinases. Science 356:eaah6345. doi: 10.1126/science.aah6345. [DOI] [PubMed] [Google Scholar]
- 56.Matilla MA, Mora DM, Krell T. 2020. The use of isothermal titration calorimetry to unravel chemotactic signaling mechanisms. Environ Microbiol 22:3005–3019. doi: 10.1111/1462-2920.15035. [DOI] [PubMed] [Google Scholar]
- 57.Xu X, Zhang H, Song Y, Lynch SV, Lowell CA, Wiener-Kronish JP, Caughey GH. 2012. Strain-dependent induction of neutrophil histamine production and cell death by Pseudomonas aeruginosa. J Leukoc Biol 91:275–284. doi: 10.1189/jlb.0711356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 59.Pineda-Molina E, Reyes-Darias JA, Lacal J, Ramos JL, Garcia-Ruiz JM, Gavira JA, Krell T. 2012. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc Natl Acad Sci U S A 109:18926–18931. doi: 10.1073/pnas.1201400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin-Mora D, Ortega A, Reyes-Darias JA, García V, López-Farfán D, Matilla MA, Krell T. 2016. Identification of a chemoreceptor in Pseudomonas aeruginosa that specifically mediates chemotaxis towards alpha-ketoglutarate. Front Microbiol 7:1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bjorkman AM, Dunten P, Sandgren MO, Dwarakanath VN, Mowbray SL. 2001. Mutations that affect ligand binding to the Escherichia coli aspartate receptor: implications for transmembrane signaling. J Biol Chem 276:2808–2815. doi: 10.1074/jbc.M009593200. [DOI] [PubMed] [Google Scholar]
- 62.Milligan DL, Koshland DE Jr. 1993. Purification and characterization of the periplasmic domain of the aspartate chemoreceptor. J Biol Chem 268:19991–19997. [PubMed] [Google Scholar]
- 63.Alvarez-Ortega C, Harwood CS. 2007. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl Environ Microbiol 73:7793–7795. doi: 10.1128/AEM.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin-Mora D, Ortega A, Perez-Maldonado FJ, Krell T, Matilla MA. 2018. The activity of the C4-dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists. Sci Rep 8:2102. doi: 10.1038/s41598-018-20283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bi S, Yu D, Si G, Luo C, Li T, Ouyang Q, Jakovljevic V, Sourjik V, Tu Y, Lai L. 2013. Discovery of novel chemoeffectors and rational design of Escherichia coli chemoreceptor specificity. Proc Natl Acad Sci U S A 110:16814–16819. doi: 10.1073/pnas.1306811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong CS, Shitashiro M, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. 2004. Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. FEMS Microbiol Lett 231:247–252. doi: 10.1016/S0378-1097(04)00009-6. [DOI] [PubMed] [Google Scholar]
- 67.Hong CS, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. 2004. The aerotaxis transducer gene aer, but not aer-2, is transcriptionally regulated by the anaerobic regulator ANR in Pseudomonas aeruginosa. J Biosci Bioeng 97:184–190. doi: 10.1016/S1389-1723(04)70188-7. [DOI] [PubMed] [Google Scholar]
- 68.Hoboth C, Hoffmann R, Eichner A, Henke C, Schmoldt S, Imhof A, Heesemann J, Hogardt M. 2009. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J Infect Dis 200:118–130. doi: 10.1086/599360. [DOI] [PubMed] [Google Scholar]
- 69.Bardy SL, Maddock JR. 2005. Polar localization of a soluble methyl-accepting protein of Pseudomonas aeruginosa. J Bacteriol 187:7840–7844. doi: 10.1128/JB.187.22.7840-7844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol 188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petrova OE, Sauer K. 2012. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ortega DR, Yang W, Subramanian P, Mann P, Kjær A, Chen S, Watts KJ, Pirbadian S, Collins DA, Kooger R, Kalyuzhnaya MG, Ringgaard S, Briegel A, Jensen GJ. 2020. Repurposing a chemosensory macromolecular machine. Nat Commun 11:2041. doi: 10.1038/s41467-020-15736-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Petrova OE, Su S, Lau GW, Panmanee W, Na R, Hassett DJ, Davies DG, Sauer K. 2014. BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog 10:e1004168. doi: 10.1371/journal.ppat.1004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang Z, Wang YH, Zhu HZ, Andrianova EP, Jiang CY, Li D, Ma L, Feng J, Liu ZP, Xiang H, Zhulin IB, Liu SJ. 2019. Cross talk between chemosensory pathways that modulate chemotaxis and biofilm formation. mBio 10:e02876-18. doi: 10.1128/mBio.02876-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim HE, Shitashiro M, Kuroda A, Takiguchi N, Ohtake H, Kato J. 2006. Identification and characterization of the chemotactic transducer in Pseudomonas aeruginosa PAO1 for positive chemotaxis to trichloroethylene. J Bacteriol 188:6700–6702. doi: 10.1128/JB.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McLaughlin HP, Caly DL, McCarthy Y, Ryan RP, Dow JM. 2012. An orphan chemotaxis sensor regulates virulence and antibiotic tolerance in the human pathogen Pseudomonas aeruginosa. PLoS One 7:e42205. doi: 10.1371/journal.pone.0042205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Babin BM, Bergkessel M, Sweredoski MJ, Moradian A, Hess S, Newman DK, Tirrell DA. 2016. SutA is a bacterial transcription factor expressed during slow growth in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:E597–E605. doi: 10.1073/pnas.1514412113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferrández A, Hawkins AC, Summerfield DT, Harwood CS. 2002. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol 184:4374–4383. doi: 10.1128/jb.184.16.4374-4383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Booth SC, Turner RJ. 2019. Phylogenetic characterization of the energy taxis receptor Aer in Pseudomonas and phenotypic characterization in Pseudomonas pseudoalcaligenes KF707. Microbiology (Reading) 165:1331–1344. doi: 10.1099/mic.0.000864. [DOI] [PubMed] [Google Scholar]
- 81.Garvis S, Munder A, Ball G, de Bentzmann S, Wiehlmann L, Ewbank JJ, Tummler B, Filloux A. 2009. Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog 5:e1000540. doi: 10.1371/journal.ppat.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garcia-Fontana C, Vilchez JI, Gonzalez-Requena M, Gonzalez-Lopez J, Krell T, Matilla MA, Manzanera M. 2019. The involvement of McpB chemoreceptor from Pseudomonas aeruginosa PAO1 in virulence. Sci Rep 9:13166. doi: 10.1038/s41598-019-49697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perez E, Stock AM. 2007. Characterization of the Thermotoga maritima chemotaxis methylation system that lacks pentapeptide-dependent methyltransferase CheR:MCP tethering. Mol Microbiol 63:363–378. doi: 10.1111/j.1365-2958.2006.05518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li M, Hazelbauer GL. 2006. The carboxyl-terminal linker is important for chemoreceptor function. Mol Microbiol 60:469–479. doi: 10.1111/j.1365-2958.2006.05108.x. [DOI] [PubMed] [Google Scholar]
- 85.Watts KJ, Taylor BL, Johnson MS. 2011. PAS/poly-HAMP signaling in Aer-2, a soluble haem-based sensor. Mol Microbiol 79:686–699. doi: 10.1111/j.1365-2958.2010.07477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garcia D, Orillard E, Johnson MS, Watts KJ. 2017. Gas sensing and signaling in the PAS-Heme domain of the Pseudomonas aeruginosa Aer2 receptor. J Bacteriol 199:e00003-17. doi: 10.1128/JB.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Airola MV, Watts KJ, Bilwes AM, Crane BR. 2010. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure 18:436–448. doi: 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Airola MV, Huh D, Sukomon N, Widom J, Sircar R, Borbat PP, Freed JH, Watts KJ, Crane BR. 2013. Architecture of the soluble receptor Aer2 indicates an in-line mechanism for PAS and HAMP domain signaling. J Mol Biol 425:886–901. doi: 10.1016/j.jmb.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sawai H, Sugimoto H, Shiro Y, Ishikawa H, Mizutani Y, Aono S. 2012. Structural basis for oxygen sensing and signal transduction of the heme-based sensor protein Aer2 from Pseudomonas aeruginosa. Chem Commun (Camb) 48:6523–6525. doi: 10.1039/c2cc32549g. [DOI] [PubMed] [Google Scholar]
- 90.Airola MV, Sukomon N, Samanta D, Borbat PP, Freed JH, Watts KJ, Crane BR. 2013. HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol 11:e1001479. doi: 10.1371/journal.pbio.1001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Greer-Phillips SE, Sukomon N, Chua TK, Johnson MS, Crane BR, Watts KJ. 2018. The Aer2 receptor from Vibrio cholerae is a dual Pas-Heme oxygen sensor. Mol Microbiol doi: 10.1111/mmi.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A 106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Briegel A, Ortega DR, Mann P, Kjær A, Ringgaard S, Jensen GJ. 2016. Chemotaxis cluster 1 proteins form cytoplasmic arrays in Vibrio cholerae and are stabilized by a double signaling domain receptor DosM. Proc Natl Acad Sci U S A 113:10412–10417. doi: 10.1073/pnas.1604693113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hyakutake A, Homma M, Austin MJ, Boin MA, Hase CC, Kawagishi I. 2005. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J Bacteriol 187:8403–8410. doi: 10.1128/JB.187.24.8403-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Biswas M, Dey S, Khamrui S, Sen U, Dasgupta J. 2013. Conformational barrier of CheY3 and inability of CheY4 to bind FliM control the flagellar motor action in Vibrio cholerae. PLoS One 8:e73923. doi: 10.1371/journal.pone.0073923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dasgupta J, Dattagupta JK. 2008. Structural determinants of V. cholerae CheYs that discriminate them in FliM binding: comparative modeling and MD simulation studies. J Biomol Struct Dyn 25:495–503. doi: 10.1080/07391102.2008.10507196. [DOI] [PubMed] [Google Scholar]
- 97.Munoz-Martinez F, Garcia-Fontana C, Rico-Jimenez M, Alfonso C, Krell T. 2012. Genes encoding Cher-TPR fusion proteins are predominantly found in gene clusters encoding chemosensory pathways with alternative cellular functions. PLoS One 7:e45810. doi: 10.1371/journal.pone.0045810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perez-Riba A, Itzhaki LS. 2019. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr Opin Struct Biol 54:43–49. doi: 10.1016/j.sbi.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 99.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signaling. Environ Microbiol 13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 100.Guvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen AI, Dolben EF, Okegbe C, Harty CE, Golub Y, Thao S, Ha DG, Willger SD, O’Toole GA, Harwood CS, Dietrich LE, Hogan DA. 2014. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog 10:e1004480. doi: 10.1371/journal.ppat.1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oku S, Hida A, Mattana T, Tajima T, Nakashimada Y, Kato J. 2017. Involvement of many chemotaxis sensors in negative chemotaxis to ethanol in Ralstonia pseudosolanacearum Ps29. Microbiology (Reading) 163:1880–1889. doi: 10.1099/mic.0.000574. [DOI] [PubMed] [Google Scholar]
- 103.Blanka A, Duvel J, Dotsch A, Klinkert B, Abraham WR, Kaever V, Ritter C, Narberhaus F, Haussler S. 2015. Constitutive production of c-di-GMP is associated with mutations in a variant of Pseudomonas aeruginosa with altered membrane composition. Sci Signal 8:ra36. doi: 10.1126/scisignal.2005943. [DOI] [PubMed] [Google Scholar]
- 104.Pu M, Sheng L, Song S, Gong T, Wood TK. 2018. Serine hydroxymethyltransferase ShrA (PA2444) controls rugose small-colony variant formation in Pseudomonas aeruginosa. Front Microbiol 9:315. doi: 10.3389/fmicb.2018.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. 2008. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol 6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, Ramos JL. 2010. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol 64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 107.De N, Navarro MV, Wang Q, Krasteva PV, Sondermann H. 2010. Biophysical assays for protein interactions in the Wsp sensory system and biofilm formation. Methods Enzymol 471:161–184. doi: 10.1016/S0076-6879(10)71010-7. [DOI] [PubMed] [Google Scholar]
- 108.Malone JG, Williams R, Christen M, Jenal U, Spiers AJ, Rainey PB. 2007. The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology (Reading) 153:980–994. doi: 10.1099/mic.0.2006/002824-0. [DOI] [PubMed] [Google Scholar]
- 109.Gellatly SL, Bains M, Breidenstein EBM, Strehmel J, Reffuveille F, Taylor PK, Yeung ATY, Overhage J, Hancock REW. 2018. Novel roles for two-component regulatory systems in cytotoxicity and virulence-related properties in Pseudomonas aeruginosa. AIMS Microbiol 4:173–191. doi: 10.3934/microbiol.2018.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pestrak MJ, Chaney SB, Eggleston HC, Dellos-Nolan S, Dixit S, Mathew-Steiner SS, Roy S, Parsek MR, Sen CK, Wozniak DJ. 2018. Pseudomonas aeruginosa rugose small-colony variants evade host clearance, are hyper-inflammatory, and persist in multiple host environments. PLoS Pathog 14:e1006842. doi: 10.1371/journal.ppat.1006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol 191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marvig RL, Sommer LM, Molin S, Johansen HK. 2015. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 113.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gloag ES, Marshall CW, Snyder D, Lewin GR, Harris JS, Santos-Lopez A, Chaney SB, Whiteley M, Cooper VS, Wozniak DJ. 2019. Pseudomonas aeruginosa interstrain dynamics and selection of hyperbiofilm mutants during a chronic infection. mBio 10:e01698-19. doi: 10.1128/mBio.01698-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Armbruster CR, Lee CK, Parker-Gilham J, de Anda J, Xia A, Zhao K, Murakami K, Tseng BS, Hoffman LR, Jin F, Harwood CS, Wong GC, Parsek MR. 2019. Heterogeneity in surface sensing suggests a division of labor in Pseudomonas aeruginosa populations. Elife 8:e45084. doi: 10.7554/eLife.45084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tognon M, Kohler T, Gdaniec BG, Hao Y, Lam JS, Beaume M, Luscher A, Buckling A, van Delden C. 2017. Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J 11:2233–2243. doi: 10.1038/ismej.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gdaniec BG, Allard PM, Queiroz EF, Wolfender JL, van Delden C, Kohler T. 2020. Surface sensing triggers a broad-spectrum antimicrobial response in Pseudomonas aeruginosa. Environ Microbiol 22:3572–3587. doi: 10.1111/1462-2920.15139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Corral-Lugo A, de la Torre J, Matilla MA, Fernandez M, Morel B, Espinosa-Urgel M, Krell T. 2016. Assessment of the contribution of chemoreceptor-based signaling to biofilm formation. Environ Microbiol 18:3355–3372. doi: 10.1111/1462-2920.13170. [DOI] [PubMed] [Google Scholar]
- 119.Lopez-Farfan D, Reyes-Darias JA, Matilla MA, Krell T. 2019. Concentration dependent effect of plant root exudates on the chemosensory systems of Pseudomonas putida KT2440. Front Microbiol 10:78. doi: 10.3389/fmicb.2019.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hueso-Gil A, Calles B, de Lorenzo V. 2020. The Wsp intermembrane complex mediates metabolic control of the swim-attach decision of Pseudomonas putida. Environ Microbiol 22:3535–3547. doi: 10.1111/1462-2920.15126. [DOI] [PubMed] [Google Scholar]
- 121.Ardre M, Dufour D, Rainey PB. 2019. Causes and biophysical consequences of cellulose production by Pseudomonas fluorescens SBW25 at the air-liquid interface. J Bacteriol 201:e00110-19. doi: 10.1128/JB.00110-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barahona E, Navazo A, Martinez-Granero F, Zea-Bonilla T, Perez-Jimenez RM, Martin M, Rivilla R. 2011. Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl Environ Microbiol 77:5412–5419. doi: 10.1128/AEM.00320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Navazo A, Barahona E, Redondo-Nieto M, Martinez-Granero F, Rivilla R, Martin M. 2009. Three independent signaling pathways repress motility in Pseudomonas fluorescens F113. Microb Biotechnol 2:489–498. doi: 10.1111/j.1751-7915.2009.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gasperotti AF, Revuelta MV, Studdert CA, Herrera Seitz MK. 2018. Identification of two different chemosensory pathways in representatives of the genus Halomonas. BMC Genomics 19:266. doi: 10.1186/s12864-018-4811-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cooper VS, Staples RK, Traverse CC, Ellis CN. 2014. Parallel evolution of small colony variants in Burkholderia cenocepacia biofilms. Genomics 104:447–452. doi: 10.1016/j.ygeno.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 126.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 127.Skerker JM, Berg HC. 2001. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A 98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Darzins A. 1994. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol 11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 129.Kearns DB, Robinson J, Shimkets LJ. 2001. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J Bacteriol 183:763–767. doi: 10.1128/JB.183.2.763-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barker AP, Vasil AI, Filloux A, Ball G, Wilderman PJ, Vasil ML. 2004. A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol Microbiol 53:1089–1098. doi: 10.1111/j.1365-2958.2004.04189.x. [DOI] [PubMed] [Google Scholar]
- 131.Miller RM, Tomaras AP, Barker AP, Voelker DR, Chan ED, Vasil AI, Vasil ML. 2008. Pseudomonas aeruginosa twitching motility-mediated chemotaxis towards phospholipids and fatty acids: specificity and metabolic requirements. J Bacteriol 190:4038–4049. doi: 10.1128/JB.00129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Beatson SA, Whitchurch CB, Sargent JL, Levesque RC, Mattick JS. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J Bacteriol 184:3605–3613. doi: 10.1128/jb.184.13.3605-3613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Berry A, Han K, Trouillon J, Robert-Genthon M, Ragno M, Lory S, Attree I, Elsen S. 2018. cAMP and Vfr control exolysin expression and cytotoxicity of Pseudomonas aeruginosa taxonomic outliers. J Bacteriol 200:e00135-18. doi: 10.1128/JB.00135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jansari VH, Potharla VY, Riddell GT, Bardy SL. 2016. Twitching motility and cAMP levels: signal transduction through a single methyl-accepting chemotaxis protein. FEMS Microbiol Lett 363:fnw119. doi: 10.1093/femsle/fnw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. 2015. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GCL, O’Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:e02456-14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kong W, Zhao J, Kang H, Zhu M, Zhou T, Deng X, Liang H. 2015. ChIP-seq reveals the global regulator AlgR mediating cyclic di-GMP synthesis in Pseudomonas aeruginosa. Nucleic Acids Res 43:8268–8282. doi: 10.1093/nar/gkv747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Whitchurch CB, Alm RA, Mattick JS. 1996. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Buensuceso RNC, Daniel-Ivad M, Kilmury SLN, Leighton TL, Harvey H, Howell PL, Burrows LL. 2017. Cyclic AMP-independent control of twitching motility in Pseudomonas aeruginosa. J Bacteriol 199:e00188-17. doi: 10.1128/JB.00188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sourjik V, Schmitt R. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327–2335. doi: 10.1021/bi972330a. [DOI] [PubMed] [Google Scholar]
- 141.Silversmith RE, Wang B, Fulcher NB, Wolfgang MC, Bourret RB. 2016. Phosphoryl group flow within the Pseudomonas aeruginosa Pil-Chp chemosensory system: differential function of the eight phosphotransferase and three receiver domains. J Biol Chem 291:17677–17691. doi: 10.1074/jbc.M116.737528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Leech AJ, Mattick JS. 2006. Effect of site-specific mutations in different phosphotransfer domains of the chemosensory protein ChpA on Pseudomonas aeruginosa motility. J Bacteriol 188:8479–8486. doi: 10.1128/JB.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Whitchurch CB, Beatson SA, Comolli JC, Jakobsen T, Sargent JL, Bertrand JJ, West J, Klausen M, Waite LL, Kang PJ, Tolker-Nielsen T, Mattick JS, Engel JN. 2005. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol Microbiol 55:1357–1378. doi: 10.1111/j.1365-2958.2005.04479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Inclan YF, Persat A, Greninger A, Von Dollen J, Johnson J, Krogan N, Gitai Z, Engel JN. 2016. A scaffold protein connects type IV pili with the Chp chemosensory system to mediate activation of virulence signaling in Pseudomonas aeruginosa. Mol Microbiol 101:590–605. doi: 10.1111/mmi.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Buensuceso RN, Nguyen Y, Zhang K, Daniel-Ivad M, Sugiman-Marangos SN, Fleetwood AD, Zhulin IB, Junop MS, Howell PL, Burrows LL. 2016. The conserved tetratricopeptide repeat-containing C-terminal domain of Pseudomonas aeruginosa FimV is required for its cyclic AMP-dependent and -independent functions. J Bacteriol 198:2263–2274. doi: 10.1128/JB.00322-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Teran-Melo JL, Pena-Sandoval GR, Silva-Jimenez H, Rodriguez C, Alvarez AF, Georgellis D. 2018. Routes of phosphoryl group transfer during signal transmission and signal decay in the dimeric sensor histidine kinase ArcB. J Biol Chem 293:13214–13223. doi: 10.1074/jbc.RA118.003910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Busch A, Guazzaroni ME, Lacal J, Ramos JL, Krell T. 2009. The sensor kinase TodS operates by a multiple step phosphorelay mechanism involving two autokinase domains. J Biol Chem 284:10353–10360. doi: 10.1074/jbc.M900521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee CK, Vachier J, de Anda J, Zhao K, Baker AE, Bennett RR, Armbruster CR, Lewis KA, Tarnopol RL, Lomba CJ, Hogan DA, Parsek MR, O’Toole GA, Golestanian R, Wong GCL. 2020. Social cooperativity of bacteria during reversible surface attachment in young biofilms: a quantitative comparison of Pseudomonas aeruginosa PA14 and PAO1. mBio 11:e02644-19. doi: 10.1128/mBio.02644-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, Parsek MR. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog 7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke Z, Wright ER, Biais N, Dalia AB, Brun YV. 2017. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358:535–538. doi: 10.1126/science.aan5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee CK, de Anda J, Baker AE, Bennett RR, Luo Y, Lee EY, Keefe JA, Helali JS, Ma J, Zhao K, Golestanian R, O’Toole GA, Wong GCL. 2018. Multigenerational memory and adaptive adhesion in early bacterial biofilm communities. Proc Natl Acad Sci U S A 115:4471–4476. doi: 10.1073/pnas.1720071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chuang SK, Vrla GD, Frohlich KS, Gitai Z. 2019. Surface association sensitizes Pseudomonas aeruginosa to quorum sensing. Nat Commun 10:4118. doi: 10.1038/s41467-019-12153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. 2012. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol 66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Colin R, Sourjik V. 2017. Emergent properties of bacterial chemotaxis pathway. Curr Opin Microbiol 39:24–33. doi: 10.1016/j.mib.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 156.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 157.Vogt SL, Green C, Stevens KM, Day B, Erickson DL, Woods DE, Storey DG. 2011. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect Immun 79:4094–4104. doi: 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pletzer D, Sun E, Ritchie C, Wilkinson L, Liu LT, Trimble MJ, Wolfmeier H, Blimkie TM, Hancock REW. 2020. Surfing motility is a complex adaptation dependent on the stringent stress response in Pseudomonas aeruginosa LESB58. PLoS Pathog 16:e1008444. doi: 10.1371/journal.ppat.1008444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chua SL, Liu Y, Yam JK, Chen Y, Vejborg RM, Tan BG, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L. 2014. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun 5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 160.Matilla MA, Krell T. 2018. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev 42. doi: 10.1093/femsre/fux052. [DOI] [PubMed] [Google Scholar]
- 161.Rossi E, Falcone M, Molin S, Johansen HK. 2018. High-resolution in situ transcriptomics of Pseudomonas aeruginosa unveils genotype independent patho-phenotypes in cystic fibrosis lungs. Nat Commun 9:3459. doi: 10.1038/s41467-018-05944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tart AH, Wolfgang MC, Wozniak DJ. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol 187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Patankar YR, Lovewell RR, Poynter ME, Jyot J, Kazmierczak BI, Berwin B. 2013. Flagellar motility is a key determinant of the magnitude of the inflammasome response to Pseudomonas aeruginosa. Infect Immun 81:2043–2052. doi: 10.1128/IAI.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kavita K, de Mets F, Gottesman S. 2018. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr Opin Microbiol 42:53–61. doi: 10.1016/j.mib.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sonnleitner E, Wulf A, Campagne S, Pei XY, Wolfinger MT, Forlani G, Prindl K, Abdou L, Resch A, Allain FH, Luisi BF, Urlaub H, Blasi U. 2018. Interplay between the catabolite repression control protein Crc, Hfq and RNA in Hfq-dependent translational regulation in Pseudomonas aeruginosa. Nucleic Acids Res 46:1470–1485. doi: 10.1093/nar/gkx1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Linares JF, Moreno R, Fajardo A, Martinez-Solano L, Escalante R, Rojo F, Martinez JL. 2010. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ Microbiol 12:3196–3212. doi: 10.1111/j.1462-2920.2010.02292.x. [DOI] [PubMed] [Google Scholar]
- 168.Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jager KE, Blasi U. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog 35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 169.Burrowes E, Baysse C, Adams C, O’Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology (Reading) 152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 170.Corona F, Reales-Calderon JA, Gil C, Martinez JL. 2018. The development of a new parameter for tracking posttranscriptional regulation allows the detailed map of the Pseudomonas aeruginosa Crc regulon. Sci Rep 8:16793. doi: 10.1038/s41598-018-34741-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu X, Yan Y, Wu H, Zhou C, Wang X. 2019. Biological and transcriptomic studies reveal hfq is required for swimming, biofilm formation and stress response in Xanthomonas axonpodis pv. citri. BMC Microbiol 19:103. doi: 10.1186/s12866-019-1476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gao M, Barnett MJ, Long SR, Teplitski M. 2010. Role of the Sinorhizobium meliloti global regulator Hfq in gene regulation and symbiosis. Mol Plant Microbe Interact 23:355–365. doi: 10.1094/MPMI-23-4-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu L, Xin L, Zeng Y, Yam JK, Ding Y, Venkataramani P, Cheang QW, Yang X, Tang X, Zhang LH, Chiam KH, Yang L, Liang ZX. 2016. A cyclic di-GMP-binding adaptor protein interacts with a chemotaxis methyltransferase to control flagellar motor switching. Sci Signal 9:ra102. doi: 10.1126/scisignal.aaf7584. [DOI] [PubMed] [Google Scholar]
- 174.Yan XF, Xin L, Yen JT, Zeng Y, Jin S, Cheang QW, Fong R, Chiam KH, Liang ZX, Gao YG. 2018. Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein. J Biol Chem 293:100–111. doi: 10.1074/jbc.M117.815704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhu Y, Yuan Z, Gu L. 2017. Structural basis for the regulation of chemotaxis by MapZ in the presence of c-di-GMP. Acta Crystallogr D Struct Biol 73:683–691. doi: 10.1107/S2059798317009998. [DOI] [PubMed] [Google Scholar]
- 176.Xin L, Zeng Y, Sheng S, Chea RA, Liu Q, Li HY, Yang L, Xu L, Chiam KH, Liang ZX. 2019. Regulation of flagellar motor switching by c-di-GMP phosphodiesterases in Pseudomonas aeruginosa. J Biol Chem 294:13789–13799. doi: 10.1074/jbc.RA119.009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sheng S, Xin L, Yam JKH, Salido MM, Khong NZJ, Liu Q, Chea RA, Li HY, Yang L, Liang ZX, Xu L. 2019. The MapZ-mediated methylation of chemoreceptors contributes to pathogenicity of Pseudomonas aeruginosa. Front Microbiol 10:67. doi: 10.3389/fmicb.2019.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Reyes-Darias JA, Garcia V, Rico-Jimenez M, Corral-Lugo A, Lesouhaitier O, Juarez-Hernandez D, Yang Y, Bi S, Feuilloley M, Munoz-Rojas J, Sourjik V, Krell T. 2015. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol Microbiol 97:488–501. doi: 10.1111/mmi.13045. [DOI] [PubMed] [Google Scholar]
- 179.Dik DA, Dominguez-Gil T, Lee M, Hesek D, Byun B, Fishovitz J, Boggess B, Hellman LM, Fisher JF, Hermoso JA, Mobashery S. 2017. Muropeptide binding and the X-ray structure of the effector domain of the transcriptional regulator AmpR of Pseudomonas aeruginosa. J Am Chem Soc 139:1448–1451. doi: 10.1021/jacs.6b12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Matsuyama BY, Krasteva PV, Baraquet C, Harwood CS, Sondermann H, Navarro MV. 2016. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:E209–E218. doi: 10.1073/pnas.1523148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kilmury SLN, Burrows LL. 2018. The Pseudomonas aeruginosa PilSR two-component system regulates both twitching and swimming motilities. mBio 9:e01310-18. doi: 10.1128/mBio.01310-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Balasubramanian D, Kumari H, Jaric M, Fernandez M, Turner KH, Dove SL, Narasimhan G, Lory S, Mathee K. 2014. Deep sequencing analyses expands the Pseudomonas aeruginosa AmpR regulon to include small RNA-mediated regulation of iron acquisition, heat shock and oxidative stress response. Nucleic Acids Res 42:979–998. doi: 10.1093/nar/gkt942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang C, Ye F, Kumar V, Gao YG, Zhang LH. 2014. BswR controls bacterial motility and biofilm formation in Pseudomonas aeruginosa through modulation of the small RNA rsmZ. Nucleic Acids Res 42:4563–4576. doi: 10.1093/nar/gku106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schulz S, Eckweiler D, Bielecka A, Nicolai T, Franke R, Dotsch A, Hornischer K, Bruchmann S, Duvel J, Haussler S. 2015. Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog 11:e1004744. doi: 10.1371/journal.ppat.1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol 51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 186.Wolfgang MC, Lee VT, Gilmore ME, Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell 4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 187.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. doi: 10.1128/jb.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Heacock-Kang Y, Sun Z, Zarzycki-Siek J, McMillan IA, Norris MH, Bluhm AP, Cabanas D, Fogen D, Vo H, Donachie SP, Borlee BR, Sibley CD, Lewenza S, Schurr MJ, Schweizer HP, Hoang TT. 2017. Spatial transcriptomes within the Pseudomonas aeruginosa biofilm architecture. Mol Microbiol 106:976–985. doi: 10.1111/mmi.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tata M, Wolfinger MT, Amman F, Roschanski N, Dotsch A, Sonnleitner E, Haussler S, Blasi U. 2016. RNASeq based transcriptional profiling of Pseudomonas aeruginosa PA14 after short- and long-term anoxic cultivation in synthetic cystic fibrosis sputum medium. PLoS One 11:e0147811. doi: 10.1371/journal.pone.0147811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cornforth DM, Dees JL, Ibberson CB, Huse HK, Mathiesen IH, Kirketerp-Moller K, Wolcott RD, Rumbaugh KP, Bjarnsholt T, Whiteley M. 2018. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci U S A 115:E5125–E5134. doi: 10.1073/pnas.1717525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Damron FH, Oglesby-Sherrouse AG, Wilks A, Barbier M. 2016. Dual-seq transcriptomics reveals the battle for iron during Pseudomonas aeruginosa acute murine pneumonia. Sci Rep 6:39172. doi: 10.1038/srep39172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cole SJ, Hall CL, Schniederberend M, Farrow Iii JM, Goodson JR, Pesci EC, Kazmierczak BI, Lee VT. 2018. Host suppression of quorum sensing during catheter-associated urinary tract infections. Nat Commun 9:4436. doi: 10.1038/s41467-018-06882-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pusic P, Tata M, Wolfinger MT, Sonnleitner E, Haussler S, Blasi U. 2016. Cross-regulation by CrcZ RNA controls anoxic biofilm formation in Pseudomonas aeruginosa. Sci Rep 6:39621. doi: 10.1038/srep39621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pusic P, Sonnleitner E, Krennmayr B, Heitzinger DA, Wolfinger MT, Resch A, Blasi U. 2018. Harnessing metabolic regulation to increase Hfq-dependent antibiotic susceptibility in Pseudomonas aeruginosa. Front Microbiol 9:2709. doi: 10.3389/fmicb.2018.02709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Boudes M, Lazar N, Graille M, Durand D, Gaidenko TA, Stewart V, van Tilbeurgh H. 2012. The structure of the NasR transcription antiterminator reveals a one-component system with a NIT nitrate receptor coupled to an ANTAR RNA-binding effector. Mol Microbiol 85:431–444. doi: 10.1111/j.1365-2958.2012.08111.x. [DOI] [PubMed] [Google Scholar]
- 197.Brewster JL, McKellar JL, Finn TJ, Newman J, Peat TS, Gerth ML. 2016. Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae. Sci Rep 6:35198. doi: 10.1038/srep35198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE Jr, Kim SH. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 199.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]