In this study, Li et al. set out to investigate the conserved genome stability factor Esc2 in budding yeast and its roles in DNA damage-induced sumoylation. Using in vitro and in vivo approaches, the authors propose that Esc2 acts as a SUMO E2 cofactor at distinct DNA structures to promote the sumoylation of specific substrates and genome maintenance.
Keywords: Esc2, homologous recombination, SUMO E2, genome maintenance
Abstract
SUMO modification regulates diverse cellular processes by targeting hundreds of proteins. However, the limited number of sumoylation enzymes raises the question of how such a large number of substrates are efficiently modified. Specifically, how genome maintenance factors are dynamically sumoylated at DNA replication and repair sites to modulate their functions is poorly understood. Here, we demonstrate a role for the conserved yeast Esc2 protein in this process by acting as a SUMO E2 cofactor. Esc2 is required for genome stability and binds to Holliday junctions and replication fork structures. Our targeted screen found that Esc2 promotes the sumoylation of a Holliday junction dissolution complex and specific replisome proteins. Esc2 does not elicit these effects via stable interactions with substrates or their common SUMO E3. Rather, we show that a SUMO-like domain of Esc2 stimulates sumoylation by exploiting a noncovalent SUMO binding site on the E2 enzyme. This role of Esc2 in sumoylation is required for Holliday junction clearance and genome stability. Our findings thus suggest that Esc2 acts as a SUMO E2 cofactor at distinct DNA structures to promote the sumoylation of specific substrates and genome maintenance.
SUMO (small ubiquitin-like modifier) regulates many cellular processes via covalent modification of a myriad of proteins. Substrate sumoylation requires the sequential action of the trio of SUMO E1, E2, and E3 enzymes, akin to ubiquitination. However, unlike the vast array of ubiquitination enzymes, all organisms examined so far contain only a single SUMO E1 and E2 enzyme and a few SUMO E3s (Pichler et al. 2017). For example, in budding yeast, the Aos1-Uba2 heterodimeric E1, the Ubc9 E2, and three mitotic E3s (Siz1, Siz2, and Mms21) are responsible for sumoylating hundreds of proteins (Albuquerque et al. 2013). How a large number of proteins are specifically and efficiently modified by a small number of sumoylation enzymes is an outstanding question. Because SUMO is directly transferred from the Ubc9 E2 active site to substrates with the help of E3s, efficient modification of large numbers of diverse substrates may require cofactors to direct E2 and E3 functions in a spatially or temporally regulated manner. Addressing this possibility could provide new insights into sumoylation mechanisms and regulation.
Here, we investigated the conserved genome stability factor Esc2 in budding yeast and its roles in DNA damage-induced sumoylation (Cremona et al. 2012; Psakhye and Jentsch 2012). The Esc2 family of proteins has been implicated in sumoylation; however, a unified mechanism of their functions has yet to be established. While the fission yeast Esc2 ortholog, Rad60, has been proposed as a general regulator of the Nse2 SUMO E3 (Mms21 ortholog) (Prudden et al. 2011), studies in budding yeast reveal a more complex scenario. Both Esc2 and Mms21 support Holliday junction (HJ) clearance in response to DNA damage (Zhao and Blobel 2005; Branzei et al. 2006; Mankouri et al. 2009; Sollier et al. 2009; Choi et al. 2010), but they were reported to affect different pathways: Mms21 promotes the sumoylation of the HJ dissolution enzyme, the Sgs1-Top3-Rmi1 complex (STR), whereas Esc2 stimulates a HJ clearance pathway mediated by the Mus81-Mms4 nuclease (Bermúdez-López et al. 2016; Bonner et al. 2016; Sebesta et al. 2017). The effect of Esc2 on the Mus81-Mms4 pathway depends on its DNA binding domain that has a strong preference for HJ and replication fork structures (Urulangodi et al. 2015; Sebesta et al. 2017). Distinct from Esc2, the Smc5/6 complex, of which Mms21 is an obligate subunit, localizes across chromosomes and affects sumoylation at a variety of DNA structures (Zhao and Blobel 2005; Lindroos et al. 2006; Takahashi et al. 2008; Bermúdez-López et al. 2016; Bonner et al. 2016; Meng et al. 2019; Winczura et al. 2019; Whalen et al. 2020). The differences described above raise the question of whether Esc2 influences sumoylation as a general regulator of the Mms21 E3 or through other mechanism(s).
Interestingly, the Esc2 family of proteins possesses two SUMO-like domains (SLD1 and SLD2), and SLD2 can bind to the Ubc9 SUMO E2 (Novatchkova et al. 2005; Prudden et al. 2009; Sollier et al. 2009; Sekiyama et al. 2010). These common features suggest a potential role for these proteins in assisting Ubc9. In this work, we tested this possibility and clarified the functional link between Esc2 and Mms21. Our targeted screen of genome stability factors reveals that Esc2 specifically influences the Mms21 substrates associating with HJs or replication fork structures. Our complementary in vivo and in vitro tests demonstrate that the role of Esc2 in sumoylation is mediated by its SLD2 binding to the so-called “backside” of Ubc9. We show that this binding contributes to HJ clearance and genome stability in cells. Our work thus uncovers Esc2 as a SUMO E2 cofactor that aids the sumoylation of Mms21 substrates located at HJs and replication forks to enhance repair completion and preserve genome integrity.
Results
Esc2 promotes the sumoylation of a subset of Mms21 substrates
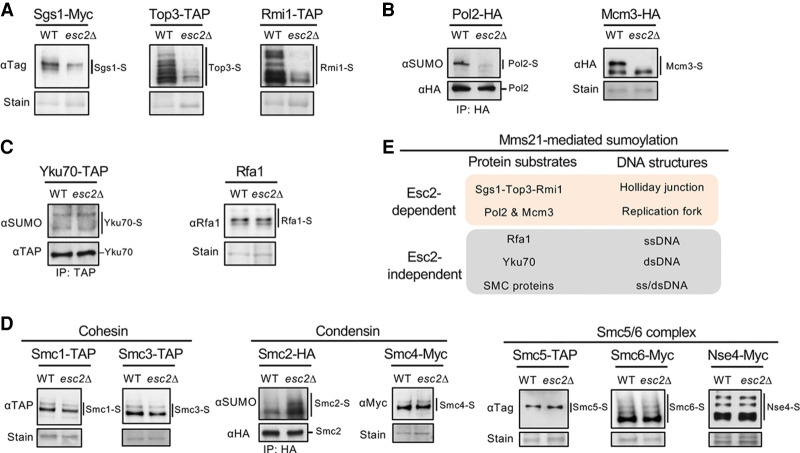
To elucidate the role(s) of Esc2 in sumoylation and its relationship with the Mms21 E3, we queried how Esc2 loss influences DNA damage-induced sumoylation of genome maintenance factors. Given that both esc2 and mms21 mutants accumulate HJ structures upon MMS treatment that are resolved by the STR complex (Branzei et al. 2006; Mankouri et al. 2009; Sollier et al. 2009), we first examined STR sumoylation. Mms21-mediated STR sumoylation is thought to occur at HJs and contribute to HJ clearance (Bermúdez-López et al. 2016; Bonner et al. 2016). We found that in esc2Δ cells, all three STR subunits had reduced sumoylation levels upon MMS treatment (Fig. 1A), suggesting that this defect could underlie the elevated HJ levels in esc2Δ cells in this condition.
Figure 1.
Esc2 promotes the sumoylation of a specific set of Mms21 substrates. (A) Sumoylation levels of the subunits of the Sgs1-Top3-Rmi1 complex are reduced in esc2Δ cells. Cells containing His8-tagged SUMO were treated with 0.03% MMS for 2 h to induce STR sumoylation. Sumoylated proteins were isolated using Ni-NTA resins and examined by immunoblotting using antibodies recognizing the tag fused to the endogenous Sgs1, Top3, or Rmi1 to visualize sumoylated forms of the proteins (-S), which are indicated by lines next to the blots. Loading is shown by Ponceau S stain (stain). (WT) Wild type. Similar methods for examining sumoylation and annotation of immunoblots are used in subsequent panels unless otherwise noted. (B) esc2Δ reduces the levels of mono-sumoylated form of Pol2 and di-sumoylated form of Mcm3. (Left) HA-tagged Pol2 was immunoprecipitated and its sumoylated form was detected by immunoblotting using anti-SUMO antibody as shown previously (Meng et al. 2019). The unmodified band was detected against the tag (HA) for equal loading control. (Right) Mcm3 sumoylation was examined as in A. (C) esc2Δ cells maintain the sumoylation levels of Yku70 and Rfa1. Yku70 sumoylation was examined as in B (left) and as shown previously (Zhao and Blobel 2005), whereas Rfa1 sumoylation was examined as in A as shown previously (Chung and Zhao 2015). (D) esc2Δ cells maintain the sumoylation levels for the subunits of three SMC complexes, namely cohesin, condensin, and the Smc5/6 complex. Sumoylation of Smc2 was examined as in B (left), and sumoylation of the other proteins was examined as in A. (E) Summary of the effects of esc2Δ on Mms21-dependent sumoylation of DNA metabolism proteins. Results in A–D show that esc2Δ reduces the sumoylation of proteins known to associate with HJ and replication fork structures but not those that mainly interact with ssDNA or dsDNA.
Since Esc2 preferentially binds to HJs and replication fork structures (Urulangodi et al. 2015; Sebesta et al. 2017), we next examined two replisome proteins whose sumoylation is regulated by Mms21. Monosumoylation of the leading strand DNA polymerase Pol2, which was suggested to occur at replication forks, and the disumoylated form of the Mcm3 subunit of the replicative helicase are largely abolished in mms21 E3 mutants (Albuquerque et al. 2016; Meng et al. 2019; Winczura et al. 2019). We found that esc2Δ phenocopied the mms21 mutant in both cases (Fig. 1B). In contrast, esc2Δ cells were proficient for the sumoylation of Mms21 substrates located at other types of DNA structures (Fig. 1C,D). These include the dsDNA end binding protein Yku70, ssDNA binding protein Rfa1, and subunits of three SMC complexes that predominantly associate with dsDNA (Zhao and Blobel 2005; Takahashi et al. 2008; Whalen et al. 2020). In addition, esc2Δ cells maintained the sumoylation of Siz E3 substrates that bind to HJ, replication fork, or DNA flap structures (Supplemental Fig. S1A–C; Hoege et al. 2002; Stelter and Ulrich 2003; Sarangi et al. 2014; Talhaoui et al. 2018). Our data (summarized in Fig. 1E) suggest that, while Esc2 has a functional partnership with Mms21, it does not act as a general regulator of this E3; rather, Esc2 specifically contributes to the sumoylation of Mms21 substrates associating with HJs and replication fork structures.
Query of Esc2 interactions with SUMO E2, SUMO E3, SUMO, and SUMO substrates
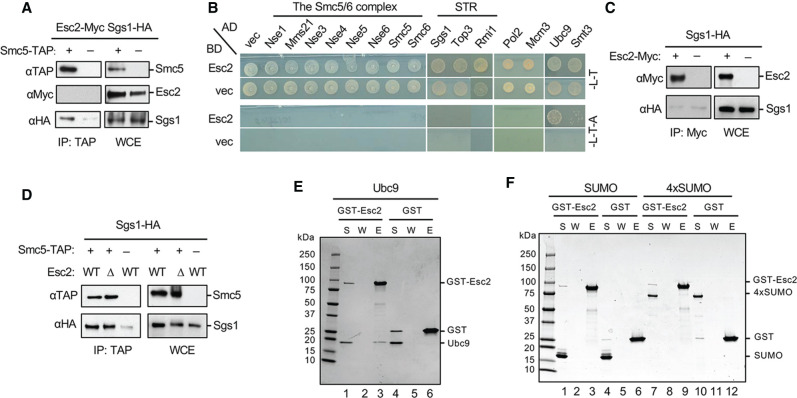
Consistent with the above notion that Esc2 is not a general Mms21 regulator, we did not detect association of Esc2 with the Smc5/6 complex, of which Mms21 is an obligate subunit, either through coimmunoprecipitation or yeast two-hybrid assays (Fig. 2A,B). We also did not find evidence that Esc2 could bind to the SUMO substrates identified above or bridge their interactions with Mms21. Yeast two-hybrid tests did not reveal interaction of Esc2 with STR subunits, Pol2, or Mcm3 (Fig. 2B). Moreover, Esc2 did not coimmunoprecipitate with Sgs1 or Pol2, whereas the association of Sgs1 with Smc5 or Pol2 with its partner protein Dpb2 was detected (Fig. 2A,C; Supplemental Fig. S2A). Purified Esc2 did not show interaction with STR or Smc5/6 in vitro (Supplemental Fig. S2B,C). Finally, Esc2 loss did not affect Smc5 association with Sgs1 or Pol2 in vivo (Fig. 2D; Supplemental Fig. S2D).
Figure 2.
Examination of Esc2 interactions with Ubc9, SUMO, STR, and the Smc5/6 complex. (A) Smc5 coimmunoprecipitates with Sgs1 but not Esc2. Cells containing endogenously Myc-tagged Esc2 and HA-tagged Sgs1, with or without TAP-tagged Smc5, were examined by co-IP tests. Representative immunoblots examining co-IP eluate (IP) and the whole cell extract (WCE) are shown. (B) Esc2 interacts with Ubc9 and SUMO (Smt3), but not subunits of the Smc5/6 complex and the STR complex nor Pol2 and Mcm3 in yeast-two-hybrid assays. (AD) Gal4 activation domain, (BD) Gal4 DNA binding domain, (vec) vector. SC-Leu-Trp media (-L-T) select for BD and AD plasmids, while SC-Leu-Trp-Ade media (L-T-A) report for positive interactions. (C) Esc2 does not coimmunoprecipitate with Sgs1. Similar levels of Sgs1 were detected in IP samples regardless of whether cells contain Myc-tagged Esc2 or not, reflecting nonspecific binding of Sgs1 to the beads. (D) Smc5 copurifies with Sgs1 regardless of the Esc2 status. Experiments were done as in A. (E,F) Protein binding assays showing that Esc2 binds to Ubc9 but not SUMO or a SUMO chain. Purified GST or GST-Esc2 proteins bound to glutathione beads were examined for their abilities to pull down Ubc9 (E), SUMO (F, lanes 1–6), or SUMO chain composed of four tandem SUMO moieties (F, lanes 7–12). Proteins were examined by SDS-PAGE, and pictures of representative gels after Coomassie blue stain are shown. (S) Supernatant, (W) wash, (E) eluate.
As Esc2 has been previously shown to interact with Ubc9 and SUMO by yeast two-hybrid assay (Sollier et al. 2009), we explored the possibility that Esc2 elicits its effects via these interactions. We found that Esc2 interacted with Ubc9 and weakly with SUMO (Smt3) in yeast two-hybrid assays (Fig. 2B). To discern direct binding from indirect association, we performed in vitro pull-down tests using purified proteins. While Esc2 interaction with Ubc9 was readily detected, we failed to detect Esc2 interaction with either the monomeric SUMO or a four-SUMO fusion construct, which can enhance SUMO-mediated interactions (Fig. 2E,F). Using microscale thermophoresis, we found that Ubc9 bound to Esc2 with a dissociation constant Kd of 372 ± 57 nM (Supplemental Fig. S2E). Taken together, the above results suggest that Esc2 can associate with the SUMO E2, which may contribute to its sumoylation function.
SLD2 binding to Ubc9 is critical for Esc2-mediated sumoylation
Esc2 family proteins contain two SUMO-like domains, SLD1 and SLD2 (Novatchkova et al. 2005). In Esc2, these domains are located C-terminal from the DNA binding domain (Fig. 3A). SLD2 has a higher degree of sequence similarity with SUMO than SLD1 and adopts a structural fold reminiscent of SUMO; however, SLD2 lacks key features required for conjugation (Prudden et al. 2009; Sekiyama et al. 2010). Structural studies show that SLD2 of the fission yeast Rad60 and the mouse Nip45 proteins bind to Ubc9 in a similar fashion as SUMO (Sekiyama et al. 2010; Prudden et al. 2011). Both SLD2 and SUMO bind to the “Ubc9 ‘backside,’” which is opposite from the Ubc9 active site that forms a thioester bond with SUMO (Supplemental Fig. S3A). While SUMO binding to the Ubc9 backside can stimulate SUMO chain formation (Knipscheer et al. 2007), it is unclear how the association of SLD2 with Ubc9 affects substrate sumoylation in cells.
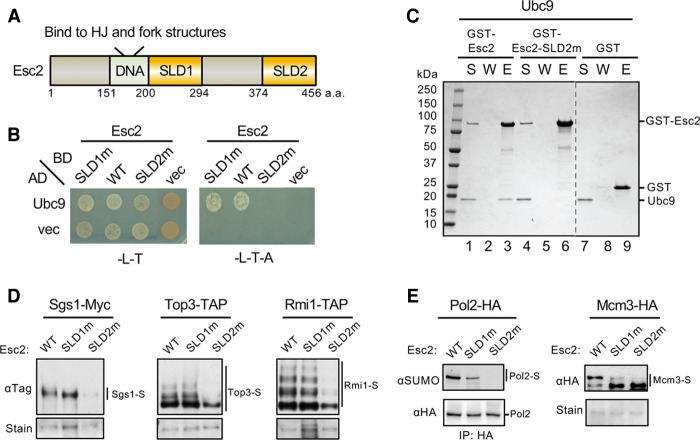
Figure 3.
Esc2 binding to Ubc9 is required for efficient sumoylation of the STR complex, Pol2 and Mcm3. (A) A schematic of Esc2 protein domains. (DNA) DNA-binding domain that prefers to bind HJ and fork structures, (SLD1) SUMO-like domain 1, (SLD2) SUMO-like domain 2. (B) Mutating the SLD2 but not SLD1 domain of Esc2 abolishes its interaction with Ubc9 in yeast two-hybrid assays. Experiments were done and data are presented as in Figure 2B. (C) Esc2-SLD2m abolished Ubc9 interaction in vitro. Experiments were done and data are presented as in Figure 2E. Dotted line denotes removal of superfluous lanes. (D) esc2-SLD2m, but not -SLD1m, reduces sumoylation levels of the STR subunits in cells. Experiments were done and data are presented as in Figure 1A. (E) esc2-SLD2m reduces levels of monosumoylation of Pol2 and disumoylation of Mcm3 in vivo. Experiments were done and data are presented as in Figure 1B.
To elucidate how SLD2 binding to Ubc9 contributes to Esc2 functions, we aimed to generate a mutant disrupting this interaction. We first built a structural model of the SLD2-Ubc9 complex using homology modeling based on the published structures of budding yeast Ubc9 and the complex of Ubc9-Rad60SLD2 (Duda et al. 2007; Sekiyama et al. 2010; Prudden et al. 2011). This model highlighted the conserved D449 residue of SLD2 at the Ubc9 interface (Supplemental Fig. S3B). We mutated D449 and the adjacent conserved residue D447 to alanine (D447A, D449A), generating the previously described esc2-SLD2m mutant that has only been examined in combination with a SLD1 mutant (D286A, I287Y or -SLD1m) (Urulangodi et al. 2015). We found that esc2-SLD2m, but not esc2-SLD1m, abrogated the interaction with Ubc9 in a yeast two-hybrid assay (Fig. 3B). In vitro pull-down tests using purified proteins validated the two-hybrid result (Fig. 3C; Supplemental Fig. S3C). Additionally, the Esc2-SLD2m mutant protein and the wild-type Esc2 protein showed similar circular dichroism profiles, indicating that the loss of interaction was not due to altered conformation of the mutant protein (Supplemental Fig. S3D). Taken together, we conclude that SLD2, but not SLD1, is responsible for the Esc2-Ubc9 interaction, and esc2-SLD2m effectively disrupts this interaction.
We then used esc2-SLD2m to evaluate the biological significance of the Esc2-Ubc9 interaction. Strikingly, esc2-SLD2m, which supported wild-type protein levels, behaved like esc2Δ in reducing the sumoylation levels of STR subunits in cells, while esc2-SLD1m had no effect (Fig. 3D; Supplemental Fig. S3E). In addition, esc2-SLD2m abolished Pol2 monosumoylation and Mcm3 disumoylation, whereas esc2-SLD1m showed milder defects (Fig. 3E). We suspect that SLD1 involvement in replication fork regulation (Urulangodi et al. 2015) and/or the compromised expression of the Esc2-SLD1m protein (Supplemental Fig. S3E) could explain the SLD1 mutant's effect on Pol2 and Mcm3 sumoylation. Collectively, our results indicate that Esc2 uses its SLD2 interaction with Ubc9 to promote substrate sumoylation in cells.
In vitro Sgs1 and Top3 sumoylation systems recapitulate in vivo requirements
To discern whether SLD2 can directly influence the sumoylation reaction, we established an in vitro sumoylation system for Sgs1 using purified proteins (Supplemental Figs. S2B, S3F). Incubation of Sgs1 with SUMO, the SUMO E1, the Ubc9 E2, and ATP produced high molecular weight Sgs1 species reminiscent of the endogenous Sgs1 sumoylation forms (Supplemental Fig. S3G; Bermúdez-López et al. 2016; Bonner et al. 2016). To confirm that these species were indeed sumoylated forms of Sgs1, we purified two Sgs1 variants known to be defective in sumoylation in vivo, namely Sgs1-K621R mutated for the main sumoylation site and Sgs1-sim mutated for its SUMO interacting motifs (Supplemental Fig. S3F; Bermúdez-López et al. 2016; Bonner et al. 2016). As expected, these variants were not sumoylated in the in vitro reactions, validating our in vitro system (Supplemental Fig. S3G).
Next, we purified the Mms21-Smc5 complex as the SUMO E3 complex, because Mms21 is not well expressed on its own (Supplemental Fig. S3F; Zhao and Blobel 2005). The addition of the Mms21-Smc5 SUMO E3 complex into the sumoylation reactions containing the STR complex greatly stimulated Sgs1 and Top3 sumoylation over time, as evidenced by their upshifted bands on immunoblots (Fig. 4A). This is in line with in vivo findings (Bermúdez-López et al. 2016; Bonner et al. 2016), demonstrating that Mms21 directly enhances Sgs1 and Top3 sumoylation. Rmi1 was not sumoylated in this system, possibly because its sumoylation requires additional factors not present in the assay. We focused on Sgs1 and Top3 sumoylation thereafter.
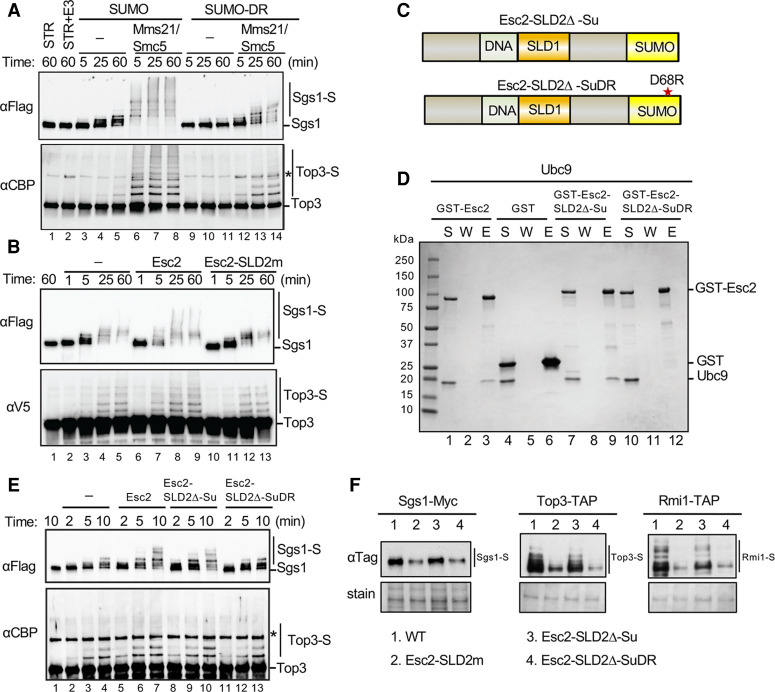
Figure 4.
Esc2 aids sumoylation via its SLD2 binding to the Ubc9 backside. (A) Mms21-Smc5 stimulates Sgs1 and Top3 sumoylation in the presence of SUMO or SUMO-D68R. In vitro sumoylation assays were performed by incubating purified STR complex with the SUMO E1, the SUMO E2, SUMO (or SUMO-DR), and ATP in the presence or absence of the Mms21-Smc5 SUMO E3 at 30°C for the indicated time (for details, see the Materials and methods). Sgs1 tagged with FLAG and Top3 tagged with CBP and their sumoylated forms were detected by immunoblotting against the tags fused to them. Asterisk indicates a cross-reaction band. (B) Esc2, but not Esc2-SLD2m, stimulates Sgs1 and Top3 sumoylation in vitro. Sumoylation assays were performed as in panel A in the presence of Mms21-Smc5 and SUMO-DR, except Top3 is tagged with V5. The inclusion of Esc2 or Esc2-SLD2m is indicated. (C) Schematics of two Esc2 variants wherein its SLD2 is replaced by SUMO (Esc2-SLD2Δ-Su) or by SUMO-D68R (Esc2-SLD2Δ-SuDR). (D) Esc2-SLD2Δ-Su, but not esc2-SLD2Δ-SuDR, protein interacts with Ubc9 in vitro. GST pull-down tests were performed and results are presented as in Figure 2E. (E) SUMO, but not SUMO-DR, can replace the SLD2 of Esc2 in stimulating Sgs1 and Top3 sumoylation. Sumoylation assays were performed as in A in the presence of Mms21-Smc5 and SUMO-DR, except with shorter time courses. (F) SLD2 can be replaced by SUMO, but not SUMO-DR, to support STR sumoylation in cells. Experiments were done and data are presented as in Figure 1A.
Esc2 stimulates Sgs1 and Top3 sumoylation and this requires SLD2 binding to Ubc9
We proceeded to test whether Esc2 directly stimulated Sgs1 and Top3 sumoylation. As in vitro sumoylation reactions require high concentrations of SUMO, we first asked whether SUMO and Esc2 compete for binding to Ubc9, which could obscure potential effects of Esc2 in the assay. Indeed, we found that Ubc9 prebound to GST-Esc2 on beads was competed off with increased levels of SUMO (Supplemental Fig. S4A). To circumvent this competition, we used a SUMO variant mutated in a key residue for binding to the Ubc9 backside, Smt3-D68R (SUMO-DR) (Duda et al. 2007; Knipscheer et al. 2007). SUMO-DR failed to compete with Esc2 for Ubc9 binding as expected (Supplemental Fig. S4B). Importantly, SUMO-DR showed wild-type competency in conjugating to Ubc9 in the presence of SUMO E1 and ATP (Supplemental Fig. S4C), indicating that it transfers proficiently from E1 to E2. Furthermore, when used in the Sgs1-Top3 sumoylation assay described above, SUMO-DR supported the sumoylation of both proteins (Fig. 4A). In these reactions, SUMO-DR gave rise to fewer sumoylated species than SUMO, particularly those of higher molecular weights, consistent with its reported impairment in SUMO chain formation (Fig. 4A; Duda et al. 2007; Knipscheer et al. 2007). Taken together, the above results validate the use of SUMO-DR in the in vitro sumoylation system.
We moved on to examine how Esc2 affected in vitro sumoylation using SUMO-DR. Esc2 did not affect E1-mediated SUMO conjugation of Ubc9 (Supplemental Fig. S4C), suggesting that Esc2 must regulate Ubc9 after it is conjugated (charged) with SUMO at its active site. Time course experiments showed that Esc2 stimulated E3-dependent sumoylation of Sgs1 and, to a lesser degree, Top3 (Fig. 4B). For instance, while a small proportion of Sgs1 was sumoylated in the absence of Esc2 in 5 min (Fig. 4B, lane 3, top), the majority of Sgs1 was sumoylated in the presence of Esc2 in the same time (Fig. 4B, lane 7, top). Importantly, the Esc2-SLD2m mutant that does not interact with Ubc9 failed to elicit the same effect, indicating that Esc2 stimulation of sumoylation hinges upon its SLD2-mediated interaction with Ubc9 (Fig. 4B). Thus, our in vivo and in vitro results demonstrate a direct role for Esc2 and its SLD2 in stimulating substrate sumoylation.
The structural mimicry of SLD2 underlies its stimulation of STR sumoylation
In addressing the mechanisms by which SLD2 binding to Ubc9 stimulates substrate sumoylation, we considered previous findings that SUMO binding to the Ubc9 backside better orients the Ubc9 active site for SUMO transfer in SUMO chain formation (Duda et al. 2007; Knipscheer et al. 2007). We tested whether SLD2 promoted substrate sumoylation via a similar structure-based mechanism by examining whether SUMO could substitute for SLD2. To this end, we generated and purified an Esc2 variant wherein its SLD2 was replaced by a SUMO moiety that lacked the diglycine motif required for conjugation (Fig. 4C). The resulting Esc2-SLD2Δ-Su fusion protein was proficient for interaction with Ubc9 (Fig. 4D; Supplemental Fig. S3C). Significantly, Esc2-SLD2Δ-Su stimulated in vitro Sgs1 and Top3 sumoylation to a similar extent as Esc2 (Fig. 4E). Moreover, this effect relied on binding to Ubc9, since the Esc2-SLD2Δ-SuDR variant containing the D68R mutation that impairs Ubc9 backside interaction could not stimulate substrate sumoylation (Fig. 4C–E). These results indicate that SLD2 exploits the SUMO binding surface on the Ubc9 backside to enhance the SUMO E2 function. We note that, unlike SUMO, Esc2 did not stimulate free SUMO chain formation, suggesting that, despite using a similar strategy for associating with the E2, SUMO and Esc2 exhibit distinct effects (Supplemental Fig. S4D).
To extend our in vitro findings, we replaced the ESC2 gene at its endogenous locus in cells with either esc2-SLD2Δ-Su or esc2-SLD2Δ-SuDR. While esc2-SLD2Δ-Su maintained wild-type sumoylation levels of STR subunits, esc2-SLD2Δ-SuDR was defective, as seen for esc2-SLD2m (Fig. 4F). Thus, complementary in vitro and in vivo data support the conclusion that SLD2 exploits the SUMO binding surface of Ubc9 to enhance substrate sumoylation.
Esc2 binding to Ubc9 limits GCRs and recombination intermediate accumulation
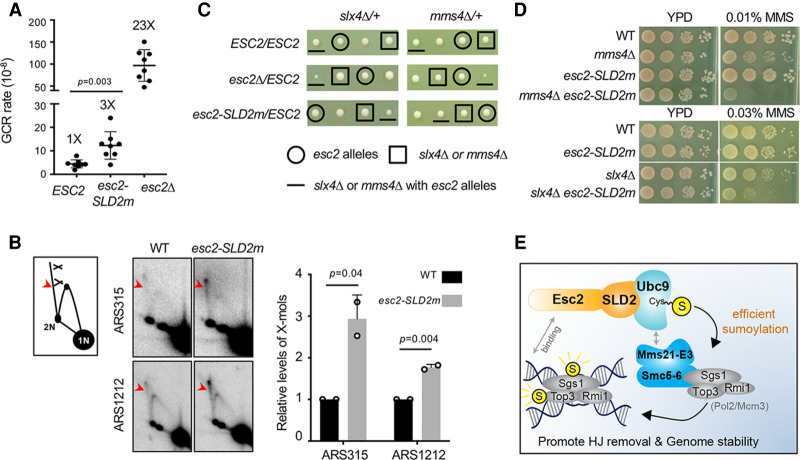
We next addressed the impact of the Esc2-Ubc9 interaction on genome maintenance. Esc2 is known to promote genome stability during growth and enhance HJ removal in the presence of MMS (Sollier et al. 2009; Albuquerque et al. 2013). We first queried genome stability using the gross chromosomal rearrangement (GCR) assay (Putnam et al. 2009). Similar to previous reports, esc2Δ cells exhibited a 23-fold increase in GCR rates compared with wild-type cells (Kanellis et al. 2007; Mankouri et al. 2009). We found that esc2-SLD2m cells showed an approximately threefold higher GCR rate than wild type (Fig. 5A), indicating that the Esc2-Ubc9 association contributes to Esc2's roles in maintaining genome stability.
Figure 5.
Esc2 binding to Ubc9 curbs levels of GCR and recombination intermediates. (A) esc2Δ and esc2-SLD2m mutants increase GCR rates. For each genotype, the median rate of at least nine cultures was calculated from two biological duplicates. Error bars are 95% confidence intervals. Two-tailed Mann–Whitney tests were performed for statistical analysis. (B) 2D gel data show that esc2-SLD2m mutants increase X-mol levels at two genomic sites. α-Factor-arrested G1 cells were released into media containing MMS for 2 h. Samples were examined by 2D gel followed by Southern blotting using probes at ARS315 or ARS1212. (Left) A schematic of 2D gel images with X-mol spike indicated by red arrowhead. (Middle) Representative 2D gel images. (Right) Quantification of relative X-mol levels from two different spore clones per genotype, with error bars representing standard deviations. WT level was set to 1.0, and the P-value is derived by Student's t-test. (C) Tetrad analyses from diploid strains with indicated genotype. Spore clones were grown for 2 d at 30°C. Spores containing different mutations are identified based on genotyping. One representative tetrad among at least nine tetrads per diploid strain is shown. (D) esc2-SLD2m worsens genotoxic sensitivity of slx4Δ and mms4Δ cells. Cells were spotted in 10-fold serial dilutions and grown for 2 d at 30°C. (E) A model for Esc2 stimulation of specific substrate sumoylation. Esc2 binding to the backside of Ubc9 (SUMO E2) through its SLD2 leads to the stimulation of sumoylation of a subset of Smc5-6-Mms21 E3 substrates, likely at HJ and replication fork sites, contributing to HJ dissolution and genome stability.
Next, we evaluated the impact of the Esc2-Ubc9 interaction on HJ removal. We used both genetic assays and 2D agarose gel electrophoresis followed by Southern blotting (2D gel) for detecting X-shaped molecules (X-mols) such as HJ structures (Fig. 5B, left). When cells replicated in MMS, esc2-SLD2m cells exhibited a two- to threefold increase in the levels of X-mols compared with wild type at loci near ARS315 or ARS1212 (Fig. 5B, middle and right). This defect likely reflects an impairment in STR-mediated HJ clearance and not the Mus81-Mms4 nuclease function, since STR is responsible for X-mol removal when cells grow in the presence of MMS (Liberi et al. 2005; Matos et al. 2011; Sebesta et al. 2017). In addition, we found that Esc2-SLD2m was proficient for Mus81-Mms4 association (Supplemental Fig. S5A). Furthermore, like STR mutants, esc2-SLD2m sensitized cells lacking Mms4 or Slx4, which acts in other HJ removal pathways (Fig. 5C,D). In both cases, esc2-SLD2m rendered these mutant cells more sensitive to MMS (Fig. 5D). In the case of slx4Δ, the double mutants also exhibited slower growth on dissection plates (Fig. 5C). As the sensitization effect of esc2-SLD2m toward mms4Δ or slx4Δ cells was less severe than esc2Δ, Esc2 must have SLD2-independent roles. Interestingly, we found that esc2-SLD2Δ-Su did not sensitize mms4Δ or slx4Δ, while esc2-SLD2Δ-SuDR did, suggesting that the observed sensitization by esc2 mutants stems from the loss of Esc2-Ubc9 interaction (Supplemental Fig. S5B,C). In summary, the corroborative data described above suggest that the Esc2-Ubc9 interaction limits recombination intermediate accumulation in cells, and this can be partly mediated by regulating STR sumoylation.
Discussion
Protein sumoylation regulates many cellular processes, yet it remains unclear how the limited number of sumoylation enzymes can efficiently modify hundreds of diverse substrates within the SUMO proteome. A previous study from the Pichler group showed that sumoylation of the SUMO E2 enzyme leads to increased modification of certain substrates in mammalian cells (Knipscheer et al. 2008). Our work suggests another means wherein an E2 cofactor can enable specific substrate sumoylation in yeast. We show that the conserved Esc2 protein aids the Ubc9 E2 in substrate sumoylation both in vivo and in vitro. Mechanistically, Esc2 uses a SUMO-like domain to bind the Ubc9 backside, stimulating the sumoylation reaction. Genetic and 2D gel results suggest that this role of Esc2 positively affects genome stability and HJ structure elimination.
We found that Esc2 regulated the sumoylation of a subset of Mms21 substrates that associate with HJ and replication fork structures. Previous studies have suggested that STR and Pol2 sumoylation likely occurs at HJ and replication fork structures, respectively (Bonner et al. 2016; Meng et al. 2019). As Esc2 preferentially binds to these DNA structures compared with dsDNA or ssDNA (Urulangodi et al. 2015; Sebesta et al. 2017), Esc2 may be a DNA structure-specific sumoylation regulator.
Our cellular and in vitro data support the conclusion that Esc2 stimulates substrate sumoylation and provide an understanding of the possible mechanisms. Building upon previous findings, we show that the Esc2-Ubc9 interaction depends on its SLD2 rather than SLD1. Our results further suggest that Esc2 likely employs its SLD2 to bind the Ubc9 backside, as seen for its homologs (Prudden et al. 2009; Sekiyama et al. 2010). Importantly, we show for the first time that esc2-SLD2m mimics esc2Δ in reducing substrate sumoylation in cells. We also established an in vitro system for assaying Sgs1 and Top3 sumoylation in detail. Using this system, we demonstrate that Esc2 directly stimulates the sumoylation reaction in a manner depending on its binding to Ubc9. Significantly, in vitro and in vivo Esc2-dependent sumoylation was supported when SLD2 was replaced by nonconjugatable SUMO, again in a manner requiring its binding to Ubc9. Thus, Esc2 likely uses its SLD2 to exploit a SUMO binding surface on Ubc9 to stimulate E2 function. A noteworthy distinction between SLD2 and SUMO binding to Ubc9 is that the former did not promote free SUMO chain formation; rather, it stimulated substrate sumoylation. Despite this difference, it is possible that Esc2 binding to the Ubc9 backside may also help to orient Ubc9 for transferring SUMO to the substrate lysine residue. This premise is consistent with our observation that Esc2 affects Ubc9 after it is charged with SUMO at its active site.
Molecular details regarding how Esc2 facilitates SUMO transfer from Ubc9 to specific Mms21 substrates is currently unclear. In principle, Esc2 could achieve this effect via binding both to Ubc9 and the Mms21 E3. However, we did not detect a stable interaction between Esc2 and Mms21, though a transient interaction that may elude our detection cannot be excluded. Also, we did not find evidence that Esc2 could stably associate with substrates or bridge substrate-E3 association. Rather, our data suggest that the Esc2-Ubc9 association is crucial for Esc2-mediated sumoylation, at least for the substrates examined here. It is likely that the association of both Esc2 and Mms21 with HJ and replication fork structures poise them for collaboration at these sites. The specific requirement of Esc2 also suggests that Mms21 action entails distinct features from the Siz E3s. While Siz E3s possess SUMO interaction regions that promote sumoylation (Streich and Lima 2016), Mms21 may use Esc2 to enhance sumoylation. For example, Esc2 binding to Ubc9 during Mms21-mediated sumoylation may better orient the donor SUMO to the target lysine. Alternatively, Esc2 binding to Ubc9 may lead to its own sumoylation, which then allows transient interactions with Mms21 and/or substrates to foster SUMO transfer. Regardless of the mechanism(s), Esc2 collaboration with Mms21 enhances sumoylation events required for HJ and replication fork control during genome stress.
We showed that the impaired sumoylation of STR in esc2-SLD2m cells correlated with more X-mols and sensitization of other HJ processing mutants. Thus, the previously documented role of Esc2 in HJ removal during DNA replication (Mankouri et al. 2009; Sollier et al. 2009) can be partly explained by Esc2-mediated STR sumoylation. Although Esc2 can stimulate Mus81-Mms4-mediated HJ cleavage, we show Esc2-SLD2m is proficient for binding to this nuclease. Thus, Esc2 appears to use distinct domains to regulate both STR and the Mus81-Mms4 pathways, likely at different cell cycle stages, thereby acting as a master regulator of HJ removal.
We found that esc2-SLD2m had moderately elevated GCR rates. Impairment of multiple sumoylation events likely contributes to this, as a pol2 sumoylation-defective mutant showed a more subtle GCR increase than esc2-SLD2m (Meng et al. 2019). While certain genome instability events, such as HJ accumulation, may not be captured by this GCR assay, the smaller increase in GCRs in esc2-SLD2m cells compared with esc2Δ cells is consistent with the milder esc2-SLD2m phenotype observed in the other assays. These differences likely reflect additional roles of Esc2 in genome regulation, including affecting the DNA helicase Srs2 at replication forks (Urulangodi et al. 2015).
Our data, in conjunction with previous findings, suggest a model wherein Esc2 and Mms21 localized at HJ and replication fork structures collaborate for timely and efficient substrate sumoylation (Fig. 5E). Esc2 binding to the Ubc9 backside can help the SUMO E2 to adopt a productive conformation required for SUMO transfer. Esc2 may also help to target Ubc9 to HJs and replication fork structures to increase the local availability of the enzyme. We note that while SLD2 binding to the Ubc9 backside is a conserved feature of the Esc2 family of proteins (Prudden et al. 2009; Sekiyama et al. 2010), variations of the above model may apply to other organisms. In particular, the fission yeast Rad60 can stably associate with the Nse2 SUMO E3, suggesting broader effects on sumoylation in this organism (Raffa et al. 2006; Prudden et al. 2009). It is also interesting to note that the mammalian Esc2 homolog Nip45 is required for coping with genotoxins, though the underlying mechanisms remain to be understood (Hurov et al. 2010). Elucidating the roles of Esc2 homologs and their SLDs in multicellular organisms will be informative.
In summary, we provide insights into how a conserved SUMO E2 interacting protein stimulates sumoylation during genome maintenance. Our work sets the foundation for future studies on other Ubc9 binding partners to better understand how sumoylation efficiency and specificity can be achieved in a variety of cellular contexts and organisms.
Materials and methods
Yeast strains, plasmids, primers, and genetic procedures
All yeast strains are derivatives of W1588-4C, a RAD5 derivative of W303 (MATa ade2-1 can1-100 ura3-1 his3-11,15 leu2-3,112 trp1-1 rad5-535). At least two strains per genotype were examined in each experiment, and only one is listed in Supplemental Table S1. Standard procedures were used for cell growth, media preparation, epitope tagging at endogenous loci, and spot assays. Plasmids and primers used are listed in Supplemental Tables S2 and S3, respectively. For Figure 5C, at least nine tetrads for each cross were dissected and analyzed, and only one tetrad was shown.
Detection of protein sumoylation in cells
Two methods were used for detecting protein sumoylation according to previous publications. In most cases, sumoylated proteins were pulled down from cells containing His8-tagged SUMO expressed from its endogenous promoter and probed for specific substrates by immunoblotting (Ulrich and Davies 2009; Wei and Zhao 2016). In brief, exponentially growing cells were treated with 0.03% MMS for 2 h before harvest. Protein extracts prepared by 55% TCA were dissolved in buffer A (6 M guanidine HCl, 100 mM sodium phosphate at pH 8.0, 10 mM Tris-HCl at pH 8.0) and incubated overnight with Ni-NTA resin after the addition of 0.05% Tween 20 and 4.4 nM imidazole. Resins were washed twice with buffer A containing 0.05% Tween 20 and four times with buffer C (8 M urea, 100 mM sodium phosphate at pH 6.3, 10 mM Tris-HCl at pH 6.3) containing 0.05% Tween 20. HU buffer (8 M urea, 200 mM Tris-HCl at pH 6.8, 1 mM EDTA, 5% SDS, 0.1% bromophenol blue, 1.5% DTT, 200 mM imidazole) was used to elute proteins to be examined by SDS-PAGE and immunoblotting analyses. Ponceau S stain was used to ensure equal loading.
Epitope-tagged Pol2, Yku70, Smc2, and Saw1 proteins were immunoprecipitated, and their sumoylated forms were detected by an anti-SUMO antibody in immunoblotting analyses (Cremona et al. 2012). In brief, cells were treated as above, and proteins were extracted by RIPA buffer (50 mM Tris-HCl at pH 7.4, 5 mM EDTA, 150 mM NaCl, 1.25% Triton-X 100, 1× protease inhibitor cocktail from Sigma, 40 mM NEM). Immunoprecipitation was carried out by incubating the extracts with sepharose beads conjugated with either IgG recognizing the TAP tag fused to Yku70 and Saw1 or anti-HA antibody for HA-tagged Pol2 and Smc2 at 4°C for 2–6 h. Beads were washed with RIPA containing 0.1% SDS and eluted with protein loading buffer. The unmodified form of the protein, detected by antibody recognizing its corresponding tag in immunoblots, was used for equal loading. Antibodies used included: anti-Myc (Bio X Cell 9E10), anti-TAP (Sigma P1291), anti-HA (Roche 3F10), anti-Rfa1 (a gift from S. Brill), anti-GST (Sigma G7781), anti-SUMO (Zhao and Blobel 2005), anti-PCNA (gift from Helle Ulrich), anti-FLAG (Sigma M2), anti-CBP (Santa Cruz Biotechnology SC33000), anti-V5 (InvitrogenR960-25), anti-Ubc9 (Santa Cruz Biotechnology yN-19 and yC-19), and anti-Dpb2 (gift from H. Araki).
Yeast two-hybrid assay
Standard assay was performed as described previously (Dhingra et al. 2019). Briefly, plasmids containing the Gal4 activation domain (AD) and Gal4 DNA binding domain (BD) with or without the fusion of genes encoding the proteins of interest were transformed in the two-hybrid host strain pJ69-4. The resultant transformants were mixed for each pair of plasmids and spotted on plates containing SC-Trp-Leu (selection of plasmids) and SC-Trp-Leu-Ade (report the expression of the ADE2 reporter) media. Pictures were taken after plates were incubated for 48 h at 30°C.
Coimmunoprecipitation
Exponentially growing yeast cells were treated with 0.03% MMS for 2 h before harvest. Cells were disrupted by glass bead beating in lysis buffer with Benzonase added to digest nucleic acid before centrifugation for 15 min at 20,000g to obtain whole-cell extract (WCE). The lysis buffer contains 25 mM K-HEPES (pH 7.6), 100 mM NaCl, 100 mM K-glutamate, 5 mM Mg (OAc)2, 0.02% NP-40, and 0.5% Triton X-100 supplemented by protease inhibitor cocktail (Sigma) and Complete Ultra EDTA-free protease inhibitor (Roche). Proteins of interest were immunoprecipitated using IgG-sepharose beads (for TAP-tagged proteins) or Protein G beads in combination with specific antibodies recognizing the tags on the proteins for 2–4 h at 4°C. Beads were washed five to six times using lysis buffer, and proteins were eluted using loading buffer. After boiling for 5 min, eluted proteins were loaded onto 4%–20% gradient gels (Bio-Rad) and subjected to SDS-PAGE and immunoblotting analyses.
Protein expression and purification
Expression and purification of most recombinant proteins were carried out following previously published protocols. These include FLAG-Sgs1 (Niu et al. 2010), V5-Top3/GST-Rmi1 (Wang et al. 2018), the Mms21/Smc5 complex (Duan et al. 2009), Myc-Smc5/His9-Strep-Tactin-Smc6 complex (Xue et al. 2014), Esc2 and its variants (Sebesta et al. 2017), Smt3, Smt3-D68R, Ubc9, Aos1-Uba2 (Zhao and Blobel 2005), 4xSmt3 (Gillies et al. 2016), and the Mus81-Mms4 complex (Matulova et al. 2009). Expression and purification of CBP-Top3 and Rmi1 were performed largely as described (Devbhandari et al. 2017). Briefly, yeast cells were lysed in extraction buffer (45 mM Hepes KOH at pH 7.6, 10% glycerol, 0.02% NP40, 300 mM NaCl, 1 mM DTT, 2 mM CaCl2). Calmodulin affinity resin (Agilent) was incubated with lysate, then washed with washing buffer (45 mM Hepes KOH at pH 7.6, 10% glycerol, 0.02% NP40, 300 mM NaCl, 1 mM DTT, 2 mM CaCl2) and eluted with elution buffer (45 mM Hepes KOH at pH 7.6, 10% glycerol, 0.02% NP40, 300 mM NaCl, 1 mM DTT, 1 mM EDTA, 2 mM EGTA). The eluate containing CBP-Top3 and Rmi1 complex was diluted with an equal volume of T buffer (25 mM Tris-Cl at pH 7.4, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal, 1 mM DTT) and loaded onto a Resource S column (GE Healthcare). The column was washed with T buffer containing 150 mM KCl and developed with a 25-mL gradient of 150–650 mM KCl. CBP-Top3/Rmi1 was eluted at ∼350 mM KCl, and the peak fractions were pooled and concentrated in an Ultracel-10K concentrator (Amicon) before storing at −80°C in aliquots.
In vitro protein binding assays
For GST pull-down assays, 5 μg of GST-tagged Esc2 or its variants was incubated with 2 μg of Ubc9 (Figs. 2E, 3C, 4D) or 2 μg of Smt3/4xSmt3 (Fig. 2F) in 30 μL of T buffer (25 mM Tris-Cl at pH 7.4, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal, 1 mM DTT) supplemented with 80 mM KCl for 30 min at 4°C. The protein mixture was incubated with 10 μL of Glutathione Sepharose 4B resin (GE Healthcare) for 30 min at 4°C. After washing the resin four times with 200 μL of T buffer with 80 mM KCl, bound proteins were eluted with 20 μL of 2% SDS. For pull-down tests shown in Supplemental Figure S2C (lanes 1–6), 0.5 µg of FLAG-Sgs1, 0.5 µg of V5-Top3/GST-Rmi1, and 1.0 µg of GST-Esc2 were incubated in 30 µL T buffer with 80 mM KCl for 30 min at 4°C. The mixture was further incubated with 10 µL of anti-FLAG agarose resin (Sigma) for 30 min at 4°C. After washing the resin four times with 200 μL of the same buffer, bound proteins were eluted with 20 μL of 2% SDS. For Strep-Tactin pull-down assays shown in Supplemental Figure S2C (lanes 7–12), 0.5 µg of Myc-Smc5/(His)9-StreptagII-Smc6 complex and 1.0 µg of GST-Esc2 were incubated in 30 µL of T buffer with 80 mM KCl for 30 min at 4°C. The mixture was incubated with 10 µL of Strep-Tactin resin that retains StreptagII-Smc6 for 30 min at 4°C. After washing the resin four times with 200 μL of the same buffer, bound proteins were eluted with 20 μL of 2% SDS. For GST pull-down assays shown in Supplemental Figure S5A, 3 μg of GST-tagged Esc2 or its variants was incubated with 1.6 μg of the Mus81-Mms4 complex in 30 μL of T buffer supplemented with 100 mM KCl for 30 min at 4°C. The protein mixture was incubated with 10 μL of Glutathione Sepharose 4B resin (GE Healthcare) for 30 min at 4°C. After washing the resin four times with 200 μL of T buffer with 100 mM KCl, bound proteins were eluted with 20 μL of 2% SDS. In all cases, 10% of the supernatant (S) and eluate (E) fractions and 2% of the wash (W) fraction were analyzed by SDS-PAGE and/or immunoblotted using anti-GST antibody (Sigma) (Supplemental Fig. S2C) or anti-His6 antibody (Sigma) (Supplemental Fig. S5A).
In vitro sumoylation assays
The in vitro sumoylation assay of the Sgs1-Top3-Rmi1 complex shown in Figure 4A was carried out by first incubating 20 nM STR complex with 2.2 μM Smt3 (or Smt3-D68R), 50 nM Aos1-Uba2 (E1), and 280 nM Ubc9 (E2), with or without 40 nM Mms21/Smc5 complex (E3) in buffer R containing 45 mM Hepes-Na (pH 7.0), 5 mM MgCl2, 72 mM KCl, and 0.1 mM DTT for 10 min on ice. The sumoylation reaction, upon the addition of 5 mM ATP, was shifted to 30°C. Samples were taken at indicated time points and mixed with sample loading buffer. Sumoylation reactions shown in Figure 4, B and E, were supplemented with or without 300 nM wild-type or mutant Esc2 in buffer R for 10 min on ice before adding ATP. In all cases, samples were taken at different time points to mix with sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting using antibodies recognizing the tags on Sgs1 and Top3, including anti-FLAG (Sigma), anti-CBP (Genscript), or anti-V5 (Rockland).
A free SUMO chain formation assay (Supplemental Fig. S4D) was performed as above except omitting STR and the SUMO E3 in the reactions. Ubc9 thioester formation assay (Supplemental Fig. S4C) was performed as previously described (Knipscheer et al. 2007). In brief, 2.2 μM Smt3 (or Smt3-D68R) was incubated with 50 nM Aos1-Uba2, 280 nM Ubc9, with or without 300 nM Esc2 in buffer R for 10 min on ice. The reaction, upon the addition of 5 mM ATP, was incubated at 30°C for the indicated time. Samples were mixed with nonreducing (without DTT) or reducing (with 50 mM DTT) loading buffer, analyzed by SDS-PAGE, and immunoblotted using an anti-Ubc9 antibody (Santa Cruz Biotechnology).
2D agarose gel electrophoresis
2D gel analysis was performed as previously described (Meng et al. 2020). Briefly, log-phase yeast cultures were treated with 5 µg/mL α factor (Bio Basic) until at least 90% of cells exhibited G1 arrest. Cells were released from arrest by pronase treatment (Millipore), and MMS was added to a final concentration 0.03% for 2 h. Cells were collected and treated with zymolyase to produce spheroplasts. After cell lysis, proteinase K (Sigma-Aldrich) was added and DNA was purified by CsCl gradient centrifugation and precipitated. Extracted DNA was digested by EcoRI and separated by agarose gel electrophoresis in two dimensions. Separated DNA was transferred onto Hybond-XL membranes (GE Healthcare) and analyzed by Southern blotting using probes hybridizing to the ARS315 or ARS1212 locus. Primers used for probe amplification are listed in Supplemental Table S3. For quantification, the signals of 1N DNA were obtained from short exposures and X-mol signals from longer exposure. Signal levels fell within the linear range of detection for the PhosphorImager.
GCR assay
GCR rate measurement was performed as described previously (Meng et al. 2019). At least nine cultures were examined for each genotype. Cells were washed and serial dilutions were plated on synthetic complete (SC) medium and FC medium containing canavanine and 5-FOA. GCR rates were calculated as m/NT using the following formula: m × (1.24 + ln[m]) − NFC = 0, where m is mutational events, NFC is the number of colonies on FC plates, and NT is the number of colonies on SC plates. The upper and lower 95% confidence intervals (95% CI) were calculated as described (Putnam and Kolodner 2010). A two-tailed Mann-Whitney test was performed as described previously (Myung et al. 2004) using GraphPad Prism version 7.
Microscale thermophoresis (MST) analyses
His-tagged Ubc9 was labeled with a His-Tag labeling kit RED-tris-NTA (NT-L118, NanoTemper Technologies). The labeling reaction was performed according to the manufacturer's instructions. Twelve doses of Ubc9-His from 4 µM–0.0039 uM were prepared by 1:1 serial dilution using MST buffer, which contains 20 mM HEPES (pH 7.5), 80 mM KCl, 0.01% Igepal, and 1% glycerol. Each dose of Ubc9-His in 10 µL of MST buffer was mixed with 40 nM Red-tris-NTA in 10 µL of MST buffer for 30 min at room temperature in the dark. The mixture was loaded into premium Monolith NT.115 Capillaries (NanoTemper Technologies, MO-AK005) for binding affinity assay. MST was measured using a Monolith NT.115 instrument (NanoTemper Technologies) at 25°C. Instrument parameters were adjusted to 70% excitation power and medium MST power. Data was analyzed by MO.Affinity Analysis software (version 2.1.3, NanoTemper Technologies) using the signal from an MST-on time of 5 sec. The Kd of Ubc9-His labeling was 4 nM. To test the binding affinity of Ubc9 with Esc2, 40 nM Red-tris-NTA in 300 µL of MST buffer was mixed with 80 nM of Ubc9-His in 300 µL of MST buffer at room temperature for 30 min in the dark for labeling. After centrifugation at 15,000g for 10 min, the supernatant was transferred to a fresh tube for the binding assay with Esc2. A total of 16 concentrations of Esc2 were serially diluted using an equal volume of MST buffer from 20 µM to 0.6 nM. Ten microliters of each dose of Esc2 was mixed with 10 µL of His-labeled Ubc9 supernatant. The mixture of Ubc9 and Esc2 was loaded into premium capillaries for MST assay with parameters set up at 80% excitation power and medium MST power. Binding affinity was analyzed by the MO.Affinity Analysis software using the signal from an MST-on time of 2.5 sec. Three individual experiments were used to calculate the standard deviation.
Circular dichroism (CD) analyses
Purified Esc2 and Esc2-SLD2m proteins were diluted to 2.9 μM in 300 μL of CD buffer (10 mM NaH2PO4, 100 mM KCl at pH 7.0). Spectra were collected by a Jasco J-710 spectropolarimeter at 20°C.
Supplementary Material
Acknowledgments
We thank Dana Branzei, Lumir Krejci, and Mark Hochstrasser for sharing yeast strains; Steve Brill, Helle Ulrich, and Hirano Araki for sharing antibodies; Weibin Wang for sharing the Top3-Rmi1 complex; Nicolas Paquet for sharing the Mus81-Mms4 complex; Koyi Choi for her early work on Esc2; and Prabha Sarangi for comments on the manuscript. We are grateful for the support of Patrick Sung, in whose laboratory in vitro work was initiated; Dirk Remus, who shared the strains for expressing the CBP-Top3 and Rmi1 complex; and Steven Whitten who helped in CD analyses. We acknowledge Dr. Matthew J. Hart and Dr. Daifeng Jiang for the MST analyses, funded in part by the University of Texas Health Science Center at San Antonio Greehey Children's Cancer Research Institute RNAi High-Throughput Screening Facility, Cancer Prevention and Research Institute of Texas grant RP160844, and Mays Cancer Center National Institutes of Health grant CA054174. This work is supported by the National Institutes of Health grants R21 ES028792 and R15 GM139135 and startup funds from Texas State University to X.X., and National Institute of General Medical Science grants GM080670 and GM131058 to X.Z.
Author contributions: S.L., J.N.B., X.X., and X.Z. designed experiments. S.L. and J.N.B. performed in vivo experiments, and X.X., S.S., A.S., and L.G. performed in vitro tests. B.W. generated structural models and purified sumoylation enzymes for in vitro tests. S.L., X.X., and X.Z. wrote the manuscript with editing by J.N.B.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.344739.120.
References
- Albuquerque CP, Wang G, Lee NS, Kolodner RD, Putnam CD, Zhou H. 2013. Distinct SUMO ligases cooperate with Esc2 and Slx5 to suppress duplication-mediated genome rearrangements. PLoS Genet 9: e1003670. 10.1371/journal.pgen.1003670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque CP, Liang J, Gaut NJ, Zhou H. 2016. Molecular circuitry of the SUMO (small ubiquitin-like modifier) pathway in controlling sumoylation homeostasis and suppressing genome rearrangements. J Biol Chem 291: 8825–8835. 10.1074/jbc.M116.716399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez-López M, Villoria MT, Esteras M, Jarmuz A, Torres-Rosell J, Clemente-Blanco A, Aragon L. 2016. Sgs1's roles in DNA end resection, HJ dissolution, and crossover suppression require a two-step SUMO regulation dependent on Smc5/6. Genes Dev 30: 1339–1356. 10.1101/gad.278275.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JN, Choi K, Xue X, Torres NP, Szakal B, Wei L, Wan B, Arter M, Matos J, Sung P, et al. 2016. Smc5/6 mediated sumoylation of the Sgs1-Top3-Rmi1 complex promotes removal of recombination intermediates. Cell Rep 16: 368–378. 10.1016/j.celrep.2016.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. 2006. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522. 10.1016/j.cell.2006.08.050 [DOI] [PubMed] [Google Scholar]
- Choi K, Szakal B, Chen YH, Branzei D, Zhao X. 2010. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol Biol Cell 21: 2306–2314. 10.1091/mbc.e10-01-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I, Zhao X. 2015. DNA break-induced sumoylation is enabled by collaboration between a SUMO ligase and the ssDNA-binding complex RPA. Genes Dev 29: 1593–1598. 10.1101/gad.265058.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, Zhao X. 2012. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol Cell 45: 422–432. 10.1016/j.molcel.2011.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devbhandari S, Jiang J, Kumar C, Whitehouse I, Remus D. 2017. Chromatin constrains the initiation and elongation of DNA replication. Mol Cell 65: 131–141. 10.1016/j.molcel.2016.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra N, Wei L, Zhao X. 2019. Replication protein A (RPA) sumoylation positively influences the DNA damage checkpoint response in yeast. J Biol Chem 294: 2690–2699. 10.1074/jbc.RA118.006006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H. 2009. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell 35: 657–668. 10.1016/j.molcel.2009.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, van Waardenburg RC, Borg LA, McGarity S, Nourse A, Waddell MB, Bjornsti MA, Schulman BA. 2007. Structure of a SUMO-binding-motif mimic bound to Smt3p-Ubc9p: conservation of a non-covalent ubiquitin-like protein-E2 complex as a platform for selective interactions within a SUMO pathway. J Mol Biol 369: 619–630. 10.1016/j.jmb.2007.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies J, Hickey CM, Su D, Wu Z, Peng J, Hochstrasser M. 2016. SUMO pathway modulation of regulatory protein binding at the ribosomal DNA locus in Saccharomyces cerevisiae. Genetics 202: 1377–1394. 10.1534/genetics.116.187252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. 10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ. 2010. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev 24: 1939–1950. 10.1101/gad.1934210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellis P, Gagliardi M, Banath JP, Szilard RK, Nakada S, Galicia S, Sweeney FD, Cabelof DC, Olive PL, Durocher D. 2007. A screen for suppressors of gross chromosomal rearrangements identifies a conserved role for PLP in preventing DNA lesions. PLoS Genet 3: e134. 10.1371/journal.pgen.0030134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. 2007. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J 26: 2797–2807. 10.1038/sj.emboj.7601711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, Johnson ES, Mann M, Sixma TK, Pichler A. 2008. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell 31: 371–382. 10.1016/j.molcel.2008.05.022 [DOI] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. 2005. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19: 339–350. 10.1101/gad.322605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos HB, Ström L, Itoh T, Katou Y, Shirahige K, Sjögren C. 2006. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell 22: 755–767. 10.1016/j.molcel.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Mankouri HW, Ngo HP, Hickson ID. 2009. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in Saccharomyces cerevisiae. Mol Biol Cell 20: 1683–1694. 10.1091/mbc.e08-08-0877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC. 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172. 10.1016/j.cell.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulova P, Marini V, Burgess RC, Sisakova A, Kwon Y, Rothstein R, Sung P, Krejci L. 2009. Cooperativity of Mus81.Mms4 with Rad54 in the resolution of recombination and replication intermediates. J Biol Chem 284: 7733–7745. 10.1074/jbc.M806192200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Meng X, Wei L, Peng XP, Zhao X. 2019. Sumoylation of the DNA polymerase ε by the Smc5/6 complex contributes to DNA replication. PLoS Genet 15: e1008426. 10.1371/journal.pgen.1008426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Wei L, Devbhandari S, Zhang T, Xiang J, Remus D, Zhao X. 2020. DNA polymerase ε relies on a unique domain for efficient replisome assembly and strand synthesis. Nat Commun 11: 2437. 10.1038/s41467-020-16095-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Smith S, Kolodner RD. 2004. Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae. Proc Natl Acad Sci 101: 15980–15985. 10.1073/pnas.0407010101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111. 10.1038/nature09318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M, Bachmair A, Eisenhaber B, Eisenhaber F. 2005. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2-NIP45) family. BMC Bioinformatics 6: 22. 10.1186/1471-2105-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Fatouros C, Lee H, Eisenhardt N. 2017. SUMO conjugation—a mechanistic view. Biomol Concepts 8: 13–36. 10.1515/bmc-2016-0030 [DOI] [PubMed] [Google Scholar]
- Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN. 2009. Molecular mimicry of SUMO promotes DNA repair. Nat Struct Mol Biol 16: 509–516. 10.1038/nsmb.1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Perry JJ, Nie M, Vashisht AA, Arvai AS, Hitomi C, Guenther G, Wohlschlegel JA, Tainer JA, Boddy MN. 2011. DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes. Mol Cell Biol 31: 2299–2310. 10.1128/MCB.05188-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S. 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820. 10.1016/j.cell.2012.10.021 [DOI] [PubMed] [Google Scholar]
- Putnam CD, Kolodner RD. 2010. Determination of gross chromosomal rearrangement rates. Cold Spring Harb Protoc 2010: pdb.prot5492. 10.1101/pdb.prot5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hayes TK, Kolodner RD. 2009. Specific pathways prevent duplication-mediated genome rearrangements. Nature 460: 984–989. 10.1038/nature08217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa GD, Wohlschlegel J, Yates JR 3rd, Boddy MN. 2006. SUMO-binding motifs mediate the Rad60-dependent response to replicative stress and self-association. J Biol Chem 281: 27973–27981. 10.1074/jbc.M601943200 [DOI] [PubMed] [Google Scholar]
- Sarangi P, Altmannova V, Holland C, Bartosova Z, Hao F, Anrather D, Ammerer G, Lee SE, Krejci L, Zhao X. 2014. A versatile scaffold contributes to damage survival via sumoylation and nuclease interactions. Cell Rep 9: 143–152. 10.1016/j.celrep.2014.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M, Urulangodi M, Stefanovie B, Szakal B, Pacesa M, Lisby M, Branzei D, Krejci L. 2017. Esc2 promotes Mus81 complex-activity via its SUMO-like and DNA binding domains. Nucleic Acids Res 45: 215–230. 10.1093/nar/gkw882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiyama N, Arita K, Ikeda Y, Hashiguchi K, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M. 2010. Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45. Proteins 78: 1491–1502. 10.1002/prot.22667 [DOI] [PubMed] [Google Scholar]
- Sollier J, Driscoll R, Castellucci F, Foiani M, Jackson SP, Branzei D. 2009. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol Biol Cell 20: 1671–1682. 10.1091/mbc.e08-08-0875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191. 10.1038/nature01965 [DOI] [PubMed] [Google Scholar]
- Streich FC Jr, Lima CD. 2016. Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature 536: 304–308. 10.1038/nature19071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Dulev S, Liu X, Hiller NJ, Zhao X, Strunnikov A. 2008. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet 4: e1000215. 10.1371/journal.pgen.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talhaoui I, Bernal M, Mullen JR, Dorison H, Palancade B, Brill SJ, Mazón G. 2018. Slx5-Slx8 ubiquitin ligase targets active pools of the Yen1 nuclease to limit crossover formation. Nat Commun 9: 5016. 10.1038/s41467-018-07364-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD, Davies AA. 2009. In vivo detection and characterization of sumoylation targets in Saccharomyces cerevisiae. Methods Mol Biol 497: 81–103. 10.1007/978-1-59745-566-4_6 [DOI] [PubMed] [Google Scholar]
- Urulangodi M, Sebesta M, Menolfi D, Szakal B, Sollier J, Sisakova A, Krejci L, Branzei D. 2015. Local regulation of the Srs2 helicase by the SUMO-like domain protein Esc2 promotes recombination at sites of stalled replication. Genes Dev 29: 2067–2080. 10.1101/gad.265629.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Daley JM, Kwon Y, Xue X, Krasner DS, Miller AS, Nguyen KA, Williamson EA, Shim EY, Lee SE, et al. 2018. A DNA nick at Ku-blocked double-strand break ends serves as an entry site for exonuclease 1 (Exo1) or Sgs1-Dna2 in long-range DNA end resection. J Biol Chem 293: 17061–17069. 10.1074/jbc.RA118.004769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Zhao X. 2016. A new MCM modification cycle regulates DNA replication initiation. Nat Struct Mol Biol 23: 209–216. 10.1038/nsmb.3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen JM, Dhingra N, Wei L, Zhao X, Freudenreich CH. 2020. Relocation of collapsed forks to the nuclear pore complex depends on sumoylation of DNA repair proteins and permits Rad51 association. Cell Rep 31: 107635. 10.1016/j.celrep.2020.107635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winczura A, Appanah R, Tatham MH, Hay RT, De Piccoli G. 2019. The S phase checkpoint promotes the Smc5/6 complex dependent SUMOylation of Pol2, the catalytic subunit of DNA polymerase ε. PLoS Genet 15: e1008427. 10.1371/journal.pgen.1008427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Choi K, Bonner J, Chiba T, Kwon Y, Xu Y, Sanchez H, Wyman C, Niu H, Zhao X, et al. 2014. Restriction of replication fork regression activities by a conserved SMC complex. Mol Cell 56: 436–445. 10.1016/j.molcel.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G. 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci 102: 4777–4782. 10.1073/pnas.0500537102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.