Mitochondrial dysfunction is one of the major pathological attributes of cardiac hypertrophy and is associated with reduced expression of PGC1α in cardiomyocytes. However, the transcriptional regulation of PGC1α remains elusive. Here, we show that parkin interacting substrate (PARIS), a KRAB zinc finger protein, prevented PGC1α transcription despite the induction of cardiomyocytes with hypertrophic stimuli. Moreover, PARIS expression and its nuclear localization are enhanced in hypertrophy both in vitro and in vivo.
KEYWORDS: cardiac hypertrophy, mitochondrial dysfunction, oxidative stress, SUMOylation, mitochondria, transcriptional repression
ABSTRACT
Mitochondrial dysfunction is one of the major pathological attributes of cardiac hypertrophy and is associated with reduced expression of PGC1α in cardiomyocytes. However, the transcriptional regulation of PGC1α remains elusive. Here, we show that parkin interacting substrate (PARIS), a KRAB zinc finger protein, prevented PGC1α transcription despite the induction of cardiomyocytes with hypertrophic stimuli. Moreover, PARIS expression and its nuclear localization are enhanced in hypertrophy both in vitro and in vivo. Knocking down PARIS resulted in mitochondrial biogenesis and improved respiration and other biochemical features that were compromised during hypertrophy. Furthermore, a PARIS-dependent proteome showed exclusive binding of a deSUMOylating protein called DJ-1 to PARIS in control cells, while this interaction is completely abrogated in hypertrophied cells. We further demonstrate that proteasomal degradation of DJ-1 under oxidative stress led to augmented PARIS SUMOylation and consequent repression of PGC1α promoter activity. SUMOylation-resistant mutants of PARIS failed to repress PGC1α, suggesting a critical role for PARIS SUMOylation in hypertrophy. The present study, therefore, proposes a novel regulatory pathway where DJ-1 acts as an oxidative stress sensor and contributes to the feedback loop governing PARIS-mediated mitochondrial function.
INTRODUCTION
A robust adaptive system usually helps cells to withstand a wide variety of stresses brought about by morphological and or molecular adjustments in the living system. Hypertrophy of cardiomyocytes is an example of one such adaptation by which the myocardium compensates for increased workload due to various kinds of stimuli, including hypertension, ischemia, or valvular diseases. However, prolonged hypertrophic stimulation leads to a pathological condition and subsequent heart failure. At the cellular level, the mitochondrion is one of the major organelles which bear the impact of all kinds of stresses. In cardiomyocytes, mitochondria undergo extensive remodeling accompanied by significant biochemical alterations. Myocyte hypertrophy is associated with impaired mitochondrial biogenesis and mitophagy, leading to a cellular energy imbalance and oxidative stress (1–3). Previous studies have shown that recovery of mitochondrial function in diseased myocardium can potentially alleviate hypertrophic manifestations, although the precise cellular mechanism remains unknown (4).
Peroxisome proliferator-activated receptor gamma coactivator 1α (PGC1α) is one of the principal regulators of mitochondrial functions as well as biogenesis, evidenced by mitochondrial abnormalities in PGC1α knockout mice (5, 6). PGC1α coactivates transcription factors such as nuclear respiratory factors 1 and 2 (NRF1 and NRF2) and mitochondrial transcriptional factor A (TFAM), to name a few, which are also important for maintaining mitochondrial physiology (7–10). PGC1α also acts as a transcriptional coactivator for several nuclear receptors, like the peroxisome proliferator-activated receptor (PPAR) family of transcription factors and estrogen-related receptor α (ERRα), thereby orchestrating several metabolic pathways (11). Downregulation of PGC1α is associated with cardiomyocyte hypertrophy. While several studies have successfully addressed the mechanism of PGC1α inactivation (12), transcriptional regulation of PGC1α during hypertrophy remains less explored. Therefore, the overall objective of this study was to investigate the plausible mechanism of transcriptional regulation of PGC1α in the context of cardiac hypertrophy.
Parkin interacting substrate (PARIS) is a zinc finger transcriptional repressor containing four zinc finger domains and one KRAB domain. The zinc finger domains regulate sequence-specific DNA binding in the promoter region of target genes, while the KRAB domain mediates transcriptional repressor activity (13, 14). Parkinson’s disease is closely associated with left ventricular hypertrophy (15). PARIS was reported as a transcriptional repressor of PGC1α in Parkinson's disease model (13). However, the role of PARIS in cardiac hypertrophy still remains unexplored.
Previous studies have shown that SUMOylation is one of the major posttranslational modifications that control PARIS activity (16). SUMOylation is a reversible process which enzymatically attaches small ubiquitin-like modifier (SUMO) to a lysine residue (17–19) in the substrate protein at a conserved SUMO motif, ψKXD/E, where ψ is any large hydrophobic residue, K is lysine, X can be any amino acid residue, and D/E represents aspartate or glutamate (20). PARIS is known to be SUMOylated at amino acid residues K189 and K286 in neurons, leading to transcriptional repression of PGC1α gene expression (14). Therefore, understanding the functional significance of the SUMOylation status of PARIS in hypertrophy might reveal a novel mechanism of regulation of mitochondrial function in myocytes.
In the present study, we report a novel role of PARIS in the regulation of mitochondrial functions in hypertrophied myocytes. It reveals a new molecular mechanism that controls PGC1α transcription by regulating the SUMOylation status of PARIS, which eventually determines the fate of mitochondria and other hypertrophic manifestations in cardiomyocytes.
RESULTS
Enhanced expression of PARIS in hypertrophied cardiomyocytes.
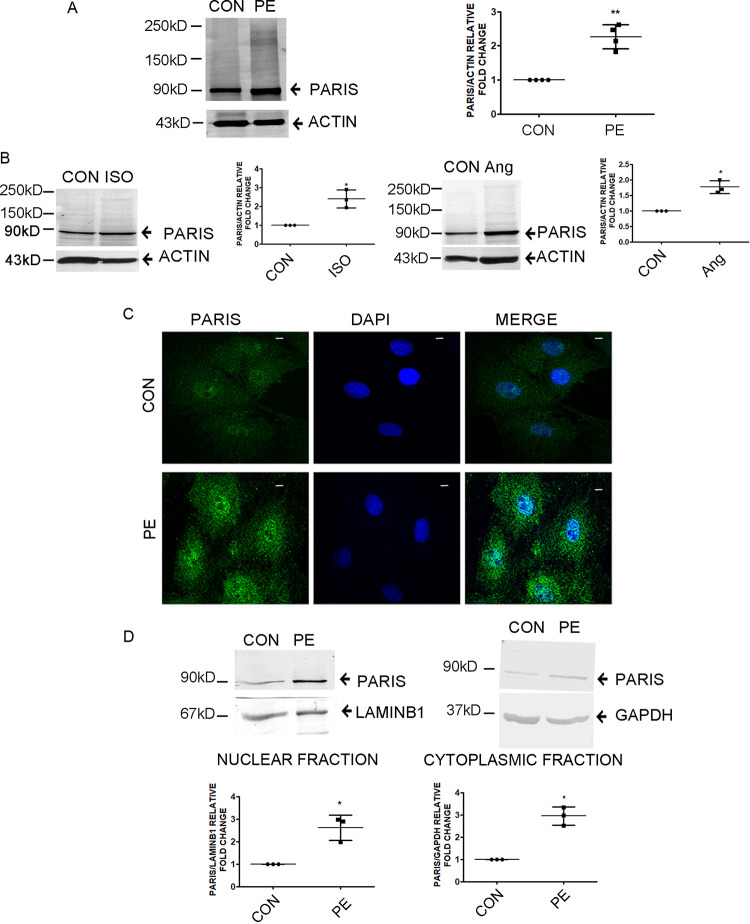
To investigate the role of PARIS in hypertrophy, the expression of PARIS was assessed by Western blot analysis in phenylephrine (PE)-treated H9C2 cardiomyocytes. Hypertrophied cardiomyocytes showed a significant increase in PARIS protein expression in comparison to control cells (Fig. 1A). PARIS protein levels were also monitored in two different in vitro models of hypertrophy. Both isoproterenol and angiotensin II (AngII)-induced hypertrophied myocytes showed elevated levels of PARIS compared to respective controls (Fig. 1B). Immunofluorescence analysis also confirmed increased expression of PARIS with enhanced nuclear localization in PE-treated H9C2 cardiomyocytes compared to controls (Fig. 1C). Immunoblotting of subcellular fractionations showed increased nuclear localization of PARIS in hypertrophied myocytes compared to controls (Fig. 1D).
FIG 1.
Enhanced PARIS expression in hypertrophied myocytes. (A and B) Immunoblots showing increased expression of PARIS in H9C2 cardiomyocytes after treatment with the hypertrophic agent PE (A) or with ISO or Ang II (B). Actin was used as an internal loading control. n = 3 or 4 for each group from 3 independent experiments. **, P < 0.01, and *, P < 0.05 compared to controls. (C) Confocal micrographs showing increased PARIS expression (green) in hypertrophied myocytes. DAPI (4′,6-diamidino-2-phenylindole) was used to counterstain the nucleus. Bars, 10 μm. (D) Immunoblot of PARIS expression in subcellular fractions in PE-treated H9C2 myocytes compared to controls. Lamin B1 and GAPDH were used as loading controls for nuclear and cytosolic fractions, respectively. The data represent 3 independent experiments. *, P < 0.05 compared to controls.
PARIS regulates cardiomyocyte hypertrophy.
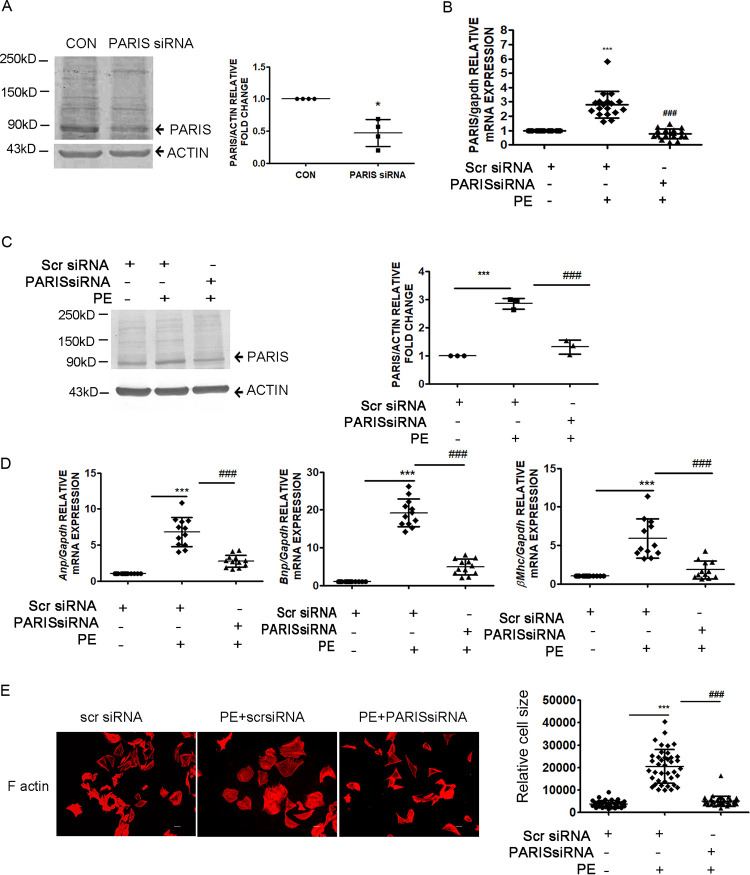
H9C2 cardiomyocytes were subjected to small interfering RNA (siRNA)-mediated knockdown of PARIS to evaluate its role in hypertrophy. Successful knockdown of PARIS expression both at baseline (Fig. 2A) and in PE-treated hypertrophied myocytes (Fig. 2C) was confirmed by decreased protein expression as well as reduced mRNA expression (Fig. 2B). Silencing PARIS in hypertrophied cardiomyocytes considerably reduced the expression of hypertrophic marker genes such as Anp, Bnp, and βMhc (21, 22) compared to PE-treated cells (Fig. 2D). Moreover, PARIS knockdown prevented PE-induced enlargement of cardiomyocytes, as shown by cell size measurement (Fig. 2E), pointing to the involvement of PARIS in hypertrophied manifestations.
FIG 2.
Regulation of hypertrophic manifestations by PARIS. (A to C) Validation of PARIS knockdown in H9C2 cardiomyocytes, as shown by decreased protein expression at baseline (A) and in the presence of PE (C) as well as decreased mRNA expression (B). Actin and GAPDH were used as internal loading controls for immunoblotting and qRT-PCR, respectively. Graphical representation of decreased PARIS expression. n = 3 or 4 for each group from 3 independent experiments. ***, P < 0.001, and *, P < 0.05 compared to controls; ###, P < 0.001 compared to PE treatment for Western blot analysis. n = 12 for each group from 4 independent experiments for qRT-PCR. ***, P < 0.001 compared to controls; ###, P < 0.001 compared to PE treatment. (D) qRT-PCR showing downregulated expression of hypertrophic marker genes Anp, Bnp, and βMhc in PARIS siRNA-treated hypertrophied myocytes. Gapdh expression was used as an internal loading control. n = 12 for each group from 4 independent experiments; ***, P < 0.001 compared to controls; ###, P < 0.001 compared to PE treatment. (E) Immunostaining of F-actin with Alexa Fluor-phalloidin in H9C2 myocytes. Bar, 10 μm. Graphical representation showing significantly decreased cell size in PARIS siRNA-treated hypertrophied myocytes, ***, P < 0.001 compared to controls; ###, P < 0.001 compared to PE treatment. n = 50 cells.
PARIS mediates mitochondrial dysfunction in hypertrophied myocytes.
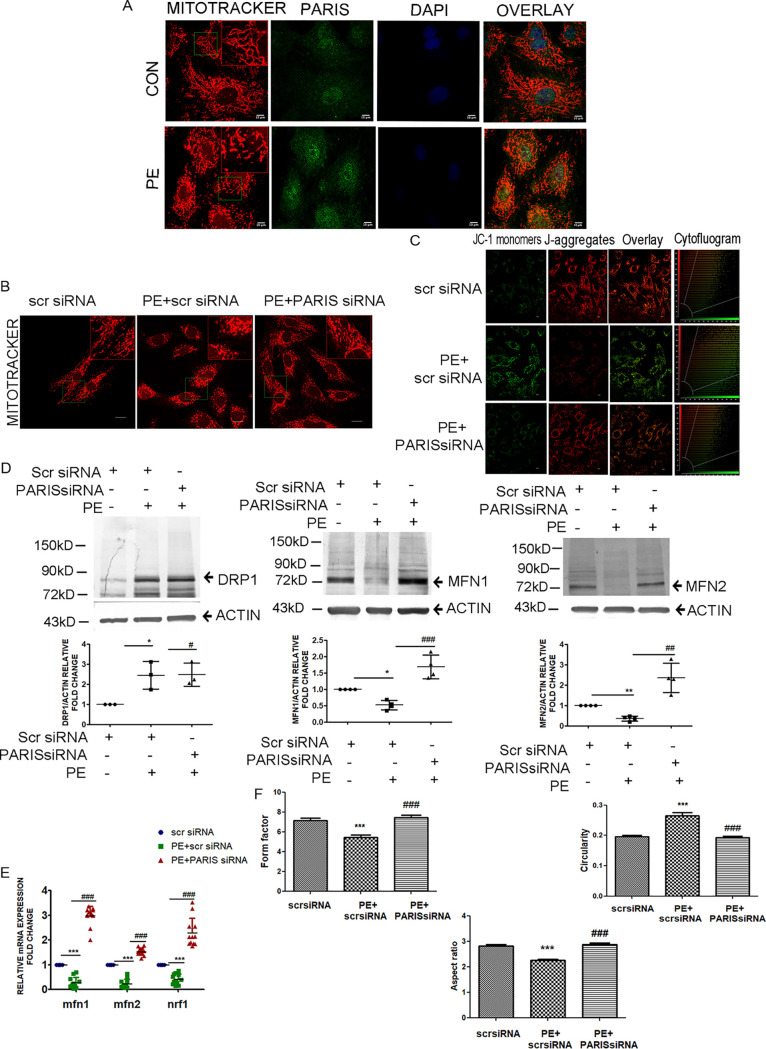
(i) PARIS regulates mitochondrial dynamics. Cardiac hypertrophy is associated with mitochondrial dysfunction (3). Therefore, the effect of PARIS knockdown on mitochondrial biology was studied in PE-treated H9C2 cardiomyocytes. PE-induced mitochondrial fragmentation was associated with increased PARIS expression (Fig. 3A). PARIS knockdown in hypertrophied H9C2 myocytes showed an overall improvement in mitochondrial function compared to PE treatment. Mitochondrial morphology was assessed by staining with MitoTracker red. Knocking down PARIS restored PE-induced mitochondrial fragmentation in hypertrophied cells (Fig. 3B) showing additional fused, elongated, and connected networks, indicating causal involvement of the protein with mitochondrial morphology.
FIG 3.
Regulation of mitochondrial dynamics by PARIS in hypertrophied myocytes. (A) Confocal micrographs showing fragmented mitochondria (red) and increased PARIS expression (green) in hypertrophied myocytes. DAPI was used to counterstain nuclei. Bars, 10 μm. n = 6 for each group. (Inset) Magnified view (×2) of the region marked by the green box. (B) MitoTracker red staining of cells transfected with either scrambled siRNA or PARIS siRNA in the presence or absence of PE treatment showing improved mitochondrial morphology in the PARIS siRNA-treated group. Bars, 20 μm. (Inset) Magnified view (×2) of the region marked by the green box. (C) Confocal micrograph of JC-1 staining displaying mitochondria with green fluorescence for compromised ΔΨm and red fluorescence for positive ΔΨm. Cytofluorogram demonstrating the red-to-green ratio in different treatment groups. Bars, 10 μm. (D) Immunoblot showing increased MFN1 and MFN2 expression with no significant change of DRP1 expression in PARIS siRNA treatment. Actin was used as an internal loading control. Graphical representation of immunoblot data showing the amount of change in expression of DRP1, MFN2, and MFN1 protein in PARIS knockdown hypertrophied myocytes. n = 3 or 4 for each group. *, P < 0.05, and **, P < 0.01 compared to controls. ##, P < 0.01, and ###, P < 0.001 compared to PE. (E) qRT-PCR showing significantly increased expression of Mfn1, Mfn2, and Nrf1 in PARIS siRNA treatment. Gapdh expression was used as the loading control. n = 12 for each group from 4 independent experiments. ***, P < 0.001 compared to controls; ###, P < 0.001 compared to PE treatment. (F) Assessment of mitochondrial morphology showing increased mitochondrial interconnectivity, represented by form factor, reduced circularity, and increased aspect ratio in the cells treated with PARIS siRNA compared to PE-treated group. n = 40 for each group from 3 independent experiments; ***, P < 0.001 compared to controls; ###, P < 0.001 compared to PE treatment.
Mitochondrial membrane potential and mitochondrial morphology are interconnected. To evaluate whether PARIS is involved in the loss of mitochondrial health, mitochondrial membrane potential (ΔΨm) was measured by staining mitochondria with the potential-dependent dye JC1. PARIS knockdown in hypertrophied myocytes showed an increase in ΔΨm (red versus green fluorescence) compared to PE treatment, signifying improved mitochondrial health by inhibiting PARIS expression in hypertrophied cells (Fig. 3C).
A dynamic balance between mitochondrial fusion and fission is important for maintaining homeostasis. PE treatment directs this balance toward more fission cycles, thus leading to mitochondrial dysfunction. To analyze the effect of PARIS inhibition on mitochondrial dynamics, expression of mitochondrial fusion (MFN1 and MFN2) and fission (DRP1) proteins was evaluated. Immunoblotting revealed elevated protein expression of MFN1 and MFN2; however, no significant change in the expression of DRP1 was observed by inhibiting PARIS in hypertrophied myocytes compared to PE treatment (Fig. 3D). A significant increase in the expression of Mfn1, Mfn2, and the mitochondrial protein gene Nrf1 (Fig. 3E) further suggested that the role of PARIS is as a negative regulator of mitochondrial dynamics.
Moreover, to confirm the PARIS-dependent regulation of mitochondrial fusion and fission, mitochondrial network connectedness was assessed. The average mitochondrial interconnectivity as shown by form factor was found to be increased, mitochondrial circularity (an indicator of roundness) was reduced, and the aspect ratio (the ratio between the major and minor axes of the mitochondria) was significantly increased by knocking down PARIS in hypertrophied myocytes compared to PE treatment (Fig. 3F), correlating with increased MFN1/2 protein expression.
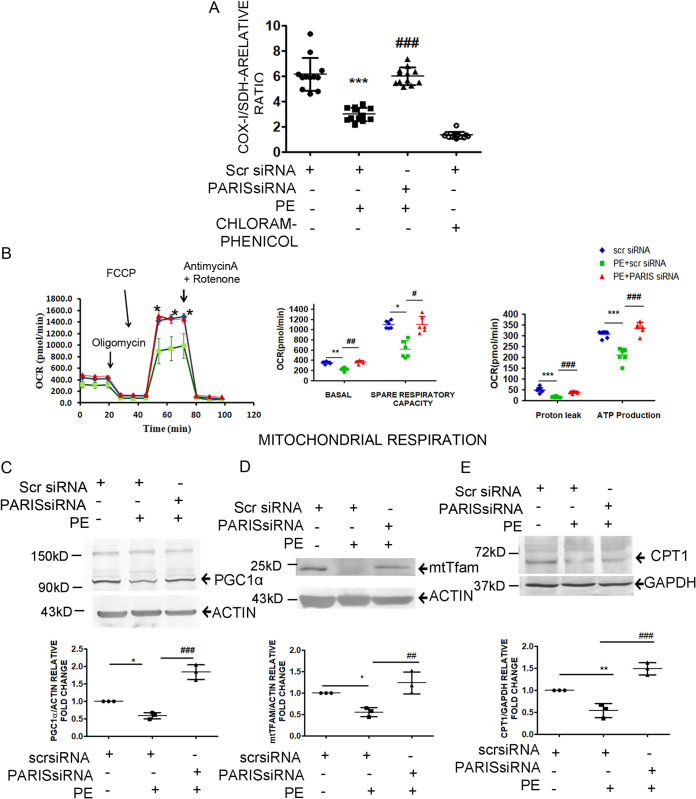
(ii) PARIS regulates mitochondrial bioenergetics. To further validate the regulation of mitochondrial biology by PARIS, mitochondrial biogenesis was assessed in PARIS knockdown hypertrophied cardiomyocytes. Mitochondrial biogenesis was measured using the ratio of mitochondrial DNA (mtDNA)-encoded COX-I to nuclear DNA (nDNA)-encoded SDH-A. Knocking down PARIS in hypertrophied H9C2 cardiomyocytes significantly enhanced mitobiogenesis compared to PE treatment, implying enhanced synthesis of new mitochondria to compensate for the loss of damaged mitochondria in myocyte, therefore supporting the role of PARIS in mitochondrial dysfunction (Fig. 4A).
FIG 4.
Regulation of mitochondrial functions by PARIS in hypertrophied myocytes. (A) Measurement of mitochondrial biogenesis using MitoBiogenesis in-cell ELISA by measuring the ratio of mtDNA-encoded COX-I and nDNA-encoded SDH-A levels showing increased mitobiogenesis in PARIS siRNA treatment. n = 12 for each group from 4 independent experiments. ***, P < 0.001 compared to controls; ###, P < 0.001 compared to PE treatment. (B) Analysis of OCR using a Seahorse XFe24 metabolic analyzer. Oligomycin, FCCP, and rotenone combined with antimycin A were added sequentially and showed significantly increased oxygen consumption in PARIS siRNA-treated hypertrophied myocytes. Graphical representation of the OCR kinetics analysis showing reduced OCR in several stages of respiration in PE-treated cardiomyocytes, including basal ATP-linked, proton leak nonmitochondrial respiration and maximal respiration, which was restored by knocking down PARIS in hypertrophied cardiomyocytes. n = 6 for each group from 3 independent experiments. *, P < 0.05, **, P < 0.01, and ***, P < 0.001 compared to controls; #, P < 0.05, ##, P < 0.01, and ###, P < 0.001 compared to PE treatment. Immunoblots showing the change in expression of PGC1α (C) and downstream genes such as mtTFAM (D) and CPT1 (E) genes in PARIS siRNA-treated hypertrophied myocytes. Actin was the internal loading control for PGC1α and mtTFAM, and GAPDH was the internal loading control for CPT1. Graphical representation of immunoblot data showing amount of change in expression of PGC1α, mtTFAM, and CPT1 proteins in PARIS knockdown hypertrophied myocytes. The data represent 3 independent experiments. *, P < 0.05, and **, P < 0.01 compared to controls. ##, P < 0.01, and ###, P < 0.001 compared to PE.
To confirm the role of PARIS in mitochondrial bioenergetics, mitochondrial stress was studied with a Seahorse XFe flux analyzer. PARIS-deficient hypertrophied cardiomyocytes showed a significantly increased oxygen consumption rate (OCR), ATP turnover, maximal respiration, and spare mitochondrial capacity compared to those subjected to PE treatment, signifying recovery of impaired mitochondrial function in the PARIS knockdown hypertrophied myocytes (Fig. 4B).
To investigate whether PARIS has any role in the downregulation of PGC1α in hypertrophied myocytes, PGC1α protein expression was observed in PARIS-silenced PE-treated cells. siRNA-mediated knockdown of PARIS prevented PE induced suppression of PGC1α protein expression as revealed by immunoblotting (Fig. 4C). This phenomenon was also associated with a significant increase in mitochondrial transcription factor TFAM (Fig. 4D) and carnitine palmitoyltransferase I (CPTI) (Fig. 4E) expression in PARIS knockdown hypertrophied cells. Therefore, all these results established the potential role of PARIS in initiating mitochondrial dysfunction during myocyte hypertrophy.
Transcriptional repression of PGC1α by PARIS in cardiomyocytes.
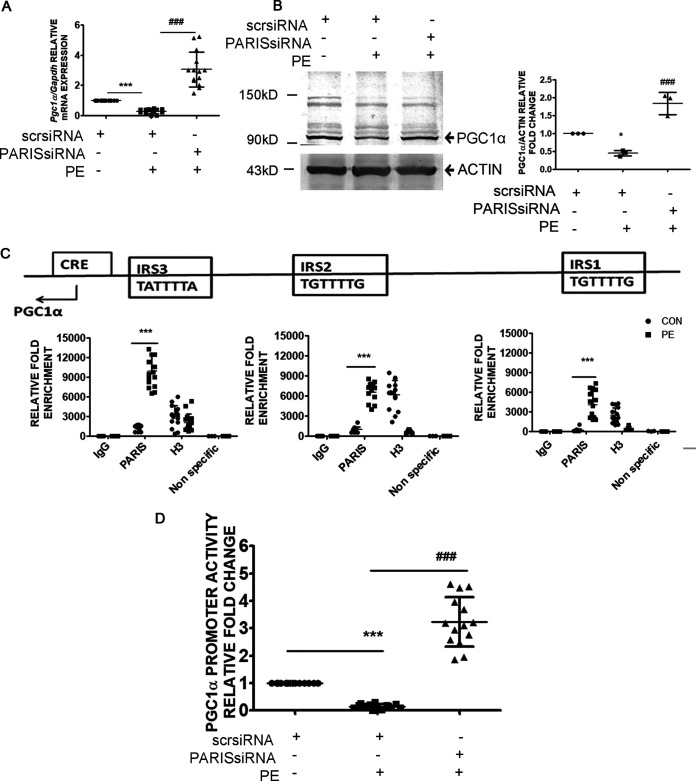
Knocking down PARIS in hypertrophied cells successfully prevented the downregulation of PGC1α at the levels of both mRNA (Fig. 5A) and protein (Fig. 5B). PARIS transcriptionally represses PGC1α by binding to the 3 conserved sequences on the rat PGC1α promoter known as insulin response sequences (IRS) (13), i.e., IRS1 (TGTTTTG), IRS2 (TGTTTTG), and IRS3 (TATTTTA) (23). To investigate whether PARIS-mediated regulation of PGC1α expression in hypertrophied myocytes was associated with its binding to PGC1α promoter, chromatin immunoprecipitation with anti-PARIS antibody was performed, followed by quantitative reverse transcription-PCR (qRT-PCR) using primers targeting predicted binding sites on the PGC1α promoter sequence. PARIS occupancy of the PGC1α promoter region was increased in hypertrophied myocytes compared to controls, indicating direct binding of PARIS to the PGC1α promoter (Fig. 5C). To determine whether the binding of PARIS modulates transcriptional activity of PGC1α promoter, a luciferase assay was performed. PE treatment reduced PGC1α promoter-driven gene expression, while knockdown of PARIS restored PGC1α promoter activity in hypertrophied myocytes, suggesting negative regulation of PGC1α transcription by PARIS (Fig. 5D).
FIG 5.
Regulation of PGC1α transcriptional activity by PARIS in hypertrophy. (A) qRT-PCR showing a significant increase in Pgc1α gene expression in PARIS siRNA-treated hypertrophied H9C2 myocytes. Gapdh expression was used as the internal loading control. n = 12 for each group from 4 independent experiments. ***, P < 0.001 compared to controls; ###, P < 0.001 compared to PE treatment. (B) Western blot showing restored PGC1α protein expression in PARIS siRNA-treated hypertrophied myocytes. Actin was used as the loading control. Graphical representation of immunoblot data showing the amount of change in expression of PGC1α. n = 3 for each group from 3 independent experiments. *, P < 0.05 compared to controls; ###, P < 0.001 compared to PE. (C) Cross-linked chromatin fragments of H9C2 cardiomyocytes of different treatment groups were immunoprecipitated with anti-PARIS, anti-rabbit IgG (−ve control), and anti-H3 (+ve control). DNA was isolated and PCR amplified using primer sets designed from the promoter regions (IRS1, IRS2, and IRS3) of PGC1α showing increased PARIS occupancy on the PGC1α promoter in PE-treated cells. A nonspecific primer was used to detect specificity of ChIP experiments. n = 15 for each group except for the nonspecific primer groups, where n = 3; ***, P < 0.001 compared to controls. Locations of IRS1, IRS2, IRS3, and CRE motifs are shown on the PGC1α promoter. (D) Relative luciferase activity of the 1-kb rat PGC1α promoter (−1000 to +40 relative to the TSS) (Pgc1αluc) in H9C2 cells of different treatment groups showing enhanced PGC1α transcriptional activity in PARIS knockdown hypertrophied H9C2 myocytes. n = 14 for each group from 5 independent experiments. ***, P < 0.001 compared to controls; ###, P < 0.001 compared to PE treatment.
PARIS is SUMOylated in hypertrophied cardiomyocytes.
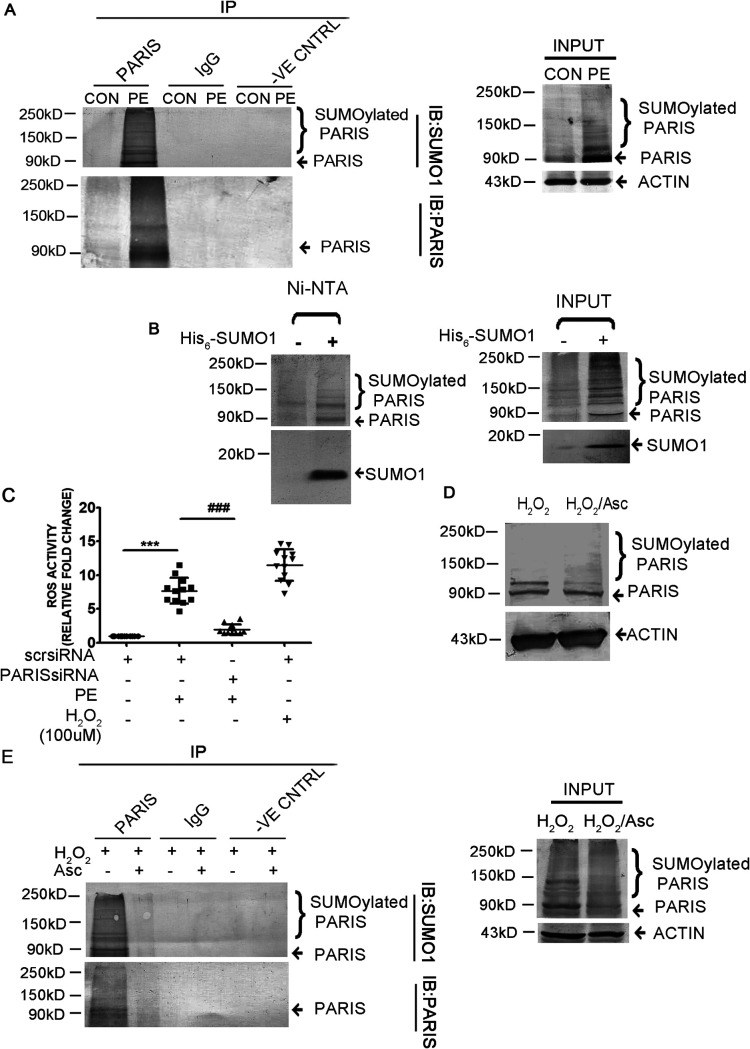
To investigate SUMOylation of PARIS in cardiac hypertrophy, PARIS was coimmunoprecipitated with SUMO1 antibody in PE-treated H9C2 cells. Immunoblotting with anti-PARIS antibody showed the presence of slowly migrating high-molecular-weight bands above PARIS in PE-treated cells, indicating SUMOylated forms of the protein. To explicitly establish that the high-molecular-weight bands correspond to sumoylated PARIS, the cell lysates were prepared under denaturing conditions using 2× SDS buffer followed by coimmunoprecipitation and were analyzed by Western blotting. Denaturing helps remove any interacting partners, and it also protects SUMOylated proteins from isopeptidases. These data strongly suggest that slower-migrating higher-molecular-weight bands of PARIS are derived from covalent linking to SUMO1 (Fig. 6A). To clearly establish that the higher-molecular-weight bands correspond to SUMOylated PARIS, HEK293 cells were transfected with His-tagged SUMO1 and then lysed under denaturing conditions using guanidine HCl buffer. The His6-SUMO1 was then purified with nickel-charged agarose beads (Ni-nitrilotriacetic acid [NTA]) followed by Western blot analysis with anti-PARIS antibody, showing SUMO1-conjugated PARIS (Fig. 6B). A band corresponding to unmodified PARIS was observed in the absence of SUMO1 (Fig. 6B). The presence of histidine in the zinc finger motif of PARIS probably retained the protein on Ni-NTA beads (24).
FIG 6.
The function of PARIS is regulated by SUMOylation in hypertrophied cardiomyocytes. (A) Coimmunoprecipitation of control and PE-treated cell lysates extracted under denaturing conditions using anti-PARIS antibody followed by immunoblotting with anti-SUMO1 antibody showing increased SUMOylation of PARIS during hypertrophy. Control beads and IgG served as negative controls. Untreated control and PE-treated cell lysates served as input. The data represent 3 independent experiments. (B) HEK293 cells were transfected with His6-SUMO1. Transfected cells were lysed under denaturing conditions, and the SUMO1-modified proteins were purified by chromatography on Ni-NTA–agarose beads followed by immunoblotting using anti-PARIS antibody, showing slower-migrating bands of SUMOylated PARIS (lane 2). n = 3 for each group from 3 independent experiments. (C) Measurement of ROS content using a DCFDA assay kit in H9C2 cells in different treatment groups showing significantly reduced ROS level in PARIS siRNA treatment; H2O2 was used as a positive control. n = 12 for each group from 4 independent experiments. ***, P < 0.001 compared to controls; ###, P < 0.001 compared to PE treatment. (D) Western blot demonstrating decreased SUMOylation of PARIS in ascorbate treatment compared to H2O2-treated H9C2 cells. Actin was used as the loading control. n = 3 for each group from 3 independent experiments. (E) Coimmunoprecipitation under denaturing conditions using anti-PARIS antibody followed by immunoblotting with anti-SUMO1 antibody in different treatment groups showing reduced SUMOylation of PARIS in cells in which ROS had been scavenged. Control beads and IgG served as negative controls. H2O2- and H2O2-Asc-treated cell lysed under denaturing condition served as input. n = 3 for each group from 3 independent experiments.
Hypertrophy-associated mitochondrial dysfunction is accompanied by generation of reactive oxygen species (ROS) (25). To investigate the effect of PARIS silencing on cellular ROS production in hypertrophied myocytes, a fluorometric assay was performed to measure cellular ROS content. The cellular ROS level, which was increased in PE-treated cells, was reduced in PARIS-silenced hypertrophied myocytes (Fig. 6C). ROS generation was mimicked in H9C2 cardiomyocytes by treatment with H2O2, and ROS accumulation was scavenged by pretreating H2O2-treated cells with ascorbate. To investigate whether accumulated ROS influences PARIS SUMOylation, Western blot analysis was performed. SUMOylation of PARIS was enhanced by H2O2 treatment, while it was reduced by scavenging the accumulated ROS (Fig. 6D). ROS-mediated PARIS SUMOylation was further confirmed by coimmunoprecipitating PARIS and SUMO1 under denaturing condition in H9C2 myocytes treated with H2O2 either alone or together with ascorbate. Immunoblotting revealed increased SUMOylation of PARIS under oxidative stress compared to cells in which ROS had been scavenged (Fig. 6E), suggesting oxidative stress-induced PARIS SUMOylation in cardiac hypertrophy.
PARIS SUMOylation regulates PGC1α transcription.
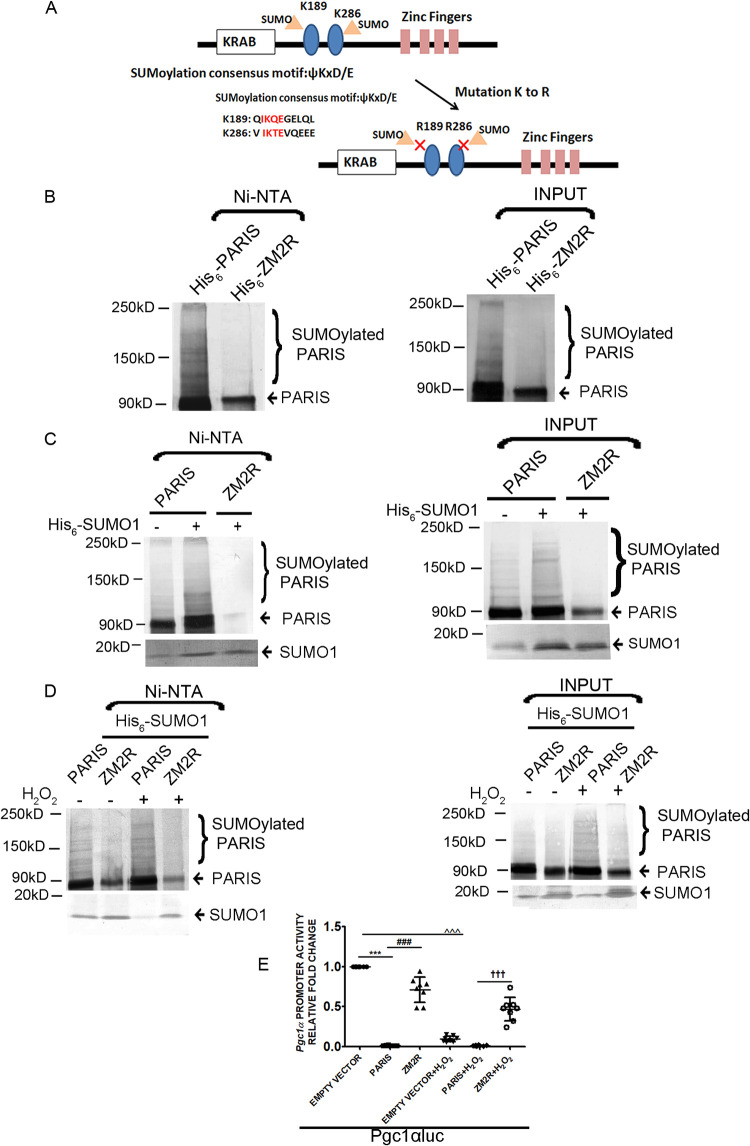
To investigate the functional significance of PARIS SUMOylation, lysine residues K189 and K286 of PARIS were mutated to arginine (ZM2R) (Fig. 7A). HEK293 cells transfected with either histidine-tagged wild-type PARIS (His6-PARIS) or mutant PARIS (His6-ZM2R) were lysed under denaturing conditions using guanidine HCl buffer. Protein lysates were purified using Ni-NTA–agarose followed by Western blotting. Immunoblotting showed slowly migrating high-molecular-weight bands of PARIS, suggesting that SUMOylation of PARIS was present in cells transfected with the wild type (His6-PARIS) but was reduced in those transfected with mutant PARIS (His6-ZM2R) (Fig. 7B). To further validate PARIS SUMOylation, HEK293 cells were cotransfected with His-tagged SUMO1 (His6-SUMO1) and PARIS or ZM2R and then lysed under denaturing conditions in guanidine HCl buffer. The His-tagged proteins were purified with Ni-NTA and analyzed by Western blotting using anti-PARIS antibody. Immunoblotting further confirmed that the slower-migrating bands are SUMOylated PARIS (Fig. 7C).
FIG 7.
SUMOylation dependent regulation of PGC1α transcription by PARIS. (A) Schematic representation of the human PARIS protein. Its KRAB and zinc finger motifs and two lysines (K189 and K286) in the consensus motif for SUMOylation are indicated. SUMOylation conserved sequences surrounding two SUMOylation sites (K189 and K286) in the human PARIS protein. K189 and K286 were mutated to arginine to make a SUMO-resistant mutant of PARIS. (B) HEK293 cells were transfected with either His6-PARIS or His6-ZM2R. Transfected cells were lysed under denaturing conditions, and then SUMO1-conjugated PARIS was purified by chromatography on Ni-NTA–agarose beads followed by immunoblotting using anti-PARIS antibody showing reduced SUMOylation in His6-ZM2R-transfected cells (lane 2). Lysates of cells transfected with either His6-PARIS or His6-ZM2R served as input. The data represent 3 independent experiments. (C) HEK293 cells were cotransfected with His6-SUMO1 along with either PARIS or ZM2R. Transfected cells were lysed under denaturing conditions; then, the SUMO1 modified proteins were purified by Ni-NTA–agarose beads followed by immunoblotting using anti-PARIS antibody showing reduced SUMOylation of ZM2R (lane 3). Total lysates from cells cotransfected with His6-SUMO1 and PARIS or ZM2R served as input. Experiments were repeated 3 times (n = 3). (D) HEK293 cells were cotransfected with His6-SUMO1 along with either PARIS or ZM2R in the presence or absence of H2O2.Transfected cells were lysed under denaturing conditions, and SUMO1-conjugated proteins were purified with Ni-NTA–agarose beads followed by immunoblotting using anti-PARIS antibody showing reduced SUMOylation of ZM2R. Wild-type PARIS showed enhanced SUMOylation under oxidative stress (lane 3), while ZM2R SUMOylation was significantly reduced (lane 4). Total lysates from cells cotransfected with His6-SUMO1 and PARIS or ZM2R in the presence or absence of H2O2 served as input. The data represent 3 independent experiments. (E) Relative luciferase activity in HEK293 cells transfected with the Pgc1αluc promoter either alone or cotransfected with wild-type or mutant PARIS in different treatment groups showing restored PGC1α transcriptional activity in ZM2R mutant-transfected cells. n = 8 for each group from 3 independent experiments; ***, P < 0.001 compared to PGC1αluc; ###, P < 0.001 compared to wild-type PARIS; ^̂̂, P < 0.001compared to PGC1αluc; †††, P < 0.001 compared to wild-type PARIS with H2O2.
To confirm that oxidative stress regulates PARIS SUMOylation, HEK293 cells were cotransfected with either wild-type PARIS or ZM2R and His6-SUMO1 in the presence or absence of H2O2. The transfected cells were lysed under denaturing condition and were purified with Ni-NTA–agarose followed by Western blotting using anti-PARIS antibody. Immunoblots showed enhanced SUMOylation of wild-type PARIS but not ZM2R under oxidative stress (Fig. 7D). These data confirm that K189 and K286 residues on PARIS are essential for SUMOylation and that this PARIS SUMOylation is modulated by oxidative stress. To check the effect of PARIS SUMOylation on PGC1α transcription activity, a luciferase assay was performed in HEK293 cells transfected with either PARIS or ZM2R in the presence or absence of H2O2. ROS accumulation significantly reduced PGC1α promoter activity in PARIS-transfected cells, while the transcriptional activity of PGC1α was not reduced extensively in ZM2R-containing cells (Fig. 7E). These data strongly suggest the significance of PARIS SUMOylation for the repression of PGC1α gene transcription during cardiac hypertrophy.
PARIS interacts with DJ-1.
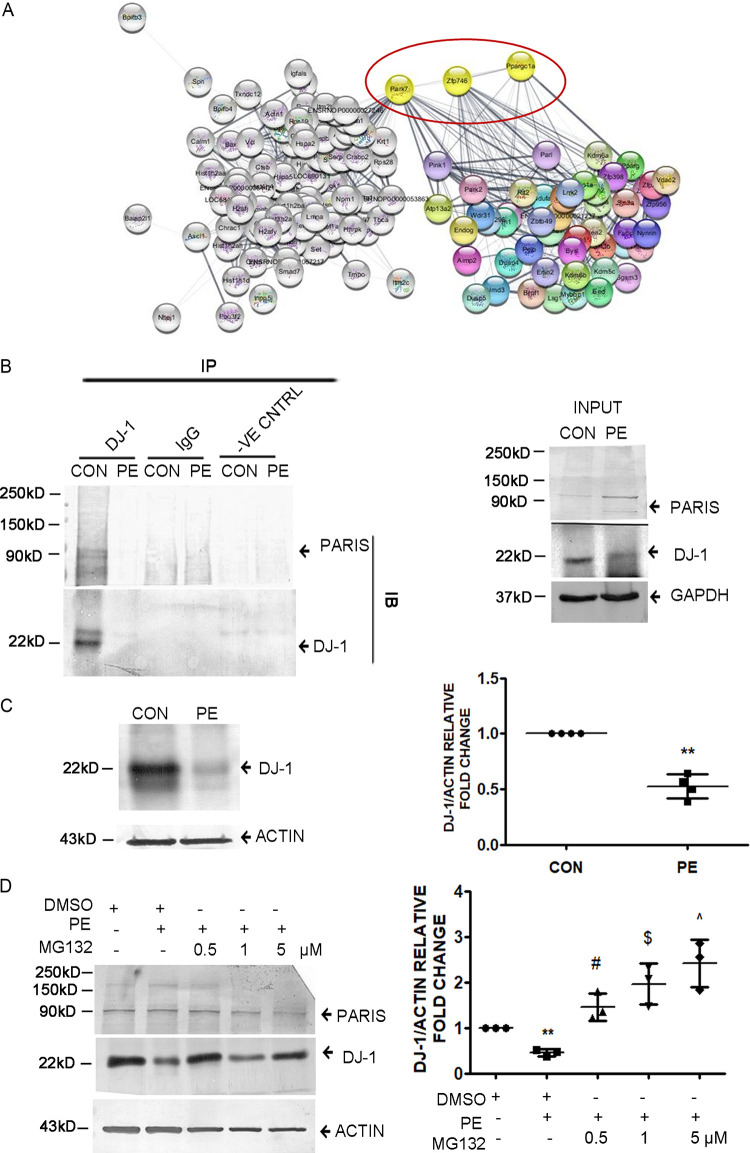
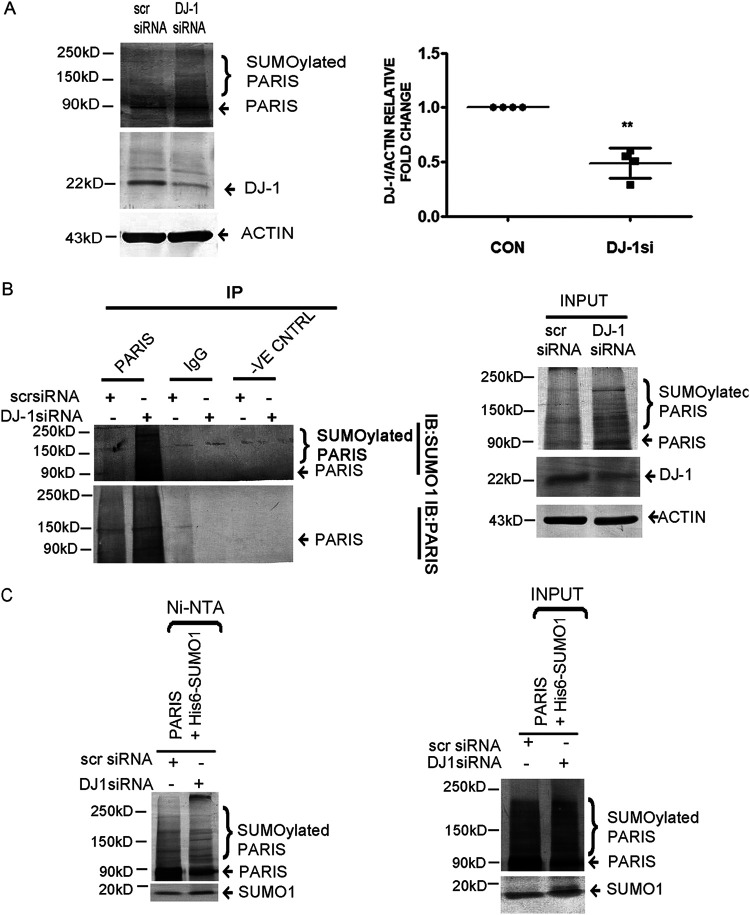
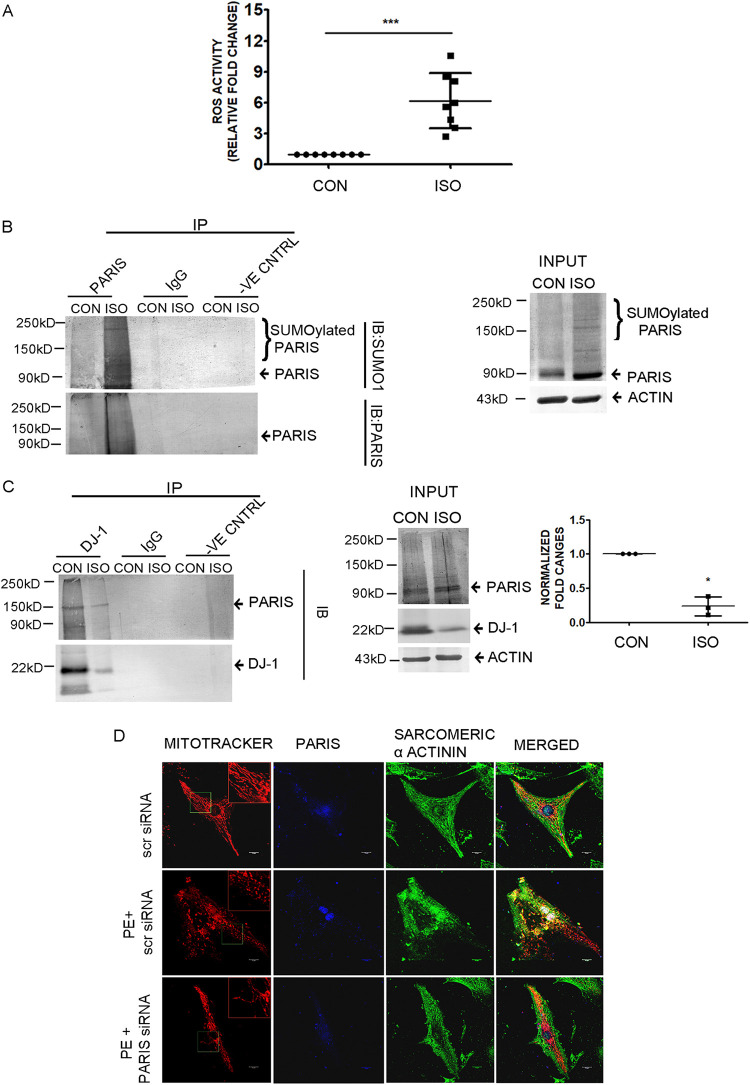
To explore the mechanism by which PARIS activation can be regulated in cells, total cellular proteins were immunoprecipitated using the anti-PARIS antibody followed by mass spectrometry. Mass spectrometry analysis revealed a list of interacting partners of PARIS (Fig. 8A), in which DJ-1 was one of the prominent proteins. Mass spectrometry data were validated by evaluating PARIS and DJ-1 interaction both under hypertrophy conditions and at baseline by coimmunoprecipitation. Coimmunoprecipitation showed reduced interaction of PARIS with DJ-1 in hypertrophied myocytes compared to controls (Fig. 8B).
FIG 8.
DJ-1 interacts with PARIS in H9C2 cardiomyocytes. (A) Interactome study showing DJ-1–PARIS–PGC1α interaction. Known interactome of PARIS from STRING merged with the PARIS interactome obtained from mass spectrometry studies. n = 3 for each group from 3 independent experiment. P < 0.05. (B) Coimmunoprecipitation assay of H9C2 cell lysate using anti-DJ-1 antibody followed by immunoblotting with anti-PARIS antibody in control and PE-treated cells showing decreased DJ-1 interaction with PARIS in PE treatment. Control beads and IgG served as negative controls. Control and PE-treated cell lysates served as input. The data represent 3 independent experiments. (C) Immunoblot showing reduced DJ-1 expression in PE-treated H9C2 myocytes compared to controls. Actin was used as the loading control. Graphical representation of immunoblot data showing the amount of change in expression of DJ-1 protein in hypertrophied myocytes. n = 4 for each group from 4 independent experiments. **, P < 0.01 compared to controls. (D) Immunoblot depicting the restoration of DJ-1 in MG132 (0.5, 1, and 5 μM)-treated hypertrophied myocytes compared to PE-treated cells. Actin was used as an internal loading control. Experiments were repeated 3 times (n = 3). Graphical representation of restored DJ-1 expression in MG132-treated hypertrophied cells. Actin was used as the loading control. n = 3 for each group from 3 independent experiments. **, P < 0.01 compared to controls. #, $, and ^, P < 0.05 compared to PE.
Interestingly, it was observed that the DJ-1 protein level was reduced in hypertrophied cardiomyocytes (Fig. 8C). To investigate whether DJ-1 is degraded in hypertrophied myocytes, H9C2 cells were treated with either PE alone or in combination with different doses of the proteasome inhibitor MG132 (500 nM, 1 μM, and 5 μM). Subsequent Western blot analysis revealed that DJ-1 expression was rescued in MG132-treated hypertrophied cells compared to PE treatment (Fig. 8D), suggesting proteasome-mediated degradation of DJ-1 during hypertrophy.
DJ-1 inhibits PARIS SUMOylation.
Next, the functional significance of DJ-1 and PARIS interaction was analyzed. The previous report showed that DJ-1 functions as a SUMO inhibitor (26). Therefore, PARIS SUMOylation was assessed in H9C2 cardiomyocytes after knocking down DJ-1 expression by siRNA treatment. Silencing DJ-1 increased PARIS SUMOylation, as revealed by Western blotting (Fig. 9A). The role of DJ-1 in PARIS SUMOylation was further confirmed by coimmunoprecipitating PARIS with SUMO1 under denaturing conditions. Coimmunoprecipitation showed that PARIS SUMOylation was enhanced by knocking down DJ-1 (Fig. 9B). DJ1-mediated inhibition of PARIS SUMOylation was further validated by cotransfecting PARIS and His6-SUMO1 in either scrambled or DJ-1 siRNA-treated HEK293cells. Cells were then lysed under denaturing conditions, and SUMOylated PARIS was purified using Ni-NTA agarose followed by Western blot analysis. These data showed enhanced SUMOylation of PARIS in DJ-1 siRNA-treated cells, indicating that DJ-1 inhibits PARIS SUMOylation (Fig. 9C).
FIG 9.
Inhibition of PARIS SUMOylation by DJ-1 in H9C2 cardiomyocytes. (A) Immunoblot showing increased PARIS SUMOylation in DJ-1 siRNA-treated H9C2 myocytes. Actin was used as an internal loading control. Graphical representation showing decreased DJ-1 expression in H9C2 cells transfected with DJ-1 siRNA. n = 4 for each group from 4 independent experiments. **, P < 0.01 compared to controls. (B) Coimmunoprecipitation of DJ-1 siRNA-treated cell lysates extracted under denaturing conditions using anti-PARIS antibody followed by immunoblotting with anti-SUMO1 antibody showing increased PARIS SUMOylation by knocking down DJ-1 in H9C2 cells. Control beads and IgG served as negative controls. Scrambled siRNA- and DJ-1 siRNA-transfected cell lysates served as input. The data represent 3 independent experiments for each group (n = 3). (C) HEK293 cells were cotransfected with His6-SUMO1and PARIS in scrambled siRNA- or DJ-1 siRNA-treated cells. Transfected cells were lysed in denaturing lysis buffer; then, the SUMO1-modified proteins were purified on Ni-NTA–agarose beads followed by immunoblotting using anti-PARIS antibody showing enhanced SUMOylation of PARIS in DJ-1 knockdown cells. Lysates of either scrambled siRNA- or DJ-1 siRNA-treated cells cotransfected with His6-SUMO1 and PARIS served as input. Each experiment was repeated 3 times for each group.
Oxidative stress affects DJ-1–PARIS interaction.
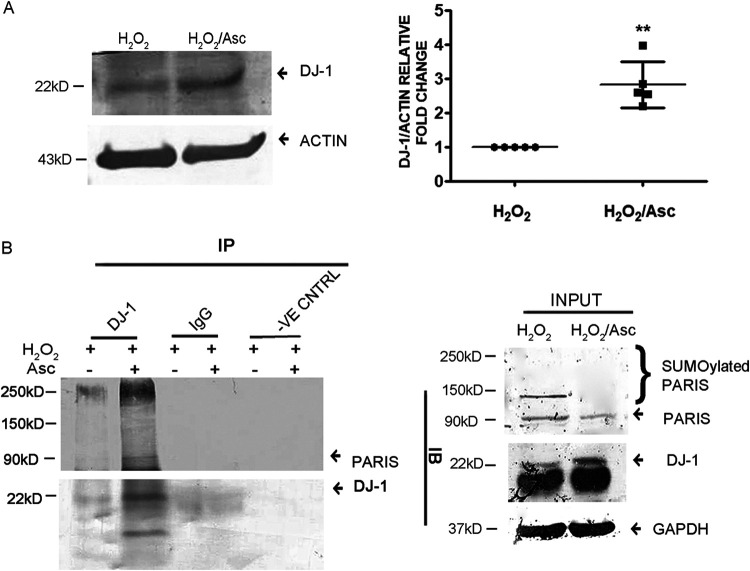
Furthermore, to analyze whether accumulated ROS in cardiac hypertrophy is responsible for DJ-1 degradation, DJ-1 expression was assessed in H2O2-treated H9C2 cells in the presence or absence of ascorbate by Western blotting. H2O2 treatment decreased the DJ-1 protein level; however, scavenging the accumulated ROS by ascorbate recovered its protein expression (Fig. 10A). To further confirm whether the downregulated interaction of DJ-1 and PARIS is due to oxidative stress, binding of PARIS with DJ-1 was analyzed both in cells with accumulated ROS and in those in which it had been scavenged. Immunoblotting revealed an increased association between these two proteins in cardiomyocytes in which ROS had been scavenged compared to those in which it had accumulated (Fig. 10B).
FIG 10.
Oxidative stress affects DJ-1 PARIS interaction. (A) Immunoblot showing restored DJ-1 protein expression in ascorbate treatment. Actin was used as an internal loading control. Graphical representation of restored DJ-1 expression. n = 5 for each group from 5 independent experiments. **, P < 0.01 compared to H2O2 treatment. (B) Coimmunoprecipitation using anti-DJ-1 antibody followed by immunoblotting with anti-PARIS antibody showing increased DJ-1 binding to PARIS in H9C2 cells in which ROS had been scavenged compared to H2O2 treatment. Control beads and IgG served as negative controls. H2O2 and H2O2-Asc treated cell lysates served as input. The data represent 3 independent experiments.
Oxidation of DJ-1 affects inhibition of PARIS SUMOylation.
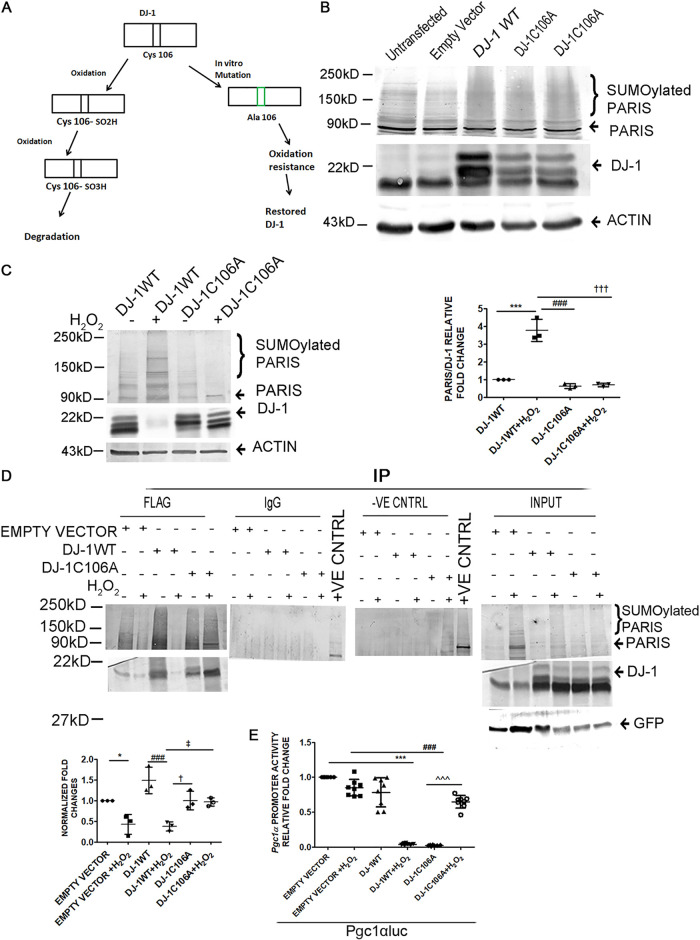
As it has been shown that DJ-1 is degraded under oxidative stress, an oxidation-resistant mutant was generated by mutating the cysteine residue at position 106 of DJ-1 to alanine (DJ-1C106A) (Fig. 11A). HEK293 cells were transfected with either wild-type DJ-1 (DJ-1WT) or the DJ-1C106A mutant, and PARIS SUMOylation was assessed. PARIS SUMOylation was reduced in the presence of DJ-1WT or DJ-1C106A compared to nontransfected cells or cells transfected with an empty vector, as revealed by Western blotting (Fig. 11B). The effect of oxidative stress on DJ-1 and its regulatory function for PARIS SUMOylation was further investigated in HEK293 cells transfected with either DJ-1WT or DJ-1C106A in the presence or absence of H2O2. While DJ-1WT was degraded, the expression of DJ-1C106A was stabilized in the presence of H2O2 (Fig. 11C). The interaction of PARIS and DJ-1 was further analyzed to confirm the hypothesis. For this, Flag-tagged DJ-1 was coimmunoprecipitated with PARIS using an anti-Flag antibody in either DJ-1WT- or DJ-1C106A-transfected HEK293 cells in the presence or absence of H2O2, followed by immunoblotting with anti-PARIS antibody. The interaction of DJ-1 and PARIS was significantly reduced in HEK293 cells transfected with either DJ-1WT or empty vector under oxidative stress, while DJ-1C106A interacted with PARIS in the presence of H2O2 (Fig. 11D). To further confirm that DJ-1-mediated inhibition of PARIS SUMOylation is necessary for the release of transcriptional repression on the PGC1α promoter, a luciferase assay using a 1-kb rat PGC1α promoter (−1000 to +40 relative to the transcription start site [TSS]) (Pgc1αluc) was performed on HEK293 cells in the presence or absence of H2O2. Our results revealed enhanced PGC1α transcriptional activity in both DJ-1WT- and DJ-1C106A-containing cells. It was also observed that stabilized DJ-1C106A successfully restored PGC1α transcriptional activity; however, DJ-1WT failed to enhance the promoter activity in the presence of H2O2 (Fig. 11E), signifying that DJ-1 deSUMOylates PARIS and thereby regulates PGC1α transcription.
FIG 11.
Oxidation of DJ-1 affects PARIS SUMOylation inhibition. (A) Schematic diagram showing the oxidation-sensitive site of DJ-1. The Cys106 residue of DJ-1 was mutated to alanine to prepare an oxidation-resistant mutant (DJ-1C106A). (B) Immunoblot of HEK293 cell lysate transfected with either wild-type DJ-1 (DJ-1WT) or the oxidation-resistant mutant DJ-1C106A showing decreased PARIS SUMOylation. n = 3 for each group from 3 independent experiments. (C, left) Western blot of PARIS and DJ-1 in HEK293 cells transfected with the WT or the mutant showing degradation of DJ-1WT and restoration of DJ-1C106A upon H2O2 treatment. (Right) Graphical representation of immunoblot data showing the amount of change in PARIS SUMOylation under oxidative stress. The data represent 3 independent experiments. ***, P < 0.001 compared to DJ-1WT; ### and †††, P < 0.001 compared to DJ-1WT with H2O2. (D, top) Coimmunoprecipitation using anti-Flag antibody and immunoblotting with anti-PARIS antibody in either DJ-1WT- or DJ-1C106A-transfected HEK293 cells resulted in decreased DJ-1/PARIS interaction in DJ-1WT or empty vector-transfected HEK293 cells in the presence of H2O2. Control beads and IgG served as negative controls. Cell lysates containing different plasmids with different treatment groups served as input. (Bottom) Graphical representation of coimmunoprecipitation. The data represent 3 independent experiments. *, P < 0.05 compared to empty vector; ###, P < 0.001 compared to DJ-1WT; † and ‡, P < 0.05 compared to DJ-1WT with H2O2. (E) Relative luciferase activity of the 1-kb rat PGC1α promoter (−1000 to +40 relative to the TSS) (Pgc1αluc) in HEK293 cells containing different plasmids with different treatment groups showing enhanced PGC1α transcriptional activity in both DJ-1WT- and DJ-1C106A-containing cells and reduced activity in the presence of H2O2. n = 8 for each group from 3 independent experiments. ***, P < 0.001 compared to Pgc1αluc; ###, P < 0.001 compared to DJ-1WT; ^̂̂, P < 0.001compared to DJ-1WT with H2O2.
Elevated cardiac expression of PARIS in hypertrophied myocardium.
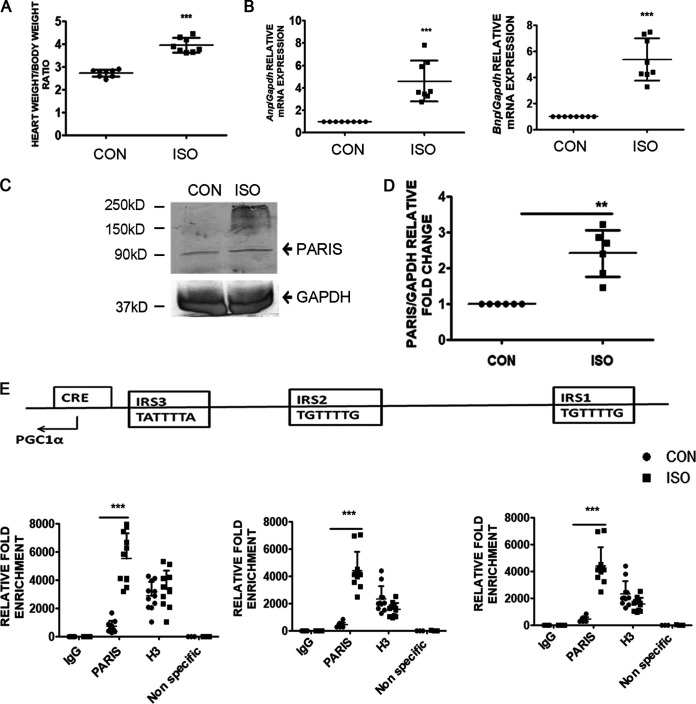
To validate the role of PARIS in cardiac hypertrophy, the expression of PARIS was estimated in isoproterenol (ISO)-induced hypertrophied myocardium in vivo. Hypertrophy was assessed by measuring the ratio of heart weight (HW; in milligrams) to body weight (BW; in grams). HW:BW was increased significantly in ISO-treated rats (Fig. 12A). Hypertrophic marker genes like Anp and Bnp were also shown to be increased significantly in hypertrophied rat hearts (Fig. 12B).
FIG 12.
Expression of PARIS is elevated in hypertrophied myocardium in vivo. (A) Assessment of hypertrophy by measuring the ratio of heart weight to body weight of rats. n = 8 for each group. ***, P < 0.001 compared to controls. (B) qRT-PCR showing increased expression of hypertrophic marker genes, i.e., Anp and Bnp, in ISO-treated hypertrophied rat hearts compared to controls. Gapdh expression was used as an internal control. n = 8 for each group. ***, P < 0.001 compared to controls. (C) Western blots depicting increased PARIS expression in ISO-treated hypertrophied rat hearts compared to controls. GAPDH expression was used as the internal loading control. (D) Graphical representation of increased PARIS expression. n = 6 for each group from 3 independent experiments. **, P < 0.01 compared to controls. (E) Cross-linked chromatin fragments of either hypertrophied heart tissues or controls were immunoprecipitated with anti-PARIS antibody, rabbit IgG (−ve control), or H3 (+ve control). DNA was PCR amplified using primer sets designed from the promoter regions (IRS1, IRS2, and IRS3) of PGC1α showing increased PARIS occupancy on the PGC1α promoter during hypertrophy. A nonspecific primer was used to ensure specificity of ChIP experiments. n = 8 for each group except for the nonspecific primer groups, where n = 3. ***, P < 0.001 compared to controls.
Hypertrophied hearts showed elevated expression of PARIS compared to controls (Fig. 12C and D). To further confirm the functional role of PARIS in cardiac hypertrophy, chromatin immunoprecipitation was performed to detect PGC1α promoter occupancy by PARIS. Hypertrophied myocardium showed increased PARIS binding to the PGC1α promoter (−1000 to +40 relative to the TSS) (Fig. 12E).
Functional significance of PARIS in hypertrophied myocardium.
Cardiac hypertrophy-associated cellular ROS production was measured by fluorometric assay showing increased ROS accumulation in hypertrophied myocardium compared to controls (Fig. 13A). Coimmunoprecipitation of PARIS with SUMO1 under denaturing condition showed increased PARIS SUMOylation, further confirming SUMOylation of PARIS in hypertrophied myocardium (Fig. 13B). An association between DJ-1 and PARIS was also observed in hypertrophied myocardium, which exhibited increased interaction between these 2 proteins at the baseline and reduced interaction during cardiac hypertrophy (Fig. 13C).
FIG 13.
Functional role of elevated PARIS expression in hypertrophied myocardium. (A) Measurement of ROS content using a DCFDA assay kit showing significantly increased ROS levels in hypertrophied myocardium. n = 8 for each group. ***, P < 0.001 compared to controls. (B) Coimmunoprecipitation using anti-PARIS antibody followed by immunoblotting with anti-SUMO1 antibody from tissue lysates under denaturing conditions showed increased SUMOylation of PARIS in hypertrophied myocardium. Control beads and IgG served as negative controls. Tissue lysates from control and ISO-treated rat heart served as input. n = 3 for each group from 3 independent experiments. (C) Coimmunoprecipitation showing DJ-1 and PARIS associated strongly in control rat myocardium compared to ISO-treated hypertrophied myocardium using an anti-DJ-1 antibody for immunoprecipitation and anti-PARIS antibody for immunoblotting. Control beads and IgG served as negative controls. Tissue lysates from control and ISO-treated rat heart served as input. Graphical representation of coimmunoprecipitation data. The data represent 3 independent experiments. *, P < 0.05 compared to controls. (D) Confocal micrographs showing mitochondrial morphology in NRVM of different treatment groups. Bar, 10 μm. (Inset) Magnified view (2×) of the region marked by the green box.
The role of PARIS in mitochondrial dysfunction in cardiac hypertrophy was monitored by confocal microscopy. To correlate PARIS expression with mitochondrial morphology and function, neonatal rat ventricular myocytes (NRVM) were transfected with either scrambled or PARIS siRNA followed by PE treatment. Immunofluorescence staining using MitoTracker red for mitochondria, anti-PARIS antibody (blue), and sarcomeric α-actinin (green) was also performed. Knocking down PARIS in neonatal rat ventricular myocytes reestablished the mitochondrial network, compared to the punctate morphology due to PE treatment (Fig. 13D), suggesting that PARIS is a prohypertrophic molecule in the myocardium and contributes to mitochondrial dysfunction in cardiac hypertrophy.
DISCUSSION
Cardiac hypertrophy is associated with energy imbalance, mitochondrial dysfunction, and oxidative stress in cardiomyocytes, leading to impaired hemodynamics. Several studies have suggested that PGC1α functions as an important signaling node that impacts several aspects of cellular abnormalities during cardiac hypertrophy (9, 27–31), including mitochondrial dysfunction. While downregulated PGC1α expression has been implicated in the progression of the disease, the precise mechanism of the repression of PGC1α remains unaddressed. The present study provides evidence of a nuanced molecular mechanism of mitochondrial dysfunction by PARIS through transcriptional control of PGC1α.
The role of PARIS in cardiac hypertrophy has remained unexplored. Our study demonstrates that PARIS expression, as well as nuclear localization, is enhanced in hypertrophied cardiomyocytes both in vitro and in vivo (Fig. 1A and B and Fig. 12C). Furthermore, knockdown of PARIS in hypertrophied myocytes significantly reduces hypertrophic marker gene expression and myocyte size (Fig. 2D and E), suggesting an important role for PARIS in the regulation of myocyte hypertrophy.
Mitochondrial dysfunction during hypertrophy is associated with the loss of the mitochondrial reticulate network and a concomitant drop in mitochondrial membrane potential (3, 4, 32). Knocking down PARIS expression maintains mitochondrial membrane potential (Fig. 3C) and also recovers fused elongated healthy mitochondrial morphology (Fig. 3B) in hypertrophied myocytes, indicating that PARIS is a negative regulator of mitochondrial function.
A dynamic balance between mitochondrial fission and fusion is necessary to maintain homeostasis (33, 34). An increase in mitochondrial fission leads to increased dysfunctional mitochondria and reduced mitobiogenesis (35–38). Increase fission leads to decreased expression of mitochondrial fusion proteins like MFN1 and MFN2. PARIS knockdown increases both MFN1 and MFN2 expression, while no significant change in Drp1 expression is observed (Fig. 3D and E). Further, the assessment of mitochondrial dynamics by measuring mitochondrial fragmentation reveals increased mitochondrial length by inhibiting PARIS in hypertrophied myocytes (Fig. 3F). While PE treatment significantly reduces functional healthy mitochondria, silencing PARIS in hypertrophied myocytes restores mitochondrial biogenesis (Fig. 4A). Furthermore, oxidative phosphorylation is one of the major functions of mitochondria, measured by oxygen consumption rate and subsequent ATP production (39). Hypertrophic stress disrupted oxidative phosphorylation, resulting in reduced ATP production and mitochondrial respiration, thus corroborating earlier reports (40). However, PARIS knockdown in hypertrophied myocytes augments mitochondrial oxidative phosphorylation (Fig. 4B), revealing a prohypertrophic role for PARIS which leads to hypertrophic manifestations by at least partially regulating mitochondrial physiology.
It is interesting that the improvement of mitochondrial function achieved by knocking down PARIS in hypertrophied cells is also associated with the restoration of PGC1α expression (Fig. 4C and Fig. 5A and B), indicating negative regulation of PGC1α by PARIS. While hypertrophic stress is associated with enhanced binding of PARIS to the PGC1α promoter region (Fig. 5C) and downregulation of PGC1α transcription, PARIS depletion restored PGC1α transcription in hypertrophied myocytes (Fig. 5D). These data suggest that binding of PARIS on PGC1α promoter is necessary for the repression of PGC1α promoter activity under hypertrophic conditions, corroborating earlier studies in the neuronal system (13, 14). However, the exact mechanism of repression of PGC1α by PARIS remains unclear. Like other KRAB zinc finger proteins, PARIS might mediate its repressor activity by interacting with corepressors and histone-modifying enzymes (41). The involvement of any of these regulatory players in PARIS-mediated PGC1α repression needs further investigation.
Previous reports demonstrated that repression of PGC1α is associated with enhanced PARIS SUMOylation. Here, we show that PARIS SUMOylation is induced in cardiac hypertrophy (Fig. 6A). Furthermore, in hypertrophied cardiomyocytes, mitochondrial dysfunction leads to oxidative stress, culminating in enhanced SUMOylation (42, 43) of PARIS. However, scavenging ROS significantly reduces PARIS SUMOylation (Fig. 6D and E), confirming ROS-mediated regulation of PARIS SUMOylation. The SUMOylation of PARIS is significantly involved in regulating PGC1α promoter activity. The SUMO-resistant mutant of PARIS (ZM2R) fails to repress PGC1α transcription in the presence of ROS, suggesting that oxidative stress-induced SUMOylation of PARIS is functionally important for the repression of PGC1α transcription (Fig. 7E) in cardiac hypertrophy. However, the mechanism of repression of PGC1α activity by PARIS SUMOylation still remains obscure. The possibilities include SUMOylation-mediated changes in DNA binding activity of PARIS and altered methylation patterns of its target promoters through interaction with DNA methyltransferases and acetyltransferases.
PARIS-dependent proteome profiling shows DJ-1 as a novel interacting partner (Fig. 8A). DJ-1 was first discovered as a protein whose gene is mutated in patients with Parkinsonism (44, 45).
The role of DJ-1 in the development of cardiac diseases is well documented. Heart-specific deletion of DJ-1 in mice showed an elevated ROS production in hypertrophied myocardium (46, 47) supporting the role of DJ-1 as an oxidative stress sensor (26, 48–51). Our study shows reduced interaction of DJ-1 and PARIS in hypertrophied cardiomyocytes along with the diminished expression of DJ-1 protein (Fig. 8B and C). Further investigation showed that proteasome blockade rescued DJ-1 protein expression, suggesting proteasome-mediated degradation of DJ-1 in cardiac hypertrophy (Fig. 8D). Interestingly, scavenging ROS in H9C2 cardiomyocytes recovered DJ-1 protein expression as well as its interaction with PARIS (Fig. 10), suggesting oxidative stress-mediated degradation of DJ-1 in cardiac hypertrophy.
Consistent with the reported role of DJ-1 as an inhibitor of SUMOylation (52, 53), knocking down DJ-1 expression in cardiomyocytes further enhances PARIS SUMOylation (Fig. 9), suggesting that DJ-1 is a SUMO inhibitor of PARIS. The observation that DJ-1 inhibits PARIS SUMOylation was further verified by generating an oxidation-resistant mutant of DJ-1 (DJ-1C106A) (Fig. 11A). DJ-1 contains 3 cysteine residues (C46, C53, and C106), among which C106 is the most vulnerable to oxidation (54, 55). We observed that DJ-1C106 remains resistant to degradation, as well as inhibiting PARIS SUMOylation, although DJ-1WT fails to inhibit PARIS SUMOylation in the presence of ROS, further confirming that DJ-1 functions as a deSUMOylating agent of PARIS (Fig. 11B and C). The interaction of DJ-1WT and PARIS is reduced after H2O2 treatment; however, mutant DJ-1 binds to PARIS effectively (Fig. 11D). Moreover, PGC1α promoter activity is enhanced by the stabilized DJ-1C106A in cells in which ROS has accumulated. In contrast, the activity is reduced in the presence of DJ-1WT in H2O2-treated cells (Fig. 11E). Therefore, we have uncovered a mechanism in which DJ-1 regulates PARIS SUMOylation and recruitment on the PGC1α promoter, thereby modulating PGC1α transcription in cardiomyocytes. However, the exact mechanism by which DJ-1 regulates PARIS SUMOylation is as yet unclear. It is possible that DJ-1 directly binds to PARIS, causing its deSUMOylation. Alternatively, DJ-1 may not have any deSUMOylase activity; instead, it might either work as a chaperone or bring other deSUMOylases to PARIS, affecting SUMOylation of PARIS.
Further validations in vivo show enhanced PARIS protein SUMOylation (Fig. 13B) under hypertrophic stress, resulting in increased PGC1α promoter binding (Fig. 12E) and transcriptional repression. Consistent with the findings in vitro (Fig. 6C), increased ROS accumulation is also observed in hypertrophied heart representing increased oxidative stress during cardiac hypertrophy (Fig. 13A). Furthermore, DJ-1 does not associate with PARIS in hypertrophied myocardium, indicating degradation of DJ-1 under oxidative stress (Fig. 13C). Silencing PARIS restores the mitochondrial network (Fig. 13D) in neonatal rat ventricular cardiomyocytes, confirming the prohypertrophic role of PARIS through regulating mitochondrial dysfunction.
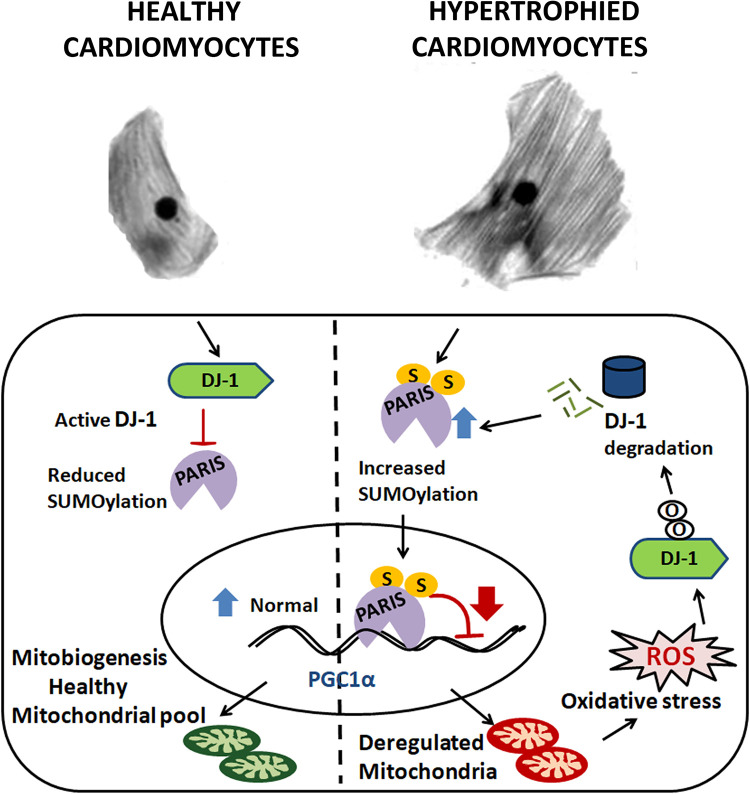
In conclusion, our data for the first time unveil a novel role for PARIS in regulating hypertrophy-associated mitochondrial dysfunction. We depict a regulatory mechanism where DJ-1 functions as an oxidative stress sensor during cardiac hypertrophy. DJ-1 undergoes proteasomal degradation in response to oxidative stress, leading to stabilization of SUMOylated PARIS. This enhanced PARIS SUMOylation subsequently represses PGC1α transcription, resulting in aggravated mitochondrial function and oxidative stress. Hence, our study depicts how mitochondrial dysfunction contributes to a feed-forward loop involving the DJ-1–PARIS axis, resulting in further deterioration and pathological manifestation (Fig. 14). Validation of this novel regulatory mechanism in a genetic animal model of cardiac hypertrophy may identify a therapeutic target to combat cardiovascular diseases.
FIG 14.
Schematic representation depicting a feed-forward loop involving PARIS-mediated mitochondrial dysfunction in cardiomyocytes, which oxidative stress generated due to mitochondrial dysfunction during cardiac hypertrophy; this in turn regulates DJ-1 and subsequent PARIS SUMOylation, resulting in downregulation of PGC1α. In healthy cardiomyocytes, active DJ-1 inhibits PARIS SUMOylation, thus maintaining normal PGC1α transcription and mitobiogenesis. In hypertrophy, enhanced PARIS SUMOylation inhibits PGC1α transcription, leading to mitochondrial dysfunction and generation of ROS in cardiomyocytes. The resulting oxidative stress in turn degrades DJ-1, which acts in a feed-forward loop relieving inhibition of PARIS SUMOylation. Thus, activated PARIS downregulates PGC1α, leading to mitochondrial dysfunction.
MATERIALS AND METHODS
Cell culture and treatment of cells.
H9C2(2-1) is a well-established model for studying the molecular mechanism of myocytes (56–58). H9C2(2-1) cardiomyocytes were acquired from the National Centre for Cell Science (Pune, India) and cultured in Dulbecco’s modified Eagle medium (DMEM) with high glucose (4.5 g/liter), sodium bicarbonate (3.7 g/liter), and fetal bovine serum (FBS; 10%) in an incubator maintained at 37°C, 5% CO2, and 80% relative humidity (RH). Cells were serum starved for 18 to 24 h before experimentation.
Since H9C2 cell lines are very difficult to transfect, HEK293 cells were used to perform most of the transfection experiments. HEK293 cells were cultured in DMEM with high glucose (4.5 g/liter), sodium bicarbonate (3.7 g/liter), and FBS (10%) in an incubator at 37°C, 5% CO2, and 80% RH.
Animal studies.
For the generation of the hypertrophic model in vivo, 24-week-old male Sprague-Dawley rats (Rattus norvegicus) were used. Sprague-Dawley rat pups (2 days old) were also utilized for the preparation of neonatal rat ventricular myocytes. The investigation conforms to the guidelines of the Instructional Animal Ethics Committee of CSIR Indian Institute of Chemical Biology.
Generation of hypertrophy in vitro and in vivo.
Hypertrophy was generated by treating serum-starved H9C2(2-1) cells with 100 μM phenylephrine (PE; Sigma-Aldrich, USA), 10 μM isoproterenol (ISO; Sigma-Aldrich, USA), or 300 nM angiotensin II (AngII; Sigma-Aldrich, USA) for 24 h (59, 60).
Left ventricular hypertrophy in 24-week-old rats was generated by ISO treatment for 14 days (61, 62). Briefly, animals were anesthetized with an intraperitoneal injection of a combination of 80 mg/kg ketamine and 9.3 mg/kg xylazine. Infusion of ISO (5 mg/kg/day) for 14 days was performed by subcutaneously implanting osmotic minipumps (Alzet, Cupertino, CA). Hypertrophy was assessed by determining the ratio of heart weight to body weight and expression of hypertrophy marker genes. The age-matched sham-treated controls were infused with 0.9% saline. At the end of the experiment, animals were euthanized via intravenous ketamine injection, and hearts were dissected. For subsequent protein and RNA extraction, cardiac tissue samples from all groups were collected and preserved at −80°C.
Isolation and culture of neonatal rat ventricular myocytes.
Neonatal rat ventricular myocytes (NRVM) from 2-day-old Sprague-Dawley rat pups were isolated as described earlier (63, 64) with few modifications. Hearts were dissected and rinsed in 1× Ads buffer (116.3 mM NaCl, 19.7 mM HEPES, 9.4 mM NaH2PO4, 5.5 mM glucose, 5.3 mM KCl, 0.83 mM MgSO4 [pH 7.4]). Atria were removed, and ventricles were minced and incubated at 37°C in a solution of enzymes containing 0.2% collagenase type II (381 U/mg) and pancreatin (0.6 mg/ml) in 1× Ads buffer in the presence of 95% O2 and 5% CO2 for 4 successive digestions of 10 min each. The supernatant of the first tissue digestion was discarded. The supernatants obtained after each digestion were centrifuged for 5 min at 1,000 rpm. The cell pellets obtained in each round of digestion were resuspended in M199 growth medium (supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin, pH 7.2). The resuspended cells were pooled, preplated, and incubated for 1 h in a CO2 incubator for cardiomyocyte enrichment. The supernatants containing cardiomyocytes were collected by brief centrifugation and resuspended in growth medium. The cells were plated onto collagen I-coated coverslips (Biocoat; BD Labware, Bedford, MA) and incubated in a CO2 incubator. The medium was replaced with growth medium after 24 h. The majority of the cells were cardiomyocytes, as confirmed by immunostaining with cardiac tissue-specific sarcomeric α-actinin antibody.
Plasmid construction and cloning.
A green fluorescent protein (GFP)-tagged plasmid (PIRES-hrGFP1α) expressing rat DJ-1 was overexpressed using a standard molecular biology cloning technique. The PARIS Cfugw construct was kindly provided by T. M. Dawson's laboratory (The Johns Hopkins Medical Institutions, Baltimore, MD). PARIS was subcloned into the PIRES-hrGFP-1α vector. The promoter region of Pgc1α (−1 to −1000 relative to the TSS) was cloned into the PGL3 vector upstream of the luciferase gene (Pgc1αluc). For the construction of His-tagged wild-type and mutant PARIS, PIRES-hrGFP-1α–PARIS was digested with BamHI and EcoRI and subcloned into the pcDNA4/myc/HisC vector. Human His6-SUMO1 was PCR amplified with a forward primer containing the 6His sequence and cloned into PIRES-hrGFP-1α using a standard molecular biology cloning technique. The primers used for plasmid construction are listed in Table 1.
TABLE 1.
Primers used for plasmid constructiona
| Primer | Sequence |
|---|---|
| Dj-1WT | F: 5′CGCGGATCCGAAATGGCATCCAAAAGAGC3′ |
| R: 5′CCGGAATTCCTCTCGTCTTTGAGAACAAGC3′ | |
| Pgc1αluc | F: 5′CGGGGTACCCAGGGCATAGGGTTGGAATC3′ |
| R: 5′AATGCTAGCTCAACTCCAATCCACTCTG3′ | |
| His6SUMO1 | F1: 5′ACTAAGCTTACCATGCATCATCACCATCACCATTCTGACCAGGAGGCAAAAC3′ |
| R1: 5′TATGGATCCCTAAACTGTTGAATGACCCCCCGTTTGTTCCTG3′ |
F, forward; R, reverse.
Site-directed mutagenesis.
A DJ-1 oxidation-resistant mutant (DJ-1C106A) and mutants with mutations of SUMO1 binding sites in PARIS (ZK189R, ZK286R, and ZM2R) were generated by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), following the manufacturer's instructions. The mutation was verified by sequencing from Bioserve Biotechnologies (India) Pvt Ltd. Primers used for plasmid construction are listed in Table 2.
TABLE 2.
Primers used for plasmid construction for site-directed mutagenesisa
| Primer | Sequence |
|---|---|
| PAR7 C106A | F: 5′GCCGTAGGACCCGCAGCGATGGCAGCTATGAG3′ |
| R: 5′CTCATAGCTGCCATCGCTGCGGGTCCTACGGC3′ | |
| ZK189R | F: 5′CACCCTCCTGCCTGATCTGCATCAAGAGGTCTG3′ |
| R: 5′CAGACCTCTTGATGCAGATCAGGCAGGAGGGTG3′ | |
| ZK286R | F: 5′GATGTAAAAATTGTAATAAGAACAGAAGTCCAGGAAGAGGAGGTG3′ |
| R: 5′CCACCTCCTCTTCCTGGACTTCTGTTCTTATTACAATTTTTACATC3′ |
F, forward; R, reverse.
Treatment of cells.
(i) Transfection of cells. HEK293 cells were transfected with DJ-1WT, DJ-1C106A, PARIS, or ZM2R constructs using Lipofectamine 2000 (Invitrogen, CA).
(ii) siRNA treatment. H9C2 cardiomyocytes were treated with either PARIS siRNA and DJ-1 siRNA, PGC1α siRNA, or scrambled siRNA (scrsiRNA) (Silencer Select siRNA; Ambion) using Lipofectamine RNAiMax (Invitrogen, CA) as per the manufacturers’ instructions.
(iii) H2O2 treatment. Cells were treated with 100 μM H2O2 (Sigma-Aldrich, USA) for 24 h. For scavenging the accumulated ROS, cells were pretreated with 50 μM ascorbate (Sigma-Aldrich, USA) for 2 h followed by H2O2 treatment for 24 h.
(iv) Proteasomal inhibition by MG132. Hypertrophied myocytes were treated with various doses of MG132 (Z-Leu-Leu-Leu-al) (500 nM to 5 μM) (Millipore) for 2 h before PE treatment. Control cells were treated with an equivalent amount of dimethyl sulfoxide (DMSO).
RNA isolation and cDNA synthesis.
Total RNA from cells was isolated using TRI reagent (Sigma-Aldrich, USA) following the manufacturer's instructions. One microgram of total RNA was reverse transcribed to cDNA using a Verso cDNA synthesis kit (Thermo Fisher Scientific, MA) using the manufacturer's protocol.
Real-time qRT-PCR.
Relative quantification of transcript levels was performed using TaqMan probes (Anp, Bnp, βMhc, Nrf1, Mfn1, Mfn2, Pgc1α, PARIS, and Gapdh) (Applied Biosystems, CA) using the manufacturer’s protocol. All reactions were performed in a total volume of 20 μl. Relative fold changes in mRNA transcript levels were calculated from cycle threshold (CT) values using 2−ΔΔCT methods. Gapdh expression was used as an internal reference control.
Protein isolation.
For in vivo experiments, hearts were excised and perfused in chilled phosphate-buffered saline (PBS). Then, the heart tissues were homogenized in 1× radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher Scientific, MA) with added protease inhibitor cocktail (Sigma-Aldrich, USA) using a homogenizer. For in vitro experiments, cells were lysed using 1× RIPA lysis buffer in the presence of a protease inhibitor cocktail. For SUMOylation detection, proteins were isolated in 1× RIPA lysis buffer in the presence of a protease inhibitor cocktail and 20 mM N-ethylmaleimide (NEM). The tissue and cell lysates were then subjected to centrifugation at 16,000 × g for 20 min at 4°C. The supernatants were collected from each sample, and concentrations of protein were estimated with a DC protein assay (Bio-Rad, CA) at 750 nm using a UV–visible-light (Vis) spectrophotometer (Bio-Rad, CA).
For isolation of proteins under denaturing condition and to detect endogenous PARIS SUMOylation, cells were harvested after the respective treatments and lysed using 2× SDS lysis buffer (150 mM Tris-HCl [pH 7.2], 4% sodium dodecyl sulfate [SDS], 20% glycerol, 20 mM NEM).The lysates were boiled at 100°C for 10 min. The lysates were subjected to high-speed centrifugation at room temperature for 15 min. The lysates were diluted (1:10) to remove SDS before immunoprecipitation was performed (65).
Nuclear fractionations were carried out using a commercially available kit (Affymetrix nuclear fractionation kit) according to the manufacturer's protocol. Actin and lamin B1 were used as cytosolic and nuclear loading controls, respectively.
Western blotting.
Isolated protein samples (40 μg) were resolved in 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were incubated in 5% bovine serum albumin (BSA)–Tris-buffered saline containing Tween (TBST) for 1 h at room temperature. The membranes were separately incubated with primary antibodies against PARIS (Sigma-Aldrich, USA), MFN2, MFN1 (Abcam, Cambridge, UK), DJ-1, Drp1, GAPDH, lamin B1, GFP, SUMO1 (Cell Signaling Technology, Danvers, MA), TFAM, CPT1, His (Santa Cruz Biotechnology, Inc., USA), PGC1α (Novus Biologicals, USA), actin, and Flag (Sigma-Aldrich, USA), diluted in 5% BSA–TBST overnight at 4°C. The membranes were incubated with secondary antibodies conjugated with alkaline phosphatase diluted in 5% BSA–TBST followed by three washes with 1× TBST at each step. The detection was performed by nitroblue tetrazolium (NBT)–5-bromo-4-chloro-3-indolylphosphate (BCIP), and subsequent densitometric quantifications were performed by using ImageJ software. Each blot was normalized with its respective loading control before calculation of the fold changes.
Coimmunoprecipitation.
Five hundred micrograms of the protein samples in IP lysis buffer (Thermo Fisher Scientific, MA) with added protease inhibitor cocktail was subjected to coimmunoprecipitation. Pierce protein A/G magnetic beads (Thermo Fisher Scientific, MA) were coupled with PARIS, DJ-1, SUMO1, and Flag antibodies or nonspecific rabbit or mouse IgG by incubating them at 4°C for 4 h. Protein lysates were then incubated with these antibody-coupled A/G agarose beads or control A/G agarose beads overnight at 4°C with end-to-end rotation to ensure homogeneous mixing. The proteins were eluted from beads using 0.1 M glycine HCl (pH 2.5 to 3), and the eluted samples were neutralized by the addition of 1/10 volume of 1 M Tris HCl (pH 8.5). The elution fraction was subjected to Western blot analysis. Ten percent of the starting material was used as the input. Blots were detected with NBT-BCIP, and subsequent densitometric quantification was performed with ImageJ software.
Ni-NTA pulldown assay for enrichment of SUMOylated PARIS.
His-tagged proteins were isolated under denaturing condition and were purified by Ni-NTA chromatography as described previously (66). HEK293 cells were either transfected with His-tagged PARIS or cotransfected with His-tagged SUMO and wild-type or mutant PARIS, followed by respective treatments. Cells were then harvested and lysed in lysis buffer (6 M guanidine HCl; 10 mM Tris-HCl, pH 8; 100 mM sodium phosphate, 10 mM imidazole, pH 8.0; 5 mM β mercaptoethanol; 0.1% Triton X-100), and viscosity was reduced by vortexing followed by centrifugation at 16,000 × g for 20 min. The supernatants were collected and protein concentrations were estimated by DC protein assay (Bio-Rad, CA) at 750 nm using a UV-Vis spectrophotometer (Bio-Rad, CA). Equal amounts of protein were mixed with 40 μl of Ni–NTA–agarose beads (Invitrogen, Chatsworth, CA) and incubated for 3 h at room temperature. The beads were successively washed with wash buffer A (8 M urea, 10 mM Tris, 100 mM sodium phosphate buffer [pH 8.0], 0.1% Triton X-100, 5 mM β-mercaptoethanol) and wash buffer B (8 M urea, 10 mM Tris, 100 mM sodium phosphate buffer [pH 6.3], 0.1% Triton X-100, 5 mM β-mercaptoethanol). After the last wash, the beads were eluted with elution buffer (200 mM imidazole, 5% SDS, 150 mM Tris-HCl [pH 6.7], 30% glycerol, 720 mM β-mercaptoethanol, 0.0025% bromophenol blue). The eluted samples were subjected to Western blot analysis for detection with anti-PARIS antibody.
Immunocytochemistry.
The cells were seeded and cultured in 6-well dishes on coverslips, fixed in 4% paraformaldehyde for 20 min, and permeabilized with 0.1% Triton X-100 for 10 min, followed by blocking with 3% BSA for an hour at room temperature. For immunofluorescence, the cells were incubated with Alexa Fluor 546-phalloidin in 1% BSA for 45 min at room temperature. The cells were stained with phalloidin, and images were taken in a fluorescence microscope using a 20× lens objective in an EVOS FL inverted microscope. The cell size was measured by calculating region of interest (ROI) using ImageJ software. Mitochondria were localized by staining with MitoTracker Red CMXROS (300 nM) (Thermo Fisher Scientific, MA) for 30 min before fixing in paraformaldehyde. The cells were also incubated with primary antibodies against PARIS overnight, followed by Alexa Fluor 488 for PARIS for 1 h. Neonatal rat ventricular myocytes were incubated with anti-PARIS antibody, followed by Alexa Fluor 350-conjugated secondary antibody and anti-sarcomeric α-actinin antibody and then by Alexa Fluor 488-conjugated secondary antibody. Nuclear counterstaining was performed with NucBlue (Invitrogen, CA). Cells were washed 3 times after each step with 1× PBS. Stained cells were mounted with ProLong Gold antifade (Invitrogen, CA), and images were captured within 24 h.
For live-cell experiments, the cells were seeded and cultured in 35-mm glass-bottom dishes (MatTek, Ashland, MA). The cells from different treatment groups were stained with JC1 (Cayman Chemical, USA), a potential-dependent dye, for 30 min and then images were captured using a confocal microscope. JC1 dye exhibits potential-dependent accumulation in mitochondria, giving rise to J aggregates, indicated by a fluorescence emission shift from green (529 nm) to red (590 nm). The fluorescence intensity ratio of red to green is a good indicator of mitochondrial membrane potential.
Mass spectrometry.
H9C2 cell lysates were coimmunoprecipitated with the anti-PARIS antibody to check the possible interacting partners of PARIS. The proteins bound to PARIS were eluted, followed by in-solution tryptic digestion for proteomic analysis. Dithiothreitol (DTT; 100 mM) was added to the eluate and kept at 60°C for 30 min. Then the samples were cooled to room temperature. Iodoacetamide (IAA; 200 mM) was added, and samples were kept at room temperature for 1 h. Next, trypsin (1 μg/100 μg of protein) was added for in-solution digestion, and the solution was held at 37°C for overnight. The reaction was stopped on the next day by adding 0.1% formic acid, and the solution was held for 5 to 10 min at room temperature. The samples were lyophilized, reconstituted in 0.1% formic acid, and analyzed using a high-resolution mass spectrometer (Orbitrap, LTQ; Thermo, USA).
Assessment of mitochondrial fragmentation.
A confocal microscope was used to measure mitochondrial morphology in response to stress and also in PARIS knockdown hypertrophied myocytes. The amount of fragmentation was quantified by measuring the length of the mitochondrial filament. H9C2 cells were seeded on coverslips. The cells were treated with either scrambled siRNA or PARIS siRNA in the presence or absence of PE. The cells were stained with MitoTracker Red CMXROS (300 nM) (Thermo Fisher Scientific, MA) for 30 min before being washed three times with PBS. Images were acquired on a Leica SP8 confocal microscope (60×).
Mitochondrial morphology was analyzed on ImageJ 1.46r using the Mito-Morphology macro (67) tool. Mitochondrial interconnectivity and elongation were measured from the epifluorescence micrograph. The form factor (perimeter2/4π · area) indicates the interconnectivity of mitochondria, and circularity indicates roundness; values (0 to 1) were measured. A circularity value of 1 indicates perfect spheroids. The aspect ratio (AR), which is the ratio of major axis to minor axis and reflects the length-to-width ratio, was measured.
Microscopy.
Confocal imaging was performed primarily (Fig. 3C and 12D) on the Leica microscope (Leica Microsystems, Mannheim, Germany); three lasers were used (405, 488, and 594 nm). Oil immersion objectives (63×, 1.4 numerical aperture) were used for image acquisition via Leica LAS-X software. Other confocal imaging (Fig. 1C and 3A) was performed on a Revolution XD system (Andor Technology, Belfast, UK) equipped with an Olympus IX81 microscope, a Yokogawa CSU-X1 spinning disc confocal unit, and three laser lines (405, 488, and 594 nm). A 60×/1.42 numerical aperture oil immersion objective was used for image acquisition via Andor iQ version 2.7 software. Images in Fig. 3B were acquired using an Olympus FVi10 microscope (Olympus Corporation, Tokyo, Japan), with one laser (562 nm) and a 0.60×/1.4 numerical aperture oil immersion objective.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed using a commercially available kit (Millipore) according to the manufacturer’s protocol. Briefly, the cells were fixed in 1% paraformaldehyde and suspended in ChIP lysis buffer. Lysates were sonicated (Qsonica, USA) to shear DNA to an average fragment size of 500 to 1,000 bp. Approximately 25 μg of sheared chromatin was immunoprecipitated using either anti-PARIS antibody or nonspecific rabbit IgG or H3 as a positive control. A 20-μl portion of the sheared chromatin without antibody incubation was used as the input sample. PARIS-bound chromatin was eluted with ChIP elution buffer. DNA was de-cross-linked from immunoprecipitated chromatin by heating overnight at 65°C. DNA was then isolated using phenol-chloroform (Sigma-Aldrich, USA) and ethanol precipitation, and samples were subsequently detected by qPCR using SYBR green with the specific primers listed in Table 3. A nonspecific primer was also used to confirm the specificity of the ChIP experiment.
TABLE 3.
Primers for ChIP qPCR using SYBR greena
| Primer | Sequence |
|---|---|
| IRS1 | F1: 5′CGGGGTACCCAGGGCATAGGGTTGGAATC3′ |
| R1: 5′AATGCTAGCTCAACTCCAATCCACTCTG3′ | |
| IRS2 | F2: 5′CGGGGTACCCAGGGCATAGGGTTGGAATC3′ |
| R2: 5′AATGCTAGCCCTTACTGAGAGTGAACTGAAG3′ | |
| IRS3 | F3: 5′TAAGCACTCGAGCAGGGCATAGGGTTGGAATC3′ |
| R3: 5′TAAGCAAAGCTTCCTTACTGAGAGTGAACTGAAG3′ | |
| Nonspecific | F: 5′AGCTGTTGGTCATCTCATTCTGT3′ |
| R: 5′GACCCTGAAAAAGGGGTCTGT3′ |
F, forward; R, reverse.
Dual-luciferase assay.
Dual-luciferase activity assays were carried out using a commercially available kit (Promega, USA) according to the manufacturer's protocol. Briefly, H9C2 cardiomyocytes were cotransfected with a Pgc1αluc vector and a Renilla vector using Lipofectamine 3000 (Invitrogen, CA). After 48 h, luciferase activity was measured using a plate reader (Thermo Fisher Scientific, MA) with the kit-provided LARII reagent and Stop and Glow reagent according to the manufacturer's instruction. All the data were normalized to those for Renilla luciferase.
Measurement of cellular ROS.
Cellular reactive oxygen species (ROS) production was quantified using a commercially available kit (DCFDA/H2DCFDA cellular reactive oxygen species detection assay kit; Abcam, Cambridge, UK). Briefly, cells were lysed using 1× lysis buffer, and 20 μg of protein from the cell lysate of each sample was incubated with 25 μM 2,7-dichloro-fluorescein (DCFDA) solution in the 1× buffer. Cellular ROS production was measured at excitation and emission wavelengths of 485 and 535 nm, respectively, using the SpectraMax Paradigm multimode detection platform (Molecular Devices, CA).
Seahorse XFe24 analysis.
The oxygen consumption rate (OCR), which is an indicator of mitochondrial respiration, was measured with a Seahorse XFe24 extracellular flux analyzer (Agilent Technologies, CA). The cells were evenly seeded (50,000 cells/well) in the XFe24 cell culture plate and allowed to attach. The cells were transfected either with scrsiRNA or PARIS siRNA in the presence or absence of PE. The cell culture media were replaced with XF base medium (Seahorse; Bioscience) and kept in a non-CO2 incubator at 37°C for 1 h. Three basal measurements of the OCR were taken before the injection of pharmacological manipulators of mitochondrial respiratory chain proteins (used for measurement of several bioenergetic parameters). After measurement of the basal OCR, 1 μM oligomycin A (Sigma-Aldrich, USA) was injected into each sample to inhibit the proton flow through ATP synthase and blocking of all ATP-linked oxygen consumption. Thereafter, 750 nM carbonyl cyanide-p-trifluoromethoxy phenylhydrazone (FCCP; Sigma-Aldrich, USA) was injected for measuring maximum OCR, followed by 1 μM antimycin A (Sigma-Aldrich, USA) and rotenone for inhibiting all mitochondrial respiratory complexes. After the injection of each inhibitor, three measurements were performed for each sample, and the measurements were normalized to the total protein content of the respective samples using a DC protein assay (Bio-Rad, CA). Basal mitochondrial OCR was derived by subtracting nonmitochondrial OCR (remaining OCR after antimycin A/rotenone addition). Maximum OCR was stimulated by FCCP injection. Maximum OCR was calculated by deducting nonmitochondrial respiratory rate from the maximum respiratory rate after FCCP injection. The difference in OCR following oligomycin A inhibition and antimycin A/rotenone inhibition is considered OCR due to proton leak. The effect of PARIS knockdown on the OCR of hypertrophied H9C2 myocytes was plotted against time.
Mitobiogenesis assay.
The mitochondrial biogenesis assay was performed in PARIS knockdown hypertrophied cardiomyocytes using a MitoBiogenesis in-cell ELISA kit (Abcam, Cambridge, UK) according to the manufacturer’s protocol. Briefly, H9C2 cardiomyocytes were seeded (50,000 cells/well) on 96-well plates. The cells were then divided into 4 groups: control cells, PE-treated cells transfected with scrsiRNA, cells transfected with PARIS siRNA (48 h) followed by PE treatment, and cells treated with chloramphenicol (10 μM) used as a positive control. The cells were fixed with 4% paraformaldehyde. The cells were treated with freshly prepared 0.5% acetic acid for 5 min to block endogenous alkaline phosphatase activity. The cells were permeabilized with 0.1% Triton X-100 for 30 min and then blocked with 2× blocking solution for 1 h. These cells were then incubated with primary antibodies against mtDNA-encoded COX-I (cytochrome c oxidase subunit 1), and nuclear-DNA-encoded SDH-A (succinate dehydrogenase complex, subunit A) proteins. Cells were then incubated with secondary antibodies, i.e., allophycocyanin (AP) for SDH-A and horseradish peroxidase (HRP) for COX-I. The reactions were sequentially developed first with AP reagent and then with HRP development solution. A 15-min kinetic reaction with a 1-min interval was recorded using the SpectraMax Paradigm multimode detection platform (Molecular Devices, CA). All the experiments were performed in triplicate and with a minimum of 3 repetitions per group. In this assay, the activities of two mitochondrial enzymes were measured simultaneously by spectrophotometry. Subunit I of mitochondrial complex IV (COX-I) is encoded by mtDNA, whereas the subunit of complex II (SDH-A) is encoded by nuclear DNA (nDNA). Thus, the ratio of COX–1 activity to SDH-A activity represents the status of mitochondrial biogenesis.
Statistical analysis.
The results are presented as means and standard deviations (SD). Each experiment was repeated at least three times. For each experiment, the fold changes were calculated in different groups relative to the control group, and all the control samples were set to a value of 1. Differences between two groups were evaluated by Student's t test (independent), and the complete data set among all treatment groups (three or more, as indicated) was evaluated by one-way analysis of variance (ANOVA). Post hoc analysis was conducted for testing statistical significance among different groups. A P value of <0.05 was considered the threshold value for statistical significance between the control and various treatment groups.
ACKNOWLEDGMENTS
We thank Ted M. Dawson (Johns Hopkins Medical Institutions, Baltimore, MD, USA) for the kind gift of PARIS Cfugw construct. We gratefully acknowledge the CSIR-IICB Central Instrument facility for allowing to use of confocal microscopes (STED, Leica, Germany; Zeiss, Germany and Olympus, Japan). We thank Sounak Bhattacharya and Banasri Das for assistance in confocal microscopy. Technical assistance provided by Santu Paul in high-resolution mass spectrometry (Orbitrap, LTQ, Thermo Fischer, USA) is gratefully acknowledged. We also thank Swapan Mandal for laboratory assistance.
This study was supported by grants (BSC 0206 and MLP 115) from the Council of Scientific and Industrial Research (CSIR), New Delhi, India. DM is a recipient of a fellowship from CSIR [31/002(1012)/2015-EMR-I].
We declare that we have no conflicts of interest with the contents of this article.
Arun Bandyopadhyay and Dibyanti Mukherjee designed the study and contributed reagents. Dibyanti Mukherjee and Vivek Chander performed experiments. Dibyanti Mukherjee, Vivek Chander, and Arun Bandyopadhyay analyzed the data. Arun Bandyopadhyay and Dibyanti Mukherjee wrote the manuscript. Arun Bandyopadhyay, Vivek Chander, Dibyanti Mukherjee edited the manuscript.
REFERENCES
- 1.Doenst T, Nguyen TD, Abel ED. 2013. Cardiac metabolism in heart failure: implications beyond ATP production. Circ Res 113:709–724. doi: 10.1161/CIRCRESAHA.113.300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosca MG, Hoppel CL. 2013. Mitochondrial dysfunction in heart failure. Heart Fail Rev 18:607–622. doi: 10.1007/s10741-012-9340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosca MG, Tandler B, Hoppel CL. 2013. Mitochondria in cardiac hypertrophy and heart failure. J Mol Cell Cardiol 55:31–41. doi: 10.1016/j.yjmcc.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siasos G, Tsigkou V, Kosmopoulos M, Theodosiadis D, Simantiris S, Tagkou NM, Tsimpiktsioglou A, Stampouloglou PK, Oikonomou E, Mourouzis K, Philippou A, Vavuranakis M, Stefanadis C, Tousoulis D, Papavassiliou AG. 2018. Mitochondria and cardiovascular diseases-from pathophysiology to treatment. Ann Transl Med 6:256. doi: 10.21037/atm.2018.06.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scarpulla RC 2011. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta 1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang H, Kang S-U, Zhang S, Karuppagounder S, Xu J, Lee Y-K, Kang B-G, Lee Y, Zhang J, Pletnikova O, Troncoso JC, Pirooznia S, Andrabi SA, Dawson VL, Dawson TM. 2016. Adult conditional knockout of PGC-1α leads to loss of dopamine neurons. eNeuro 3:ENEURO.0183-16.2016. doi: 10.1523/ENEURO.0183-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 8.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. 2000. Peroxisome proliferator–activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest 106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. 2006. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-γ coactivator 1α. Proc Natl Acad Sci U S A 103:10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ventura-Clapier R, Garnier A, Veksler V. 2008. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1α. Cardiovasc Res 79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 11.Lehman JJ, Boudina S, Banke NH, Sambandam N, Han X, Young DM, Leone TC, Gross RW, Lewandowski ED, Abel ED, Kelly DP. 2008. The transcriptional coactivator PGC-1α is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. Am J Physiol Heart Circ Physiol 295:H185–H196. doi: 10.1152/ajpheart.00081.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Marcos PJ, Auwerx J. 2011. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr 93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin J-H, Ko HS, Kang H, Lee Y, Lee Y-I, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. 2011. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson's disease. Cell 144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishida T, Yamada Y. 2016. SUMOylation of the KRAB zinc-finger transcription factor PARIS/ZNF746 regulates its transcriptional activity. Biochem Biophys Res Commun 473:1261–1267. doi: 10.1016/j.bbrc.2016.04.051. [DOI] [PubMed] [Google Scholar]
- 15.Piqueras-Flores J, López-García A, Moreno-Reig Á, González-Martínez A, Hernández-González A, Vaamonde-Gamo J, Jurado-Román A. 2018. Structural and functional alterations of the heart in Parkinson’s disease. Neurol Res 40:53–61. doi: 10.1080/01616412.2017.1390933. [DOI] [PubMed] [Google Scholar]
- 16.Escobar-Ramirez A, Vercoutter-Edouart A-S, Mortuaire M, Huvent I, Hardivillé S, Hoedt E, Lefebvre T, Pierce A. 2015. Modification by SUMOylation controls both the transcriptional activity and the stability of delta-lactoferrin. PLoS One 10:e0129965. doi: 10.1371/journal.pone.0129965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P, Costanzo P. 2013. KRAB-zinc finger proteins: a repressor family displaying multiple biological functions. Curr Genomics 14:268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sampson DA, Wang M, Matunis MJ. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem 276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson KA, Henley JM. 2010. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pichler A, Fatouros C, Lee H, Eisenhardt N. 2017. SUMO conjugation–a mechanistic view. Biomol Concepts 8:13–36. doi: 10.1515/bmc-2016-0030. [DOI] [PubMed] [Google Scholar]
- 21.Gardner DG 2003. Natriuretic peptides: markers or modulators of cardiac hypertrophy? Trends Endocrinol Metab 14:411–416. doi: 10.1016/S1043-2760(03)00113-9. [DOI] [PubMed] [Google Scholar]
- 22.Mair J, Friedl W, Thomas S, Puschendorf B. 1999. Natriuretic peptides in assessment of left-ventricular dysfunction. Scan J Clin Lab Invest 59:132–142. doi: 10.1080/00365519909168337. [DOI] [PubMed] [Google Scholar]
- 23.Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. 2003. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes 52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 24.Sentis S, Le Romancer M, Bianchin C, Rostan MC, Corbo L. 2005. Sumoylation of the estrogen receptor α hinge region regulates its transcriptional activity. Mol Endocrinol 19:2671–2684. doi: 10.1210/me.2005-0042. [DOI] [PubMed] [Google Scholar]
- 25.Dikalov SI, Nazarewicz RR. 2013. Angiotensin II-induced production of mitochondrial reactive oxygen species: potential mechanisms and relevance for cardiovascular disease. Antioxid Redox Signal 19:1085–1094. doi: 10.1089/ars.2012.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu Y, Lambert JP, Nicholson CK, Kim JJ, Wolfson DW, Cho HC, Husain A, Naqvi N, Chin L-S, Li L, Calvert JW. 2016. DJ-1protects the heart against ischemia–reperfusion injury by regulating mitochondrial fission. J Mol Cell Cardiol 97:56–66. doi: 10.1016/j.yjmcc.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu P-H, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM. 2005. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab 1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Duncan JG, Finck BN. 2008. The PPAR-PGC-1 axis controls cardiac energy metabolism in healthy and diseased myocardium. PPAR Res 2008:253817. doi: 10.1155/2008/253817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang H, Ward WF. 2006. PGC-1α: a key regulator of energy metabolism. Adv Physiol Educ 30:145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 30.Lu Z, Xu X, Hu X, Fassett J, Zhu G, Tao Y, Li J, Huang Y, Zhang P, Zhao B. 2010. PGC-1α regulates expression of myocardial mitochondrial antioxidants and myocardial oxidative stress after chronic systolic overload. Antioxid Redox Signal 13:1011–1022. doi: 10.1089/ars.2009.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DI W, Lv J, Jiang S, Lu C, Yang Z, Ma Z, Hu W, Yang Y, Xu B. 2018. PGC-1: the energetic regulator in cardiac metabolism. Curr Issues Mol Biol 28:29–46. doi: 10.21775/cimb.028.029. [DOI] [PubMed] [Google Scholar]
- 32.Schwarzer M, Osterholt M, Lunkenbein A, Schrepper A, Amorim P, Doenst T. 2014. Mitochondrial reactive oxygen species production and respiratory complex activity in rats with pressure overload-induced heart failure. J Physiol 592:3767–3782. doi: 10.1113/jphysiol.2014.274704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorn GW II 2013. Mitochondrial dynamics in heart disease. Biochim Biophys Acta 1833:233–241. doi: 10.1016/j.bbamcr.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legros F, Lombès A, Frachon P, Rojo M. 2002. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell 13:4343–4354. doi: 10.1091/mbc.e02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishihara N, Jofuku A, Eura Y, Mihara K. 2003. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem Biophys Res Commun 301:891–898. doi: 10.1016/s0006-291x(03)00050-0. [DOI] [PubMed] [Google Scholar]
- 36.Knowlton AA, Chen L, Malik ZA. 2014. Heart failure and mitochondrial dysfunction: the role of mitochondrial fission/fusion abnormalities and new therapeutic strategies. J Cardiovasc Pharmacol 63:196–206. doi: 10.1097/01.fjc.0000432861.55968.a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pennanen C, Parra V, López-Crisosto C, Morales PE, Del Campo A, Gutierrez T, Rivera-Mejías P, Kuzmicic J, Chiong M, Zorzano A, Rothermel BA, Lavandero S. 2014. Mitochondrial fission is required for cardiomyocyte hypertrophy mediated by a Ca2+-calcineurin signaling pathway. J Cell Sci 127:2659–2671. doi: 10.1242/jcs.139394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu C, Huang Y, Li L. 2017. Drp1-dependent mitochondrial fission plays critical roles in physiological and pathological progresses in mammals. Int J Mol Sci 18:144. doi: 10.3390/ijms18010144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sansbury BE, Riggs DW, Brainard RE, Salabei JK, Jones SP, Hill BG. 2011. Responses of hypertrophied myocytes to reactive species: implications for glycolysis and electrophile metabolism. Biochem J 435:519–528. doi: 10.1042/BJ20101390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DA Costa RM, Fais RS, Dechandt CR, Louzada-Junior P, Alberici LC, Lobato NS, Tostes RC. 2017. Increased mitochondrial ROS generation mediates the loss of the anti- contractile effects of perivascular adipose tissue in high-fat diet obese mice. Br J Pharmacol 174:3527–3541. doi: 10.1111/bph.13687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu C, Zhang S, Gao X, Gao X, Xu X, Lv Y, Zhang Y, Zhu Z, Zhang C, Li Q, Wong J, Cui Y, Zhang W, Ma L, Wang C. 2012. Roles of Kruppel-associated Box (KRAB)-associated co-repressor KAP1 Ser-473 phosphorylation in DNA damage response. J Biol Chem 287:18937–18952. doi: 10.1074/jbc.M111.313262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsutsui H, Kinugawa S, Matsushima S. 2009. Mitochondrial oxidative stress and dysfunction in myocardial remodelling. Cardiovasc Res 81:449–456. doi: 10.1093/cvr/cvn280. [DOI] [PubMed] [Google Scholar]
- 43.Lu B, Christensen IT, Yu T, Wang C, Yan Q, Wang X. 2019. SUMOylation evoked by oxidative stress reduced lens epithelial cell antioxidant functions by increasing the stability and transcription of TP53INP1 in age-related cataracts. Oxid Med Cell Longev 2019:7898069. doi: 10.1155/2019/7898069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonifati V, Rizzu P, Squitieri F, Krieger E, Vanacore N, van Swieten JC, Brice A, van Duijn CM, Oostra B, Meco G, Heutink P. 2003. DJ-1(PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol Sci 24:159–160. doi: 10.1007/s10072-003-0108-0. [DOI] [PubMed] [Google Scholar]
- 45.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MCJ, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. 2003. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 46.Aleyasin H, Rousseaux MW, Phillips M, Kim RH, Bland RJ, Callaghan S, Slack RS, During MJ, Mak TW, Park DS. 2007. The Parkinson's disease gene DJ-1 is also a key regulator of stroke-induced damage. Proc Natl Acad Sci U S A 104:18748–18753. doi: 10.1073/pnas.0709379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Billia F, Hauck L, Grothe D, Konecny F, Rao V, Kim RH, Mak TW. 2013. Parkinson-susceptibility gene DJ-1/DJ-1 protects the murine heart from oxidative damage in vivo. Proc Natl Acad Sci U S A 110:6085–6090. doi: 10.1073/pnas.1303444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinat C, Shendelman S, Jonason A, Leete T, Beal MF, Yang L, Floss T, Abeliovich A. 2004. Sensitivity to oxidative stress in DJ-1-deficient dopamine neurons: an ES-derived cell model of primary Parkinsonism. PLoS Biol 2:e327. doi: 10.1371/journal.pbio.0020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taira T, Saito Y, Niki T, Iguchi-Ariga SMM, Takahashi K, Ariga H. 2004. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep 5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim RH, Smith PD, Aleyasin H, Hayley S, Mount MP, Pownall S, Wakeham A, You-TEN AJ, Kalia SK, Horne P, Westaway D, Lozano AM, Anisman H, Park DS, Mak TW. 2005. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc Natl Acad Sci U S A 102:5215–5220. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y, Gehrke S, Haque ME, Imai Y, Kosek J, Yang L, Beal MF, Nishimura I, Wakamatsu K, Ito S, Takahashi R, Lu B. 2005. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc Natl Acad Sci U S A 102:13670–13675. doi: 10.1073/pnas.0504610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong N, Xu J. 2008. Synergistic activation of the human MnSOD promoter by DJ-1 and PGC-1α: regulation by SUMOylation and oxidation. Hum Mol Genet 17:3357–3367. doi: 10.1093/hmg/ddn230. [DOI] [PubMed] [Google Scholar]
- 53.Zhong N, Kim CY, Rizzu P, Geula C, Porter DR, Pothos EN, Squitieri F, Heutink P, Xu J. 2006. DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor. J Biol Chem 281:20940–20948. doi: 10.1074/jbc.M601935200. [DOI] [PubMed] [Google Scholar]
- 54.Yanagida T, Kitamura Y, Yamane K, Takahashi K, Takata K, Yanagisawa D, Yasui H, Taniguchi T, Taira T, Honda T, Ariga H. 2009. Protection against oxidative stress-induced neurodegeneration by a modulator for DJ-1, the wild-type of familial Parkinson’s disease-linked PARK7. J Pharmacol Sci 109:463–468. doi: 10.1254/jphs.08323sc. [DOI] [PubMed] [Google Scholar]
- 55.Fernandez-Caggiano M, Schröder E, Cho H-J, Burgoyne J, Barallobre-Barreiro J, Mayr M, Eaton P. 2016. Oxidant-induced interprotein disulfide formation in cardiac protein DJ-1 occurs via an interaction with peroxiredoxin 2. J Biol Chem 291:10399–10410. doi: 10.1074/jbc.M115.699850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hescheler J, Meyer R, Plant S, Krautwurst D, Rosenthal W, Schultz G. 1991. Morphological, biochemical, and electrophysiological characterization of a clonal cell (H9C2) line from rat heart. Circ Res 69:1476–1486. doi: 10.1161/01.res.69.6.1476. [DOI] [PubMed] [Google Scholar]
- 57.Frentzou GA, Collier ME, Seymour A-ML, Ettelaie C. 2010. Differential induction of cellular proliferation, hypertrophy and apoptosis in H9C2 cardiomyocytes by exogenous tissue factor. Mol Cell Biochem 345:119–130. doi: 10.1007/s11010-010-0565-8. [DOI] [PubMed] [Google Scholar]
- 58.Watkins SJ, Borthwick GM, Arthur HM. 2011. The H9C2 cell line and primary neonatal cardiomyocyte cells show similar hypertrophic responses in vitro. In Vitro Cell Dev Biol Anim 47:125–131. doi: 10.1007/s11626-010-9368-1. [DOI] [PubMed] [Google Scholar]
- 59.Banerjee P, Bandyopadhyay A. 2014. Cytosolic dynamics of annexin A6 trigger feedback regulation of hypertrophy via atrial natriuretic peptide in cardiomyocytes. J Biol Chem 289:5371–5385. doi: 10.1074/jbc.M113.514810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banerjee P, Chander V, Bandyopadhyay A. 2015. Balancing functions of annexin A6 maintain equilibrium between hypertrophy and apoptosis in cardiomyocytes. Cell Death Dis 6:e1873. doi: 10.1038/cddis.2015.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khatua TN, Dinda AK, Putcha UK, Banerjee SK. 2016. Diallyl disulfide ameliorates isoproterenol induced cardiac hypertrophy activating mitochondrial biogenesis via eNOS- Nrf2-Tfam pathway in rats. Biochem Biophys Rep 5:77–88. doi: 10.1016/j.bbrep.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou R, Ma P, Xiong A, Xu Y, Wang Y, Xu Q. 2017. Protective effects of low-dose rosuvastatin on isoproterenol-induced chronic heart failure in rats by regulation of DDAH- ADMA-NO pathway. Cardiovasc Ther 35:e12241. doi: 10.1111/1755-5922.12241. [DOI] [PubMed] [Google Scholar]
- 63.Gomez J, Potreau D, Raymond G. 1994. Intracellular calcium transients from newborn rat cardiomyocytes in primary culture. Cell Calcium 15:265–275. doi: 10.1016/0143-4160(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 64.Bandyopadhyay A, Shin DW, Ahn JO, Kim DH. 2000. Calcineurin regulates ryanodine receptor/Ca2+-release channels in rat heart. Biochemical J 352:61–70. doi: 10.1042/bj3520061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao Y, Pollack D, Nieves E, Winchell A, Callaway M, Vigodner M. 2015. Can your protein be sumoylated? A quick summary and important tips to study SUMO-modified proteins. Anal Biochem 477:95–97. doi: 10.1016/j.ab.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. 2009. Detection of protein SUMOylation in vivo. Nat Protoc 4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- 67.Dagda RK, Cherra SJ, Kulich SM, Tandon A, Park D, Chu CT. 2009. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem 284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]