Abstract
The molecular process of transcription by RNA Polymerase II is highly conserved among eukaryotes (“classic model”). A distinct way of locating transcription start sites (TSSs) has been identified in a budding yeast Saccharomyces cerevisiae (“scanning model”). Herein, we applied genomic approaches to elucidate the origin of the scanning model and its underlying genetic mechanisms. We first identified TSSs at single-nucleotide resolution for 12 yeast species using the nAnT-iCAGE technique, which significantly improved the annotations of these genomes by providing accurate 5′ boundaries for protein-coding genes. We then inferred the initiation mechanism of each species based on its TSS maps and genome sequences. We discovered that the scanning model likely originated after the split of Yarrowia lipolytica and the other budding yeasts. Species that use the scanning model showed an adenine-rich region immediately upstream of the TSS that might facilitate TSS selection. Both initiation mechanisms share a strong preference for pyrimidine–purine dinucleotides surrounding the TSS. Our results suggest that the purine is required to accurately recruit the first nucleotide, thereby increasing the chances of a messenger RNA of being capped during mRNA maturation, which is critical for efficient translation initiation during protein biosynthesis. Based on our findings, we propose a model for TSS selection in the scanning-model species, as well as a model for the stepwise process responsible for the origin and evolution of the scanning model.
Transcription of protein-coding genes is an essential process in the “central dogma” of molecular biology that describes the conversion of genetic codes from the DNA into functional products. A crucial step of transcriptional regulation occurs at the level of transcription initiation, as it determines not only the number of transcripts produced but also the locations of the transcription start site (TSS). Therefore, transcription initiation has been a focus of many studies of gene regulation (Roeder 1996). Genome-wide studies in eukaryotic organisms revealed that transcription initiation is highly dynamic (The FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group 2005; Carninci et al. 2006; Hoskins et al. 2011; The ENCODE Project Consortium 2012; Lu and Lin 2019). Transcription can be initiated from multiple TSSs in most genes, and alternative usage of TSSs in different cell types or growth conditions is prevalent in mammals, the fruit fly, and yeast (Davuluri et al. 2008; Batut et al. 2013; Lu and Lin 2019). Limited TSS shift was observed in studies based on different yeast strains or growth conditions, suggesting a role of genetic and environmental factors in controlling alternative TSS usage (Börlin et al. 2019; Policastro et al. 2020). Switching between TSSs appears to be associated with differential gene expression (Lu and Lin 2019). Transcript isoforms produced by alternative TSS usage tend to have different translation efficiencies (Cheng et al. 2018). From an evolutionary perspective, changes in TSSs were thought to be associated with the divergence of gene expression patterns and phenotypic traits (Lin and Li 2012).
Most studies of the RNA Polymerase II (Pol II) transcription machinery have focused on promoters containing a TATA box (Patikoglou et al. 1999), which was the first identified core promoter element (Smale and Kadonaga 2003). The process of transcription initiation from TATA-containing promoters is highly conserved from archaea to eukaryotes. The first step in transcription initiation is the binding of TATA binding protein (TBP) to the TATA box. Other general transcription factors (GTFs) bind to the TBP–TATA complex in a defined order and recruit Pol II to form a preinitiation complex (PIC) that allows Pol II to reach a TSS directly (Bernard et al. 2010; Li et al. 2015; Blombach et al. 2016). Therefore, most TSSs locate ∼30 bp downstream from the TATA box, in what we refer to as the “classic model” herein. The model budding yeast species Saccharomyces cerevisiae appears to use a distinct mechanism of transcription initiation (Choi et al. 2002; Hahn and Young 2011). Specifically, the PIC in S. cerevisiae scans for favorable TSSs and initiates transcription mainly 40–120 bp downstream from the TATA box, designated here as the “scanning model” (Giardina and Lis 1993; Kuehner and Brow 2006; Fishburn and Hahn 2012). A recent study revealed that the scanning model is used at all promoters in S. cerevisiae, suggesting that it serves as a global transcriptional initiation mechanism (Qiu et al. 2020). In contrast, Schizosaccharomyces pombe, a fission yeast that is distantly related to S. cerevisiae, follows an initiation pattern similar to that of classic-model species in TATA-containing promoters (Choi et al. 2002). Thus, the most parsimonious explanation posits that the scanning model may have originated during the evolution of S. cerevisiae after it diverged from S. pombe more than 500 million years ago (MYA) (Rhind et al. 2011). However, more accurate timing of the origin of the scanning model, as well as its underlying genetic basis, has yet to be determined.
Studying the evolution of transcription initiation will provide a better understanding of the molecular mechanisms underlying the identification of TSSs by the PIC. For instance, in both classic-model and scanning-model species, transcription is mostly initiated from a purine (the +1 site) on the coding strand, with a pyrimidine immediately upstream of it (the −1 site), which forms a pyrimidine–purine (PyPu) dinucleotide (The FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group 2005; Hoskins et al. 2011). It has been shown that the −1 pyrimidine facilitates the stacking of the first nucleotide by Pol II (Zhang et al. 2014). However, it remains unclear why a purine serves as the first transcribed nucleotide. In addition, most TSSs show an adenine nucleotide 8 bp upstream of their position (abbreviated as −8A hereafter) in S. cerevisiae (Zhang and Dietrich 2005; Lu and Lin 2019). A structural study showed that the B-reader helix of TFIIB recognizes the −8A and that −8A is critical for TSS selection (Kostrewa et al. 2009). Whether the preference for −8A exists in other scanning-model species is not known. A better understanding of the sequence context required for the identification of favorable TSSs in scanning-model species should provide insights into the genetic mechanisms of transcription initiation.
Transcription within a core promoter is commonly initiated from a cluster of nearby TSSs, instead of a single TSS (The FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group 2005). The width of a TSS cluster (TC) and the distribution of transcription activities among its TSSs may vary substantially from one core promoter to another, forming different shapes of distribution of transcription signals, which are called promoter shapes (Carninci et al. 2006). Core promoters are generally divided into “sharp” and “broad” classes based on their promoter shapes (Carninci et al. 2006; Hoskins et al. 2011). Transcription initiation at sharp core promoters mainly occurs from a single predominant TSS, whereas initiation is more dispersed at broad core promoters. In mammals, sharp core promoters are often associated with genes showing tissue-specific expression, whereas broad core promoters are enriched in ubiquitously expressed genes, suggesting that the promoter shape reflects the different regulatory needs of a gene (Carninci et al. 2006). It was found that sharp core promoters are more likely to contain a TATA box (Carninci et al. 2006; Hoskins et al. 2011), indicating an influence of the presence of TATA box on promoter shape. By examining the TSS maps for 81 lines of Drosophila melanogaster, Schor et al. (2017) identified thousands of genetic variants that may influence transcription level and core promoter shape. However, the major genetic determinants behind promoter shape are not entirely understood.
In this study, we conducted comparative analyses of the TSS maps and genomic sequences for 12 yeast species. We have pinpointed the origin of the scanning model, inferred key genetic innovations associated with its evolution, identified the sequence context required for transcription initiation, and hypothesized their functional consequences on transcription initiation and promoter shape. These findings contributed to our understanding of the evolutionary divergence of transcription initiation mechanisms and the functional roles of sequencing elements in this key process of the central dogma of molecular biology.
Results
Evolutionary dynamics of transcription initiation landscapes in yeasts
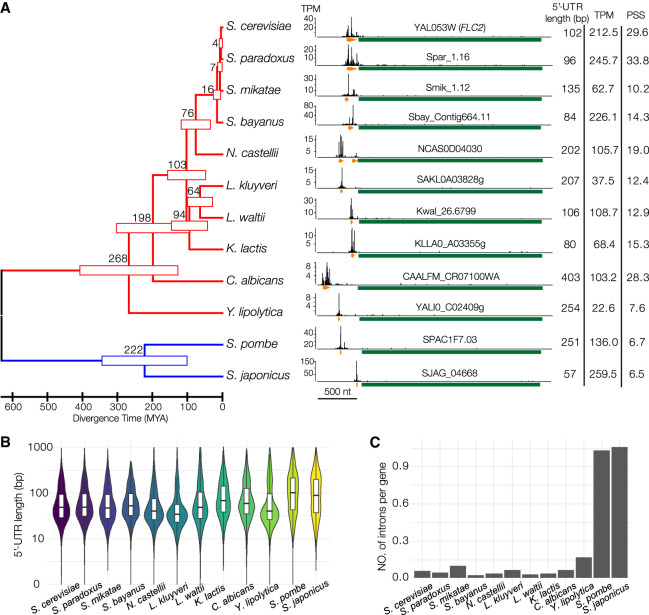
We generated high-resolution TSS maps for 10 budding yeast species and two fission yeast species, including S. cerevisiae, S. pombe, and other important species, with estimated divergence times ranging from four million years to more than 500 million years (Fig. 1A; Supplemental Table S1). We obtained these TSS maps using the no-amplification nontagging cap analysis of gene expression (nAnT-iCAGE) technique (Murata et al. 2014). A total number of 838 million CAGE tags from the 12 yeast species were produced, providing an ultrahigh sequencing depth (Supplemental Table S2). We applied the Poisson model to remove candidate TSSs that were likely because of technical artifacts, or stochastic transcription from non–bona fide core promoters (see Methods). On average, each species used 286,433 TSSs when grown in rich medium (Supplemental Table S3), supporting the pervasive nature of transcription initiation in yeasts, given their small genome size (∼12 million bp) and gene numbers (approximately 5000−6000).
Figure 1.
Generation of TSS maps at single-nucleotide resolution for 12 yeast species. (A) An example of TSS maps of orthologous FLC2 genes from 12 yeast species. We inferred the phylogenetic tree of the 12 yeast species based on RPB2 protein sequences, the largest subunit of the Pol II complex, using the maximum likelihood method. The full species names are provided in Supplemental Table S1. The estimated divergence times and 95% confidence intervals for all branching points in the tree are provided as numbers and horizontal bars, respectively. The tree was drawn to scale. In the TSS map of each species, the top track illustrates the distributions of TSS signals. The second track shows the locations and boundaries of core promoters (orange arrow) and the locations of gene coding regions (green bar). (B) Violin plot showing the distribution of 5′-UTR lengths in each species. (C) Number of introns per gene in each of the 12 yeast genomes.
We developed a peak-based clustering method, called “Peakclu,” to identify TCs, representing core promoters (Supplemental Fig. S1). We assigned TCs to Pol II transcribed genes as their core promoters based on their position proximity (see Methods). We identified core promoters for 83.7% protein-coding genes for these species (ranging from 4571 genes in Schistosomiasis japonicus to 5348 genes in S. cerevisiae). We defined the representative TSS of a protein-coding gene as the TSS with the highest CAGE signal within the promoter region. Our core promoter and TSS data improve the annotations of these genomes by providing 5′ boundaries for most genes at single-nucleotide resolution. We provided the updated genomes annotation files for the 12 yeast species in general feature format (GFF) as Supplemental Datasets S1−S12. Only TCs with tags per million (TPM) greater than one were considered as qualified core promoters for subsequent analyses (Supplemental Dataset S13).
To better characterize the evolutionary patterns of core promoters, we delineated protein-coding genes of the 12 yeast species into 6614 orthologous groups using OrthoDB (Supplemental Dataset S14; Kriventseva et al. 2019). We observed that core promoters between orthologous genes tended to have distinct features, including transcription activities, length of 5′ UTR, and core promoter shape, as illustrated by the orthologous group of FLC2 as an example (Fig. 1A). These features are most similar between the closely related species, such as S. cerevisiae and Saccharomyces paradoxus, which have diverged ∼4 MYA. In contrast, these features showed reduced similarity with increasing divergence times. For example, we detected larger differences between S. cerevisiae and its second-most related species Saccharomyces mikatae, suggesting that these features may be related to genetic factors. However, these differences do not show a simple linear correlation with their divergence times on a broader scale, probably because changes in these features were not directional, or they might have diverged at a much higher rate than their respective genomic sequences.
One of the most significant differences related to TSSs between budding yeasts and fission yeasts is the 5′-UTR length. The budding yeasts have a shorter median length of 5′ UTR than that of fission yeasts (Fig. 1B; Supplemental Dataset S15). The median 5′-UTR length in fission yeasts is more similar to that of higher eukaryotes, such as 106 bp in tomato and 111 bp in cow (Leppek et al. 2018). Exonization from introns in 5′ UTR regions has been observed (Hooks et al. 2014). Elongation of 5′ UTR by exonization is more likely to occur in genomes with a higher intron density. Because budding yeasts show a significantly lower intron density than fission yeasts (Fig. 1C), we hypothesize that the massive loss of introns in budding yeast genomes largely eliminates the possibility of 5′ UTR elongation through the exonization process.
In each examined species, 5′-UTR lengths vary greatly among individual genes. We measured the coefficient of variation (CV) of 5′-UTR lengths among orthologous genes for each KEGG pathway to quantify their evolutionary divergence. We determined that KEGG groups with the most divergent 5′-UTR length are generally related to metabolism pathways, such as ether lipid metabolism and riboflavin metabolism (Supplemental Fig. S2A). The median lengths of 5′ UTR among genes in the riboflavin metabolism pathway range from 21–294 bp across our 12 species (Supplemental Fig. S2B). In contrast, the KEGG groups with the most conserved 5′-UTR length are enriched in essential cellular function pathways, such as ribosome and RNA transport (median 5′-UTR lengths ranging from 37–63 bp) (Supplemental Fig. S2C). These results support the hypothesis that 5′-UTR length, which is primarily determined by the location of TSSs, relates to gene functions and their expression profiles (Lin and Li 2012), although the underlying mechanism remains to be investigated further.
The scanning model emerged during the evolution of budding yeasts
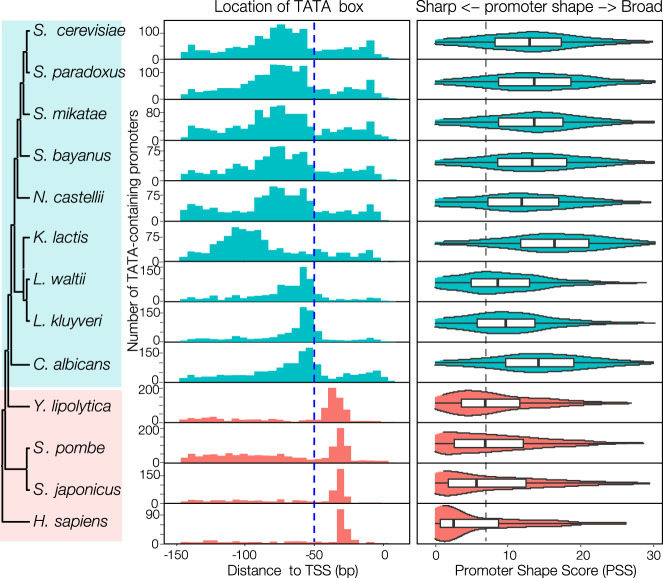
A distinct feature distinguishing the classic model from the scanning model is the distance between the TATA box and TSS. Therefore, we used this feature to infer the transcription initiation mechanism of a given species. We searched for TATA box motifs using the consensus sequence TATAWAWR (Basehoar et al. 2004; Rhee and Pugh 2012) in promoter regions in each species examined. We observed a binary pattern of TATA box positioning among the 12 species (Fig. 2). In one group, including a known classic-model species S. pombe, as well as the other fission yeast examined S. japonicus and a budding yeast Yarrowia lipolytica, TATA box motifs are well positioned ∼30 bp upstream of TSS, similar to that of human, supporting the notion that they all use the “classic model.” In the other group, which includes the known scanning-model species S. cerevisiae and all other budding yeasts except Y. lipolytica, TATA boxes are mainly distributed over a broad range from 50–120 bp upstream of the TSS, suggesting that these species use the scanning model for transcription initiation (Fig. 2). Y. lipolytica, which diverged ∼200 MYA, is the earliest branching species among budding yeasts examined. These results support the hypothesis that the origin of the scanning model occurred after the divergence of Y. lipolytica during the evolution of budding yeasts.
Figure 2.
Inference of transcription initiation mechanisms using TATA-containing promoters. The left panel displays phylogenetic relationships of the 12 yeast species with human as an outgroup. The middle panel shows the distributions of distances between the TATA box (TATAWAWR) and TSS in each species. The numbers of TATA-containing promoters and genes in each species are provided in Supplemental Table S4. The blue dashed line indicates the location of the −50 position. The names of the scanning-model species are shaded in cyan, and classic-model species are shaded in salmon. The right panel shows the distribution of promoter shape score (PSS) of TATA-containing promoters in each species. The gray dashed line indicates the median PSS value of Y. lipolytica, S. pombe, and S. japonicus.
In classic-model species, transcription from TATA-containing promoters tends to be initiated from a narrow range of TSSs, resulting in core promoters with a sharper shape than those from TATA-less promoters (Carninci et al. 2006). We calculated the promoter shape score (PSS) for both classes of core promoters in all examined species (see Methods). The sharpest promoters have a PSS of zero, whereas the PSS increases as core promoters become broader. This analysis revealed two significant findings. First, PSS values for TATA-containing promoters in the three classic-model species are significantly lower (sharper) than those from scanning-model species (Fig. 2). Such a difference is probably because of different mechanisms of locating TSSs between the two classes of species: Indeed, in scanning-model species, the PIC scans DNA sequences downstream from a TATA box to select favorable TSSs instead of starting from a fixed distance, leading to a broader distribution of TSSs. These results also corroborate our robust inference of transcription initiation mechanisms based on distributions of TATA box locations. Second, the PSS values of TATA box–containing promoters are significantly lower than those of TATA-less promoters in classic-model species, but we saw no such differences in scanning-model species (Supplemental Fig. S3). This observation suggests that the scanning mechanism is used for promoters with and without a TATA box in scanning-model species, which is consistent with the results of a separate study (Qiu et al. 2020). Therefore, our findings based on core promoter shape further support that the scanning model originated after the split of Y. lipolytica during the evolution of budding yeasts.
Because the eighth position of the TATA box consensus sequence TATAWAWR minimally affects its interaction with TBP (Patikoglou et al. 1999), we repeated our analyses by searching for TATA box motifs using only the sequence TATAWAW. Although we identified ∼30% more TATA box motifs from promoter regions using this shorter consensus sequence, the distribution patterns of both TATA-TSS distances and promoter shapes remain unchanged in all species (Supplemental Table S4; Supplemental Fig. S4), highlighting the robustness of our conclusions. As we obtained the TSS maps for these species from cells grown in rich medium and given that TSS activities can significantly change under different growth conditions (Lu and Lin 2019), we sought to determine whether physiological regulation might influence transcription initiation. Therefore, we generated a set of distributions for TATA-TSS distances based on published TSS maps derived from nine distinct growth conditions in S. cerevisiae (Lu and Lin 2019) and from five growth conditions in S. pombe (Thodberg et al. 2019). We observed highly similar distributions across samples within each species (Supplemental Fig. S5). These observations support that the molecular machinery of transcription initiation is independent of physiological regulation or growth conditions in both scanning-model and classic-model species.
Purine as the first recruited nucleotide is critical for accurate transcription initiation and efficient 5′ capping
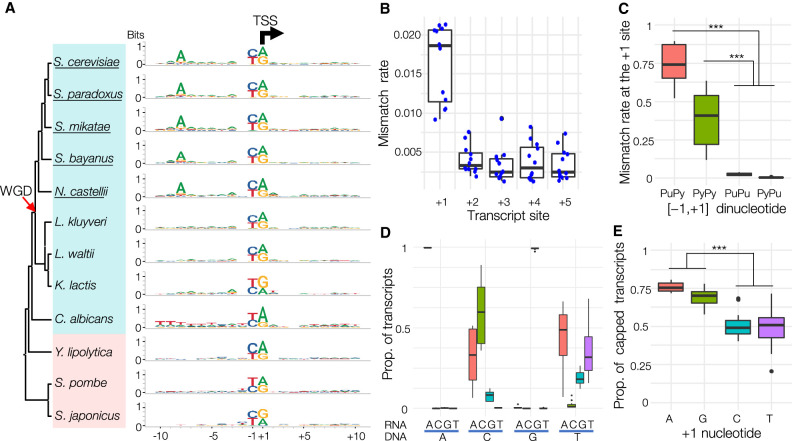
We retrieved sequences surrounding the dominant TSS (±10 bp) of all core promoters from all yeast species (Supplemental Dataset S13). The consensus sequence showed a strong preference of PyPu dinucleotide at TSSs in all species examined (Fig. 3A; Supplemental Fig. S6). Unlike the pyrimidine at the −1 site, the functional role of purine at the +1 site remains elusive. In contrast to DNA replication, transcription initiation does not require an RNA primer, and Pol II adds the first nucleotide to the template strand without the formation of a phosphodiester bond. Owing to the difference between transcription initiation and extension, we speculated that the mismatch rate at the +1 site might be higher than at other sites. The strong preference for a single type of nucleotide for transcription initiation may theoretically reduce the probability of transcription errors.
Figure 3.
Functional roles of PyPu dinucleotide in transcription initiation. (A) Consensus sequences of core promoters in the 12 yeast species. The sequence logo was generated using sequences from −10 to +10 bp surrounding the dominant TSS of all core promoters in each species (Supplemental Dataset S13). The black arrow indicates the TSS position (the +1 site) and the transcription direction. The red arrow indicates the occurrence of WGD, and the names of WGD species are underlined. (B) Boxplot of the distributions of mismatched rates at the first five sites of transcripts in the 12 species. Each blue dot represents the mismatch rate at each site in a species. (C) Mismatch rates in transcripts initiated from different −1/+1 dinucleotides: PuPy, PyPy, PuPu, and PyPu. (*) P < 0.01; (**) P < 0.001; (***) P = 0. (D) Proportion of each type of nucleotides added by Pol II at the +1 site of RNA transcripts. On the x-axis, the type of recruited nucleotides (RNA) is shown above the blue lines, and the nucleotides on the sense strand (DNA) are shown under the blue lines. (E) Boxplot illustrates proportions of transcripts with a detected G-cap at the 5′ end among transcripts with different starting nucleotides in the 12 yeast species.
The raw CAGE sequencing reads from the 12 yeast species allowed us to test our hypotheses by comparing transcript sequences and their genomic templates (see Methods). As predicted, we discovered that the mismatch rate at the +1 site was about six times higher than at the next four sites (Fig. 3B). We then examined the consequences of different dinucleotides at the [−1,+1] sites on initiation fidelity. Transcripts initiated from PyPu (0.003) and PuPu (0.026) dinucleotides showed drastically lower proportions of mismatched nucleotides than from PyPy (0.405) or PuPy (0.740) dinucleotides. This result showed that a purine at the +1 site is critical for recruiting the correct nucleoside triphosphate to the first site, whereas the nucleotide at the −1 site has a minimal effect (Fig. 3C; Supplemental Fig. S7A). We noticed that a purine, particularly adenine, was frequently incorporated by Pol II among those mismatches initiated from a pyrimidine site (Fig. 3D; Supplemental Fig. S7B). For instance, if the +1 site on the coding strand was a thymine, Pol II then tended to recruit an adenine rather than a thymine, suggesting that purines are strongly preferred by Pol II for the +1 site, regardless of the template nucleotide.
The next question raised by our results was why Pol II would prefer purines, especially adenines, as the initiation nucleotide. The first nucleotide of the primary mRNA receives a cap structure (e.g., N7-methylated guanosine [m7G]) as a post-transcriptional modification, which then allows for cap-dependent initiation of protein synthesis and prevention of exonuclease cleavage (Both et al. 1975; Muthukrishnan et al. 1975). To determine the effect of different nucleotides at the 5′ end of mRNAs on 5′ capping, we examined the raw CAGE sequencing reads to calculate the proportion of m7G caps for transcripts with different types of nucleotides at the 5′ end (see Methods). Our analysis revealed that transcripts with a purine at the 5′ end had much higher rates of being capped by m7G (73.2%) compared with those with a pyrimidine (49.2%) (Fig. 3E; Supplemental Fig. S7C).
Some uncapped transcripts could be generated by premature reverse transcription (RT) stops or 5′−3′ decay of mRNA, generating “false TSSs.” If uncapped transcripts have more pyrimidines than purines at their first site, it could introduce bias in quantifying 5′ capping rates. To minimize the impacts of these truncated transcripts, we calculated 5′ capping rates using TSSs with more stringent support. First, we examined 5′ capping rates for TSSs identified by different techniques, such as transcript leader sequencing (TL-seq), which replaces m7G with a linker by 5′ ligation (Arribere and Gilbert 2013). A recent study (Spealman et al. 2018) used TL-seq to generate TSS maps for four budding yeast species, two of which (S. cerevisiae and S. paradoxus) we included in this study. As TL-seq does not sequence the m7G cap, we could not infer capping rates based on TL-seq reads. Instead, we used our CAGE reads to calculate capping rates for TSSs identified by both techniques, which were considered as “high-confidence” TSSs. To further filter those possibly originating from technical artifacts, we selected TSSs with TPM greater than one in TL-seq data for subsequent analyses (46,514 and 33,506 TSSs from S. cerevisiae and S. paradoxus, respectively; see Methods). We obtained consistent results from studies, as the capping rates of TSSs with adenine and guanine were significantly higher than those with cytosine and thymine in both species (ANOVA, P < 0.01) (Supplemental Fig. S8).
Because transcripts created by RT stop or mRNA decay lack a 5′ cap, if a TSS is not supported by reads with a G-cap (noncapped site), we can consider it as a potential “false TSS.” We found that only ∼6% of reads belong to noncapped sites, suggesting a limited impact of RT stop or mRNA decay on TSS calling in promoter regions. By excluding those reads of noncapped sites, our calculation of 5′ capping rates based on TSSs that are supported by at least one G-capped transcript and located within assigned core promoters yielded a similar result (Supplemental Fig. S8C). Therefore, quantifications of 5′ capping rates based on three different sets of TSSs reached the same conclusion. Our results suggested that the strong preference for a purine at the +1 site of transcripts provides their best chance to be capped by m7G, thereby increasing the probability of successful protein biosynthesis and reducing mRNA cleavage by exonucleases.
The gain of an adenine-rich region immediately upstream of TSS during the evolution of scanning-model yeasts may have facilitated TSS selection
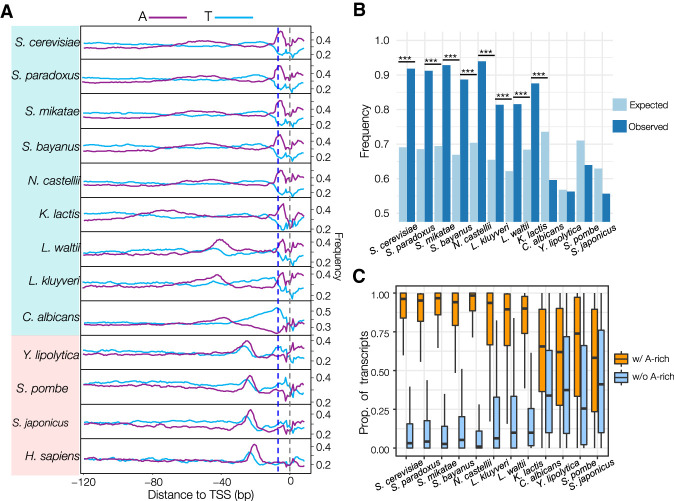
An adenine is present at 8 bp upstream of most TSSs (−8A) in S. cerevisiae (Zhang and Dietrich 2005; Lu and Lin 2019). We confirmed the predominance of −8A in other budding yeast species that have experienced an ancestral whole-genome duplication (WGD) (Fig. 3A; Supplemental Fig. S9). By conducting a sliding-window analysis of nucleotide frequency, we determined the existence of an adenine-rich (A-rich) region immediately upstream of the TSS, with a peak close to the −8 position, in all scanning-model yeasts except Candida albicans (Fig. 4A; Supplemental Fig. S9). We also detected a similar A-rich region at the same location in S. cerevisiae in all growth conditions examined based on published TSS maps (Lu and Lin 2019), suggesting its independence from physiological regulations (Supplemental Fig. S10). As an opportunistic pathogen of humans, C. albicans is the earliest diverging lineage among the scanning-model species. All classic-model species lacked the TSS-proximity A-rich region. Instead, they showed an AT-rich region ∼30 bp upstream of the TSS, corresponding to the location of the TATA box (Fig. 4A; Supplemental Fig. S9). Based on the phylogenetic distribution of the A-rich region, the most parsimonious explanation suggests that the enrichment of the TSS-proximity A-rich region originated after the divergence of C. albicans during the evolution of budding yeasts. In addition, the common ancestor of WGD yeasts gained a strong preference for adenine at a specific location (−8A) within the A-rich region. The gain of the −8A in WGD species suggests that these species may show a more stringent requirement for the distance between adenine and TSS for transcription initiation.
Figure 4.
Presence of an adenine-rich (A-rich) region in the scanning-model species and its functions. (A) Sliding-window analysis of A/T frequencies in the region from −120 to +10 bp surrounding the dominant TSS of all core promoters in the 12 yeast species and human (Supplemental Dataset S13). The window size is 5 bp with a step size of 1 bp. Blue dashed line refers to the −8 site. The gray dashed line refers to the TSS position (the +1 site). (B) Frequency of expected and observed A-rich (at least two adenines) region in the window from −9 to −3 bp. (***) P = 0, chi-square test. (C) Proportions of transcripts initiated from TSSs with and without an A-rich region. This figure was generated based on all core promoters in the 12 species.
If the A-rich region indeed plays a key role in transcription initiation in scanning-model species, we hypothesized that it should be overrepresented immediately upstream of TSSs in scanning-model species but not in classic-model species. We first aimed to enumerate the number of adenines in the 7-bp window (from −9 to −3) between the two types of species. We then expressed the results as frequencies for minimum adenine numbers from one to seven between the two types of species (Supplemental Fig. S11). We determined that promoters with at least two adenines showed significantly higher frequencies in scanning-model species compared with classic-model species. Therefore, we defined A-rich as the presence of two or more adenines within the 7-bp window. Based on this definition, we observed the presence of the A-rich region in 91.80% of TSSs in S. cerevisiae, compared with an expected frequency of 69.1% based on nucleotide frequencies, supporting the claim that A-rich regions are overrepresented in the 7-bp window (P = 1.8 × 10−227, chi-square test) (Fig. 4B). We detected such enrichment in all scanning-model species, excluding C. albicans. In contrast, the A-rich region was not enriched, or even underrepresented, in all classic-model species (Fig. 4B). Because one PyPu dinucleotide should be detected in every 5-bp window by chance, this observation also explains why TSSs locate a few base pairs downstream from A-rich regions in scanning-model species.
Transcription initiation from a core promoter may occur from an array of nearby TSSs, and some TSSs may lack an upstream A-rich region. If the A-rich region is required for efficient transcription initiation in the scanning-model species, we might expect that most transcripts within a core promoter would be initiated from TSSs associated with an A-rich region. As shown in Figure 4C, we discovered that the proportion of transcripts initiated from A-rich-associated TSSs was much higher than that from A-rich-less TSSs in all scanning-model species except C. albicans, supporting our hypothesis. In C. albicans, we observed a thymine-rich region upstream of TSSs (Supplemental Fig. S9), suggesting that the molecular mechanisms of transcription initiation in C. albicans might differ from other scanning-model species.
Genetic basis underlying the evolutionary conservation and divergence of core promoters
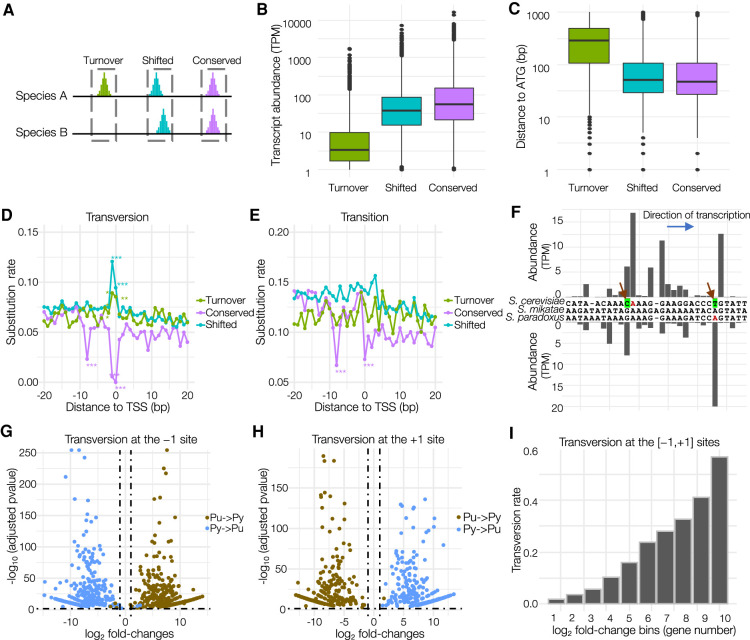
Our results suggest the critical roles of PyPu and the A-rich region (or −8A in WGD species) in transcription initiation. We then examined the evolutionary patterns associated with these sequence elements and how they may influence the divergence of TSSs and core promoters. We focused on the three closely related species, S. cerevisiae, S. paradoxus, and S. mikatae, which allowed us to align their entire genomes for the accurate identification of orthologous core promoters. By using S. mikatae as an outgroup because it diverged from the other two species earlier, we divided orthologous core promoters into three groups (see Methods) (Fig. 5A; Supplemental Dataset S16). The Conserved group included orthologous core promoters present in both S. cerevisiae and S. paradoxus, with the same positions for the dominant TSSs. We defined the Shifted group as those core promoters in which the dominant TSS changed in one or both species. We classified lost or newly gained core promoters in S. cerevisiae or S. paradoxus as the Turnover group.
Figure 5.
Genetic basis underlying the evolutionary divergence of core promoters and TSSs in budding yeasts. (A) A schematic diagram of three types of core promoters based on their evolutionary patterns. (B) Different transcriptional activities among the three types of core promoters. (C) The Turnover core promoters tended to locate more upstream from the ATG codon than the other groups. (D) The rate of transversion substitution at each site in the region surrounding the dominant TSS (from −20 to +20 bp), which were calculated between orthologous core promoters in S. cerevisiae and S. paradoxus. The sites with significantly higher or lower substitution rates are indicated by asterisks (chi-square test). (*) P < 0.01; (**) P < 0.001; (***) P = 0. (E) The rate of transition substitution at each site in the region centered around the TSS (from −20 to +20 bp). (F) An example of Shifted core promoters (upstream of YDL182W) and their genomic sequences in three closely related budding yeasts. New mutations in S. cerevisiae and S. paradoxus are indicated by arrows. (G) Volcano plot illustrating that mutations from pyrimidine to purine (Py → Pu) and from purine to pyrimidine (Pu → Py) at the −1 site have the opposite impacts on transcriptional activities of TSSs. The horizontal dashed line refers to the adjusted P-value of 0.05, and the vertical dashed lines indicate ≥1 or ≤−1 log2 fold-changes. (H) Volcano plot illustrates that mutations from pyrimidine to purine (Py → Pu) and from purine to pyrimidine (Pu → Py) at the +1 site have the opposite impacts on transcriptional activities of TSSs. (I) The TSSs with larger fold-changes in transcriptional activities are more likely to be associated with transversion mutation at the [−1,+1] sites.
Overall, we discovered the prevalence of core-promoter turnover. S. cerevisiae gained 670 and lost 229 core promoters since its divergence from S. paradoxus, accounting for 10.3% and 3.5% of its core promoters, respectively (Supplemental Fig. S12A). We detected similar patterns in S. paradoxus. When compared with Conserved and Shifted core promoters, Turnover core promoters tended to show lower transcriptional activity (Fig. 5B) and usually were located at further upstream of the translation start codons (Fig. 5C).
We examined the genomic sequences from −20 to +20 bp surrounding the dominant TSSs for each group of core promoters to infer their associated genetic changes. We observed distinct patterns of genetic divergence at the −8, −1, and +1 sites between the three types of core promoters. In Conserved core promoters, the rates of nucleotide substitutions, particularly transversions, were nearly depleted at the −8, −1, and +1 sites (Fig. 5D–E), suggesting that the nucleotide type at these positions is critical for maintaining core promoter activities. In contrast, we observed elevated transversion rates at the −1, +1 sites in the Shifted and Turnover groups (P < 0.001, chi-square test) (Fig. 5D). For example, the core promoter of YDL182W locus in S. cerevisiae changed the position of its dominant TSS since its divergence from S. paradoxus (Fig. 5F). S. cerevisiae lost the ancestral dominant TSS owing to a transversion mutation that replaced adenine with thymine at the +1 site, converting PyPu to PyPy. Concomitantly, S. cerevisiae gained a new dominant TSS 13 bp upstream of the ancestral TSS by replacing guanine to cytosine, generating a new PyPu dinucleotide (Fig. 5F). Moreover, active core promoters in the Turnover group had a significantly higher frequency of PyPu and −8A than their inactive counterparts (silent core promoters) (Supplemental Fig. S12B). However, both groups showed similar frequencies of TATA box, supporting that nucleotide turnovers at the −8, −1, and +1 sites played an important role in the evolutionary divergence of core promoter activities.
We then evaluated the consequence of transversions at the −1 and +1 sites on transcription initiation activity. When the nucleotide at the −1 site changed from the preferred pyrimidine to purine, most core promoters experienced significantly reduced transcriptional activity. We observed the opposite pattern in core promoters where pyrimidine replaced purine (Fig. 5G). In contrast, a change from a purine to pyrimidine at the +1 site significantly reduced promoter transcriptional activities because purine is the preferred nucleotide, and again, we noted the opposite pattern when purine replace pyrimidine (Fig. 5H). These results further support the importance of PyPu dinucleotides in transcription initiation. When we sorted TSSs based on their fold-changes in transcription activity between S. cerevisiae and S. paradoxus, we discovered that the proportion of TSSs with transversion mutations at the [−1,+1] sites increased as the fold-change in transcriptional activity increased. Within the group of TSSs with the largest fold-changes, 56.7% showed an association with transversion mutations at the [−1,+1] sites (Fig. 5I), suggesting that the TSSs with the most significant evolutionary divergence in transcriptional activities are more likely owing to transversion mutations at the [−1,+1] positions.
Other common motifs in yeast core promoters
A GA element (GAAAAA) was identified as a conserved promoter element in most TATA-less promoters in S. cerevisiae (Seizl et al. 2011). Notably, the GA element was enriched in promoter regions in all scanning-model yeast species, at a position similar to that of the TATA box in TATA-containing promoters (Supplemental Fig. S13). These findings suggest that the GA element might function as binding sites for GTF in scanning-model species, supporting the presence of two distinct Pol II transcription initiation machinery in yeasts.
Our TSS maps allowed us to predict other putative core promoter motifs in yeasts using de novo motif discovery methods. We showed that, besides the TATA box, eight motifs were significantly enriched in promoter regions in at least three yeast species (Supplemental Fig. S14A). These shared motifs generally mapped to similar locations relative to the TSS within each type of transcription initiation mechanism (Supplemental Fig. S14B), further supporting the presence of two models of Pol II initiation. For example, motifs that match to the binding sites of REB1p, ABF1p, and TOD6p were detected in most scanning-model species, and they located at similar positions as the TATA box. These motifs, as well as the GA element, manifested very little co-occurrence with the TATA box (Supplemental Fig. S15), indicating that these motifs might play an important role in transcription initiation from TATA-less promoters.
Discussion
In this study, we generated quantitative TSS maps for 10 budding yeasts and two fission yeasts, representing the most comprehensive TSS atlas in yeasts to date. These TSS maps improve genome annotations for these species by providing 5′ boundaries for most protein-coding genes at single-nucleotide resolution. Most importantly, our study contributed to the field of transcription regulation by providing a better understanding of the functions of several key sites surrounding the TSS and by unraveling the origin and evolutionary process that led to the scanning-model of transcription initiation.
Functional roles of the PyPu and A-rich region in transcription initiation
Our study improves our understanding of the role of PyPu during transcription initiation. The strong preference for purine as the first base of transcripts likely stems from the intrinsic preference of Pol II and from positive influences subsequent post-transcriptional modification and protein biosynthesis. We determined that transversions at the [−1,+1] sites, which disrupt PyPu dinucleotides, result in remarkable changes in TSS activities and TSS shifts, supporting their importance in transcription initiation. Furthermore, these results uncover a key genetic mechanism underlying the evolutionary divergence of TSSs and core promoters. We discovered that disruption of the PyPu sites was sufficient to eliminate its transcriptional initiation activities (Fig. 5D). However, other factors likely play a more important role in gaining a new TSS. In most cases, the birth of a new TSS in a promoter region does not require mutations to obtain a PyPu dinucleotide owing to its prevalence (1 PyPu in every 5 bp). A study on the human genome showed that most new TSSs emerged from transposable elements owing to retrotransposon activities (Li et al. 2018). However, yeasts are known for their scarcity in active transposable elements (Bleykasten-Grosshans and Neuvéglise 2011). We speculate that evolutionary innovations in trans- or cis-regulatory factors probably play a more important role in the birth of new TSS in yeasts.
Another sequence signature of the TSS in scanning-model species is the presence of an A-rich region in the −9 to −3 window. Particularly, WGD species displayed a strong preference for adenine at the −8 position (−8A). Structural studies indicated that the −8A in S. cerevisiae is recognized by the B-reader helix of TFIIB, which is required for TSS selection (Kostrewa et al. 2009; Sainsbury et al. 2013). It is reasonable to postulate that adenines in the A-rich region might serve as binding sites for a PIC component, probably TFIIB, for the scanning model of transcription initiation. The interaction between TFIIB and the A-rich region might temporarily pause the scanning process and direct Pol II to initiate transcription from its downstream PyPu. Therefore, the A-rich region in scanning-model species might have a similar role as the TATA box in classic-model species, serving as an anchor point for PIC to initiate transcription. In addition, because one PyPu is expected to be present by chance in each 5-bp window, a requirement for the presence of an A-rich region immediately upstream of PyPu largely eliminates initiation from many other PyPu sites, reducing the production of undesired transcript isoforms. This hypothesis is consistent with a previous study in which mutation at the −8A in S. cerevisiae led to almost complete loss of its corresponding transcription activity (Kostrewa et al. 2009). However, how PIC components interact with the A-rich region requires further interrogations.
A proposed model for transcription initiation in the scanning-model species
Based on what we learned about the functional roles of PyPu and the A-rich region, we propose a model to describe how transcription is initiated in scanning-model species (Fig. 6A). In brief, the assembly of PIC on the TATA box occurs similarly in both classic-model and scanning-model species. In the classic model, transcription initiation would occur if PyPu is present near the location of the Pol II catalytic center. In the scanning model, the PIC scans downstream from the TATA box for the combination of an A-rich region and PyPu in a 10-bp window (from −9 to +1 bp). The A-rich region might serve as an anchor point for PIC. If a PyPu is available within a few base pairs downstream from the A-rich region, transcription can be initiated by Pol II. Otherwise, the PIC continues to scan the promoter sequence until it reaches a favorable sequence combination.
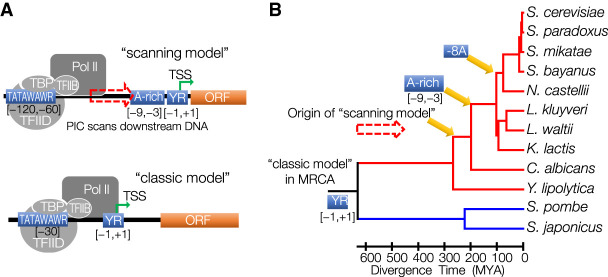
Figure 6.
The origin and evolution of the scanning model of transcription initiation mechanism. (A) Schematic illustration of two distinct mechanisms of transcription initiation: classic model and scanning model. Both initiation mechanisms share a strong preference for PyPu dinucleotides at the [−1,+1] sites. In the classic model, transcription initiates at ∼30 bp downstream from the TATA box, where PIC is assembled. In contrast, the PIC in scanning-model species scans DNA downstream from the TATA box for TSSs with favorable genomic context, which includes a PyPu and an A-rich region immediately upstream of it. (B) Schematic illustration of the origin of the scanning model and associated genetic innovations during the evolution of budding yeasts. The most recent common ancestor (MRCA) of the budding yeasts and fission yeasts was the classic-model species. The scanning model originated after the split between Y. lipolytica from the other budding yeasts. An A-rich region in the region from −9 to −3 bp upstream of the TSS appeared during the evolution of scanning-model species after the divergence of C. albicans. The gaining of specific −8A preference occurred in ancestral WGD species.
It is worth mentioning that the presence of a favorable sequence combination appears to be a necessary, but not sufficient, for efficient transcription initiation. Multiple lines of evidence suggested that other factors are also required for active transcription initiation from a potential TSS. A recent study showed the presence of distance constraint between TSS and PIC (Qiu et al. 2020). If TSSs are too close to where the PIC assembles, transcription initiation will be repressed. One possible factor is the Mot1–Ino80–NC2 (MINC) complex, which is involved ATP-dependent nucleosome sliding (Shen et al. 2000; True et al. 2016). It has been shown that MINC binds to TBP and suppresses transcription of cryptic transcripts and upstream antisense RNAs (uaRNAs) (Xue et al. 2017). Gene-specific transcription factors may also play key roles in the regulation of TSS activity once the PIC is assembled at the promoter, because our previous study showed that only a portion of TSSs are active under one given growth condition (Lu and Lin 2019). From an evolutionary perspective, our data showed that TSS turnover does not necessarily require mutations at these sites (Fig. 5D,E), supporting the involvement of other factors for gain or loss of transcription activities from a potential TSS.
Our results suggest that the scanning model is likely used in both TATA-containing and TATA-less promoters in these budding yeast species, which is consistent with the findings of another study (Qiu et al. 2020). In metazoans, the assembly of PIC on TATA-less promoters was thought to be mediated by bindings of TFIID to other promoter elements such as INR, MTE, and DPE (Theisen et al. 2010). However, these TFIID recognition elements appear to be absent in S. cerevisiae. It was found that TBP occupies both TATA-containing and TATA-less promoters, suggesting other factors might stabilize TBP binding (Basehoar et al. 2004). Our de novo prediction of motifs in promoter regions provides several candidates for future identification of such factors, which would be of great potential interest, as ∼80% of S. cerevisiae genes lack a TATA box in their promoters.
The origin and stepwise evolution of the scanning model in budding yeasts
One of the most significant findings of this study is that the shift of transcription initiation from the classic model to the scanning model occurred after the split of Y. lipolytica during the evolution of budding yeasts. Our study indicates that the transition from the classic model to the scanning model was likely a stepwise process that involved multiple genetic innovations in both PIC genes and promoter sequences that occurred at different evolutionary stages (Fig. 6B).
The first major evolutionary event was the switch of the TSS selection process in the ancestral budding yeast after its split from the Y. lipolytica lineage. This switch may be caused by genetic innovations in GTFs, such as TFIIB, and PoI II, resulting in a distinct transcription initiation machinery. Li et al. (1994) constructed an RNA Pol II transcription system that was reconstituted from S. pombe extracts. By swapping its TFIIB and Pol II with their counterparts of S. cerevisiae, transcription initiation of the in vitro transcription system shifted to 40−120 bp downstream from the TATA box, in contrast to 30 bp downstream as in S. pombe, suggesting that TFIIB and Pol II play key roles in determining the TSS (Li et al. 1994). Another contributing factor may be the divergence of nucleosome occupancy patterns in promoter regions. S. cerevisiae showed a wider nucleosome depletion region (NDR) immediately upstream of the TSS than S. pombe does (Moyle-Heyrman et al. 2013). We observed a similar pattern for the group of TATA box–containing core promoters between the two species based on published nucleosome occupancy data (Supplemental Fig. S16; Brogaard et al. 2012; Moyle-Heyrman et al. 2013). Therefore, the wider NDR in S. cerevisiae provides a longer naked DNA that would facilitate the scanning process. Comparative studies of sequences of each PIC component will be necessary to infer other critical genetic changes associated with the origin of the scanning model.
The second evolutionary event is the gain of the A-rich region in ancestral budding yeasts after their divergence from C. albicans. In the scanning model, a different mechanism of TSS selection should be involved for the PIC to pause the scanning process and initiate transcription. Here, we showed that an A-rich region sits immediately upstream of TSSs in all scanning-model species after their divergence from C. albicans.
The most recent evolutionary event is the origin of the preference of −8A in the A-rich region in the WGD species. Our results show that the −8 position was nearly depleted of any types of substitutions in the group of conserved core promoters (Fig. 5D,E). The positional preference of adenine in WGD species might be because of the divergence in a PIC component that directly interacts with adenines. Comparative analyses of sequence and structural features for each PIC component between the WGD and non-WGD species will be critical to better understand the function of the A-rich region and the genetic mechanism underlying the changes of positional preference of adenine in the A-rich region.
Methods
Yeast strains and CAGE sequencing
Twelve yeast species, including 10 budding yeast species and two fission yeast species, were used in this study (Supplemental Table S1). Strains were grown to log-phase in rich medium (YPD liquid medium) at 30°C. We collected samples in two biological duplicates, and total RNA was extracted with TRIzol (Invitrogen) from each sample. Two biological replicate sequencing libraries were constructed for each yeast species following the nAnT-iCAGE protocol from total RNA (Murata et al. 2014), and all nAnT-iCAGE libraries were sequenced using the Illumina NextSeq system (single-end, 75-bp reads) at DNAFORM.
Inference of phylogenetic relationships and divergence times for the 12 yeast species
The phylogenetic relationships of the 12 species were inferred using the maximum likelihood method based on the LG model with the largest subunit of Pol II RPB2 protein sequences. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 0.4419]). The divergence times for all branching points in the phylogenetic tree were calculated by the RelTime method (Tamura et al. 2012) and were calibrated by the estimated divergence times obtained from TimeTree (Kumar et al. 2017).
TSS calling and identification of core promoters based on CAGE data
The sequenced tags were mapped to each respective reference genome (Supplemental Table S1) using HISAT2 (Kim et al. 2015) with the “--no-softclip” option to avoid false TSSs. We identified reads mapping to rRNA sequences (28S, 18S, 5.8S, and 5S) with rRNAdust (http://fantom.gsc.riken.jp/5/sstar/Protocols:rRNAdust), which changes the FLAG column in SAM files to “not passing filters.” The modified SAM files were then converted into BAM format and sorted using SAMtools (Li et al. 2009) for subsequent TSSs calling. We merged the CAGE reads from biological replicates to calculate the numbers of reads supporting each TSS for all species.
We calculated the probability of observing k number of CAGE reads from one site given the sequencing depth in a species using the Poisson distribution by applying the formula
where λ is the expected number of CAGE reads per site. As transcription initiation can be initiated from both strands of DNA, λ was calculated as the total number of uniquely mapped reads divided by 2 × genome size in base pairs. We considered TSSs with a significantly larger than expected number of supporting CAGE reads (P < 0.01) as biologically significant TSSs for use in subsequent analyses. We normalized the transcriptional activity of each TSS as TPM uniquely mapped tags.
TSSs in a proximate region are likely regulated by the same set of promoter elements and give rise to a functionally equivalent set of transcripts, which can be grouped into a single TC, representing a candidate core promoter. We developed the Peakclu algorithm to identify TCs in each species. Briefly, we first applied a sliding-window approach (window size = 100 bp with step size = 1) to scan CAGE signals from the 5′ end for both strands of each chromosome. In each window, the TSS with the highest TPM value was identified as a peak, representing the dominant TSS of a TC. We grouped the surrounding TSSs with the peak into the same TC, unless a TSS was ≥30 bp away from the nearest one. To infer the width of TC, we first calculated the cumulative distribution of CAGE signals within the TC. Because some outliers may significantly increase the width, we used the positions of the 10th and 90th percentile of CAGE signals as its boundaries and designated the distance between the two positions as its width. We assigned a TC to its immediate downstream gene as its core promoter if the distance of its dominant TSS and the translation start codon of that gene was ≤1000 bp. The detailed descriptions of TC assignments are available in our previous study (Lu and Lin 2019).
We reanalyzed CAGE sequencing data from other studies following the same criteria as this study. The CAGE sequencing data for S. cerevisiae (nine growth conditions) and for S. pombe (five growth conditions) were retrieved from Lu and Lin (2019) and Thodberg et al. (2019), respectively. Human CAGE sequencing data was downloaded from Adiconis et al. (2018). We obtained raw TL-seq data for S. cerevisiae and S. paradoxus from Spealman et al. (2018). TL-seq reads were aligned to their respective reference genomes using HISAT2, with the “softclip” option to ignore the 17-bp linker sequence at the 5′ end.
Calculation of 5′-UTR length and core promoter shape
Transcription is usually initiated from an array of TSSs, instead of a single TSS, and gives rise to a set of functionally equivalent transcripts with slightly different 5′-UTR lengths (Kodzius et al. 2006). In many cases, multiple core promoters are concurrently used that generate significantly different lengths of 5′ UTRs (Lu and Lin 2019). Using one TSS to calculate 5′-UTR length therefore cannot accurately represent the uncertainty and complexity of transcription initiation. We thus calculated the 5′-UTR length of a gene X (LX) as the weighted average 5′-UTR lengths in all its transcripts based on the formula
where n is the total number of TSSs identified for gene X, ti is the number of CAGE tags mapped to the ith TSS, and di is the length of 5′ UTR in transcripts generated from the ith TSS.
We revised the equation for calculating PSS described in our previous study (Lu and Lin 2019) by reversing the negative values to positive:
where p is the probability of observing a TSS at ith TSS within a core promoter, L is the total number of TSSs that pass filtrations by the Poisson distribution, and w is the core promoter width defined as the distance between 10th and 90th quantiles. According to the revised equation, the sharpest promoter has a PSS of zero, and the PSS increases as a core promoter becomes broader.
Analysis of consensus transcription initiation sequence and estimation of capping rate
We plotted sequence motifs with the seqLogo package in R (Bembom 2019; R Core Team 2020). We calculated the frequency of mismatched nucleotide at the TSS position (+1 site) with G-capped reads only. CAGE was designed to only capture transcripts with an m7G cap through biotinylation and binding to magnetic beads, which yields a G at the first position of sequencing reads. However, ∼25% of CAGE reads start without a G. This is mainly because the wash step during library preparation does not completely remove cDNA fragments that are not captured by streptavidin beads. Another source of uncapped transcripts could be products of premature RT stops or 5′−3′ decay of mRNA, resulting in “false TSSs.” These false TSSs are expected to be located downstream from bona fide TSS or core promoters. We therefore applied two filtering steps to minimize these technical artifacts and to increase the accuracy of capping rate calculation. First, we filtered TSSs with supporting reads that are not significantly more than expected based on the Poisson model (P < 0.01). Second, we excluded TSSs locate outside the boundaries of assigned core promoters for our analysis of capping rates.
Analysis of orthologous core promoters
Orthologous core promoter analyses were conducted among S. cerevisiae, S. paradoxus, and S. mikatae. We performed pairwise genome alignments with wgVISTA (Frazer et al. 2004). To minimize background noise, we used only the sequences of core promoters with TPM > 1 as queries to search for their orthologous core promoters. We later discarded orthologous core promoter groups if they were not associated with protein-coding genes (Supplemental Dataset S16). We also excluded Turnover core promoters in subsequent analyses when generated by insertion and deletions.
Identification of TATA box motifs and de novo motif discovery
To determine the presence of the TATA box in a promoter region, we first generated a TATA box matrix based on the consensus sequences TATAWAWR and TATAWAW by seq2profile.pl in the HOMER package (Heinz et al. 2010) with zero mismatches allowed. We then used each generated TATA box matrix to search against promoter sequences (from −150 to +10 bp) for TATA box motifs in all yeast species using findMotifs.pl in HOMER. To identify novel sequence motifs enriched in promoter regions, we performed de novo motif discovery for the same set of promoter sequences by HOMER. We identified the occurrence and locations of the predicted motifs from each species using findMotifs.pl.
Data access
The raw sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA510689. The quantitative maps of TSSs and core promoters generated in this study can be visualized and downloaded from the YeasTSS database (McMillan et al. 2019; http://www.yeastss.org). The source code for analyses of CAGE data in this study is available on GitHub (https://github.com/Linlab-slu/TSSr) and as Supplemental Code.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This study was supported by the President's Research Fund from Saint Louis University and the U.S. National Science Foundation (NSF 1951332) to Z. Lin. We thank Dr. Genevieve Fourel and three anonymous reviewers for constructive comments that have significantly improved this manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.264325.120.
References
- Adiconis X, Haber AL, Simmons SK, Levy Moonshine A, Ji Z, Busby MA, Shi X, Jacques J, Lancaster MA, Pan JQ, et al. 2018. Comprehensive comparative analysis of 5′-end RNA-sequencing methods. Nat Methods 15: 505–511. 10.1038/s41592-018-0014-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Gilbert WV. 2013. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res 23: 977–987. 10.1101/gr.150342.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709. 10.1016/S0092-8674(04)00205-3 [DOI] [PubMed] [Google Scholar]
- Batut P, Dobin A, Plessy C, Carninci P, Gingeras TR. 2013. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res 23: 169–180. 10.1101/gr.139618.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembom O. 2019. seqLogo: Sequence logos for DNA sequence alignments. R package version 1520 10.18129/B9.bioc.seqLogo [DOI]
- Bernard V, Brunaud V, Lecharny A. 2010. TC-motifs at the TATA-box expected position in plant genes: a novel class of motifs involved in the transcription regulation. BMC Genomics 11: 166 10.1186/1471-2164-11-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleykasten-Grosshans C, Neuvéglise C. 2011. Transposable elements in yeasts. C R Biol 334: 679–686. 10.1016/j.crvi.2011.05.017 [DOI] [PubMed] [Google Scholar]
- Blombach F, Smollett KL, Grohmann D, Werner F. 2016. Molecular mechanisms of transcription initiation-structure, function, and evolution of TFE/TFIIE-like factors and open complex formation. J Mol Biol 428: 2592–2606. 10.1016/j.jmb.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börlin CS, Cvetesic N, Holland P, Bergenholm D, Siewers V, Lenhard B, Nielsen J. 2019. Saccharomyces cerevisiae displays a stable transcription start site landscape in multiple conditions. FEMS Yeast Res 19 10.1093/femsyr/foy128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both GW, Furuichi Y, Muthukrishnan S, Shatkin AJ. 1975. Ribosome binding to reovirus mRNA in protein synthesis requires 5′ terminal 7-methylguanosine. Cell 6: 185–195. 10.1016/0092-8674(75)90009-4 [DOI] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang JP, Widom J. 2012. A map of nucleosome positions in yeast at base-pair resolution. Nature 486: 496–501. 10.1038/nature11142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635. 10.1038/ng1789 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Otto GM, Powers EN, Keskin A, Mertins P, Carr SA, Jovanovic M, Brar GA. 2018. Pervasive, coordinated protein-level changes driven by transcript isoform switching during meiosis. Cell 172: 910–923.e16. 10.1016/j.cell.2018.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WS, Yan M, Nusinow D, Gralla JD. 2002. In vitro transcription and start site selection in Schizosaccharomyces pombe. J Mol Biol 319: 1005–1013. 10.1016/S0022-2836(02)00329-7 [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH. 2008. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet 24: 167–177. 10.1016/j.tig.2008.01.008 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Fishburn J, Hahn S. 2012. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Mol Cell Biol 32: 12–25. 10.1128/MCB.06242-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–W279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Lis JT. 1993. DNA melting on yeast RNA polymerase II promoters. Science 261: 759–762. 10.1126/science.8342041 [DOI] [PubMed] [Google Scholar]
- Hahn S, Young ET. 2011. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189: 705–736. 10.1534/genetics.111.127019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks KB, Delneri D, Griffiths-Jones S. 2014. Intron evolution in Saccharomycetaceae. Genome Biol Evol 6: 2543–2556. 10.1093/gbe/evu196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Landolin JM, Brown JB, Sandler JE, Takahashi H, Lassmann T, Yu C, Booth BW, Zhang D, Wan KH, et al. 2011. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res 21: 182–192. 10.1101/gr.112466.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, Sasaki D, Imamura K, Kai C, Harbers M, et al. 2006. CAGE: cap analysis of gene expression. Nat Methods 3: 211–222. 10.1038/nmeth0306-211 [DOI] [PubMed] [Google Scholar]
- Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. 2009. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 462: 323–330. 10.1038/nature08548 [DOI] [PubMed] [Google Scholar]
- Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simão FA, Zdobnov EM. 2019. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res 47: D807–D811. 10.1093/nar/gky1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Brow DA. 2006. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J Biol Chem 281: 14119–14128. 10.1074/jbc.M601937200 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34: 1812–1819. 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- Leppek K, Das R, Barna M. 2018. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 19: 158–174. 10.1038/nrm.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Flanagan PM, Tschochner H, Kornberg RD. 1994. RNA polymerase II initiation factor interactions and transcription start site selection. Science 263: 805–807. 10.1126/science.8303296 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Hou J, Bai L, Hu C, Tong P, Kang Y, Zhao X, Shao Z. 2015. Genome-wide analysis of core promoter structures in Schizosaccharomyces pombe with DeepCAGE. RNA Biol 12: 525–537. 10.1080/15476286.2015.1022704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lenhard B, Luscombe NM. 2018. Integrated analysis sheds light on evolutionary trajectories of young transcription start sites in the human genome. Genome Res 28: 676–688. 10.1101/gr.231449.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Li WH. 2012. Evolution of 5′ untranslated region length and gene expression reprogramming in yeasts. Mol Biol Evol 29: 81–89. 10.1093/molbev/msr143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Lin Z. 2019. Pervasive and dynamic transcription initiation in Saccharomyces cerevisiae. Genome Res 29: 1198–1210. 10.1101/gr.245456.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J, Lu Z, Rodriguez JS, Ahn TH, Lin Z. 2019. YeasTSS: an integrative web database of yeast transcription start sites. Database (Oxford) 2019 10.1093/database/baz048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle-Heyrman G, Zaichuk T, Xi L, Zhang Q, Uhlenbeck OC, Holmgren R, Widom J, Wang JP. 2013. Chemical map of Schizosaccharomyces pombe reveals species-specific features in nucleosome positioning. Proc Natl Acad Sci 110: 20158–20163. 10.1073/pnas.1315809110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Nishiyori-Sueki H, Kojima-Ishiyama M, Carninci P, Hayashizaki Y, Itoh M. 2014. Detecting expressed genes using CAGE. Methods Mol Biol 1164: 67–85. 10.1007/978-1-4939-0805-9_7 [DOI] [PubMed] [Google Scholar]
- Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 1975. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature 255: 33–37. 10.1038/255033a0 [DOI] [PubMed] [Google Scholar]
- Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. 1999. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev 13: 3217–3230. 10.1101/gad.13.24.3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policastro RA, Raborn RT, Brendel VP, Zentner GE. 2020. Simple and efficient profiling of transcription initiation and transcript levels with STRIPE-seq. Genome Res 30: 910–923. 10.1101/gr.261545.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Jin H, Vvedenskaya I, Llenas JA, Zhao T, Malik I, Visbisky AM, Schwartz SL, Cui P, Čabart P, et al. 2020. Universal promoter scanning by Pol II during transcription initiation in Saccharomyces cerevisiae. Genome Biol 21: 132 10.1186/s13059-020-02040-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Rhee HS, Pugh BF. 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301. 10.1038/nature10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936. 10.1126/science.1203357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 21: 327–335. 10.1016/0968-0004(96)10050-5 [DOI] [PubMed] [Google Scholar]
- Sainsbury S, Niesser J, Cramer P. 2013. Structure and function of the initially transcribing RNA polymerase II–TFIIB complex. Nature 493: 437–440. 10.1038/nature11715 [DOI] [PubMed] [Google Scholar]
- Schor IE, Degner JF, Harnett D, Cannavo E, Casale FP, Shim H, Garfield DA, Birney E, Stephens M, Stegle O, et al. 2017. Promoter shape varies across populations and affects promoter evolution and expression noise. Nat Genet 49: 550–558. 10.1038/ng.3791 [DOI] [PubMed] [Google Scholar]
- Seizl M, Hartmann H, Hoeg F, Kurth F, Martin DE, Söding J, Cramer P. 2011. A conserved GA element in TATA-less RNA polymerase II promoters. PLoS One 6: e27595 10.1371/journal.pone.0027595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406: 541–544. 10.1038/35020123 [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. 2003. The RNA polymerase II core promoter. Annu Rev Biochem 72: 449–479. 10.1146/annurev.biochem.72.121801.161520 [DOI] [PubMed] [Google Scholar]
- Spealman P, Naik AW, May GE, Kuersten S, Freeberg L, Murphy RF, McManus J. 2018. Conserved non-AUG uORFs revealed by a novel regression analysis of ribosome profiling data. Genome Res 28: 214–222. 10.1101/gr.221507.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. 2012. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci 109: 19333–19338. 10.1073/pnas.1213199109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen JW, Lim CY, Kadonaga JT. 2010. Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol Cell Biol 30: 3471–3479. 10.1128/MCB.00053-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thodberg M, Thieffry A, Bornholdt J, Boyd M, Holmberg C, Azad A, Workman CT, Chen Y, Ekwall K, Nielsen O, et al. 2019. Comprehensive profiling of the fission yeast transcription start site activity during stress and media response. Nucleic Acids Res 47: 1671–1691. 10.1093/nar/gky1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JD, Muldoon JJ, Carver MN, Poorey K, Shetty SJ, Bekiranov S, Auble DT. 2016. The modifier of transcription 1 (Mot1) ATPase and Spt16 histone chaperone co-regulate transcription through preinitiation complex assembly and nucleosome organization. J Biol Chem 291: 15307–15319. 10.1074/jbc.M116.735134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Pradhan SK, Sun F, Chronis C, Tran N, Su T, Van C, Vashisht A, Wohlschlegel J, Peterson CL, et al. 2017. Mot1, Ino80C, and NC2 function coordinately to regulate pervasive transcription in yeast and mammals. Mol Cell 67: 594–607.e4. 10.1016/j.molcel.2017.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dietrich FS. 2005. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res 33: 2838–2851. 10.1093/nar/gki583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Degen D, Ho MX, Sineva E, Ebright KY, Ebright YW, Mekler V, Vahedian-Movahed H, Feng Y, Yin R, et al. 2014. GE23077 binds to the RNA polymerase ‘i’ and ‘i+1’ sites and prevents the binding of initiating nucleotides. eLife 3: e02450 10.7554/eLife.02450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.