Abstract
The National Center for Biotechnology Information (NCBI) is an archive providing free access to a wide range and large volume of biological sequence data and literature. Staff scientists at NCBI analyze user-submitted data in the archive, producing gene and SNP annotation and generating sequence alignment tools. NCBI's flagship genome browser, Genome Data Viewer (GDV), displays our in-house RefSeq annotation; is integrated with other NCBI resources such as Gene, dbGaP, and BLAST; and provides a platform for customized analysis and visualization. Here, we describe how members of the biomedical research community can use GDV and the related NCBI Sequence Viewer (SV) to access, analyze, and disseminate NCBI and custom biomedical sequence data. In addition, we report how users can add SV to their own web pages to create a custom graphical sequence display without the need for infrastructure investments or back-end deployments.
The completion of human and model organism genome assemblies at the turn of the century exposed the need for visualization and analysis tools that would allow researchers to view DNA, RNA, or protein sequence side-by-side with gene models and additional empirical sequence-related data. Unlike text-based viewers, a graphical sequence viewer allows researchers to investigate genome sequences efficiently at multiple levels and make different types of discoveries: for instance, megabase-scale views to examine higher-level structural patterns and gene clusters or base-pair-level views to analyze sequence polymorphisms. Graphical sequence viewers supporting linear sequence representations can display alignments of genomic or protein sequence in parallel with annotated features and experimental data rendered in the form of boxes, line graphs, histograms, or heatmaps. Genome sequence viewers have been created independently by different groups, including UCSC, the Broad Institute Integrative Genomics Viewer (IGV), and others (Kent et al. 2002; Skinner et al. 2009; Robinson et al. 2011; Lee et al. 2020). These viewers can help researchers discern patterns in expression or molecular state among different genes or genomic regions, facilitating biological insights and aiding in hypothesis generation for further research.
The National Center for Biotechnology Information (NCBI) has served as the major US-based repository of biological sequence data for 30 yr. A wealth of experimental data is stored in the GenBank (INSDC) (Sayers et al. 2020b), Sequence Read Archive (SRA), Gene Expression Omnibus (GEO) (Barrett et al. 2013), and database of Genotypes and Phenotypes (dbGaP) (Tryka et al. 2014) archives, much of which is associated or aligned to a genome assembly. NCBI staff have analyzed the data submitted to our repositories and generated reference representations (RefSeq) for genomes, transcripts, and proteins (O'Leary et al. 2016), as well as curated reference SNPs (refSNPs) (Sherry et al. 2001) and clinical (ClinVar) variants (Landrum and Kattman 2018). Users of NCBI can also conduct BLAST searches of the NCBI database (Boratyn et al. 2013) and generate sequence–sequence alignments that can be used to inform their own sequence annotations. The abundance and uniqueness of aligned or annotated sequence data generated at NCBI, as well as the opportunity to provide users with an integrated view of and access to content from our diverse resources, led us to develop our own graphical sequence viewers to aid in the analysis of this information.
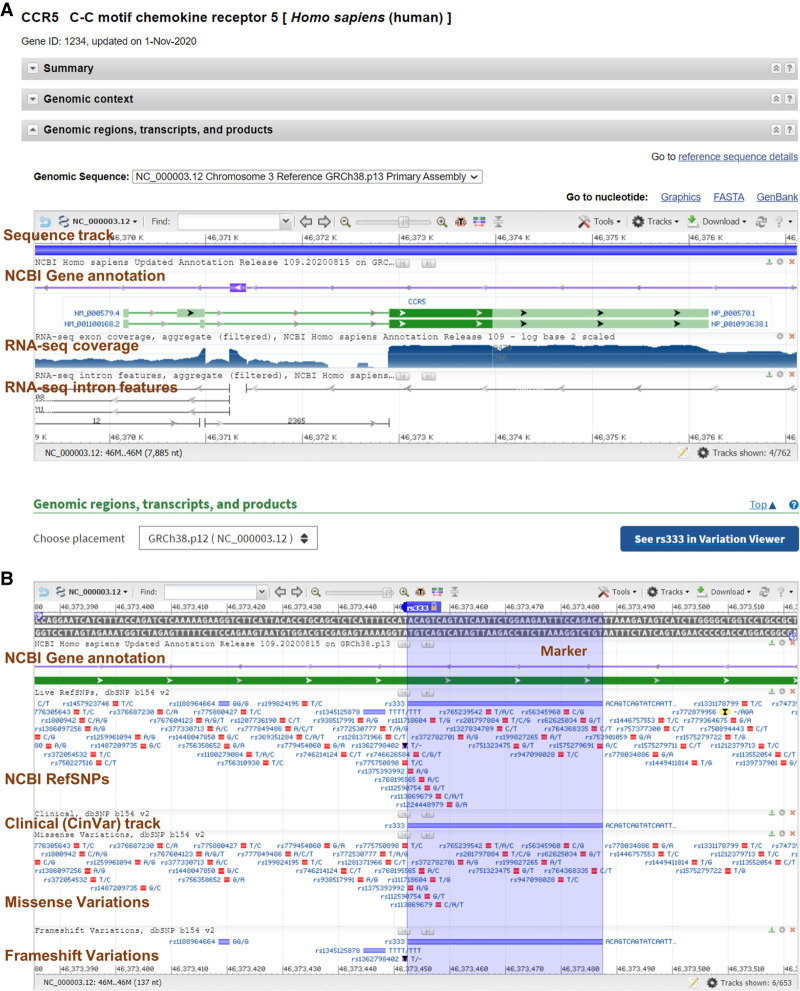
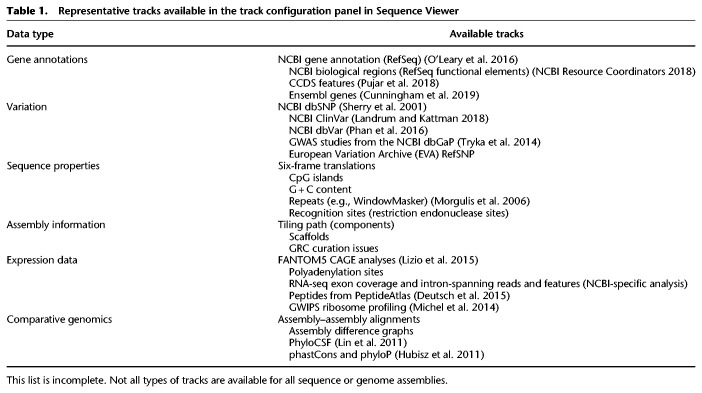
We initially developed the Sequence Viewer (SV) to provide users browsing the NCBI Nucleotide and Protein databases with graphical displays of GenBank and RefSeq sequence records and their accompanying annotations. SV is also available as an independent web application (https://www.ncbi.nlm.nih.gov/projects/sviewer/) in which users can specify the GenBank or RefSeq accession they wish to view. SV can also now be found embedded on record pages in many other NCBI resources, offering tailored graphical views of the data, with the display and data tracks configured to highlight resource-relevant content. For instance, a researcher examining the NCBI gene record page for CCR5 (Fig. 1A) will see an instance of SV configured to show the RefSeq transcript models along with RNA-seq data for the CCR5 gene region. If the researcher then goes to the NCBI SNP resource page to view a polymorphism (e.g., rs333) within CCR5, they will see an instance of SV preconfigured with a selection of NCBI SNP data tracks (Fig. 1B). In addition, the NCBI Primer-BLAST (Ye et al. 2012) and ORF Finder tools (Wheeler et al. 2003) use an embedded instance of SV to display the results of analysis relative to the query sequence. Outside of the NCBI, SV can be added to third-party pages as an embedded application (more information on this later) (https://www.ncbi.nlm.nih.gov/projects/sviewer/embedded.html).
Figure 1.
NCBI Sequence Viewer (SV) is the graphical component of many NCBI pages and is configured to display data appropriate to the resource page. (A) SV located on an NCBI gene record page. Brown labels indicate tracks for sequence, gene annotation, and RNA-seq data. (B) SV located on an NCBI SNP record page. Brown labels indicate tracks for gene annotation, RefSNPs, Clinical (ClinVar), missense, and frameshift variants. A marker is automatically placed over the variant described on the page.
Because SV displays a single designated sequence molecule, it is appropriate for viewing the data specific to a particular NCBI gene or nucleotide record or for analyzing a single-molecule bacterial or viral genome, but it does not easily allow navigation to different molecules in a multichromosome eukaryotic genome assembly. To better serve researchers working with the hundreds of diverse eukaryotic assemblies annotated by RefSeq, NCBI adapted SV to create a whole-genome browsing tool, Genome Data Viewer (GDV).
GDV is composed of an embedded instance of SV that displays sequence and track data, along with additional page elements that allow a user to search within an entire genome assembly and efficiently narrow in on their chromosome, sequence, region, or gene of interest. GDV replaced the NCBI Map Viewer (Dombrowski and Maglott 2003), NCBI's previous tool for whole-genome display. Researchers using GDV can go directly to the NCBI BLAST service (Boratyn et al. 2013) from the browser and load BLAST results as alignment tracks that can be viewed side by side with gene annotation and other data. Variation Viewer, a related browser associated with NCBI's variation resources, is functionally similar to GDV and also incorporates an instance of SV but is configured with features specifically intended for analyzing human variation data. GDV and Variation Viewer can both display the same types of NCBI variation track data.
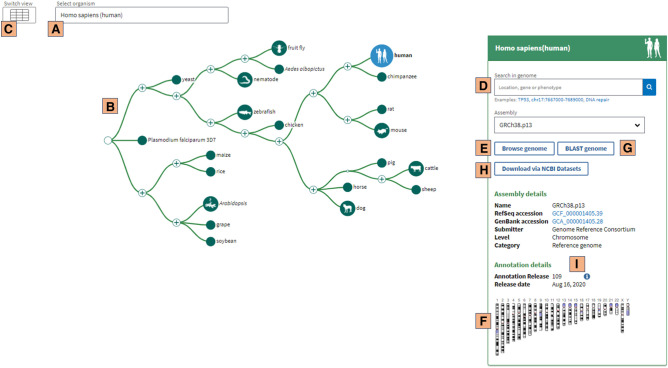
The GDV can be accessed from its own home page (Fig. 2; https://www.ncbi.nlm.nih.gov/genome/gdv/) and can also be found via links from other NCBI resources, including gene, assembly, GEO, and dbGaP record pages. GDV provides users a graphical gateway to data at the NCBI, especially RefSeq and refSNP annotation. Below, we highlight some of the functions of GDV and other instances of the NCBI SV and provide context for GDV's features with respect to the broader collection of publicly available genome browsers, including the UCSC and Ensembl genome browsers, JBrowse, and IGV.
Figure 2.
Genome Data Viewer (GDV) home page (https://www.ncbi.nlm.nih.gov/genome/gdv/). Users can search for an organism (A) or browse the taxonomic tree (B). There is also an option to navigate the available genome assemblies in table form (C). The panel on the right side provides information and links for the selected assembly. Adding a location or gene symbol in the search box (D), clicking on the “browse genome” button (E), or clicking on a chromosome in the ideogram (F) will take the user directly to the genome browser view. There are also options to BLAST the genome (G), download information using the NCBI Datasets interface (H), and obtain information about the annotation (I).
Results and discussion
GDV home page
The GDV home page (Fig. 2; https://www.ncbi.nlm.nih.gov/genome/gdv/) serves as a gateway to the RefSeq annotated eukaryotic assemblies available at NCBI (O'Leary et al. 2016). Users can browse organisms using the taxonomic tree (Fig. 2B) or search directly for species of interest by common or scientific taxonomic name using the free text search (Fig. 2A). When the user first arrives on this page, the taxonomic tree highlights a set of popular research organisms, including human, mouse, zebrafish, fruit fly, Arabidopsis thaliana, and yeast. The user can click on nodes on the tree to browse within subtrees showing different taxonomic groupings. GDV also provides the option to browse species and assemblies in a table view (Fig. 2C).
Our available list of organisms and assemblies is growing constantly and stands at more than 900 as of this writing. This list includes single-celled yeasts and protists, plants, fungi, arthropods, livestock, and all common model organisms. Multiple assemblies are available for many species; for instance, users have the choice of four human assemblies (GRCh38, GRCh37, HuRef, and CHM1_1.1), two zebrafish assemblies, and four cattle (Bos taurus) assemblies. Researchers are able to navigate among different assemblies in order to compare annotation changes between different versions and load aligned data specific to a particular assembly. In addition, because NCBI is a major source of genome annotation, the NCBI GDV browser includes a broader range of organisms and assemblies than can be found on other public browsers, such as the UCSC Genome Browser. For instance, GDV provides annotated assemblies for more than 100 flowering plants that users can access directly from our browser home page without having to load assembly sequence or hubs manually. GDV therefore supports analysis for many nonmodel organisms that may not have alternative sources for viewing genome-wide sequence data.
The panel on the right side of this page provides a centralized hub for accessing information for a selected assembly. Users are provided with the RefSeq identifier for the assembly as well as related information, including the genome submission source and annotation release and date. Links in this section take users to the NCBI Assembly resource page, NCBI annotation report (Fig. 2I), sequence and annotation downloads via NCBI Datasets (Fig. 2H), and the NCBI BLAST page (Fig. 2G) for the selected organism. Users can choose an assembly from the dropdown menu and navigate directly to the graphical browser view of the assembly by using the browse button (Fig. 2E), searching for a gene or location in the search box (Fig. 2D), or clicking on a chromosome in the ideogram (Fig. 2F).
Genome browser
Navigation
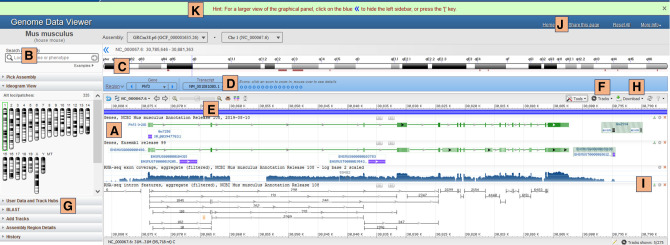
The genome browser provides users with numerous tools and panels that can aid in navigation of the genome (Fig. 3). The core element in the page is an embedded SV application (Fig. 3A). Different page elements interact with SV and with one another dynamically, so that changes in one element are incorporated by other elements (e.g., changing the chromosome in the ideogram panel on the left results in updating the region selector tool [Fig. 3C] and SV to show the corresponding chromosome). Users can hide the left sidebar in order to see a wider view of the track and sequence data in the SV.
Figure 3.
GDV. (A) NCBI SV embedded application; (B) search box; (C) region selector ideogram; (D) exon navigator; (E) zoom and pan options from SV; (F) tracks configuration menu from SV to add NCBI-provided tracks; (G) user data and track hubs and BLAST widgets to add custom tracks; (H) download data options from SV; (I) buttons to download track data, change track settings, or hide tracks (x); (J) share this page link; and (K) news banner.
Users can navigate within the browser by providing sequence or cytogenetic location coordinates or searching with free text in the main browser search box (Fig. 3B). To support the different entry points to genomic analysis, GDV accepts searches for gene symbols or known aliases, gene names, phenotypes, refSNP IDs, and RefSeq transcript, protein, and assembly component accessions.
Because genomic analyses often involve comparisons between different assemblies or sequences within an assembly, we implemented several tools to facilitate navigation between and within genomes. The pick assembly panel beneath the search box allows users to switch among different assemblies for the same organism (where available), whereas the ideogram panel supports navigation to different chromosomes. Users can select chromosomes or scaffolds from the assembly using a drop-down menu on the top center of the page.
GDV, like other genome viewers such as IGV and the UCSC and Ensembl genome browsers, provides an interactive region selector ideogram display for genome assemblies containing chromosome representations (Fig. 3C). Users can go directly to a region by highlighting within this ideogram using click and drag. In addition, for genomes with chromosome banding, clicking inside a band will zoom the display directly to that part of the sequence.
The region selector ideogram also provides access to information about regions in the GRCh37 and GRCh38 assemblies that contain alternate sequences (ALTs) or patch scaffolds curated by the Genome Reference Consortium (GRC) (Church et al. 2011). When viewing an assembly region with aligned ALT or patch scaffolds, an assembly region details panel is also available in the left sidebar region, listing all ALTs and patches aligned to the displayed region. Clicking on a sequence listed in this panel updates the display to the selected ALT or patch scaffold, enabling researchers to navigate between the chromosome and different alternate representations and exposing regions of assembly updates and known human sequence diversity. Assembly–assembly alignment tracks showing ALT or patch scaffolds aligned to the chromosomes, as well as those with pairwise alignments for different assemblies of the same organism, are also available to add to the graphical view to facilitate comparative analyses (see below for more information about unique track data provided in the NCBI viewers).
Zooming the view to gene level activates the blue exon navigator element (Fig. 3D). Here, users can select a gene and transcript and browse or zoom directly to the exons within the transcript. This tool allows users to find a particular exon within a transcript variant of a gene, for instance, in order to find a position described in the research literature (e.g., “mutation in exon 6”). The drop-down menu in this element also allows users to quickly recenter the view around the selected gene or transcript. To our knowledge, GDV's option to navigate directly to exons within an annotated transcript is unique among genome browsers.
Users can return to previously viewed regions using the back button in their web browser. The SV component also includes options to pan, zoom, and go back to prior views on its toolbar (Fig. 3E). A news banner appears on the top of the browser view, alerting returning users to a recent update to this tool (Fig. 3K).
Data provided in the NCBI SV and GDV genome browser
GDV was designed specifically to support visualization and analysis of the wide range of genomes and assemblies annotated at the NCBI (O'Leary et al. 2016). RefSeq gene annotation data tracks are shown by default in the graphical view for these assemblies. NCBI refSNP data tracks (Sherry et al. 2001) are also shown by default for human assemblies. Gene and SNP tracks are automatically updated in GDV and SV embedded instances upon new releases of the NCBI databases, so that users of the NCBI graphical viewers always have immediate access to the latest versions of RefSeq and SNP annotation.
The NCBI does not generate annotation for model organisms with their own active and well-established annotation groups, such as Drosophila melanogaster (FlyBase) and Saccharomyces cerevisiae (SGD), or for unicellular eukaryotes whose genomes are not compatible with the NCBI's eukaryotic genome annotation pipeline. However, such assemblies are available in the GDV browser, including currently hundreds of assemblies for fungi species. For these assemblies, GDV displays gene annotation tracks that copy (propagate) annotation submitted to INSDC by the assembly provider or from the model organism authority.
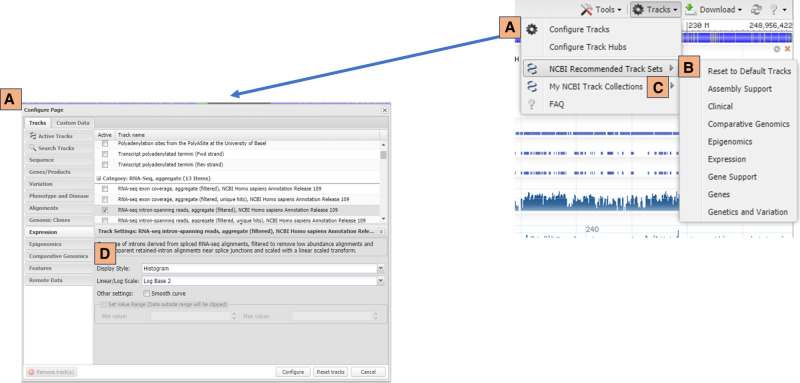
Users of GDV and other instances of the NCBI SV, such as the graphical views on the NCBI gene record pages, can further customize their view by adding additional track data. The NCBI provides tracks containing gene annotation from RefSeq (O'Leary et al. 2016) and Ensembl (Cunningham et al. 2019), variation data, assembly information, sequence properties, comparative genomics, and more. Some of these tracks are similar to the track choices provided by the UCSC or Ensembl browsers, whereas others, such as our in-house analysis of RNA-seq data, are unique to the NCBI's sequence visualization tools. These tracks can be added to the view using the tracks menu (Fig. 4A), which is found on the toolbar of the SV component (Fig. 3F). Table 1 contains examples of the tracks available in GDV.
Figure 4.
Tracks supplied by NCBI can be added using the track configuration panel (A), which can be accessed from the tracks menu located in the SV toolbar. The tracks menu on the toolbar also provides the option to add predetermined sets of tracks from the NCBI recommended track sets menu (B) or private track sets saved in “My NCBI Track Collections” (C). Track display settings can be changed within the track configuration panel (D).
Table 1.
Representative tracks available in the track configuration panel in Sequence Viewer
NCBI provides track sets that contained predefined tracks configured to support different types of analyses, such as clinical, assembly support, and gene support (Fig. 4B). These track sets provide a one-click option for naïve users looking for a set of potentially helpful tracks. Additionally, users with a MyNCBI account can save their own customized track displays for future use in their MyNCBI account (Fig. 4C; https://www.ncbi.nlm.nih.gov/tools/sviewer/faq/#tracksets). User-defined track collections saved in a MyNCBI account can be accessed from any instance of the SV application, such as the graphical sequence display on NCBI gene record pages, and can also be shared with collaborators or laboratory members.
GDV's integration with other NCBI resources, such as the GEO, SRA, and dbGaP databases, can facilitate browser-based genomic analyses of these types of data relative to analysis at other public genome browsers. A user viewing a publication describing a GEO, SRA, or dbGaP study aligned to an assembly can add the reported accession number (e.g., GSM4308119, SRR12003862, pha002856.1) in the input box in the add tracks panel in the GDV (Fig. 3G) and quickly see the content as data tracks in the genome browser. The user can also provide the identifier in the URL parameters (https://www.ncbi.nlm.nih.gov/genome/gdv/browser/help/#URLParams). Although other browsers may also support viewing of data from these databases, the NCBI GDV is unique in that these data can be added directly to the display using the study identifier.
To facilitate genome-wide analyses, the GDV is also directly accessible from the NCBI gene, assembly, and nucleotide record pages and from select aligned studies in the GEO and dbGaP databases. Links from GEO database records and the dbGaP advanced search interface (https://www.ncbi.nlm.nih.gov/gap/advanced_search/) automatically open a GDV session with the corresponding GEO or dbGaP track shown (e.g., https://www.ncbi.nlm.nih.gov/genome/gdv/browser/GEO/?id=GSE145181). Users can also navigate from GDV to additional NCBI resource pages, such as gene, SNP, and ClinVar, using links provided in the tooltips in the NCBI gene and SNP annotation tracks. Researchers can take advantage of these interconnections within GDV to perform genomic analysis requiring information found in multiple NCBI databases.
Similar to the UCSC and Ensembl genome browsers, GDV and SV support displays of data tracks provided by external sources. External data can be added to the GDV display using the user data and track hubs panel (Fig. 3F). Tracks can be uploaded in common bioinformatics formats such as alignment FASTA, GFF3, GTF, VCF, BED, and WIG. Users can also stream data hosted on remote URLs provided in bigBed, bigWig (Kent et al. 2010), multiWig, tabix VCF, and indexed BAM formats. At the time of this writing, GDV does not yet support all of the UCSC-defined display formats (e.g., bigGenePred, PSL), nor does it support display of 3D-interaction data. We welcome community input on adding support for additional data formats.
NCBI graphical sequence viewers, including GDV and the NCBI SV instances, also support data organized in the form of UCSC-browser style track hubs (Raney et al. 2014; Sayers et al. 2019). GDV's track hubs search interface provides direct access to tracks archived in the EBI Track Hub Registry (https://trackhubregistry.org/). In addition, GDV, like the UCSC and Ensembl genome browsers, can be accessed directly from the track hub records in the EBI registry.
Track data and display settings
GDV offers users the ability to customize the displays of individual tracks. Users can hide or configure tracks from the track configuration panel (Fig. 4D) or by using the icons at the right end of each track (Fig. 3I). Different public genome browsers provide conceptually similar, but somewhat distinct options, for visualizing gene, graphical, and alignment data. In this section, we highlight track data visualizations in the GDV browser and other instances of the SV graphical view component that support various analysis scenarios.
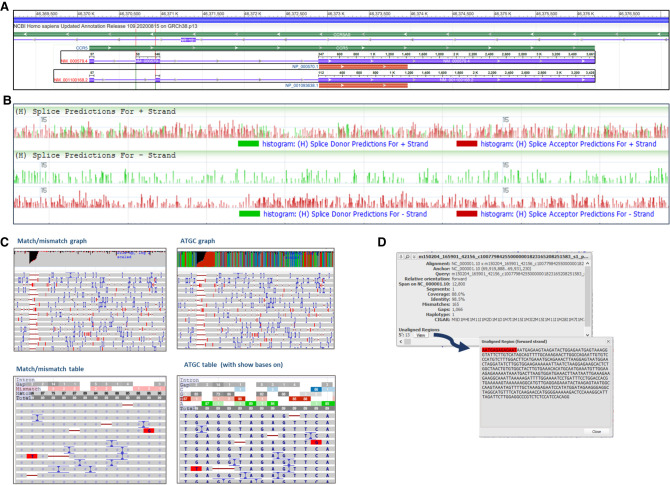
Because NCBI visualization tools are designed help users take maximum advantage of NCBI RefSeq annotation, we have developed multiple ways of configuring the gene feature tracks depending on the needs of a user. Gene tracks can be configured to show a simple gene bar or merged exon–intron model defining the genomic span of the gene, which may be useful for researchers wanting to see a general overview of all the genes in a chromosomal region of interest. For users interested in more details about the different transcript variants at a particular gene, gene annotation tracks can also be adjusted to show the different transcript models or expanded to display all transcript and CDS features along with annotated predicted protein domains or other information (https://www.ncbi.nlm.nih.gov/tools/sviewer/legends/#anchor_2). When all transcript variants are shown, the user can select (via mouse click) two transcript variants from the same gene and see differences between the variants highlighted in the red vertical hairlines (Fig. 5A). This function can be used to choose a transcript variant of interest from a gene with many annotated alternative transcripts. Additional information about transcript and CDS annotations can be obtained from the tooltips, including positional information (e.g., HGVS cDNA coordinates) and options to view the FASTA or GenBank flat file (GBFF) sequence.
Figure 5.
Track data displays for different track types. (A) Gene annotation track showing two transcript variants in a gene selected using mouse clicks. Green indicates gene; purple, transcript; red, coding sequence (CDS). Red hairlines denote positions where the transcript variants show differences in splicing. (B) MultiWig files displayed as overlay (top) or stacked (bottom set) graphs. (C) Pile-up graphs and tables for alignment data, for example, BAM files. (D) Example of data available in the tooltip of an alignment, along with a pop-up view showing sequence of unaligned nucleotides.
More continuous data, such as RNA-seq expression data formatted as a graph, can be configured on a linear or logarithmic scale and displayed as a histogram, line graph, or heatmap (https://www.ncbi.nlm.nih.gov/tools/sviewer/legends/#anchor_12), according to the preference of the user and the nature of the underlying data. Consistent with other leading browsers, including the UCSC Genome Browser and IGV, the range value (minimum and maximum) of the track can be adjusted manually in order to directly compare the signal of data in different tracks or different locations in the genome. This may be helpful in, for instance, determining which gene in a related gene family has the highest expression in a particular RNA-seq tissue sample. External track hub data formatted in a multiWig file can be viewed as a graph overlay, which may aid in directly comparing different data subsets, or as separate stacked tracks (Fig. 5B).
GDV and SV provide multiple display options for dense alignment data (https://www.ncbi.nlm.nih.gov/tools/sviewer/legends/#anchor_7), such as data coming from BLAST searches or whole-genome high-throughput sequencing projects. GDV's track display options for this type of data are conceptually similar to those found in other viewers such as IGV. At the lowest zoom levels, aligned data are shown as a coverage graph: Alignments come into view dynamically as the zoom level progresses to the sequence level. Users can choose to show or hide unaligned tails, hide duplicate reads, show gene features annotated on an alignment (such as an assembly–assembly alignment), and sort by strand or haplotype tag. We also provide the option to display a summary pile-up graph or table view, in which researchers can get an overview of the read data in a genomic region (Fig. 5C). Additional information may be found in the tooltips for individual reads, including coverage and identity information relative to the genome assembly, the alignment CIGAR string, and the nucleic acid sequence of any unaligned portions (Fig. 5D). The complete nucleotide sequence of each aligned read can also be obtained for further analysis.
Adding BLAST tracks and the BLAST Alignment Inspector
The NCBI GDV is interconnected with the NCBI BLAST service (Boratyn et al. 2013; NCBI Resource Coordinators 2018) to facilitate the analysis of genome alignments coming from BLAST analysis. Users can initiate genome BLAST searches from within the SV application or from the BLAST panel found on the GDV left sidebar (Fig. 3F; https://www.ncbi.nlm.nih.gov/genome/gdv/browser/help/#BLAST). Additionally, users can enter RIDs that they have for previously run genome BLASTs into this BLAST panel. BLAST results appear as tracks in the graphical SV application (Fig. 6C).
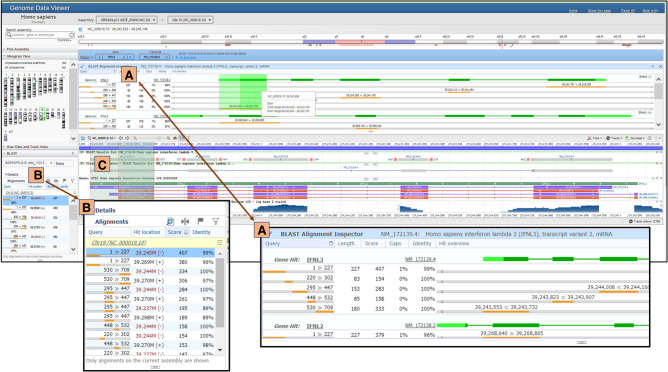
Figure 6.
GDV session displaying the BLAST Alignment Inspector. When the cursor hovers over an aligned region in the BLAST Alignment Inspector (A), the corresponding region in the SV is highlighted as well. Note the full list of alignments is reported in a table in the BLAST panel (B). BLAST alignments can also be viewed as tracks added to the SV (C).
Although other browsers also support the alignment of sequence data to a genome assembly, GDV provides a unique view tool, the BLAST Alignment Inspector, to aid in the analysis of NCBI BLAST results (Fig. 6A). The Alignment Inspector visualizes a BLAST alignment relative to a NCBI gene model. Clicking within the Alignment Inspector recenters the SV graphical view to show the corresponding region of the genome assembly, thereby allowing users to see concordances between BLAST results and annotation data. Along with the BLAST results panel on the left side (Fig. 6B), the BLAST Alignment Inspector can facilitate interpretation of results in which a query has multiple hits to an assembly. For example, the inspector makes it easier to determine the “best” hit and also to understand what elements of a query are unique and which may be repetitive, especially in the context of alignment results involving a gene family, paralogous genes, or pseudogenes.
Sharing, exporting, and printing
The communication of results is central to research. To support this need, GDV provides users with several ways to share and export data to show to colleagues or include in presentations.
GDV's “share this page” button (Fig. 3J) creates a temporary URL that captures the genome assembly, track content, and display settings of the genome browser view. For users interested in a publication-quality images of SV or GDV graphical displays, NCBI provides a download option (Fig. 3H) to export a PDF or SVG file. These vector graphics file formats are compatible with most third-party image editors.
Like other popular genome browsers, such as the UCSC Genome Browser and the Ensembl browser, GDV and SV provide users the ability to export data as files for analysis and use in other applications. There are options to download selected regions of the assembly sequence in FASTA or GBFF format. Researchers can also export annotation data for discrete sequence ranges from selected gene, feature, and SNP tracks (Fig. 3I). Currently, these data are available in BED, CSV, VCF, or GFF3 file formats, as appropriate for the data type.
Adding the NCBI SV to third-party pages
As described above, the NCBI SV is embedded on many NCBI resource pages and forms the core component of the GDV genome browser (Fig. 3). SV can also be added to third party (non-NCBI) web sites. Figure 7 diagrams how third-party web pages, such as researcher laboratory websites, can incorporate SV in a similar way as the NCBI GDV. Although other public sequence and genome viewers, including the UCSC Genome Browser, JBrowse, and IGV (IGV.js), can also be added to third-party pages, a major advantage to embedding SV is that the user will have access to the NCBI's latest set of data tracks through SV's connectivity with NCBI databases and services.
Figure 7.
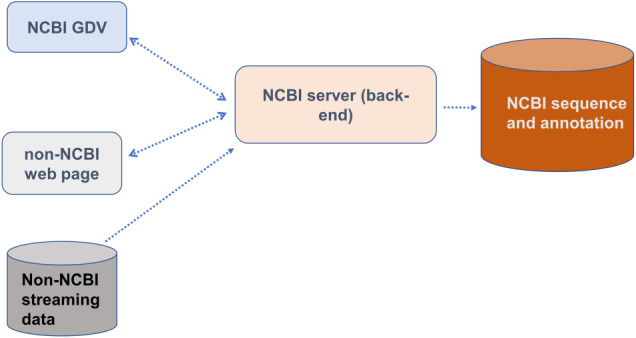
Organization of data flow for SV to NCBI in-house tools (such as the NCBI GDV browser) and non-NCBI web pages. Both GDV and non-NCBI web pages communicate with the NCBI back-end to obtain sequence and annotation data hosted at NCBI. Non-NCBI streaming data must also access NCBI back-end servers in order to be displayed in the NCBI graphical viewer application.
SV instances embedded on third-party sites can display any nucleic acid or protein sequence that has been submitted to the INSDC databases (GenBank, ENA, DDBJ). Embeds can access NCBI-provided track data aligned to the displayed sequence and can also show user-provided aligned data and stream data hosted on publicly accessible remote URLs or organized in track hubs.
The SV application can be embedded in plain HTML/JavaScript or with another framework as selected by the user. The latest version of the software, including all its dependencies, is dynamically loaded from the NCBI. Therefore, SV may be easier for less-experienced programmers to add to a web page than other viewers, such as JBrowse (Buels et al. 2016), because the embedding is performed fully on the front-end with all back-end support provided by the NCBI's data center.
Once embedded, the NCBI SV embedding API (https://www.ncbi.nlm.nih.gov/tools/sviewer/embedding-api/) allows users to customize the toolbar content, the track order, and the track display options. The embedded application can also be configured to track the number and type of interactions with the SV application by web page visitors so that researchers can analyze how often data are accessed on their web page.
SV can be embedded with the ability to automatically reinitialize it to change any settings (including the displayed molecule), so that the page can change dynamically, for instance, to incorporate release updates to annotation tracks. Multiple instances of the NCBI SV can be embedded on a single web page. Please refer to our video tutorial (https://youtu.be/JC10DCKAfyM) for an overview of how members of the scientific community can add SV to their own web pages.
Displays for unsubmitted data
NCBI SV and GDV cannot display data without exposing it to NCBI shared back-end services. For those who wish to view experimental data before public database submission or who need to keep their data secure, the NCBI provides an alternative sequence analysis software suite, Genome Workbench (GBench) (https://www.ncbi.nlm.nih.gov/tools/gbench/). GBench is a desktop software package that can be used for visualization and analysis of nonpublic data behind a firewall. GBench contains a number of different interconnected molecular analysis tools, including not just a graphical sequence viewer similar to SV but also tools for sequence alignment, a phylogenetic tree viewer, and a multiple sequence alignment viewer. This suite also includes a sequence editing package that can produce files for submission to the NCBI GenBank database (Sayers et al. 2020a; Kuznetsov and Bollin 2021).
Future directions
The GDV and other NCBI sequence visualization tools, including the NCBI SV and GBench software suite, are under active development. The NCBI actively seeks input from the biological research community. In addition to doing outreach through NCBI webinars and attendance at several meetings per year, we are conducting regular user surveys on our resource pages. We encourage those with suggestions to use the “feedback” button located at the bottom right of GDV and other NCBI resources and to leave their contact information if they would like to talk with us further. Feature updates are communicated to the public through the NCBI Insights blog (subscribe at https://ncbiinsights.ncbi.nlm.nih.gov), and we also encourage users to follow our YouTube channel (https://www.youtube.com/channel/UCvJHVo5xGSKejBbBj0A5AyQ) for news and tutorials covering the many features and functions of SV and other NCBI data analysis tools. Please also refer to the help documentation at https://www.ncbi.nlm.nih.gov/genome/gdv/browser/help/ and https://www.ncbi.nlm.nih.gov/tools/sviewer/.
As we look to the next decade of data visualization at NCBI, we note that the appreciation of intraspecies diversity has spurred a revolution in pan-genomes (Computational Pan-Genomics 2018). This revolution has been aided by advances in sequencing technology and read mapping, which have allowed researchers to more easily generate genome assemblies from any person, organism, strain, or biotype. Researchers are now able to ask questions about biological function and evolution in the context of simultaneously examining multiple similar genome sequences, for instance, multiple sequences from an evolving bacterial or viral pathogen. These questions uncover the need for better visualization tools that are able to integrate views of custom and archived data to show comparative relationships among different sequences and genome assemblies.
In the coming years, we will be working on ways to support users interested in visualizing and interpreting data from newly assembled genomes and population samples, including SRA data stored in a cloud environment. New data sets also offer the opportunity to further engage with the research community via open source software and novel web development technologies. As we refine GDV and develop new visualization platforms, we will continue to focus our efforts on providing a seamless and efficient experience for researchers interested in access and analysis of NCBI-provided data.
Methods
Graphical sequence viewer architecture
The design of the NCBI SV and genome browsers is based on a two-tier model, with a web front-end based on HTML/JavaScript and the back-end (server side) running at the NCBI's data center. The server side is built using the NCBI C++ Toolkit (https://ncbi.github.io/cxx-toolkit/). SV uses a software as a service (SaaS) model in which the front-end software component is delivered over the internet as opposed to being hosted on a company's own local computing resources. The latest version of the SV, including all its dependencies, is dynamically loaded to all embedded instances using cross-origin resource sharing (CORS).
Tiled image approach
Genomic data are rendered into tile images by high-performance back-end servers, thereby allowing for more efficient rendering of particular sequence ranges in view. When users search, pan, or zoom inside the interface, the JavaScript front-end makes a series of calls to the back-end and transmits genomic coordinates and display options to render the images and obtain tooltips and meta information about the current viewed range. PNG compressed tile images with JSON image annotations are downloaded and stitched by the front-end, providing the effect of browsing a large prerendered image.
Back-end computing
The NCBI SV back-end implements a Common Gateway Interface (CGI) directly bridged into parallel distributed rendering services running at the NCBI data center. Server-side rendering services are written in C++ and use a distributed scalable grid framework to process requests. The back-end grid framework uses game theory to optimize multiple concurrent requests coming from different users for similar data. Holistic use of this optimization results in the ability to get a back-end response asynchronously, thereby minimizing delays or disruption of service. The NCBI rendering grid processes millions of back-end calls a day, scaling for high daytime peaks and seasonal variations.
The performance profile of the back-end graphical workload is defined by data intensive algorithms aggregating multiple NCBI and third-party data sources (https://www.ncbi.nlm.nih.gov/tools/gbench/third_party_tools/). Back-end graphical algorithms use OpenGL (https://www.opengl.org/) CPU without hardware acceleration for raster images. The use of CPU rendering only marginally impacts performance but allows us to use off-the-shelf servers without the need for server-grade GPUs.
GDV components
The GDV home page and browser are developed in JavaScript. The GDV browser is composed of multiple independent components, such as the search box, SV, ideogram panel, BLAST panel, and exon navigator. Components are associated with document object model (DOM) element(s) that show the HTML representation of the component state. Components communicate with each other and synchronize their state via notifications. For example, the search component posts a notification when the search is completed, so that other components, such as the ideogram and region selector, can update the display.
NCBI-provided track data
NCBI-provided data tracks available in SV and GDV are generated in-house and stored in internal databases. These tracks are derived from publicly-accessible data in the NCBI RefSeq, Gene, GenBank, SNP, ClinVar, dbVar, dbGaP, GEO, and SRA databases or from external databases (e.g., Ensembl) (Cunningham et al. 2019) as indicated in the track title description within the application.
Third-party data
NCBI SV accesses track hub data by running UCSC Browser utilities (https://genome.ucsc.edu/util.html) on our back-end with a custom layer of compressed caching and in memory succinct data structures (https://github.com/tlk00/BitMagic) to improve performance. The SV data streaming model relies on data availability of non-NCBI data sources, which is not guaranteed by the NCBI.
Browser and code availability
The NCBI SV application can be accessed at https://www.ncbi.nlm.nih.gov/tools/sviewer/. Instructions for embedding SV on third-party pages can be found at https://www.ncbi.nlm.nih.gov/tools/sviewer/embedding-api/.
The bulk of the SV codebase is available as part of the NCBI GBench source code package. The most recent version of this code, as of this writing, is available as Supplemental Code. The current version of this code can be downloaded at https://www.ncbi.nlm.nih.gov/tools/gbench/downloads/.
NCBI's GDV browser can be accessed from its home page at https://www.ncbi.nlm.nih.gov/genome/gdv/.
All NCBI software and data are public domain under the terms of the United States Copyright Act (https://www.ncbi.nlm.nih.gov/home/about/policies/).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Arkadi Doubintchik for aid in project management and the NCBI QA team and Marina Omelchenko for testing of our graphical viewers. We thank Ray Anderson III, Hsiu-Chuan Chen, Cliff Clausen, and Vamsi Kodali for aiding in the management and retrieval of track and assembly data at NCBI. We acknowledge Deanna Church and Peter Meric for their contributions to the early design and implementation of GDV. We also thank our many internal and external users, especially Terence Murphy from the RefSeq/Gene group, as well as members of the NCBI Customer Service Division for consultation in product development. This work was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Author contributions: S.H.R., V.A.S., and A.K. planned software development and supervised work; R.P. advised software priorities and coordinated outreach to users; V.A., A.A., E.B., V.E., V.J., V.L., D.R., A.S., and E.M.W. contributed code development; S.H.R. drafted the manuscript with help from A.K., V.A.S., V.A., and A.S.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.266932.120.
References
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. 2013. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41: D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41: W29–W33. 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L, et al. 2016. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol 17: 66 10.1186/s13059-016-0924-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, et al. 2011. Modernizing reference genome assemblies. PLoS Biol 9: e1001091 10.1371/journal.pbio.1001091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Computational Pan-Genomics Consortium. 2018. Computational pan-genomics: status, promises and challenges. Brief Bioinform 19: 118–135. 10.1093/bib/bbw089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, et al. 2019. Ensembl 2019. Nucleic Acids Res 47: D745–D751. 10.1093/nar/gky1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Sun Z, Campbell D, Kusebauch U, Chu CS, Mendoza L, Shteynberg D, Omenn GS, Moritz RL. 2015. State of the human proteome in 2014/2015 as viewed through peptideAtlas: enhancing accuracy and coverage through the atlasProphet. J Proteome Res 14: 3461–3473. 10.1021/acs.jproteome.5b00500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski SM, Maglott D. 2003. Using the Map Viewer to explore genomes. In The NCBI Handbook (ed. McEntyre J, Ostell J). National Center for Biotechnology Information (US), Bethesda, MD. [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A. 2011. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform 12: 41–51. 10.1093/bib/bbq072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The Human Genome Browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26: 2204–2207. 10.1093/bioinformatics/btq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov A, Bollin C. 2021. NCBI Genome Workbench: desktop software for comparative genomics, visualization, and GenBank data submission. In Multiple sequence alignment. Methods in molecular biology, Vol. 2231 (ed. Katoh K), pp. 261–295. Humana, New York: 10.1007/978-1-0716-1036-7_16 [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Kattman BL. 2018. ClinVar at five years: delivering on the promise. Hum Mutat 39: 1623–1630. 10.1002/humu.23641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Barber GP, Casper J, Clawson H, Diekhans M, Gonzalez JN, Hinrichs AS, Lee BT, Nassar LR, Powell CC, et al. 2020. UCSC Genome Browser enters 20th year. Nucleic Acids Res 48: D756–D761. 10.1093/nar/gkz1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. 2011. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282. 10.1093/bioinformatics/btr209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato S, et al. 2015. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 16: 22 10.1186/s13059-014-0560-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Fox G, Kiran AM, De Bo C, O'Connor PB, Heaphy SM, Mullan JP, Donohue CA, Higgins DG, Baranov PV. 2014. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res 42: D859–D864. 10.1093/nar/gkt1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis A, Gertz EM, Schaffer AA, Agarwala R. 2006. WindowMasker: window-based masker for sequenced genomes. Bioinformatics 22: 134–141. 10.1093/bioinformatics/bti774 [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46: D8–D13. 10.1093/nar/gkx1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Hsu J, Tri LQ, Willi M, Mansour T, Kai Y, Garner J, Lopez J, Busby B. 2016. dbVar structural variant cluster set for data analysis and variant comparison. F1000Res 5: 673 10.12688/f1000research.8290.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujar S, O'Leary NA, Farrell CM, Loveland JE, Mudge JM, Wallin C, Girón CG, Diekhans M, Barnes I, Bennett R, et al. 2018. Consensus coding sequence (CCDS) database: a standardized set of human and mouse protein-coding regions supported by expert curation. Nucleic Acids Res 46: D221–D228. 10.1093/nar/gkx1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, Wang T, Nguyen N, Paten B, Zweig AS, Karolchik D, et al. 2014. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 30: 1003–1005. 10.1093/bioinformatics/btt637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Agarwala R, Bolton EE, Brister JR, Canese K, Clark K, Connor R, Fiorini N, Funk K, Hefferon T, et al. 2019. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 47: D23–D28. 10.1093/nar/gky1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Beck J, Brister JR, Bolton EE, Canese K, Comeau DC, Funk K, Ketter A, Kim S, Kimchi A, et al. 2020a. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 48: D9–D16. 10.1093/nar/gkz899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch-Mizrachi I. 2020b. Genbank. Nucleic Acids Res 48: D84–D86. 10.1093/nar/gkz956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311. 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. 2009. JBrowse: a next-generation genome browser. Genome Res 19: 1630–1638. 10.1101/gr.094607.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, Lee M, Popova N, Sharopova N, Kimura M, et al. 2014. NCBI's Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res 42: D975–D979. 10.1093/nar/gkt1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, et al. 2003. Database resources of the National Center for Biotechnology. Nucleic Acids Res 31: 28–33. 10.1093/nar/gkg033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.