Preeclampsia (PE) is a hypertensive disorder of uncertain etiology that is the leading cause of maternal and fetal morbidity or mortality. The dysregulation of microRNAs (miRNAs) has been highlighted as a potential factor involved in the development of PE. Therefore, our study investigated a novel miRNA, miRNA 183 (miR-183), and its underlying association with PE. Expression of miR-183, forkhead box P1 (FOXP1), and G protein subunit gamma 7 (GNG7) in placental tissues of patients with PE was determined.
KEYWORDS: Preeclampsia, microRNA-183, forkhead box P1, G protein subunit gamma 7, mechanistic target of rapamycin kinase
ABSTRACT
Preeclampsia (PE) is a hypertensive disorder of uncertain etiology that is the leading cause of maternal and fetal morbidity or mortality. The dysregulation of microRNAs (miRNAs) has been highlighted as a potential factor involved in the development of PE. Therefore, our study investigated a novel miRNA, miRNA 183 (miR-183), and its underlying association with PE. Expression of miR-183, forkhead box P1 (FOXP1), and G protein subunit gamma 7 (GNG7) in placental tissues of patients with PE was determined. Gain- and loss-of-function experiments were conducted to explore modulatory effects of miR-183, FOXP1, and GNG7 on the viability, invasion, and angiogenesis of trophoblast cells in PE. Finally, we undertook in vivo studies to explore effects of FOXP1 in the PE model. The results revealed suppressed expression of FOXP1 and significant elevations in miR-183 and GNG7 expression in placental tissues of PE patients. FOXP1 was observed to promote proliferation, invasion, and angiogenesis in human chorionic trophoblastic cells. miR-183 resulted in depletion of FOXP1 expression, while FOXP1 was capable of restraining GNG7 expression and promoting the mTOR pathway. The findings confirmed the effects of FOXP1 on PE. In conclusion, miR-183 exhibits an inhibitory role in PE through suppression of FOXP1 and upregulation of GNG7.
INTRODUCTION
Preeclampsia (PE) is a severe complication of pregnancy, occurring after the 20th week of gestation and characterized by hypertension and proteinuria, leading to perinatal and maternal morbidity and mortality worldwide (1). PE is characterized by restricted angiogenesis in early pregnancy and shallow invasion of maternal spiral arteries, resulting in placental hypoperfusion (2). PE can result in impaired function of blood vessels of the fetal placenta and increase the risk of cardiovascular diseases in the newborn, indicating that PE can interfere with fetal blood vessel development in utero (3). Mounting evidence has revealed that inadequate trophoblast migration and invasiveness associated with an impaired remodeling of the spiral arteries are crucial pathological features of PE (4). However, to date, the etiology and pathogenesis of PE remain unclear. An understanding of the underlying molecular mechanisms that are involved in trophoblast migration/invasiveness is required.
The G protein signaling pathways are known to participate in various physiological activities, including blood pressure regulation (5). Moreover, it has been reported that different α components of G-protein αβγ subunit combinations serve different physiological functions (6). G protein alpha subunit 14 (a member of the G protein family) is involved in the regulation of hypertension and tumor vascularization (7). It has also been demonstrated that G protein subunit gamma 7 (GNG7) silencing can suppress apoptosis and promote the viability and differentiation of trophoblasts by activating the mammalian target of the rapamycin (mTOR) signaling pathway in PE (8). Meanwhile, another study demonstrated that the depletion of forkhead box P1 (FOXP1) regulates the expression of genes that are significantly involved with and enriched in chemokine pathways, i.e., GNG7 (9). Interestingly, a significantly high expression of FOXP1 has been reported in embryonic neural stem cells. Regulation of FOXP1 expression has also been observed to modulate the differentiation of neurons and astrocytes (10). Moreover, highly expressed FOXP1 can effectively inhibit the Akt/mTOR signaling in a model of diabetic renal pathology (11). Meanwhile, FOXP1 expression is significantly downregulated in cardiac endothelial cells during pathological cardiac remodeling (12). A previous study reported that low expression of microRNA 183 (miR-183) regulates the Jak/Stat signaling pathway by targeting FOXP1, thus reducing hippocampal neuron injury in rats with epilepsy-like behaviors (13). A multitude of studies have reported abnormal expression profiles of placental microRNAs (miRNAs) in PE (14, 15). As demonstrated in numerous studies, miRNA-mediated gene expression in women with PE may induce endoplasmic reticulum stress, oxidative stress in PE, and endothelial reprogramming, any of which effects could enhance the risk of developing cardiovascular disease later in life (16). The miRNA expression profile of patients with PE is significantly different from that of women experiencing healthy pregnancy, in particular with respect to the effects of miRNAs in regulating the secretion of angiogenic factors and anti angiogenic factors (17). Mounting evidence has elaborated the significance of angiogenesis modulators in PE (18, 19). More importantly, multiple miRNAs have been documented to exert effects in PE by regulating trophoblast cellular biological behaviors (20–22). Considering the high expression of miRNA 152 (miR-152), miR-183, and miR-210 observed during the second trimester of women with PE (23), we hypothesized that miR-183 could potentially regulate FOXP1 during the progression of PE by mediating trophoblast cellular functions and angiogenesis. Therefore, the present study aimed to investigate whether miR-183 plays an inhibitory role in the trophoblast cell proliferation, invasion, and angiogenesis through the suppression of FOXP1 expression and upregulation of GNG7 expression.
RESULTS
miR-183 regulates expression of FOXP1 and GNG7, thereby affecting PE progression.
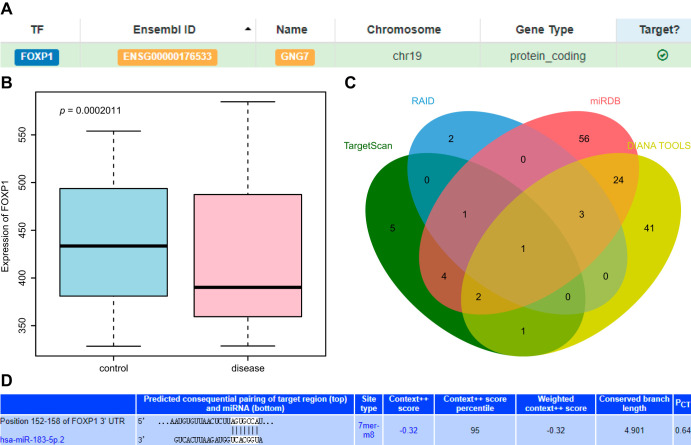
FOXP1 can regulate the expression of GNG7 (9), leading us to speculate that FOXP1 may play an important role in PE via GNG7. Consistent with the previous study, our scrutiny of the hTFtarget database revealed the targeting regulation of FOXP1 on GNG7 (Fig. 1A). Placental tissues obtained from PE patients had low expression of FOXP1, as shown in the box plot of FOXP1 expression data in the microarray data GSE6573, retrieved from the Gene Expression Omnibus (GEO) database (Fig. 1B). The upstream miRNAs of FOXP1 were predicted by the TargetScan, RAID, miRDB, and DIANA TOOLS databases, which yielded 14, 7, 91, and 72 miRNAs, respectively. The intersection of the Venn diagram identified miR-183-5p as the most critical upstream miRNA (Fig. 1C). However, there has hitherto been no evidence regarding the target relationship between miR-183 and FOXP1, which could entail a novel regulatory pathway for PE. Moreover, we next obtained the binding site map of the targeting relationship from the TargetScan database (Fig. 1D). Based on the aforementioned findings, we concluded that miR-183 might affect PE through FOXP1-mediated changes in GNG7.
FIG 1.
Bioinformatics analysis predicting miR-183, FOXP1, and GNG7 and their molecular interactions in PE. (A) Targeting relationship between FOXP1 and GNG7 retrieved in the hTFtargetScan database. (B) Box plot of FOXP1 expression data in microarray data GSE6573. The box on the left represents the expression in the placental tissues of normal samples, and the box on the right represents the expression in the placental tissues of patients with PE. (C) Venn diagram of upstream miRNA results of FOXP1 predicted by TargetScan, RAID, miRDB, and DIANA TOOLS databases. (D) The binding site map of miR-183 and FOXP1 obtained from the TargetScan database.
Low expression of FOXP1 is observed in placental tissues of patients with PE.
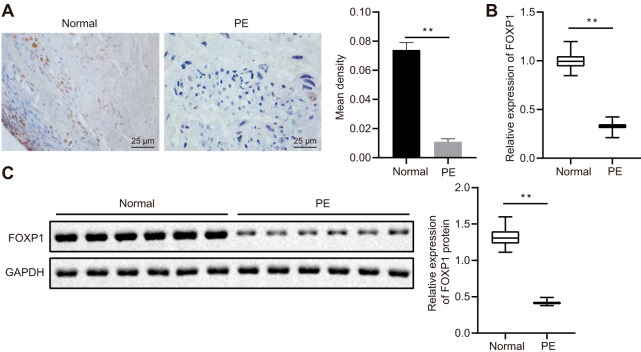
Manifestations of PE in the rodent model group, including systolic blood pressure at 18 days of gestation higher than normal and proteinuria higher than normal, confirmed the successful establishment of the PE mouse model. The clinical characteristics of patients with PE and healthy pregnant women selected for study at the Second Xiangya Hospital, Central South University, are shown in Table 1. The patients with PE presented with increased systolic and diastolic blood pressure and their newborns were observed to have lower body weight. Immunohistochemistry (IHC) and Western blot analysis were conducted to detect FOXP1 protein expression in the placental tissues of patients with PE and healthy pregnant women. Our results indicated that FOXP1-positive areas were significantly decreased in patients with PE (Fig. 2A), while mRNA and protein expression of FOXP1 was reduced, as determined by reverse transcription quantitative PCR (RT-qPCR) and Western blot analysis (Fig. 2B and C). These results highlighted the poor expression of FOXP1 in the placental tissues of patients with PE.
TABLE 1.
Clinical features of patients with preeclampsia (PE)
| Variable | Normal pregnancy (n = 24) | PE (n = 24) | P value |
|---|---|---|---|
| Age (yr) ± SD | 28.21 ± 3.73 | 27.54 ± 3.35 | 0.516 |
| Mean maternal wt (kg) ± SD | 52.96 ± 3.42 | 67.05 ± 5.96 | <0.001 |
| Mean systolic blood pressure (mm Hg) ± SD | 114.83 ± 4.66 | 160.88 ± 9.83 | <0.001 |
| Mean diastolic blood pressure (mm Hg) ± SD | 75.29 ± 4.68 | 105.21 ± 12.57 | <0.001 |
| Mean urine protein (μg/ml) ± SD | 0 | 1.21 ± 0.38 | <0.001 |
| Mean birth wt (g) ± SD | 3,014.73 ± 503.34 | 2,329.41 ± 614.21 | <0.001 |
| Mean gestation age (wk) ± SD | 38.92 ± 1.44 | 37.67 ± 1.63 | 0.007 |
FIG 2.
Reduced expression of FOXP1 is observed in the placental tissues of patients with PE. (A) IHC staining analysis of FOXP1 protein in placental tissues of patients with PE and healthy pregnant women (×400 magnification). (B) The expression of FOXP1 in the placental tissues of patients with PE and healthy pregnant women as identified by RT-qPCR. **, P < 0.01. (C) FOXP1 expression in the placental tissues of patients with PE and healthy pregnant women normalized to GAPDH as determined by Western blotting; n = 24. **, P < 0.01. The experiment was repeated 3 times independently.
FOXP1 overexpression facilitates human chorionic trophoblast cell (HTR-8/SVneo) proliferation, invasion, and angiogenesis.
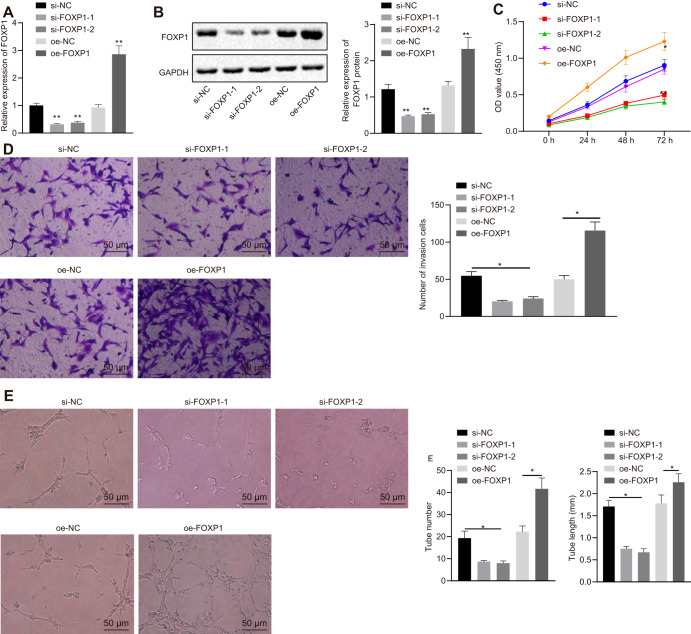
Human chorionic trophoblast cells (HTR-8/SVneo) were selected to verify the function of FOXP1. HTR-8/SVneo cells were transfected with small interfering RNA-negative control (si-NC), si-FOXP1-1, si-FOXP1-2, overexpression (oe)-NC, and oe-FOXP1, whereupon FOXP1 expression was detected by RT-qPCR and Western blot analysis. The results suggested that, compared to HTR-8/SVneo cells transfected with si-NC, FOXP1 expression was downregulated in HTR-8/SVneo cells transfected with si-FOXP1-2 or si-FOXP1-1, among which si-FOXP1-1 showed a more pronounced decline of FOXP1 and was therefore selected for subsequent experiments. In contrast to the transfection of oe-NC, FOXP1 expression was enhanced in HTR-8/SVneo cells transfected with oe-FOXP1 (Fig. 3A and B).
FIG 3.
Overexpression of FOXP1 promotes HTR-8/SVneo cell proliferation, invasion, and angiogenesis. (A) The expression of FOXP1 in HTR-8/SVneo cells after the overexpression of FOXP1 or knockdown of FOXP1, as detected by RT-qPCR. (B) The expression of FOXP1 normalized to GAPDH in HTR-8/SVneo cells after the overexpression of FOXP1 or knockdown of FOXP1, as determined by Western blotting. (C) Cell activity as measured by cell counting kit 8 (CCK-8) at 0, 24, 48, and 72 h following FOXP1 silencing or overexpression. (D) Cell invasion identified by Transwell, where the number of invasion cells were counted following FOXP1 silencing or overexpression (×200 magnification). (E) Formation of the lumen and the number and length of lumen measured by human umbilical vein endothelial cell angiogenesis assay following FOXP1 silencing or overexpression (×200 magnification). *, P < 0.05 versus treatment with si-NC or oe-NC. The experiment was repeated 3 times independently.
Cell proliferation, invasion, and angiogenesis were measured by assays of cell counting kit 8 (CCK-8), Transwell, and in vitro angiogenesis assays. Compared to the transfection of oe-NC, cell viability, invasion, and angiogenesis were increased in HTR-8/SVneo cells following transfection with oe-FOXP1, while a decrease was observed in HTR-8/SVneo cells upon the transfection of si-FOXP1 in contrast to the transfection of si-NC (Fig. 3C to E). Collectively, the above findings suggested that overexpression of FOXP1 could promote HTR-8/SVneo cell proliferation, invasion, and angiogenesis, thereby inhibiting the progression of PE.
miR-183 specifically binds FOXP1 and inhibits its expression.
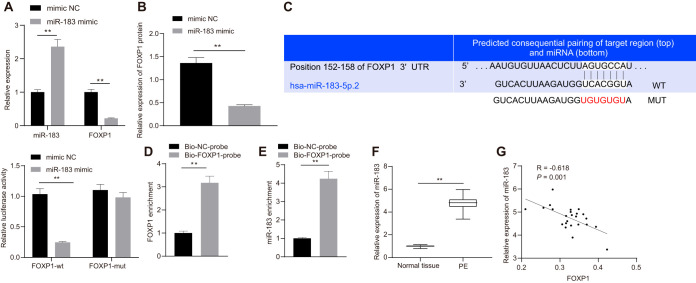
Following transfection with the miR-183 mimic, miR-183 and FOXP1 expression in HTR-8/SVneo cells was detected by RT-qPCR, the results of which suggested that miR-183 expression was significantly upregulated while that of FOXP1 was inhibited (Fig. 4A and B). Dual-luciferase reporter gene assay was used to detect luciferase activity, which showed the luciferase activity was decreased in wild-type FOXP1 3′ untranslated region (FOXP1-3′-UTRW) after transfection with miR-183 mimic, but no such difference was observed in mutant-type FOXP1 3′ untranslated region (FOXP1-3′-UTRM) expression after transfection with miR-183 mimic (Fig. 4C). RNA pulldown assay was performed, followed by RT-qPCR, the results of which revealed a large amount of FOXP1 in microspheres pulled down by FOXP1 and that miR-183 expression was elevated relative to the Bio-NC-probe group (Fig. 4D and E). The expression of miR-183 in placental tissues of patients with PE and normal placental tissues was analyzed by RT-qPCR, which showed elevated expression of miR-183 in placental tissues of patients with PE (Fig. 4F) and a negative correlation between miR-183 expression and FOXP1 expression (Fig. 4G). These results suggested that miR-183 was highly expressed in the placental tissues of patients with PE and could suppress FOXP1 expression.
FIG 4.
miR-183 negatively regulates the expression of FOXP1. (A) Expression of miR-183 and FOXP1 in Htr-8/SVneo cells measured by RT-qPCR after the transfection of miR-183 mimic. *, P < 0.05. (B) Expression of FOXP1 in Htr-8/SVneo cells after the transfection of miR-183 mimic normalized to GAPDH, as assessed by Western blotting. (C) Sequence alignment of FOXP1 3′ UTR and hsa-miR-183-5p.2 and relative luciferase activity detected by dual-luciferase reporter gene assay. **, P < 0.01. (D) FOXP1 expression detected by RNA pulldown assay and RT-qPCR using biotinylated FOXP1 and NC probes. *, P < 0.05. (E) miR-183b-5p expression relative to input detected by RNA pulldown assay and RT-qPCR using biotinylated FOXP1 and NC probes. *, P < 0.05. (F) The expression of miR-183 in the placental tissues of patients with PE and healthy pregnant women was tested by RT-qPCR. (G) Pearson correlation analysis of miR-183 expression with FOXP1 expression in placental tissues of patients with PE. The experiment was repeated 3 times independently.
Inhibition of miR-183 promotes trophoblast cell proliferation, invasion, and angiogenesis by upregulating FOXP1 expression.
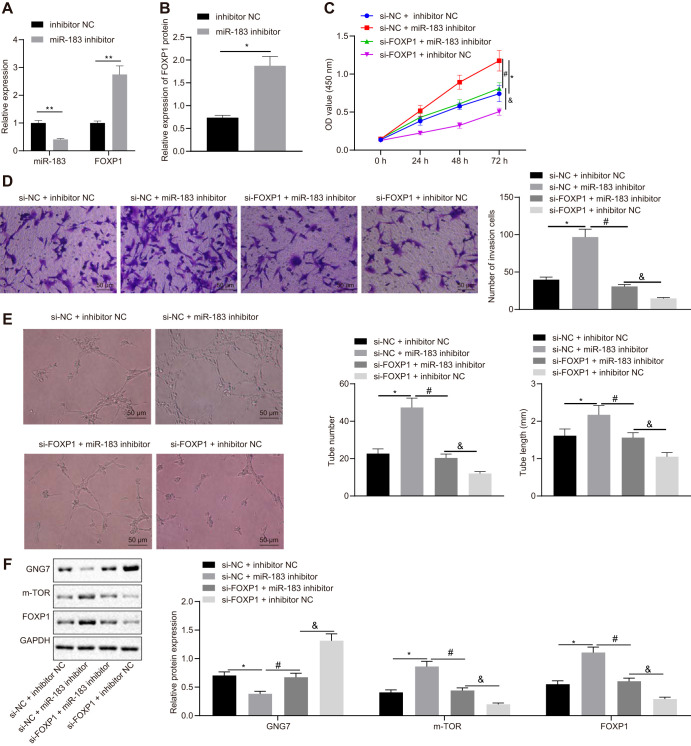
RT-qPCR was conducted to evaluate miR-183 expression in HTR-8/SVneo cells following transfection with miR-183 inhibitor, revealing that miR-183 expression was remarkably decreased. Meanwhile, results of RT-qPCR and Western blot analysis indicated that FOXP1 expression was increased in HTR-8/SVneo cells upon transfection of miR-183 inhibitor (Fig. 5A and B). To further study the effects of miR-183 and FOXP1 on biological functions of HTR-8/SVneo cells, cell proliferation, invasion, and angiogenesis were detected by CCK-8, Transwell, and in vitro angiogenesis assays. The results revealed that cell viability, invasion, and angiogenesis were significantly elevated in HTR-8/SVneo cells after the transfection of miR-183 inhibitor. Moreover, in other studies we silenced FOXP1 to conduct rescue experiments, which showed that the effects of miR-183 inhibition on HTR-8/SVneo cell proliferation, invasion, and angiogenesis were reversed by transfection of si-FOXP1 (Fig. 5C to E). Additionally, Western blot analysis showed the expression of GNG7 was reduced, while that of FOXP1 and mTOR was elevated, in HTR-8/SVneo cells transfected with miR-183 inhibitor. However, concomitant inhibition of miR-183 and FOXP1 had the opposite effects (Fig. 5F). These results suggested that inhibition of miR-183 promoted FOXP1 expression and thus stimulated proliferation, invasion, and angiogenesis in HTR-8/SVneo cells.
FIG 5.
Inhibition of miR-183 elevates FOXP1 expression, thereby promoting trophoblast cell proliferation, invasion, and angiogenesis. (A) Expression of miR-183 and FOXP1 in HTR-8/SVneo cells identified by RT-qPCR after the transfection of miR-183 inhibitor or in combination with si-FOXP1. (B) Expression of FOXP1 protein in HTR-8/SVneo cells normalized to GAPDH and detected by Western blotting after the transfection of miR-183 inhibitor or in combination with si-FOXP1. (C) Cell activity as assessed by CCK-8 assay after the transfection of miR-183 inhibitor or in combination with si-FOXP1. (D) Cell invasion as determined by Transwell, where the number of invasion cells was counted after the transfection of miR-183 inhibitor or in combination with si-FOXP1 (×200 magnification). (E) Formation of microvascular branches and the number and length of microvascular branches as tested by an in vitro angiogenesis assay after the transfection of miR-183 inhibitor (×200 magnification). (F) Expression of GNG7, FOXP1, and mTOR as determined by Western blotting in HTR-8/SVneo cells transfected with miR-183 inhibitor or in combination with si-FOXP1. *, P < 0.05 versus treatment with NC; #, P < 0.05 versus treatment with si-NC + miR-183 inhibitor; &, P < 0.05 versus treatment with si-FOXP1 + miR-183 inhibitor. The experiment was repeated 3 times independently.
FOXP1 inhibits GNG7 expression and activates the mTOR signaling pathway, promoting trophoblast cell proliferation, invasion, and angiogenesis.
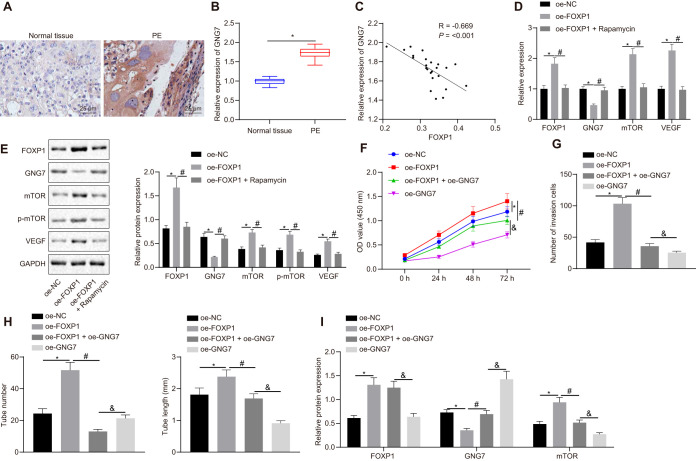
The expression of GNG7 in the placental tissues of patients with PE was detected by RT-qPCR and IHC. Our data revealed high expression of GNG7 in placental tissues of patients with PE (Fig. 6A and B) and that GNG7 expression was negatively correlated with FOXP1 expression (Fig. 6C). Furthermore, RT-qPCR was utilized to assess the expression of GNG7, mTOR, and vascular endothelial growth factor (VEGF) in HTR-8/SVneo cells. Overexpression of FOXP1 was found to suppress GNG7 expression and promote mTOR and VEGF expression in HTR-8/SVneo cells (Fig. 6D). The expression of mTOR, and VEGF along with mTOR phosphorylation levels, was subsequently investigated by Western blotting, which showed that the overexpression of FOXP1 led to an increase in the expression of mTOR, GNG7, and VEGF proteins, and mTOR phosphorylation levels, all of which changes were reversed following rapamycin treatment (Fig. 6D and E). Meanwhile, CCK-8, Transwell, and in vitro angiogenesis assays were employed to detect HTR-8/SVneo cell proliferation, invasion, and angiogenesis. The overexpression of FOXP1 was observed to promote trophoblast cell proliferation (Fig. 6F), invasion (Fig. 6G), and angiogenesis (Fig. 6H), all of which effects were impeded by restored GNG7 expression. Additionally, Western blot analysis showed the expression of GNG7 was reduced while that of FOXP1 and mTOR was elevated in HTR-8/SVneo cells transfected with oe-FOXP1. However, concomitant overexpression of FOXP1 and GNG7 provoked opposite effects (Fig. 6I). Collectively, these results supported the model that FOXP1 inhibited GNG7 expression and activated the mTOR signaling pathway, thereby promoting proliferation, invasion, and angiogenesis of trophoblast cells.
FIG 6.
FOXP1 inhibits GNG7 expression and promotes the mTOR signaling pathway and trophoblast cell proliferation, invasion, and angiogenesis. (A) Expression of GNG7 in the placental tissues of patients with PE and healthy pregnant women measured by IHC (×400 magnification). (B) Expression of GNG7 in the placental tissues of patients with PE and healthy pregnant women identified by RT-qPCR; n = 24; **, P < 0.01. (C) Pearson’s correlation analysis of GNG7 expression with FOXP1 expression in the placental tissues of patients with PE. (D) Expression of FOXP1, GNG7, mTOR, and VEGF in HTR-8/SVneo cells identified by RT-qPCR after the treatment of oe-FOXP1 or in combination with rapamycin. (E) Expression of FOXP1, GNG7, mTOR, VEGF, and p-mTOR normalized to GAPDH and assessed by Western blotting in HTR-8/SVneo cells identified by RT-qPCR after the treatment of oe-FOXP1 or in combination with rapamycin. (F) Cell viability measured by CCK-8 after the transfection of oe-FOXP1 or in combination with oe-GNG7. (G) Cell invasion rate calculated by Transwell after the transfection of oe-FOXP1 or in combination with oe-GNG7 (×200 magnification). (H) Angiogenesis and the number and length of microvascular branches determined by in vitro angiogenesis assay after the transfection of oe-FOXP1 or in combination with oe-GNG7 (×200 magnification). (I) Expression of GNG7, FOXP1, and mTOR as determined by Western blotting in HTR-8/SVneo cells transfected with oe-FOXP1 or in combination with oe-GNG7. *, P < 0.05 versus treatment with oe-NC; #, P < 0.05 versus treatment with oe-FOXP1; &, P < 0.05 versus treatment with oe-FOXP1 + oe-GNG7. The experiment was repeated 3 times independently.
miR-183 elicits PE through regulation of the FOXP1/GNG7/mTOR axis in mice.
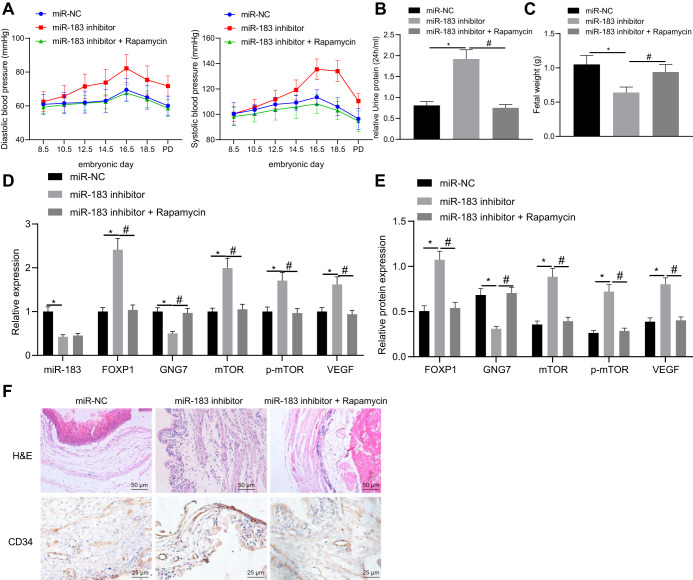
To identify the effect of miR-183 on PE in vivo, we constructed a nude mouse xenograft model of endometrial cancer (EC). Mice were injected with cells transfected with miR-183 inhibitor or in combination with rapamycin treatment. To determine whether PE-like physiological changes were induced in mice, maternal blood pressure was measured at the following embryonic days: E0, E8.5, E10.5, E12.5, E14.5, E16.5, and E18.5. Systolic and diastolic blood pressures were significantly increased in mice upon the infection of miR-183 inhibitor at gestational day 16.5 and continued to rise throughout the remaining course of the gestation (P < 0.05). Further rapamycin treatment caused no changes in systolic and diastolic blood pressure during the gestation (P > 0.05) (Fig. 7A). Postpartum blood pressure quickly returned to normal, as likewise occurs in human postpartum recovery. Maternal urine was collected and its protein content measured on E18.5, the results of which showed distinctly elevated urine protein content in mice with the injection of miR-183 inhibitor compared to miR-NC, which was reduced following further rapamycin treatment (Fig. 7B). These findings indicated that miR-183 inhibitor could potentially induce PE symptoms in mice similar to human symptoms, also with respect to lower fetal weight compared to the healthy group (Fig. 7C). RT-qPCR was used to detect the expression of miR-183, FOXP1, GNG7, mTOR, and VEGF in the placental tissues of mice treated with miR-183 inhibitor. The results suggested that expression of miR-183 and GNG7 was decreased while that of FOXP1, mTOR, and VEGF was increased in the presence of miR-183 inhibitor; however, opposite results were noted in response to rapamycin treatment (Fig. 7D). Moreover, Western blot analysis revealed that GNG7 protein expression was downregulated while the expression of FOXP1, mTOR, and VEGF proteins, along with the mTOR phosphorylation levels, were upregulated in the placental tissues of mice following the knockdown of miR-183, and that rapamycin treatment rescued these effects (Fig. 7E). Hematoxylin-eosin staining of the placental tissue sections showed that the placental structure was disordered in mice treated with miR-183 inhibitor, while rapamycin treatment normalized the placental structure (Fig. 7F). Subsequently, the placental tissue sections were stained by the CD34 antibody, which showed significant reductions in vascular compactness and branching following miR-183 inhibitor treatment, which were negated by rapamycin treatment (Fig. 7F). These results suggested that miR-183 could promote the progression of PE through regulation of the FOXP1/GNG7/mTOR axis in mice.
FIG 7.
miR-183 accelerates the progression of PE through regulation of the FOXP1/GNG7/mTOR axis in mice. (A) Systolic blood pressure and diastolic blood pressure of mice upon injection of miR-183 inhibitor or in combination with rapamycin at E0, E8.5, E10.5, E12.5, E14.5, E16.5, and E18.5; n = 20. (B) Urine protein content of mice at E18.5 following treatment with miR-183 inhibitor alone or in combination with rapamycin; n = 20. (C) Embryo weight of mice following treatment with miR-183 inhibitor or in combination with rapamycin; n = 20. (D) The expression of miR-183, FOXP1, GNG7, mTOR, p-mTOR, and VEGF in the placental tissues of mice treated with miR-183 inhibitor alone or in combination with rapamycin as measured by RT-qPCR; n = 15. (E) Expression of FOXP1, GNG7, mTOR, p-mTOR, and VEGF proteins in mouse placenta tissues normalized to GAPDH, as measured by Western blotting, following treatment with miR-183 inhibitor or in combination with rapamycin; n = 6. (F) Hematoxylin-eosin staining (upper) of placental tissues of mice following treatment with miR-183 inhibitor alone or in combination with rapamycin (×200 magnification); n = 4 to 6. CD34 staining (lower) of placental tissues of mice following treatment with miR-183 inhibitor alone or in combination with rapamycin (×400 magnification); n = 15; *, P < 0.05 versus treatment with miR-NC; #, P < 0.05 versus treatment with miR-183 inhibitor. The experiment was repeated 3 times independently.
DISCUSSION
PE is a hypertensive disorder occurring during pregnancy, which is characterized by the presence of placental and endothelial dysfunction (24). An aberrant cytotrophoblastic infiltration of the maternal spiral arteriole has been suggested as the initiating factor in the pathophysiology of PE, which results in structurally abnormal placental blood vessels, and subsequent hypoperfusion, and ischemia in the placenta (25). PE, together with other hypertensive pregnancy disorders, is consistently related to several markers of vascular injury (26). Insufficient or defective endometrial maturation can also cause PE with severe symptoms (27). Moreover, miRNAs have been demonstrated as remarkably potent factors involved in the process of maintaining homeostasis in many organs, including the placenta, by regulating a plethora of genes and participating in a variety of cellular events (28). The objective of the current study was to investigate the effects of miR-183 on trophoblast cell proliferation, invasion, and angiogenesis in PE. We thus tested our hypothesis that silencing miR-183 would play an inhibitory role in the progression of PE by elevating the expression of FOXP1 and downregulating GNG7 expression.
Initially, our findings revealed that FOXP1 was poorly expressed in PE samples and that overexpression of FOXP1 promoted proliferation, invasion, and angiogenesis of human chorionic trophoblastic cells (HTR-8/SVneo). Consistent with these findings, a previously reported study also demonstrated the crucial physiological roles of FOXP1 in macrophage function and monocyte differentiation (29). Similarly, FOXP1 was reported to be involved in the transcriptional network regulating B lymphopoiesis (30). The repression of enhanced expression of FOXP1 on cell proliferation and oxidative stress induced by high glucose treatment was also noted in a related study (11). FOXP1 was further implicated in the regulation of the transforming growth factor β1/transforming growth factor β receptor 1 (TGF-β1/TGF-βR1) in cardiac endothelial cells, and was shown to mediate the proliferation and transformation of cardiac fibroblasts into myofibroblasts during cardiac remodeling (12). In contrast, abnormal expression of miRNAs is known to be involved in the development of PE. Here, we found a high expression of miR-183 in PE while showing that downregulation of miR-183 promoted trophoblast cell proliferation, invasion, and angiogenesis by elevating VEGF via upregulation of FOXP1.
Consistent with present results, a previous study showed that high expression of miR-152, miR-183, and miR-210 exacerbated the onset of PE in the second trimester of pregnancies (23). Moreover, other research documented that miR-183 expression was upregulated following training with an elevated plus-maze in environment-enrichment mice but not in control mice (31). Accordingly, another relevant study demonstrated that restrained miR-183 expression could lead to an increase in FOXP1 expression, thus inactivating the Jak/Stat signaling pathway, promoting cell proliferation, and inhibiting the apoptosis of hippocampal neurons, by which means the injury of hippocampal neurons and progression of epilepsy were suppressed (13). Wan et al. showed that FOXP1 expression was closely related to VEGF expression in association with progression of renal clear cell carcinoma (32). VEGF also has a significant role in the abnormal vascularization of PE, such that low expression of VEGF was indicative of aggravated endothelial damage (33, 34). Our present results also showed that the overexpression of FOXP1 contributed to placental angiogenesis by increasing expression of VEGF, whereas si-FOXP1 induced opposite results in pregnant mice with PE-like symptoms.
GNG7 could serve as a potential predictor in independent cohorts, especially for the majority of emergency patients presenting with pain, while GNG7 levels also provided cross diagnostic evidence for certain psychiatric disorders (35). Moreover, the effects of GNG7 on the actin cytoskeleton, regulation of the mTOR pathway, inhibition on cell division and promotion on cell death, and autophagy were also reported in a previous study (36). The silencing of the GNG7 gene could reduce cell apoptosis and promote cell viability and differentiation of placental trophoblast cells through the activation of the mTOR signaling pathway in PE (8). Notably, silencing of GNG7 lowered apoptosis while facilitating proliferation and differentiation of placental cytotrophoblasts in rats with PE-like phenotypes through the activation of the mTOR signaling pathway (8). Moreover, downregulated FOXP1 reportedly mediates expression of multiple genes, including the CCR1, GNG7, and PLCB1 genes, which are prominently enriched in chemokine pathways, suggesting that chemokine pathways play a crucial role in FOXP1-mediated effects (9). These findings are partially inconsistent with our results, which suggested that FOXP1 could restrain the GNG7 expression to activate the mTOR signaling pathway and promote trophoblast cell proliferation, invasion, and angiogenesis.
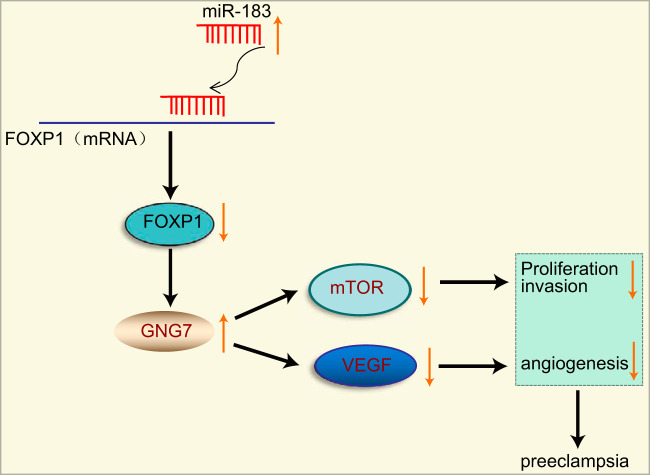
In conclusion, our study revealed that the overexpression of miR-183 could potentially upregulate GNG7 expression to inactivate the mTOR signaling pathway, thereby suppressing trophoblast cell proliferation, invasion, and angiogenesis and promoting PE progression by lowering FOXP1 expression (Fig. 8). Thus, a thorough investigation of the miR-183/FOXP1/GNG7 axis in PE and an investigation of associated gene networks are of great importance to understanding the pathophysiology of PE, and these inquiries have the potential to provide pioneering diagnostic and therapeutic approaches. Therefore, further studies are required to unveil other crucial factors involved in regulating these pathways, which can potentially aid the development of novel therapeutics for PE.
FIG 8.
Schematic of the regulatory network and function of miR-183 in PE. miR-183 targets FOXP1 and elevates the expression of GNG7 while inhibiting that of mTOR and VEGF, consequently impairing trophoblast cell proliferation, invasion, and angiogenesis and thus ultimately aggravating PE.
MATERIALS AND METHODS
Ethics statement.
The experimental design was approved by the Ethics Committee and Experimental Animal Ethics Committee of Second Xiangya Hospital, Central South University (no. 201907092). Written informed consent, including ethical agreements, were obtained from the donors or their relatives. The experimental procedures involving human beings were conducted in accordance with the principles outlined in the Declaration of Helsinki. All animal experiments were performed in strict agreement with the guidelines of animal care and use. Extensive efforts were made to ensure minimal suffering of the animals used in the study.
Microarray-based gene expression profiling.
The important regulation pathways and genes related to PE were selected according to existing research results. The upstream gene transcription factors were determined from the hTFtarget (http://bioinfo.life.hust.edu.cn/hTFtarget#!/) database and related literature. PE-related microarray data GSE6573 was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds). The expression data of selected transcription factors were extracted to draw box plots by R language. The upstream miRNAs of FOXP1 were predicted by the TargetScan database (context++ score < 0.3; weighted context++ score < 0.3) (http://www.targetscan.org/vert_71/), RNA Interactome Database (score < 0.9) (http://www.rna-society.org/raid2/index.html), miRDB (target score > 85) (http://www.mirdb.org/), and DIANA TOOLS (miTG score > 0.95) (http://diana.imis.athena-innovation.gr/DianaTools). Subsequently, the key miRNA was identified by intersection plotting and the target-binding site map of miRNA, while transcription factors were obtained from TargetScan.
Patient enrollment.
PE was diagnosed according to guidelines recommended by the American College of Obstetricians and Gynecologists. Placental tissues from patients with severe maternal complications and fetal malformations were excluded in this study. Twenty-four patients with PE and twenty-four healthy women in the third trimester of pregnancy were enrolled in this study. Placental tissue specimens (0.5 by 0.5 cm) were separated from the central part of the maternal side of the placenta immediately after delivery. The blood cells of tissues were then washed with sterile phosphate-buffered saline (PBS). Part of each specimen was fixed by immersion in 4% paraformaldehyde for 24 to 48 h and paraffin-embedded for IHC, while the remaining tissues were stored at −80°C for protein and RNA extraction.
Cell culture.
Human trophoblast HTR8/SVneo cells (CRL-3271; American Type Culture Collection, Manassas, VA) were cultured in 1640 medium supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA) and 1% penicillin and streptomycin (Invitrogen Inc., Carlsbad, CA). The HEK293A/17 SF cells (ACS-4500; American Type Culture Collection, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 1% penicillin and streptomycin (Invitrogen Inc., Carlsbad, CA) with 5% CO2 at 37°C. When growing at the logarithmic growth phase, the cells were trypsinized and seeded into 6-well plates at a density of 1 × 105 cells per well for 24 h of conventional culture. Upon reaching approximately 75% confluence, cells were transfected according to the instructions of the Lipofectamine 2000 reagent kit (Invitrogen Inc., Carlsbad, CA) with miR-NC (5′-TAACACGTCTATACGCCCA-3′), miR-183 inhibitor (5′-AGTGAATTCTACCAGTGCCAT-3′), si-NC (5′-CCTAAGGTTAAGTCGCCCTCG-3′), si-FOXP1 (5′-GCTAACACTAAACGAAATCTA-3′), oe-NC, oe-FOXP1 + oe-GNG7, or oe-GNG7. After 48 h, RT-qPCR and Western blot analysis were used for detection or subsequent functional experiments. These expression plasmids (miR-NC, miR-183 inhibitor, si-NC, si-FOXP1, oe-NC, oeFOXP1 + oe-GNG7, and oe-GNG7) were purchased from Shanghai GenePharma Co. Ltd. (Shanghai, China) with a concentration of 50 ng/ml.
IHC staining.
Paraffin-embedded sections were deparaffinized by xylene and hydrated with gradient ethanol. These sections were retrieved in citric acid solution in a pressure cooker and boiled for 1.5 min and then cooled to room temperature. Afterward, the sections were sealed with Tris-buffered saline solution containing 10% normal serum and 1% bovine serum albumin at room temperature for 2 h. The sections were then incubated with rabbit anti-FOXP1 (ab16645, 1:200; Abcam Inc., Cambridge, MA) and GNG7 (sc-377; 1:200; Santa Cruz Biotechnology Inc., Santa Cruz, CA) in normal serum as NC at 4°C overnight. Following overnight incubation, each section was further incubated for 20 min, this time with the addition of 50 μl of 3% H2O2 to eliminate the activity of endogenous peroxidase, and each section was then incubated at 37°C for 20 min with 50 μl polymer intensifier. Subsequently, the sections were further incubated with 50 μl secondary goat anti-rabbit immunoglobulin G (IgG) antibody (ab205718, 1:2,000; Abcam Inc., Cambridge, MA) at 37°C for 30 min. After incubation with secondary antibody, each section was added with 100 μl of freshly prepared diaminobenzidine tetrahydrochloride solution and observed under a microscope for 3 to 10 min, with a brown staining regarded as positive. Afterward, the sections were washed with distilled water, stained with hematoxylin, and dehydrated in an ascending series of alcohol. Finally, the sections were cleared and sealed with neutral resin and observed under a microscope. The experiment was conducted three times independently.
Western blot analysis.
Total protein in tissues and cells was extracted with radioimmunoprecipitation assay lysis buffer containing phenylmethylsulfonyl fluoride. The concentration of each protein sample was subsequently determined using a bicinchoninic acid kit (P0012; Beyotime Institute of Biotechnology, Shanghai, China). Protein (50 μg) was dissolved in 2× sodium dodecyl sulfate (SDS) sample loading buffer and boiled for 10 min. The protein lysates were separated by SDS polyacrylamide gel electrophoresis and transferred to the polyvinylidene fluoride (PVF) membrane under wet conditions. After being sealed in 5% skimmed milk for 1 h, the membrane was incubated with diluted primary rabbit anti-FOXP1 (ab16645, 1:5,000; Abcam Inc., Cambridge, MA), rabbit antibody to phosphorylated mTOR (p-mTOR) (ab109268, 1:10,000; Abcam Inc., Cambridge, MA), rabbit anti-mTOR (ab2732, 1:2,000; Abcam Inc., Cambridge, MA), mouse anti-VEGF (sc-7269, 1:2,000; Santa Cruz Biotechnology Inc., Santa Cruz, CA), rabbit anti-GNG7 (sc-377, 1:2,000; Santa Cruz Biotechnology Inc., Santa Cruz, CA), and mouse antibody to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ab8245, 1:10,000; Abcam Inc., Cambridge, MA) at 4°C overnight. Afterward, the membrane was washed three times with Tris-buffered saline with Tween (TBST). Following incubation with secondary goat anti-rabbit IgG (ab205718, 1:20,000, Abcam Inc., Cambridge, MA) and goat anti-mouse IgG (ab205719, 1:20,000; Abcam Inc., Cambridge, MA) at room temperature for 1 h, the proteins were visualized with enhanced chemiluminescence substrate (WBKLS0100; Merck Millipore, Billerica, MA). The gray value of Western blot protein analysis was determined by Image J software (National Institutes of Health, Bethesda, MD). The experiment was conducted three times independently.
Dual-luciferase reporter gene assay.
HEK293A/17 SF cells were used for dual-luciferase reporter gene assay. Synthetic FOXP1-3′-UTRW and FOXP1-3′-UTRM fragments were constructed in the pMIR-reporter plasmid. Next, FOXP1-3′-UTRW or FOXP1-3′-UTRM was transfected with miR-183 mimic or mimic NC into cells according to Lipofectamine 2000 kit instructions. After 24 h of culture at 37°C with 5% CO2, luciferase activity in each group was measured by the Dual-Luciferase reporter assay system (Promega Corp., Madison, WI). The relative luciferase activity was considered the ratio of relative light unit of firefly luciferase to that of renilla luciferase (internal reference). The luciferase activity was determined by considering the average from three replicates.
Transwell assay.
A Transwell filter was used for cell invasion assays (8 μm; Corning Incorporated, Corning, NY). The apical chamber of the basement membrane was coated with a matrix gel of 50 μl (BD Biosciences, San Jose, CA). Human trophoblast HTR8/SVneo cell line was obtained from the American Type Culture Collection (Manassas, VA) (CRL-3271). After 48 h of transfection with HTR8/SVneo cells with control or siRNA, the cells were detached into the single-cell suspension, rinsed with PBS, and placed in the apical chamber after undergoing centrifugation. Subsequently, the cells were added to the basolateral chamber with culture medium containing 20% fetal bovine serum at 5% CO2 at 37°C for 48 h. The cells that migrated to the basolateral chamber were fixed with 4% paraformaldehyde and stained with crystal violet. Finally, 5 representative fields were randomly selected for microscopic observation to obtain the total number of invasive cells (37). All experiments were conducted three times independently.
RNA isolation and quantitation.
Total RNA was extracted from tissues and cells using TRIzol reagents (15596018; Invitrogen Inc.) in strict accordance with the provided instructions and RNA concentration was measured. RNA was then reverse transcribed into cDNA using the ImProm-II reverse transcription system kit (Promega Corp., Madison, WI). After dilution with 12 μl of water, 1 μg of total RNA was incubated at 85°C for 5 min and cooled rapidly on ice for another 5 min. All the primers used in this study are shown in Table 2, with β-actin serving as the internal reference. The experiment was repeated three times independently.
TABLE 2.
Primer sequences for RT-qPCRa
| Target | Primer sequences |
|---|---|
| VEGF | F, 5′-TTCAGAGCGGAGAAAGCAT-3′; R, 5′-TAGTTCCCGAAACCCTGAG-3′ |
| mTOR | F, 5′-TTGGAGAACCAGCCCATAAGA-3′; R, 5′-ATGAGATGTCGCTTGCTTGATAA-3′ |
| VEGF | F, 5′-GGAGCAGAGATGTGGGAATGGA-3′; R, 5′-GCAAGGAGTGGGTCTCTAGGTCAA-3′ |
| GNG7 | F, 5′-CGTCTGACCTCATGAGCTACTGTGA-3′; R, 5′-CAAGGTTTCTTGTCCTTAAAGGGGTTC-3′ |
| FOXP1 | F, 5′-CTGAATCTGGTATCAAGTGTCACCCTCT-3′; R, 5′-GATTCGAGAATGGCCTGCCTGA-3′ |
| GAPDH | F, 5′-CCCACTCCTCCACCTTTGAC-3′; R, 5′-CATACCAGGAAATGAGCTTGACAA-3′ |
| miR-183 | F, 5′-CTATGGCACTGGTAGAATTCACT-3′; R, 5′-TCGTATCCAGTGCAGGGTC-3′ |
| U6 | F, 5′-CTCGCTTCGGCAGCACA-3′; R, 5′-AACGCTTCACGAATTTGCGT-3′ |
RT-qPCR, reverse transcription-quantitative PCR; F, forward; R, reverse; VEGF, vascular endothelial growth factor; GNG7, G protein subunit gamma 7; FOXP1, forkhead box P1; mTOR, mechanistic target of rapamycin kinase; miR-183, microRNA 183; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
CCK-8 assay.
The cells were cultured in 96-well plates with 5% CO2 at 37°C for 24 h. After culture at 0, 24, 48, and 72 h, cells in each group were added with 10 μl of CCK-8 solution (Beyotime Institute of Biotechnology) and further cultured for 4 h. The absorbance value was measured at 450 nm with a microplate reader. All experiments were conducted three times independently.
In vitro angiogenesis assay.
HTR8/SVneo cells were transfected with silencing or overexpression plasmids and the conditioned supernatant was collected and added to 96-well plates coated with matrix gel containing 1.5 × 104 human uterine microvascular endothelial cells (HUtMEC; c-12296; Promo Cell GmbH, Heidelberg, Germany). After 16 h of incubation, HUtMEC branches were observed with an optical microscope and photographed. The total number of branch points and branch tubes was calculated manually with the use of a photomicrograph. All experiments were conducted three times independently.
RNA pulldown assay.
DNA fragments containing FOXP1 full-length or NC sequences were amplified by PCR using primers containing t7 and cloned into plasmid vector GV394 (Shanghai Genechem Co., Ltd., Shanghai, China). The plasmids were linearized after restriction enzyme digestion using XhoI. The biotin-labeled RNA was then reverse transcribed using T7 RNA polymerase (TaKaRa Biomedical Technology, Kusatsu, Shiga, Japan) and biotin RNA labeling set (Roche Life Science, Basel, Switzerland). Following treatment by means of RNase-free DNase I (Roche) and RNeasy minikit (Qiagen, Dusseldorf, Germany), RNA was isolated for subsequent RT-qPCR. All experiments were conducted three times independently (38).
Animal model development.
CD-1 mice (aged 6 to 8 weeks, specific-pathogen-free grade) were purchased from Vital River Lab Animal Technology Co., Ltd. (Beijing, China). All mice were housed at a constant temperature of 25 to 26°C with a humidity of 70% under artificial 12-h/12-h light-dark cycles and regular UV disinfection with good ventilation, as well as free access to food and water for 1 week. Female mice (n = 30) were allowed to mate with fertile or vasectomized males (n = 15) at night, with 1 male mouse assigned to 2 female mice in a cage to increase the probability of a pregnancy. The formation of vaginal plugs served as indicators of successful mating. The morning of the vaginal plug formation was designated the 1st day of gestation or pseudopregnancy. The placenta was collected immediately after the mice were anesthetized and obtained tissues were rapidly frozen and stored at −80°C.
HIV-1-based vector plasmid pLV-EGFP has previously been reported (39), and other miR-NC or miR-183 inhibitors were prepared by substituting enhanced green fluorescent protein (EGFP) cDNA. Lentivirus packaging plasmids were transfected into HEK293A/17 SF cells and lentivirus particles were collected 2 days later. Afterward, the particles were centrifuged at 5,000 × g for 2 h and resuspended in potassium simplex optimization medium (KSOM) with the concentration adjusted to 8 × 106 TU/ml. Wild-type CD-1 female mice were intraperitoneally injected with pregnant mare serum gonadotrophin (5 units) and 48 h later injected with human chorionic gonadotropin (5 units) to stimulate ovulation, followed by mating with wild-type CD-1 males. After 4 days, the embryos of female blastocysts were collected and the zona pelluca was removed in acidic Tyrode solution (Sigma-Aldrich Chemical Company, St. Louis, MO). Blastocysts without zona pelluca were cultured in 5 μl medium containing lentivirus vector of miR-NC or miR-183 inhibitor, or both miR-183 inhibitor and rapamycin for 8 h. The transferred blastocysts were washed 3 times with KSOM, transplanted into a pseudopregnant CD-1 female, or cultured for 48 h and examined under a confocal microscope.
Statistical analysis.
All data were processed by SPSS 21.0 statistical software (IBM Corp., Armonk, NY). Measurement data were presented as mean ± standard deviation. Data between normal and model mice with normal distribution and homogeneity of variance were compared using an unpaired t test. Data among multiple groups were analyzed by one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test. Data of cell activity among multiple groups at different time points were measured by two-way ANOVA followed by Bonferroni's post hoc test. The relationship between the two indicators was identified by Pearson correlation analysis. A value of P <0.05 was considered statistically significant.
Data availability.
The data sets generated/analyzed during the current study are available upon request.
ACKNOWLEDGMENTS
We acknowledge the helpful comments on this paper received from the reviewers.
Ling Yu and Weisi Lai designed the study. Ling Yu and Weisi Lai collated the data, carried out data analyses, and produced the initial draft of the manuscript. Ling Yu contributed to drafting the manuscript. Both of us read and approved the final submitted manuscript.
We declare no conflicts of interest.
REFERENCES
- 1.Liu J, Zhuang T, Pi J, Chen X, Zhang Q, Li Y, Wang H, Shen Y, Tomlinson B, Chan P, Yu Z, Cheng Y, Zheng X, Reilly M, Morrisey E, Zhang L, Liu Z, Zhang Y. 2019. Endothelial forkhead box transcription factor P1 regulates pathological cardiac remodeling through transforming growth factor-beta1-endothelin-1 signal pathway. Circulation 140:665–680. doi: 10.1161/CIRCULATIONAHA.119.039767. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal S, Shinar S, Cerdeira AS, Redman C, Vatish M. 2019. Predictive performance of PlGF (placental growth factor) for screening preeclampsia in asymptomatic women: a systematic review and meta-analysis. Hypertension 74:1124–1135. doi: 10.1161/HYPERTENSIONAHA.119.13360. [DOI] [PubMed] [Google Scholar]
- 3.Zhou C, Yan Q, Zou QY, Zhong XQ, Tyler CT, Magness RR, Bird IM, Zheng J. 2019. Sexual dimorphisms of preeclampsia-dysregulated transcriptomic profiles and cell function in fetal endothelial cells. Hypertension 74:154–163. doi: 10.1161/HYPERTENSIONAHA.118.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaufmann P, Black S, Huppertz B. 2003. Endovascular trophoblast invasion: implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol Reprod 69:1–7. doi: 10.1095/biolreprod.102.014977. [DOI] [PubMed] [Google Scholar]
- 5.Kvehaugen AS, Melien O, Holmen OL, Laivuori H, Oian P, Andersgaard AB, Dechend R, Staff AC. 2013. Single nucleotide polymorphisms in G protein signaling pathway genes in preeclampsia. Hypertension 61:655–661. doi: 10.1161/HYPERTENSIONAHA.111.00331. [DOI] [PubMed] [Google Scholar]
- 6.Schwindinger WF, Mirshahi UL, Baylor KA, Sheridan KM, Stauffer AM, Usefof S, Stecker MM, Mirshahi T, Robishaw JD. 2012. Synergistic roles for G-protein gamma3 and gamma7 subtypes in seizure susceptibility as revealed in double knock-out mice. J Biol Chem 287:7121–7133. doi: 10.1074/jbc.M111.308395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou QY, Zhao YJ, Zhou C, Liu AX, Zhong XQ, Yan Q, Li Y, Yi FX, Bird IM, Zheng J. 2019. G protein alpha subunit 14 mediates fibroblast growth factor 2-induced cellular responses in human endothelial cells. J Cell Physiol 234:10184–10195. doi: 10.1002/jcp.27688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai WS, Ding YL. 2019. GNG7 silencing promotes the proliferation and differentiation of placental cytotrophoblasts in preeclampsia rats through activation of the mTOR signaling pathway. Int J Mol Med 43:1939–1950. doi: 10.3892/ijmm.2019.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheng H, Li X, Xu Y. 2019. Knockdown of FOXP1 promotes the development of lung adenocarcinoma. Cancer Biol Ther 20:537–545. doi: 10.1080/15384047.2018.1537999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braccioli L, Vervoort SJ, Adolfs Y, Heijnen CJ, Basak O, Pasterkamp RJ, Nijboer CH, Coffer PJ. 2017. FOXP1 promotes embryonic neural stem cell differentiation by repressing Jagged1 expression. Stem Cell Rep 9:1530–1545. doi: 10.1016/j.stemcr.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang H, Xue W, Wu X, Zheng J, Ding C, Li Y, Dou M. 2019. FOXP1 inhibits high glucose-induced ECM accumulation and oxidative stress in mesangial cells. Chem Biol Interact 313:108818. doi: 10.1016/j.cbi.2019.108818. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Zhang M, Guallar E, Wang G, Hong X, Wang X, Mueller NT. 2019. Trace minerals, heavy metals, and preeclampsia: findings from the Boston Birth Cohort. J Am Heart Assoc 8:e012436. doi: 10.1161/JAHA.119.012436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng X, Xiong W, Yuan M, Zhan J, Zhu X, Wei Z, Chen X, Cheng X. 2019. Down-regulated microRNA-183 mediates the Jak/Stat signaling pathway to attenuate hippocampal neuron injury in epilepsy rats by targeting Foxp1. Cell Cycle 18:3206–3222. doi: 10.1080/15384101.2019.1671717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frazier S, McBride MW, Mulvana H, Graham D. 2020. From animal models to patients: the role of placental microRNAs, miR-210, miR-126, and miR-148a/152 in preeclampsia. Clin Sci (Lond) 134:1001–1025. doi: 10.1042/CS20200023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemmatzadeh M, Shomali N, Yousefzadeh Y, Mohammadi H, Ghasemzadeh A, Yousefi M. 2020. MicroRNAs: small molecules with a large impact on pre-eclampsia. J Cell Physiol 235:3235–3248. doi: 10.1002/jcp.29286. [DOI] [PubMed] [Google Scholar]
- 16.Cronqvist T, Tannetta D, Morgelin M, Belting M, Sargent I, Familari M, Hansson SR. 2017. Syncytiotrophoblast derived extracellular vesicles transfer functional placental miRNAs to primary human endothelial cells. Sci Rep 7:4558. doi: 10.1038/s41598-017-04468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagana AS, Vitale SG, Sapia F, Valenti G, Corrado F, Padula F, Rapisarda AMC, D'Anna R. 2018. miRNA expression for early diagnosis of preeclampsia onset: hope or hype? J Matern Fetal Neonatal Med 31:817–821. doi: 10.1080/14767058.2017.1296426. [DOI] [PubMed] [Google Scholar]
- 18.Mundim GJ, Paschoini MC, Araujo Junior E, Da Silva Costa F, Rodrigues JV. 2016. Assessment of angiogenesis modulators in pregnant women with pre-eclampsia: a case-control study. Arch Gynecol Obstet 293:369–375. doi: 10.1007/s00404-015-3823-x. [DOI] [PubMed] [Google Scholar]
- 19.Cerdeira AS, Agrawal S, Staff AC, Redman CW, Vatish M. 2018. Angiogenic factors: potential to change clinical practice in pre-eclampsia? BJOG 125:1389–1395. doi: 10.1111/1471-0528.15042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi Z, She K, Li H, Yuan X, Han X, Wang Y. 2019. MicroRNA-454 contributes to sustaining the proliferation and invasion of trophoblast cells through inhibiting Nodal/ALK7 signaling in pre-eclampsia. Chem Biol Interact 298:8–14. doi: 10.1016/j.cbi.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Gao Y, She R, Wang Q, Li Y, Zhang H. 2018. Up-regulation of miR-299 suppressed the invasion and migration of HTR-8/SVneo trophoblast cells partly via targeting HDAC2 in pre-eclampsia. Biomed Pharmacother 97:1222–1228. doi: 10.1016/j.biopha.2017.11.053. [DOI] [PubMed] [Google Scholar]
- 22.Jiang L, Long A, Tan L, Hong M, Wu J, Cai L, Li Q. 2017. Elevated microRNA-520g in pre-eclampsia inhibits migration and invasion of trophoblasts. Placenta 51:70–75. doi: 10.1016/j.placenta.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, Long A, Jiang L, Cai L, Xie LI, Gu J, Chen X, Tan L. 2015. Quantification of preeclampsia-related microRNAs in maternal serum. Biomed Rep 3:792–796. doi: 10.3892/br.2015.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han C, Wang C, Chen Y, Wang J, Xu X, Hilton T, Cai W, Zhao Z, Wu Y, Li K, Houck K, Liu L, Sood AK, Wu X, Xue F, Li M, Dong JF, Zhang J. 2020. Placenta-derived extracellular vesicles induce preeclampsia in mouse models. Haematologica 105:1686–1694. doi: 10.3324/haematol.2019.226209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zen M, Padmanabhan S, Zhang K, Kirby A, Cheung NW, Lee VW, Alahakoon TI. 2020. Urinary and serum angiogenic markers in women with preexisting diabetes during pregnancy and their role in preeclampsia prediction. Diabetes Care 43:67–73. doi: 10.2337/dc19-0967. [DOI] [PubMed] [Google Scholar]
- 26.Amor AJ, Vinagre I, Valverde M, Pane A, Urquizu X, Meler E, Lopez E, Quiros C, Gimenez M, Codina L, Conget I, Barahona MJ, Perea V. 2020. Preeclampsia is associated with increased preclinical carotid atherosclerosis in women with type 1 diabetes. J Clin Endocrinol Metab 105:dgz031. doi: 10.1210/clinem/dgz031. [DOI] [PubMed] [Google Scholar]
- 27.Rabaglino MB, Conrad KP. 2019. Evidence for shared molecular pathways of dysregulated decidualization in preeclampsia and endometrial disorders revealed by microarray data integration. FASEB J 33:11682–11695. doi: 10.1096/fj.201900662R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu P, Zhao Y, Liu M, Wang Y, Wang H, Li YX, Zhu X, Yao Y, Wang H, Qiao J, Ji L, Wang YL. 2014. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension 63:1276–1284. doi: 10.1161/HYPERTENSIONAHA.113.02647. [DOI] [PubMed] [Google Scholar]
- 29.Shi C, Sakuma M, Mooroka T, Liscoe A, Gao H, Croce KJ, Sharma A, Kaplan D, Greaves DR, Wang Y, Simon DI. 2008. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood 112:4699–4711. doi: 10.1182/blood-2008-01-137018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, Tucker PW, Rao A. 2006. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol 7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- 31.Ragu Varman D, Marimuthu G, Rajan KE. 2013. Environmental enrichment upregulates micro-RNA-183 and alters acetylcholinesterase splice variants to reduce anxiety-like behavior in the little Indian field mouse (Mus booduga). J Neurosci Res 91:426–435. doi: 10.1002/jnr.23165. [DOI] [PubMed] [Google Scholar]
- 32.Wan L, Huang J, Chen J, Wang R, Dong C, Lu S, Wu X. 2015. Expression and significance of FOXP1, HIF-1a and VEGF in renal clear cell carcinoma. J BUON 20:188–195. [PubMed] [Google Scholar]
- 33.Ali LE, Salih MM, Elhassan EM, Mohmmed AA, Adam I. 2019. Placental growth factor, vascular endothelial growth factor, and hypoxia-inducible factor-1alpha in the placentas of women with pre-eclampsia. J Matern Fetal Neonatal Med 32:2628–2632. doi: 10.1080/14767058.2018.1443066. [DOI] [PubMed] [Google Scholar]
- 34.Xu X, Yang XY, He BW, Yang WJ, Cheng WW. 2016. Placental NRP1 and VEGF expression in pre-eclamptic women and in a homocysteine-treated mouse model of pre-eclampsia. Eur J Obstet Gynecol Reprod Biol 196:69–75. doi: 10.1016/j.ejogrb.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Niculescu AB, Le-Niculescu H, Levey DF, Roseberry K, Soe KC, Rogers J, Khan F, Jones T, Judd S, McCormick MA, Wessel AR, Williams A, Kurian SM, White FA. 2019. Towards precision medicine for pain: diagnostic biomarkers and repurposed drugs. Mol Psychiatry 24:501–522. doi: 10.1038/s41380-018-0345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Ji X, Li Z, Yang X, Wang W, Zhang X. 2016. G protein gamma subunit 7 induces autophagy and inhibits cell division. Oncotarget 7:24832–24847. doi: 10.18632/oncotarget.8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He B, Yang X, Li Y, Huang D, Xu X, Yang W, Dai Y, Zhang H, Chen Z, Cheng W. 2018. TLR9 (Toll-like receptor 9) agonist suppresses angiogenesis by differentially regulating VEGFA (vascular endothelial growth factor A) and sFLT1 (soluble vascular endothelial growth factor receptor 1) in preeclampsia. Hypertension 71:671–680. doi: 10.1161/HYPERTENSIONAHA.117.10510. [DOI] [PubMed] [Google Scholar]
- 38.Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, Lu Z, Bai C, Watabe K, Mo YY. 2013. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res 23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada Y, Ueshin Y, Isotani A, Saito-Fujita T, Nakashima H, Kimura K, Mizoguchi A, Oh-Hora M, Mori Y, Ogata M, Oshima RG, Okabe M, Ikawa M. 2007. Complementation of placental defects and embryonic lethality by trophoblast-specific lentiviral gene transfer. Nat Biotechnol 25:233–237. doi: 10.1038/nbt1280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated/analyzed during the current study are available upon request.