Abstract
The Cre/loxP system is a powerful tool for gene function study in vivo. Regulated expression of Cre recombinase mediates precise deletion of genetic elements in a spatially– and temporally–controlled manner. Despite the robustness of this system, it requires a great amount of effort to create a conditional knockout model for each individual gene of interest where two loxP sites must be simultaneously inserted in cis. The current undertaking involves labor-intensive embryonic stem (ES) cell–based gene targeting and tedious micromanipulations of mouse embryos. The complexity of this workflow poses formidable technical challenges, thus limiting wider applications of conditional genetics. Here, we report an alternative approach to generate mouse loxP alleles by integrating a unique design of CRISPR donor with the new oviduct electroporation technique i-GONAD. Showing the potential and simplicity of this method, we created floxed alleles for five genes in one attempt with relatively low costs and a minimal equipment setup. In addition to the conditional alleles, constitutive knockout alleles were also obtained as byproducts of these experiments. Therefore, the wider applications of i-GONAD may promote gene function studies using novel murine models.
Our understanding of the genetic mechanisms of human diseases has been largely expanded by loss-of-function studies using engineered mouse models. Two types of gene knockout mouse models are commonly used: global and conditional, each with unique advantages. Ubiquitous deletion of a gene from all tissues in a global knockout model can mimic the genetic condition of human disease, thus permitting a quick and thorough evaluation of gene function in vivo (Cheon and Orsulic 2011; Doyle et al. 2012; Wang et al. 2013; Amoasii et al. 2017; Gurumurthy and Lloyd 2019). Given the flexible design of gene inactivation, for example, frame shift mutation caused by a small insertion or deletion (indel) or targeted removal of exon(s), the design and creation of a global knockout mouse model is relatively easy. However, genetic studies using this type of model may have inherent limitations. First, for a gene that is widely expressed, pleiotropic effects from its deletion in all tissues can obscure the cell type–specific gene functions. Second, an early onset lethality or gross abnormality of a mutant will prevent its application for studying gene function at adult stages or during aging conditions.
Cre/loxP-mediated conditional knockout models can circumvent these difficulties. Cre can delete the flanked gene sequence between two loxP sites through DNA recombination (Sauer and Henderson 1988). Built on this principle, more sophisticated models of hormone-sensitive or tetracycline-inducible conditional knockouts were developed that allowed precise temporal control of gene disruption (Danielian et al. 1998; Jaisser 2000; Schonig et al. 2002; Belteki et al. 2005; Feil et al. 2009). These models have afforded valuable opportunities to interrogate the context-dependent gene function, thus providing clinically relevant information to treat genetic disease. The Cre/loxP system requires the creation of a conditional allele for the gene of interest. Although many tissue-specific/hormone-inducible Cre-expressing mouse strains are readily available, generation of the loxP-flanked (floxed) alleles is challenging and labor-intensive due to the lack of an efficient method for their generation (Lewandoski 2001; Skarnes et al. 2011; Bouabe and Okkenhaug 2013).
The common approach for generating floxed alleles was established in the 1980s (Thomas and Capecchi 1987; Mansour et al. 1988; Capecchi 1989; Zijlstra et al. 1989; te Riele et al. 1992; Limonta et al. 1995; Skarnes 2015). It utilizes homologous recombination in embryonic stem cells and requires technically challenging embryo-manipulation procedures. Such practice is largely restricted to transgenic core facilities, making the approach costly, time-consuming, and lacking in guaranteed success. Although this method has cumulatively created many invaluable mouse models over the past decades (Skarnes et al. 2011), scaling this approach up to functionally characterize the vast majority of the genome, with its rapidly expanding gene list (Chen et al. 2020) and associated genetic elements, is a challenging endeavor. Therefore, an alternative method that is inexpensive and easy to implement is awaited by the mouse genetic research community. Ideally, such methodology can be performed by regular laboratory personnel, for example, graduate students, with necessary technical training. Toward this goal, both the gene targeting strategy and the delivery method should be streamlined.
CRISPR genome-editing technologies have revolutionized genetic studies (Hsu et al. 2014; Irion et al. 2014; Platt et al. 2014; Aida et al. 2015; Square et al. 2015; Wang et al. 2016a; Zu et al. 2016; Jiang and Doudna 2017; Adli 2018; Miura et al. 2018; Gurumurthy and Lloyd 2019; Rasys et al. 2019; Yuan et al. 2019). However, successful applications of CRISPR-based mutagenesis have been largely restricted to creation of global loss-of-function models. Compared with random indels, the efficiency of precise editing through homology-mediated repair (HDR) is low (Doudna and Charpentier 2014; Jiang and Marraffini 2015; Richardson et al. 2016; Zhang et al. 2017; Aird et al. 2018). Because the generation of conditional alleles requires simultaneous integration of two loxP sites precisely in the same chromosomal region (in cis), the chance of success is significantly lower when compared with other HDR projects.
Single-stranded oligo DNA (ssODNA) that contains homologous sequences flanking the nuclease-induced dsDNA break site emerged as an ideal form of HDR template that delivers higher knock-in efficiency and specificity (Miura et al. 2015; Yoshimi et al. 2016). A recent study of the interactions between the Cas9 protein with its DNA substrate provided a rational to improve the HDR design (Richardson et al. 2016). Specifically, asymmetric target-strand ssODNAs were shown to be highly effective in introducing point mutations (Richardson et al. 2016). Using this type of ssODNA, we previously generated two conditional alleles for the Mymx gene by microinjection with an overall 12% efficiency (Bi et al. 2018). It should be noted, however, that an independent test of loxP insertions for 30 genes showed a nonsignificant impact of homology arm symmetry on HDR efficiency (Lanza et al. 2018). Reported in this large-scale test, the efficiency of loxP insertion by using short ssODNA, either symmetric or asymmetric designs, is around 9% (Lanza et al. 2018). More recently, utilizing various types of short ssODNA to separately insert loxP sites, extensive efforts from a consortium of core facilities and laboratories reported an overall 1% efficiency on 56 loci (Gurumurthy et al. 2019a). These studies revealed large disparities of the gene targeting efficiency through microinjection.
In addition to microinjection, alternative CRISPR delivery methods were reported (Kaneko and Mashimo 2015; Takahashi et al. 2015; Gurumurthy et al. 2016; Wang et al. 2016b; Modzelewski et al. 2018; Ohtsuka et al. 2018; Teixeira et al. 2018). Of special interest, a mouse zygote-stage embryo transfection strategy called improved-Genome editing via Oviductal Nucleic Acids Delivery (i-GONAD) was reported (Ohtsuka et al. 2018). This method delivers a gene editing cocktail into mouse zygotes through oviduct electroporation. Operation of i-GONAD is easier than the ES-cell-based approach or microinjection because mouse zygotes no longer need to be individually handled and transferred into pseudopregnant mice (Ohtsuka et al. 2018; Gurumurthy et al. 2019b), yet the HDR efficiency of i-GONAD was comparable to that achieved through microinjection (Ohtsuka et al. 2018). For the creation of conditional alleles, one recent study proposed a two-step i-GONAD workflow that sequentially inserts two loxP sites, one at a time (Sato et al. 2020). However, the proof-of-principle test of this approach was not satisfactory, that is, only one loxP was integrated (Sato et al. 2020). Therefore, both the CRISPR targeting strategy and the logistics of i-GONAD for creation of conditional alleles require improvement. Here, we aim to develop and test a new approach by integrating a unique design of asymmetric loxP-ssODNA with the i-GONAD delivery method to create mouse conditional alleles.
Results
Exploiting the i-GONAD method to create a conditional allele: a proof-of-principle test
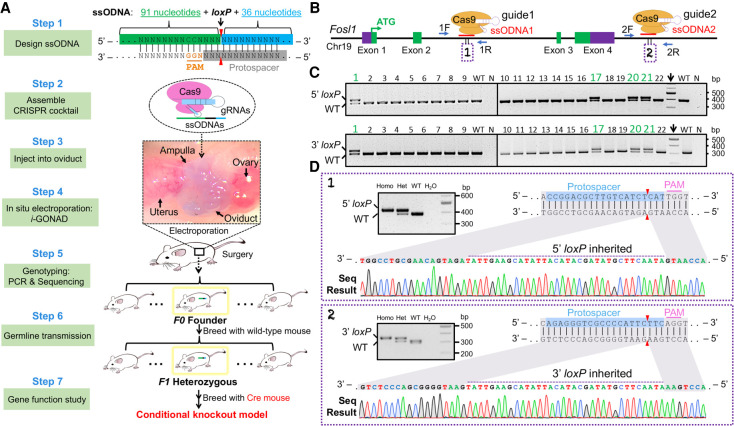
The principle of HDR template design, major experimental procedures, and milestones for this method are schematized in Figure 1A. It involves using two guide RNAs (gRNAs) and two short ssODNAs as HDR donors for loxP insertions. Each ssODNA is 161 nt long, composed of 91 nt of the 5′ homology arm from the PAM-proximal side, 34 nt of loxP sequence, and 36 nt of the 3′ homology arm from the PAM-distal side.
Figure 1.
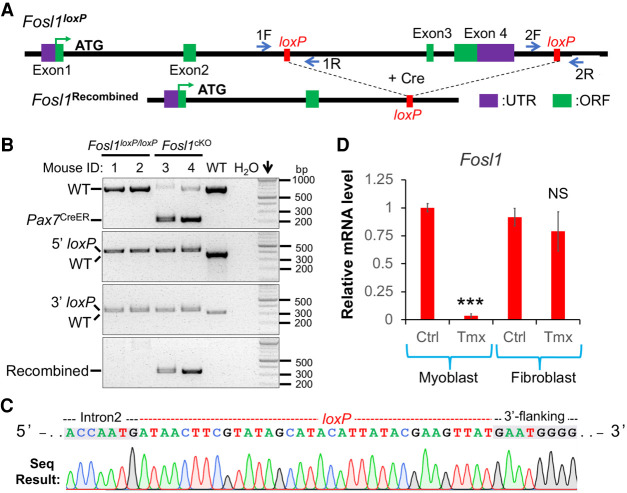
Generation of a conditional allele by i-GONAD: design and a proof-of-principle test. (A) Schematic illustration of the strategy to generate a conditional allele by i-GONAD. The necropsy image was taken to show the anatomical location of the oviduct with a blue ink indicator. (B) Fosl1 gene structure and relative positions of ssODNA, gRNA, and genotyping primer. (ssODNA) Single-stranded oligo DNA. (C) Fosl1 genotyping results for the F1 generation. Top row detects 5′-loxP insertion by primer pair 1F&1R; bottom row detects 3′-loxP insertion by primer pair 2F&2R; 5′-loxP band size is 426 bp; 3′-loxP is 351 bp. Mouse that inherited both 5′- and 3′-loxP is highlighted in green. (Arrow) DNA marker lane, (N) negative control of PCR (water). (D) Validations of loxP sites by Sanger sequencing.
We first tested the i-GONAD method by generating a mouse conditional allele for the Fosl1 (fos-like antigen 1) gene for which the expression at both mRNA and protein levels was induced during muscle regeneration (Supplemental Fig. S1). The function of the Fosl1 gene during this biological process remains unknown. Because Fosl1 global knockout mice die as embryos (Schreiber et al. 2000), studying the postnatal muscle-specific function of this gene requires a conditional allele.
The mouse Fosl1 gene contains four exons, with the last two being close to each other and representing 64% of the coding sequence including critical domains of the FOSL1 protein (Matsuo et al. 2000). Therefore, deletions of exons 3 and 4 can unequivocally abolish gene function. We designed a pair of gRNAs and the corresponding ssODNAs to insert loxP sites flanking these exons (Fig. 1B). Different from microinjection, which directly delivers CRISPR components into the zygotes, i-GONAD involves a two-step transfer of the gene editing cocktail: first, the cocktail is injected into the lumen of the oviduct, followed by a second step of oviduct electroporation that transfers the cocktail into zygotes (Fig. 1A; Ohtsuka et al. 2018). Owing to the volume restriction of the oviduct and inevitable dilutions of the gene editing cocktail by the much larger volume of oviduct fluid, we used concentrated Cas9 protein, ssODNAs, and gRNAs in molar ratios of 1:6:10, as we previously used in microinjection experiments (Bi et al. 2018).
CD-1 female mice in estrus were mated with C57BL/6J males. We performed i-GONAD on two females that showed copulation plugs. To avoid false detection of loxP sites in scenarios of random ssODNA integration, genotyping primers were designed in the regions outside the homology arms of donors. Successful incorporation of loxP was identified by a 34-base-pair (bp) increase in PCR amplicon size. Our results revealed the simultaneous 5′- and 3′-loxP insertions in one mouse (#2) (Supplemental Fig. S2). We also detected 5′- or 3′-loxP integrations in other mice: #1, #10, #11, #14, #15, and #20 (Supplemental Fig. S2). Because transgenic founders are commonly mosaic, we tested germline editing of the #2 mouse by breeding it with wild-type (WT) mice. Out of a total of 22 filial 1 (F1) progenies obtained, four showed simultaneous inheritance of 5′- and 3′-loxP sites (Fig. 1C). The fidelity of these loxP sites was also validated by sequencing (Fig. 1D). Intercrossing of the heterozygous mice generated homozygous Fosl1loxP/loxP mutants at expected Mendelian ratios. These mutants appeared phenotypically normal, indicating that loxP insertions do not alter gene function, a prerequisite for conditional alleles.
More tests of the i-GONAD method to create conditional alleles for another four genes
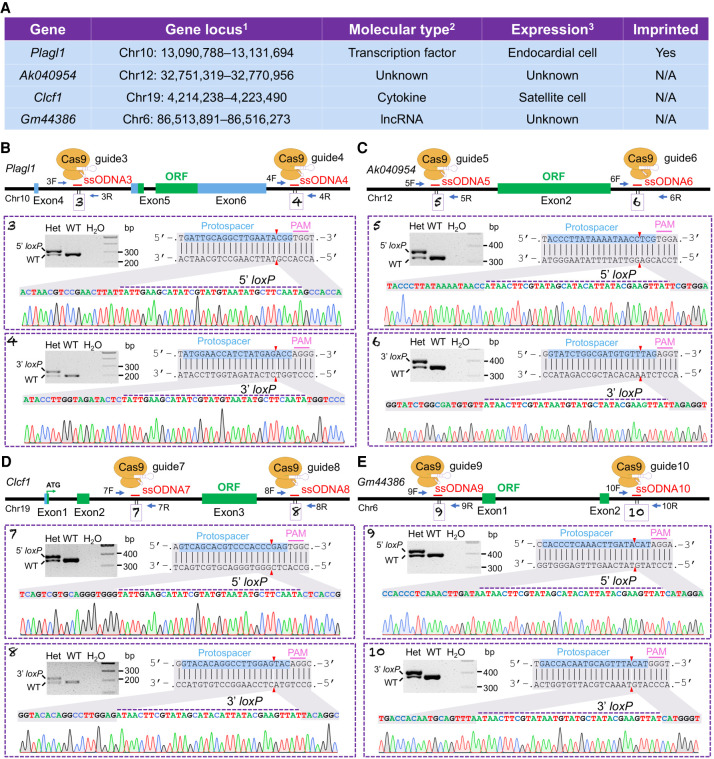
To our knowledge, the Fosl1loxP allele is the first conditional allele created by the i-GONAD protocol in one step, which is easier and faster than two-step approaches. To better evaluate the efficiency, we performed additional tests on four other genes: Plagl1 (pleiomorphic adenoma gene-like 1), Ak040954, Clcf1 (cardiotrophin-like cytokine factor 1), and Gm44386. In addition to having different genomic locations, these genes were chosen because of our research interests, their distinct functions, and patterns of epigenetic regulations (Fig. 2A). As such, tests on these loci may demonstrate the broader utility of this method.
Figure 2.
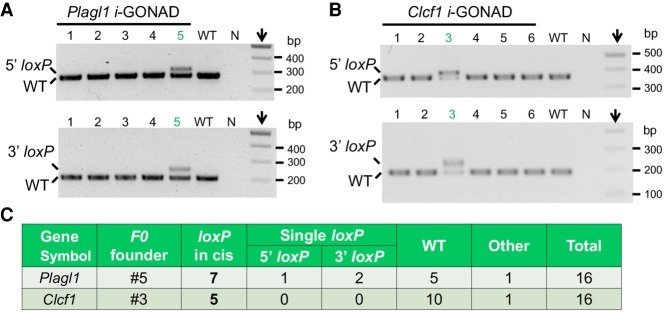
Generation of conditional alleles for another four genes by the i-GONAD method. (A) Gene information: 1coordinates as of mouse GRCm38/mm10 genome assembly; 2NCBI annotation; 3query of Tabula Muris single-cell RNA sequencing data. (N/A) Data not available. (B–E) Gene structures and relative positions of ssODNA, gRNA, and genotyping primer for Plagl1 (B), Ak040954 (C), Clcf1 (D), and Gm44386 (E) genes. For Plagl1, 5′- and 3′-loxP bands are 312 bp and 234 bp; for Ak040954, 5′- and 3′-loxP bands are 361 bp and 391 bp; for Clcf1, 5′- and 3′-loxP bands are 389 bp and 222 bp; for Gm44386, 5′- and 3′-loxP bands are 431 bp and 415 bp. Boxed panels showed sequencing verifications of loxP sites in F1 generations. (Het) Heterozygous.
We first tested the synergy and editing efficiency of gRNAs in mouse fibroblasts (Supplemental Fig. S3). The gRNA pair that can generate large deletions between the gRNAs was chosen for i-GONAD. The loxP sequence contains an 8-bp asymmetric core spacer that defines the orientation of the loxP cassette (Sternberg et al. 1981; Sauer and Henderson 1988). Deletion of the flanked sequence requires that the two loxP sites are aligned in the same direction (Guo et al. 1997). We therefore adjusted the orientation of the loxP sequence within the HDR donors when gRNAs targeted opposite DNA strands.
For the Plagl1 gene, we aimed to generate a conditional allele by flanking the coding exons 5 and 6 with loxP sites (Fig. 2B). Four females were used for the i-GONAD procedure. Among 28 pups that were born, three (#8, #10, #11) showed simultaneous 5′- and 3′-loxP insertions, and another eight mice showed either 5′- or 3′-loxP insertions (Supplemental Fig. S4A). Among 15 progeny obtained from breeding the #10 founder with C57BL/6J WT mice, eight pups showed successful germline transmission of both loxP sites (Supplemental Fig. S4B). Correct targeting in these mice was also confirmed by sequencing (Fig. 2B, boxed panels). Of note, all other progeny showed the insertion of only the 3′-loxP site (Supplemental Fig. S4B). This reveals the mosaicism of genome editing.
The long noncoding gene Ak040954 contains two exons. We targeted the major exon 2 which represents 91% of the transcript (Fig. 2C). Three female mice were used for i-GONAD. Among 11 mice that were born, two pups (#5, #7) showed simultaneous 5′- and 3′-loxP insertions, whereas another two pups (#8, #9) showed only 5′-loxP insertions (Supplemental Fig. S4C). Among nine progeny obtained from breeding of #5 founder with C57BL/6J WT, three pups showed successful germline transmissions of floxed alleles that contained both the 5′- and 3′-loxP sites (Supplemental Fig. S4D). The fidelity of loxP sequences was validated by sequencing (Fig. 2C, boxed panels). We also observed the inheritance of other types of mutations, showing up as >34 bp insertions (#2, #7) (Supplemental Fig. S4D), which mirrored the genotype of their F0 parent (Supplemental Fig. S4C).
The Clcf1 gene contains three exons that together encode a 225-amino acid cytokine. We generated a conditional knockout allele by targeting exon 3 (Fig. 2D) that encodes the majority of the protein. Among eight pups produced, one mouse (#2) showed simultaneous 5′- and 3′-loxP insertions, and another two mice (#1, #4) showed either 3′- or 5′-loxP insertion, respectively (Supplemental Fig. S4E). Among 13 progeny of the #2 founder, eight mice showed successful germline transmissions of both 5′- and 3′-loxP insertions (Supplemental Fig. S4F). The fidelity of loxP sites was also confirmed by sequencing (Fig. 2D, boxed panels). All other pups showed only 5′-loxP insertions (Supplemental Fig. S4F). This is consistent with the genotype of the F0 founder for which the 5′-loxP insertion was nearly homozygous (Supplemental Fig. S4E).
Using the i-GONAD method, we also generated a conditional allele for the Gm44386 gene, whereby the coding exons were flanked by two loxP sites (Fig. 2E). Among nine pups produced, two (#1, #5) showed simultaneous 5′- and 3′-loxP insertions, whereas another two (#3, #9) showed only 3′-loxP insertions (Supplemental Fig. S4G). Successful transmissions of the two loxP sites were also confirmed by PCR (Supplemental Fig. S4H) and sequencing (Fig. 2E, boxed panels).
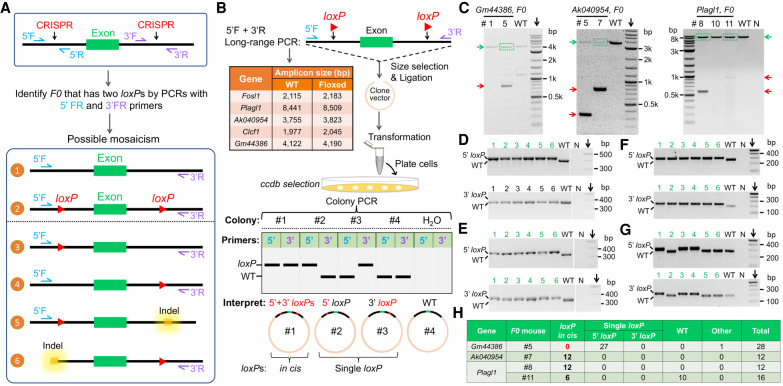
A cloning-based strategy to rapidly identify founders with loxP insertions in cis
By Mendel's law of inheritance, the simultaneous transmission of 5′- and 3′-loxP sites into the F1 generation validated loxP insertions in cis for five founders, that is, one for each gene. In addition to these mice, we also obtained other F0 founders that showed both 5′- and 3′-loxP sites. This includes the #5 mouse for the Gm44386 gene (Supplemental Fig. S4G), the #7 mouse for the Ak040954 gene (Supplemental Fig. S4C), and the #8 and #11 mice for the Plagl1 gene (Supplemental Fig. S4A). To deconvolute the potential mosaicism in these F0 founders (Fig. 3A) and identify more floxed alleles, we devised a cloning-based strategy of genotyping.
Figure 3.
Validations of floxed alleles by long-range genotyping PCR. (A) Illustration of the potential mosaicism of F0 founders. Although founders with 5′- and 3′-loxP insertions can be separately identified by genotyping PCR using 5′F + 5′R and 3′F + 3′R primers, this cannot distinguish whether loxP insertions are in cis or in trans. (B) Schematic to show the design and major steps of genotyping strategy. (C) DNA electrophoresis results of long-range PCR. The DNA bands enclosed in the green box were purified and cloned into vectors. (D–G) Representative genotyping results of bacterial colonies from cloning of the purified long-range PCR products for founder #5 of the Gm44386 gene (D), founder #7 for the Ak040954 gene (E), and founder #8 (F) and #11 (G) for the Plagl1 gene. (H) Summary of bacterial colony genotyping results.
As illustrated in Figure 3B, long-range genotyping PCR was performed with the forward primer from the 5′ gRNA region (5′F) and the reverse primer from 3′ gRNA region (3′R). Because the sizes of these PCR products were large (Fig. 3B), gel electrophoresis cannot reliably identify floxed alleles from others, for example, single loxP, WT, or small indels. We leveraged the principle of molecular cloning to genotype a single DNA molecule amplified by the long-range PCR (Fig. 3B). During transformation, the plasmid incompatibility ensures that each bacterium only maintains one vector that hosts one DNA insert. This provides an opportunity to verify the presence of two loxP sites in a single DNA molecule through genotyping the bacterial colony using 5′F&5′R and 3′F&3′R primers, which give rise to much shorter amplicons that can be analyzed by regular gel electrophoresis (Fig. 3B). In rare cases where a competent cell takes more than one vector, such heterogeneity can also be detected by PCR and will be excluded from analyses.
As expected, the separation of long-range PCR products by electrophoresis is poor (green arrows, Fig. 3C). To reduce the cloning background, we purified the large amplicons (green boxes, Fig. 3C) for ligations. Genotyping results of bacterial colonies for the #5 mouse of the Gm44386 gene showed only 5′ loxP, whereas the 3′ gRNA region was WT-size (Fig. 3D). Mimicking the bacterium genotyping results, the progeny of this mouse only showed 5′ loxP (Supplemental Fig. S4I). Compared with the genotype of this founder (Supplemental Fig. S4G), these observations indicate a mosaicism that possibly includes scenarios #3 and #6 illustrated in Figure 3A (lower panel). In comparison, floxed alleles were detected for all other founder mice (Fig. 3E–H). As a validation, breeding of the #7 mouse (Ak040954) produced F1 progeny that contained both loxP sites (three out of 10 pups). Together, these experiments proved the accuracy of the cloning-based method to identify floxed alleles.
Byproducts of the i-GONAD method can serve as global knockout models
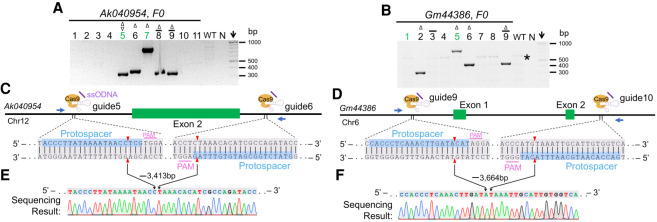
The long-range PCR revealed large-deletion alleles from these i-GONAD experiments (red arrows, Fig. 3C). Indeed, deletions of various sizes were observed for all five genes (Fig. 4A–D; Supplemental Figs. S5–S7). In total, 20 out of 54 (37%) F0 generation mice showed large deletions. As a consequence, critical exons for these genes were removed as confirmed by sequencing. This includes deletions of 7489 bp for the Plagl1 gene (Supplemental Fig. S5C), 3413 bp for the Ak040954 gene (Fig. 4E), 3664 bp for the Gm44386 gene (Fig. 4F), 1812 bp for the Fosl1 gene (Supplemental Fig. S6B), and 1755 bp for the Clcf1 gene (Supplemental Fig. S7B). Note that the amplicons for WT or floxed alleles were too large and thus not detected in these PCR conditions. Collectively, these “byproduct” mutants obtained from loxP projects may serve as global knockout models.
Figure 4.
Null alleles of Ak040954 and Gm44386 genes generated by the i-GONAD method. (A,B) Genotyping results of Ak040954 (A) and Gm44386 (B) for the F0 generation using primer pairs shown in C and D. The mice #5 to #9 in A and mice #2, #5, #6, and #9 in B showed large deletions. The star in B indicates a faint, nonspecific band. Note that WT or floxed alleles are too large to be detected in current PCR conditions. (C,D) Gene structures and positions of gRNA and genotyping primers for Ak040954 (C) and Gm44386 (D) genes. (E,F) Sanger sequencing results of founder #9 and #6 as shown in A and B, respectively. For all panels: (WT) wild-type, (N) negative control (water), green ID highlights founders with floxed alleles, (arrow) DNA marker lane, mice with single-side loxP integration are indicated by a bar over the ID, (Δ) deletions, (▿) large insertions.
Other types of mutations from the F0 generation included smaller indels produced by either 5′- or 3′-gRNA, for example, mice #15 and #20 for the Fosl1 gene (Supplemental Fig. S2), and mice #1, #2, #6, and #8 for the Clcf1 gene (Supplemental Fig. S4E). In scenarios where intron-exon splicing sites were destroyed, abnormal splicing may join incompatible exons, making these mutations potentially useful as knockout or hypomorphic alleles.
Examination of off-target mutagenesis by i-GONAD
The high efficiency of i-GONAD prompted us to examine the genome editing specificity in F0 founders that contained floxed alleles (Supplemental Fig. S8A). We used the polyacrylamide gel electrophoresis (PAGE) method (Zhu et al. 2014) to quickly and sensitively detect small indels. Among 10 gRNAs, #2 gRNA produced off-target mutagenesis in the predicted site (Supplemental Fig. S8A–C). Coincidently, only this off-target site had identical sequence with the 10-bp PAM-proximal “seed” region of the gRNA (Supplemental Fig. S8A). This aligns with the notion that a single nucleotide mismatch within the PAM-proximal region is not tolerated by the CRISPR-Cas9 (Qi et al. 2013; Fortin et al. 2019). Together, our results showed that genome editing delivered by i-GONAD is largely specific, though caution is also warranted when gRNAs with predicted low-specificity were used.
Test of i-GONAD on the Mecp2 gene
We continued to examine whether the short symmetric ssODNA donors can work for i-GONAD in generation of conditional alleles. For this purpose, we chose the Mecp2 (methyl CpG binding protein 2) gene because multiple groups have attempted to insert loxP by microinjecting symmetric ssODNA donors (Yang et al. 2013; Gurumurthy et al. 2019a). As such, using the same designs of gRNA and ssODNA, it provides an indirect comparison of i-GONAD with the microinjection approach.
Three CD-1 female mice that showed copulation plugs after mating with C57BL/6J males were used for the i-GONAD procedure. Embryos at day 12.5 postconception (E12.5) were collected for genotyping analysis. The high efficiency of gRNAs was revealed by big truncations and possible elimination of primer binding sites in multiple samples (#1, #2, #4, #6, #9, #15) (Supplemental Fig. S9A,B). Among 19 total embryos, two showed a 5′-loxP site; four showed a 3′-loxP site, with one embryo (#3) showing both 5′- and 3′-loxP sites. Long-range PCR (Supplemental Fig. S9B) and cloning-based genotyping analysis (Supplemental Fig. S9C,D) confirmed in cis loxP insertions in this sample. These results were consistent with a previous report that the efficiency of loxP insertions in this locus was relatively low (Gurumurthy et al. 2019a).
Repeating i-GONAD on Plagl1 and Clcf1 genes using C57BL/6J females
i-GONAD can efficiently deliver CRISPR cocktails for a variety of hybrid or inbred mouse strains (Ohtsuka et al. 2018). We used CD-1 females because of their good postsurgery performance and generally large litter size. In our experience, performing i-GONAD on CD-1 females is also easier than C57BL/6J females thanks to larger volumes of the oviduct. However, with technical proficiency gained from these practices, we continued to determine whether conditional alleles could also be produced from the inbred C57BL/6J strain.
Plag1 and Clcf1 genes were selected for these tests. C57BL/6J females that showed copulation plugs after mating with C57BL/6J males were used for i-GONAD. As previously observed, the pregnancy rate of C57BL/6J females was low (Ohtsuka et al. 2018). From multiple breeding pairs, only two females produced a total of five embryos for the Plagl1 experiment, and another two females produced six for the Clcf1 experiment. Nevertheless, we identified one embryo that showed a floxed allele for each gene (Fig. 5A,B), which was also confirmed by analyzing the long-range PCR products (Fig. 5C; Supplemental Fig. S10). Together, these results are in agreement with previous observations that inbred strains can be used for genome editing by the i-GONAD method (Ohtsuka et al. 2018).
Figure 5.
Testing i-GONAD for generation of conditional alleles using C57BL/6J mice. (A) Plagl1 genotyping results of E12.5 embryos. For the 5′ gRNA region, WT band is 278 bp, loxP band is 312 bp; for the 3′ gRNA region, WT band is 200 bp, loxP band is 234 bp. (B) Clcf1 genotyping results for E12.5 embryos. For the 5′ gRNA region, WT band is 355 bp, loxP band is 389 bp; for the 3′ gRNA region, WT band is 188 bp, loxP band is 222 bp. (C) Summary of bacteria colony genotyping results shown in Supplemental Figure S10C,D.
Validation of the Fosl1 conditional allele
We continued to validate the design and utility of the conditional allele that we generated by i-GONAD. Through serial crossing of Fosl1loxP/loxP with Pax7CreER mice (Lepper et al. 2009), a widely used muscle stem cell–specific Cre deleter, we obtained Pax7CreER:Fosl1loxP/loxP conditional knock out mouse model, indicated by Fosl1cKO. Tamoxifen was administered into adult mutants to activate CreER and thus the removal of exons 3 and 4 of the Fosl1 gene (Fig. 6A). Two days after the last dosage of tamoxifen, muscle tissues were collected for genotyping analyses. As expected, the recombined allele can be specifically detected in muscle samples from Fosl1cKO but not littermate Fosl1loxP/loxP mice (Fig. 6B). Sequencing confirmed the correct recombination between 5′- and 3′-loxP sites that excised the targeted exons and joined intron 2 with the 3′ region of the Fosl1 gene (Fig. 6C). Because muscle stem cells account for less than 5% of total nuclei in intact muscle tissues (Snow 1981), the intact floxed allele was also readily detected in Fosl1cKO muscle samples (Fig. 6B).
Figure 6.
Successful recombination of the Fosl1 floxed allele mediated by Cre. (A) Gene structures of the Fosl1loxP and recombined alleles following Cre-mediated DNA recombination. (B) Genotyping results of adult muscle tissues from Fosl1loxP/loxP and littermate Fosl1cKO mice. 5′- and 3′-loxP were detected by primers 1F + 1R and 2F + 2R, respectively. The floxed allele was detected by primers 1F + 2R. (C) Sanger sequencing result that validated DNA recombination in Fosl1cKO muscle sample. (D) qPCR that measured Fosl1 expression in myoblasts and fibroblasts isolated from the Fosl1cKO mice. qPCR primers are located in exon 4 which is floxed. (Ctrl) Vehicle control, (Tmx) 4-OH tamoxifen. (***) P < 0.001, (NS) not significant. Data are mean ± SEM.
We then isolated muscle precursor cells from Fosl1cKO mouse and confirmed the robust inactivation of Fosl1 gene detected by qPCR using primers from exon 4 of this gene (Fig. 6D). As a negative control, fibroblasts that were also isolated from Fosl1cKO mouse showed a normal level of expression for the Fosl1 gene (Fig. 6D). We did not examine Fosl1 gene expression in whole muscle tissues because muscle precursor cells only account for a small portion of the tissue, whereas the remaining cell types (Pax7–) also abundantly express the Fosl1 gene, as shown by single-cell RNA sequencing analyses of muscle tissues (Supplemental Fig. S11; The Tabula Muris Consortium 2018). These results validated the utility of the conditional allele produced by the i-GONAD method.
Discussion
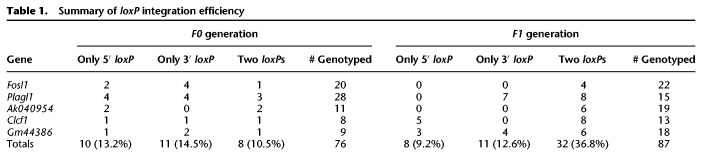
The overall targeting efficiency of producing floxed alleles by i-GONAD was 10% (eight out of 76) (Table 1). In addition to these desired mutants, the frequency of obtaining F0 mice with either 5′- or 3′-loxP insertion was 28% (21 out of 76). Therefore, the combined loxP-insertion efficiency was 38%, close to the HDR efficiency that was previously reported using the i-GONAD method (49%) or through microinjection (52%) (Ohtsuka et al. 2018). In F1 generations, the chance of inheriting two loxP sites (in cis) was 37% (32 out of 87 pups) (Table 1), indicating efficient germline editing by i-GONAD. These tests demonstrate that our approach, using the i-GONAD method and the HDR template design, is robust, fast, and efficient for the generation of mouse conditional alleles.
Table 1.
Summary of loxP integration efficiency
One recent large-scale test of microinjection reported an 11% loxP-insertion efficiency by using asymmetric ssODNAs and a 7% efficiency by using symmetric ssODNAs (Lanza et al. 2018). Compared with these dual-ssODNA approaches, long ssODNA, composed of loxP–Exon(s)–loxP sequences, was shown to be more efficient in generating floxed alleles (Quadros et al. 2017; Lanza et al. 2018; Miura et al. 2018; Miyasaka et al. 2018). Because both loxP sites were synthesized in one piece, this predicts simultaneous integrations of two loxP sites. However, technical barriers do exist for the preparations of long ssODNA (Lanza et al. 2018). Depending on the length of the floxed area, production of long ssODNA that meets the required yield, purity, and fidelity by either chemical synthesis or enzymatic reactions is still challenging. For instance, even without considering the homology arms, the floxed regions for our genes measured 2–7 kb. In comparison, the maximum size of ssODNA (megamer) that can be ordered from IDT is 2 kb. Therefore, broader applications of long ssODNA await improvement of DNA synthesis technology.
In comparison with the conventional floxing method, an alternative strategy of conditional gene-inactivation by leveraging the exon-splicing machinery (Guzzardo et al. 2017) could make the use of single-piece ssODNA more realistic. This conditional knockout strategy involves the insertion of a small artificial intron that harbors two loxP sites into a coding exon of the target gene. In the absence of Cre, this foreign sequence can be fully excised by splicing machinery without affecting gene function. In the presence of Cre activity, recombination of loxP sites will destroy the artificial intron which causes translational termination, thus abrogating gene function. It remains to be tested whether the artificial-intron ssODNA can produce higher targeting efficiency when delivered by i-GONAD.
In addition to the formats of donors, the HDR efficiency can also be affected by donor concentration at the editing site (Liu et al. 2019). Consistent with this notion, HDR frequency was improved when ssODNA was chemically linked to Cas9 protein (Ma et al. 2017; Aird et al. 2018; Ling et al. 2020). In a simpler form, one can test whether the more stable ssODNA, improved by chemical modifications, can enhance i-GONAD efficiency. Other methods to improve HDR frequency include chemical inhibition of the nonhomologous end-joining pathway (Maruyama et al. 2015), engineering of Cas9 protein (Charpentier et al. 2018; Jayavaradhan et al. 2019), and the timing of gene targeting in S/G2 phases of the cell-cycle (Lin et al. 2014). The i-GONAD procedure is performed around 4:00 p.m. of the day when the vaginal plug is observed (Ohtsuka et al. 2018; Gurumurthy et al. 2019b). This marks approximately 16 h post-insemination (hpi), a stage when zygotes transit from the S to the G2 phase of the first cell-cycle (Luthardt and Donahuc 1973; Howlett and Bolton 1985; Debey et al. 1989). In comparison, microinjection or ex vivo zygote electroporation was commonly performed at an earlier time that corresponds to 10–12 hpi (Lanza et al. 2018; Teixeira et al. 2018). The timing differences of these CRISPR delivery methods may affect the efficiency of loxP insertions.
One limitation of the current study is the relatively smaller number of genes tested, as compared with the larger-scale microinjection experiments (Lanza et al. 2018) or multicenter consortium studies (Gurumurthy et al. 2019a). Because we do not have access to microinjection core or ex vivo zygote electroporation setups, we cannot directly compare the performance of these delivery methods. However, the initial report of i-GONAD confirmed that its HDR efficiency was comparable with that achieved through microinjection (Ohtsuka et al. 2018). Similarly, the 10% efficiency that we observed for loxP insertions by i-GONAD was also comparable with that from microinjection (11%–12%) (Bi et al. 2018; Lanza et al. 2018), though locus-to-locus variability was observed. For instance, the efficiency of creating a floxed allele for the Fosl1 gene is 5% (one out 20) versus 18% (two out 11) for the Ak040954 gene (Table 1). This could be caused by different levels of chromatin accessibility, a major determinant of Cas9 binding and activity (Wu et al. 2014; Chari et al. 2015). Nevertheless, successful delivery of CRISPR reagents is required but certainly cannot guarantee the same editing efficiency for different loci.
To provide an indirect comparison with the microinjection method, we repeated i-GONAD on the Mecp2 gene. We selected this locus because it was previously tested by the microinjection method, though a large discrepancy of targeting efficiency, ranging from 0% to 16%, was reported (Yang et al. 2013; Gurumurthy et al. 2019a). The i-GONAD efficiency of producing a floxed allele for this locus is 5%. Of note, the gRNAs did show high editing activity in producing null alleles. Therefore, i-GONAD is a reliable method to deliver the CRISPR cocktail into zygotes, and the efficiency of producing conditional alleles can vary largely at different loci.
We referred to our previous experience of microinjection (Bi et al. 2018) for the molar ratios of Cas9, gRNA, and ssODNA. It remains unknown whether other concentrations of these reagents could enhance the success rate of i-GONAD. Although i-GONAD does not require direct handling of the mouse embryo, its success depends on many factors. First, similar to microinjection, i-GONAD requires steady hand-control of the microcapillary needle for oviduct injection. Second, a thorough understanding of the mouse reproductive system is essential. In our experience, oviduct injections visualized by dye solution have afforded valuable training opportunities. In addition, we recommend testing gRNAs in cultured mouse cells, for example, fibroblasts, before applying them for i-GONAD. Last, one should monitor the off-target mutations in picked founders, especially when the predicted off-target site is located near the gene of interest. In summary, our study provides a complete gene targeting workflow to create, analyze, and authenticate conditional alleles. This may promote gene function studies in vivo by providing an inexpensive alternative to generate custom mouse models.
Methods
Oviduct electroporation by the i-GONAD protocol
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Georgia. Mouse oviduct electroporation was performed as previously reported (Ohtsuka et al. 2018). Briefly, 6- to 10-wk-old CD-1 or C57BL/6J female mice were mated with C57BL/6J stud males the day before electroporation. The copulated female mice were used for surgery to expose the oviduct. CRISPR gene editing cocktails were freshly assembled and contained 6 μM Cas9 protein (IDT 1081058, Lot # 0000405530), 30 μM gRNA (Alt-R crRNA annealed with tracrRNA, IDT 1072534, Lot # 0000403961), and 18 μM ssODNA (IDT Ultramer DNA Oligo, standard desalting). This cocktail was delivered into the oviduct through microcapillary injection. Oviduct electroporation was performed using a CUY21EDIT II electroporator with the following protocol: Pd A: 100 mA, Pd on: 5 msec, Pd off: 50 msec, three cycles, decay 10%. The sequences for gRNA and ssODNA are provided in Supplemental Materials.
Mouse genotyping analysis
Genotyping PCR was performed using genomic DNA extracted from the toe clipping with the primers listed in Supplemental Materials. For Sanger sequencing, PCR products were first gel-purified and cloned into the pCRII Topo vector (Thermo Fisher Scientific K460001) and sequenced with T7 or SP6 primers. Long-range PCR was performed using LongAmp Hot Start Taq 2X Master Mix (NEB M0533S) to examine genomic DNA that was purified by a Monarch Genomic DNA Purification kit (NEB T3010). The large amplicons were gel-purified and cloned into the pCR-XL-2-TOPO vector (Thermo Fisher Scientific K8050-10). The top off-targeting sites were predicted by Cas-OFFinder (Bae et al. 2014).
Tamoxifen and muscle injury
Tamoxifen (Sigma-Aldrich T5648) was dissolved in ethanol (10 mg/mL). This stock solution was diluted in sesame oil (Sigma-Aldrich S3547) with a ratio of 1:9 before injection. Two milligrams of tamoxifen were administered by intraperitoneal injection. Muscle injury was induced by injecting 1.2% barium chloride (50 μL) into the tibialis anterior muscle.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Junfei Wen and Hector Romero-Soto for technical assistance. We also thank the Center for Molecular Medicine and the Complex Carbohydrate Research Center for administrative support and the IACUC and URAR team from the University of Georgia for mouse colony management. We thank Dr. Shihuan Kuang and Jun Wu from Purdue University and Dr. Xiaochun Li from the University of Texas Southwestern Medical Center for sharing reagents and Dr. Jonathan Eggenschwiler for critical reading of this manuscript. We also thank Dr. Masato Ohtsuka from Tokai University and Dr. Channabasavaiah Gurumurthy from the University of Nebraska Medical Center for consultations on the oviduct electroporation method and patient instructions and other members from the Bi lab for technical advice. This work was supported by start-up funding from the University of Georgia.
Author contributions: R.S., H.Z., and P.B. designed and performed the research; R.S. and P.B. analyzed the data; and P.B. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.265439.120.
References
- Adli M. 2018. The CRISPR tool kit for genome editing and beyond. Nat Commun 9: 1911 10.1038/s41467-018-04252-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R, Wada Y, Tanaka KF, Sakuma T, Yamamoto T, Tanaka K. 2015. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol 16: 87 10.1186/s13059-015-0653-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird EJ, Lovendahl KN, St Martin A, Harris RS, Gordon WR. 2018. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun Biol 1: 54 10.1038/s42003-018-0054-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoasii L, Long CZ, Li H, Mireault AA, Shelton JM, Sanchez-Ortiz E, McAnally JR, Bhattacharyya S, Schmidt F, Grimm D, et al. 2017. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med 9: eaan8081 10.1126/scitranslmed.aan8081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS. 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30: 1473–1475. 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. 2005. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res 33: e51 10.1093/nar/gni051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi P, McAnally JR, Shelton JM, Sánchez-Ortiz E, Bassel-Duby R, Olson EN. 2018. Fusogenic micropeptide Myomixer is essential for satellite cell fusion and muscle regeneration. Proc Natl Acad Sci 115: 3864–3869. 10.1073/pnas.1800052115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouabe H, Okkenhaug K. 2013. Gene targeting in mice: a review. Methods Mol Biol 1064: 315–336. 10.1007/978-1-62703-601-6_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. 1989. The new mouse genetics: altering the genome by gene targeting. Trends Genet 5: 70–76. 10.1016/0168-9525(89)90029-2 [DOI] [PubMed] [Google Scholar]
- Chari R, Mali P, Moosburner M, Church GM. 2015. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods 12: 823–826. 10.1038/nmeth.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Khedher AHY, Menoret S, Brion A, Lamribet K, Dardillac E, Boix C, Perrouault L, Tesson L, Geny S, et al. 2018. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat Commun 9: 1133 10.1038/s41467-018-03475-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Brunner AD, Cogan JZ, Nuñez JK, Fields AP, Adamson B, Itzhak DN, Li JY, Mann M, Leonetti MD, et al. 2020. Pervasive functional translation of noncanonical human open reading frames. Science 367: 1140–1146. 10.1126/science.aay0262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon DJ, Orsulic S. 2011. Mouse models of cancer. Annu Rev Pathol-Mech 6: 95–119. 10.1146/annurev.pathol.3.121806.154244 [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. 1998. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol 8: 1323–1326. 10.1016/S0960-9822(07)00562-3 [DOI] [PubMed] [Google Scholar]
- Debey P, Renard JP, Coppey-Moisan M, Monnot I, Geze M. 1989. Dynamics of chromatin changes in live one-cell mouse embryos: a continuous follow-up by fluorescence microscopy. Exp Cell Res 183: 413–433. 10.1016/0014-4827(89)90401-1 [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Doyle A, McGarry MP, Lee NA, Lee JJ. 2012. The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res 21: 327–349. 10.1007/s11248-011-9537-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil S, Valtcheva N, Feil R. 2009. Inducible Cre mice. Methods Mol Biol 530: 343–363. 10.1007/978-1-59745-471-1_18 [DOI] [PubMed] [Google Scholar]
- Fortin JP, Tan J, Gascoigne KE, Haverty PM, Forrest WF, Costa MR, Martin SE. 2019. Multiple-gene targeting and mismatch tolerance can confound analysis of genome-wide pooled CRISPR screens. Genome Biol 20: 21 10.1186/s13059-019-1621-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Gopaul DN, Van Duyne GD. 1997. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389: 40–46. 10.1038/37925 [DOI] [PubMed] [Google Scholar]
- Gurumurthy CB, Lloyd KCK. 2019. Generating mouse models for biomedical research: technological advances. Dis Model Mech 12: dmm029462 10.1242/dmm.029462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy CB, Takahashi G, Wada K, Miura H, Sato M, Ohtsuka M. 2016. GONAD: a novel CRISPR/Cas9 genome editing method that does not require ex vivo handling of embryos. Curr Protoc Hum Genet 88: 15.18.1–15.18.12. 10.1002/0471142905.hg1508s88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy CB, O'Brien AR, Quadros RM, Adams J, Alcaide P, Ayabe S, Ballard J, Batra SK, Beauchamp MC, Becker KA, et al. 2019a. Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: a multi-center evaluation. Genome Biol 20: 171 10.1186/s13059-019-1776-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy CB, Sato M, Nakamura A, Inui M, Kawano N, Islam MA, Ogiwara S, Takabayashi S, Matsuyama M, Nakagawa S, et al. 2019b. Creation of CRISPR-based germline-genome-engineered mice without ex vivo handling of zygotes by i-GONAD. Nat Protoc 14: 2452–2482. 10.1038/s41596-019-0187-x [DOI] [PubMed] [Google Scholar]
- Guzzardo PM, Rashkova C, Dos Santos RL, Tehrani R, Collin P, Bürckstümmer T. 2017. A small cassette enables conditional gene inactivation by CRISPR/Cas9. Sci Rep 7: 16770 10.1038/s41598-017-16931-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett SK, Bolton VN. 1985. Sequence and regulation of morphological and molecular events during the first cell cycle of mouse embryogenesis. J Embryol Exp Morphol 87: 175–206. [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, Krauss J, Nusslein-Volhard C. 2014. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 141: 4827–4830. 10.1242/dev.115584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaisser F. 2000. Inducible gene expression and gene modification in transgenic mice. J Am Soc Nephrol 11(Suppl. 16): S95–S100. [PubMed] [Google Scholar]
- Jayavaradhan R, Pillis DM, Goodman M, Zhang F, Zhang Y, Andreassen PR, Malik P. 2019. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat Commun 10: 2866 10.1038/s41467-019-10735-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang FG, Doudna JA. 2017. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46: 505–529. 10.1146/annurev-biophys-062215-010822 [DOI] [PubMed] [Google Scholar]
- Jiang W, Marraffini LA. 2015. CRISPR-Cas: new tools for genetic manipulations from bacterial immunity systems. Annu Rev Microbiol 69: 209–228. 10.1146/annurev-micro-091014-104441 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Mashimo T. 2015. Simple genome editing of rodent intact embryos by electroporation. PLoS One 10: e0142755 10.1371/journal.pone.0142755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza DG, Gaspero A, Lorenzo I, Liao L, Zheng P, Wang Y, Deng Y, Cheng C, Zhang C, Seavitt JR, et al. 2018. Comparative analysis of single-stranded DNA donors to generate conditional null mouse alleles. BMC Biol 16: 69 10.1186/s12915-018-0529-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM. 2009. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460: 627–631. 10.1038/nature08209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M. 2001. Conditional control of gene expression in the mouse. Nat Rev Genet 2: 743–755. 10.1038/35093537 [DOI] [PubMed] [Google Scholar]
- Limonta J, Pedraza A, Rodríguez A, Freyre FM, Barral AM, Castro FO, Lleonart R, Gracía CA, Gavilondo JV, de la Fuente J. 1995. Production of active anti-Cd6 mouse-human chimeric antibodies in the milk of transgenic mice. Immunotechnology 1: 107–113. 10.1016/1380-2933(95)00010-0 [DOI] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. 2014. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3: e04766 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling X, Xie B, Gao X, Chang L, Zheng W, Chen H, Huang Y, Tan L, Li M, Liu T. 2020. Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates. Sci Adv 6: eaaz0051 10.1126/sciadv.aaz0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, Ma W. 2019. Methodologies for improving HDR efficiency. Front Genet 9: 691 10.3389/fgene.2018.00691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthardt FW, Donahuc RP. 1973. Pronuclear DNA synthesis in mouse eggs. An autoradiographic study. Exp Cell Res 82: 143–151. 10.1016/0014-4827(73)90256-5 [DOI] [PubMed] [Google Scholar]
- Ma M, Zhuang F, Hu X, Wang B, Wen XZ, Ji JF, Xi JJ. 2017. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res 27: 578–581. 10.1038/cr.2017.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. 1988. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336: 348–352. 10.1038/336348a0 [DOI] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. 2015. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33: 538–542. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, Wagner EF. 2000. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat Genet 24: 184–187. 10.1038/72855 [DOI] [PubMed] [Google Scholar]
- Miura H, Gurumurthy CB, Sato T, Sato M, Ohtsuka M. 2015. CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Sci Rep 5: 12799 10.1038/srep12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura H, Quadros RM, Gurumurthy CB, Ohtsuka M. 2018. Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat Protoc 13: 195–215. 10.1038/nprot.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka Y, Uno Y, Yoshimi K, Kunihiro Y, Yoshimura T, Tanaka T, Ishikubo H, Hiraoka Y, Takemoto N, Tanaka T, et al. 2018. CLICK: one-step generation of conditional knockout mice. BMC Genomics 19: 318 10.1186/s12864-018-4713-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewski AJ, Chen S, Willis BJ, Lloyd KCK, Wood JA, He L. 2018. Efficient mouse genome engineering by CRISPR-EZ technology. Nat Protoc 13: 1253–1274. 10.1038/nprot.2018.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka M, Sato M, Miura H, Takabayashi S, Matsuyama M, Koyano T, Arifin N, Nakamura S, Wada K, Gurumurthy CB. 2018. i-GONAD: a robust method for in situ germline genome engineering using CRISPR nucleases. Genome Biol 19: 25 10.1186/s13059-018-1400-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen SD, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. 2014. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159: 440–455. 10.1016/j.cell.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T, Aida T, Redder R, Richardson GP, Inagaki Y, Sakai D, et al. 2017. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol 18: 92 10.1186/s13059-017-1220-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasys AM, Park S, Ball RE, Alcala AJ, Lauderdale JD, Menke DB. 2019. CRISPR-Cas9 gene editing in lizards through microinjection of unfertilized oocytes. Cell Rep 28: 2288–2292.e3. 10.1016/j.celrep.2019.07.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. 2016. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34: 339–344. 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- Sato M, Miyagasako R, Takabayashi S, Ohtsuka M, Hatada I, Horii T. 2020. Sequential i-GONAD: an improved in vivo technique for CRISPR/Cas9-based genetic manipulations in mice. Cells 9: 546 10.3390/cells9030546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N. 1988. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA 85: 5166–5170. 10.1073/pnas.85.14.5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonig K, Schwenk F, Rajewsky K, Bujard H. 2002. Stringent doxycycline dependent control of CRE recombinase in vivo. Nucleic Acids Res 30: 134e 10.1093/nar/gnf134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M, Wang ZQ, Jochum W, Fetka I, Elliott C, Wagner EF. 2000. Placental vascularisation requires the AP-1 component Fra1. Development 127: 4937–4948. [DOI] [PubMed] [Google Scholar]
- Skarnes WC. 2015. Is mouse embryonic stem cell technology obsolete? Genome Biol 16: 109 10.1186/s13059-015-0673-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342. 10.1038/nature10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow MH. 1981. Satellite cell distribution within the soleus muscle of the adult mouse. Anat Rec 201: 463–469. 10.1002/ar.1092010303 [DOI] [PubMed] [Google Scholar]
- Square T, Romasek M, Jandzik D, Cattell MV, Klymkowsky M, Medeiros DM. 2015. CRISPR/Cas9-mediated mutagenesis in the sea lamprey Petromyzon marinus: a powerful tool for understanding ancestral gene functions in vertebrates. Development 142: 4180–4187. 10.1242/dev.125609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Hamilton D, Hoess R. 1981. Bacteriophage-P1 site-specific recombination. II. Recombination between loxP and the bacterial chromosome. J Mol Biol 150: 487–507. 10.1016/0022-2836(81)90376-4 [DOI] [PubMed] [Google Scholar]
- The Tabula Muris Consortium. 2018. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562: 367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi G, Gurumurthy CB, Wada K, Miura H, Sato M, Ohtsuka M. 2015. GONAD: Genome-editing via Oviductal Nucleic Acids Delivery system: A novel microinjection independent genome engineering method in mice. Sci Rep 5: 11406 10.1038/srep11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M, Py BF, Bosc C, Laubreton D, Moutin MJ, Marvel J, Flamant F, Markossian S. 2018. Electroporation of mice zygotes with dual guide RNA/Cas9 complexes for simple and efficient cloning-free genome editing. Sci Rep 8: 474 10.1038/s41598-017-18826-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H, Maandag ER, Berns A. 1992. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci USA 89: 5128–5132. 10.1073/pnas.89.11.5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem-cells. Cell 51: 503–512. 10.1016/0092-8674(87)90646-5 [DOI] [PubMed] [Google Scholar]
- Wang HY, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HF, La Russa M, Qi LS. 2016a. CRISPR/cas9 in genome editing and beyond. Annu Rev Biochem 85: 227–264. 10.1146/annurev-biochem-060815-014607 [DOI] [PubMed] [Google Scholar]
- Wang W, Kutny PM, Byers SL, Longstaff CJ, DaCosta MJ, Pang C, Zhang Y, Taft RA, Buaas FW, Wang H. 2016b. Delivery of Cas9 protein into mouse zygotes through a series of electroporation dramatically increases the efficiency of model creation. J Genet Genomics 43: 319–327. 10.1016/j.jgg.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. 2014. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32: 670–676. 10.1038/nbt.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang HY, Shivalila CS, Cheng AW, Shi LY, Jaenisch R. 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T. 2016. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun 7: 10431 10.1038/ncomms10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TT, Zhong Y, Wang YG, Zhang T, Lu R, Zhou MY, Lu YY, Yan KN, Chen YJ, Hu ZH, et al. 2019. Generation of hyperlipidemic rabbit models using multiple sgRNAs targeted CRISPR/Cas9 gene editing system. Lipids Health Dis 18: 69 10.1186/s12944-019-1013-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JP, Li XL, Li GH, Chen WQ, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu YW, et al. 2017. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol 18: 35 10.1186/s13059-017-1164-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Xu Y, Yu S, Lu L, Ding M, Cheng J, Song G, Gao X, Yao L, Fan D, et al. 2014. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci Rep 4: 6420 10.1038/srep06420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra M, Li E, Sajjadi F, Subramani S, Jaenisch R. 1989. Germ-line transmission of a disrupted β2-microglobulin gene produced by homologous recombination in embryonic stem cells. Nature 342: 435–438. 10.1038/342435a0 [DOI] [PubMed] [Google Scholar]
- Zu Y, Zhang XS, Ren JF, Dong XH, Zhu Z, Jia L, Zhang QH, Li WM. 2016. Biallelic editing of a lamprey genome using the CRISPR/Cas9 system. Sci Rep-Uk 6: 23496 10.1038/srep23496 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.