Abstract
Recent evidence from proteomics and deep massively parallel sequencing studies have revealed that eukaryotic genomes contain substantial numbers of as-yet-uncharacterized open reading frames (ORFs). We define these uncharacterized ORFs as novel ORFs (nORFs). nORFs in humans are mostly under 100 codons and are found in diverse regions of the genome, including in long noncoding RNAs, pseudogenes, 3′ UTRs, 5′ UTRs, and alternative reading frames of canonical protein coding exons. There is therefore a pressing need to evaluate the potential functional importance of these unannotated transcripts and proteins in biological pathways and human disease on a larger scale, rather than one at a time. In this study, we outline the creation of a valuable nORFs data set with experimental evidence of translation for the community, use measures of heritability and selection that reveal signals for functional importance, and show the potential implications for functional interpretation of genetic variants in nORFs. Our results indicate that some variants that were previously classified as being benign or of uncertain significance may have to be reinterpreted.
Recent evidence from proteomics, proteogenomics, ribosome profiling, and massively parallel sequencing studies have revealed that prokaryotic and eukaryotic genomes contain a substantial number of as-yet-uncharacterized and unannotated open reading frames (ORFs) (Firth and Brierley 2012; Andrews and Rothnagel 2014; Prabakaran et al. 2014; Albuquerque et al. 2015; Saghatelian and Couso 2015; Hellens et al. 2016; Brunet et al. 2018; Miravet-Verde et al. 2019). These ORFs have largely evaded detection owing to the original conservative definition of a gene with annotation criteria of one coding sequence (CDS) per transcript, a minimum of 100 codons for each CDS, “ATG” as the only start codon, and conservative definitions of Kozak sequences (Plaza et al. 2017; Brunet et al. 2018). There have also been recent advances in the sensitivity of proteomics methods such as ribosome profiling and mass spectrometry (MS), the ability to sequence genomes and transcripts at a deeper depths, and the ability to integrate these two data types, which our laboratory specializes in and calls systems proteogenomics (Prabakaran et al. 2014). These advances have revealed that we have underestimated the genome's coding potential, with many unannotated ORFs showing evidence of translation in humans alone (Ma et al. 2014; Chen et al. 2020; Erady et al. 2021). These ORFs are mostly under 100 codons and found in diverse regions of the genome, including in long noncoding RNAs (lncRNAs), pseudogenes, 3′ UTRs, 5′ UTRs, and alternative reading frames of canonical protein coding exons (Prabakaran et al. 2014; Plaza et al. 2017; Brunet et al. 2018).
Based on their genomic location and their size, these unannotated ORFs have been defined in numerous ways including as short ORFs (sORFs), small ORFs (smORFs), alternative ORFs (altORFs), and upstream/downstream ORFs (u/dORFs). The definitions for these labels, like those for original gene annotations, again set arbitrary bounds and even tend to vary between reports and species (Olexiouk et al. 2018). In this study, we have attempted to collate and reclassify all of these observed ORFs, and we refer here to any unannotated ORF as a “novel ORF” (nORF), which encompasses all of the above definitions. Specifically, a nORF is any ORF that can encode a not-yet-classified transcript or protein product, or an isoform of one, with no bounds on the number of codons, location, number of ORFs per transcript, or start codon. Although nORFs may appear by chance in the genome (Olexiouk et al. 2018), in this study we have exclusively focused on nORFs with experimental evidence of translation from MS or ribosome profiling studies and have attempted to interpret their potential functional consequences.
For humans, two published databases of note—OpenProt (Brunet et al. 2019) and sORFs.org (Olexiouk et al. 2018)—have compiled and analyzed translation data from ribosome profiling and MS studies to identify and share novel proteins. Evidence from these two databases has helped challenge conventional gene annotations to provide critical data for the field of nORFs, but both still have important limitations because of ambiguous definitions of these nORFs. sORFs.org, for instance, only considers ORFs under 100 codons, presents many duplicate or highly similar entries, and shares data in formats difficult to use in downstream analyses. OpenProt, although more accessible, has far fewer entries with experimental evidence, partly owing to only considering ORFs above 30 codons and limiting ORFs to ATG start codons and canonical transcripts. Additionally, annotation pipelines differ between the databases and are somewhat outdated, making comparisons difficult. Overall, the field lacks a consensus definition of what a nORF is and an accessible central resource with nORF data consolidated in a consistent manner. In this study, we address these needs with the curation and redefinition of nORFs.
Although mechanisms by which uORFs influence the translation of nearby canonical genes have been investigated (McGillivray et al. 2018; Whiffin et al. 2020), the functional consequence of nORFs more generally remains largely unexplored. The functional evidence that does exist has implicated nORFs in roles both related to and independent of nearby canonical genes (Brunet et al. 2018), including mRNA decapping (Pueyo et al. 2016), muscle regeneration (Matsumoto et al. 2017), and insulin secretion (Hu et al. 2016). Additionally, we have shown previously that nORF encoded protein-like products can form structures with potential biological functions (Erady et al. 2021), can be regulated by post-translational modifications (Erady et al. 2021), are biologically regulated in mouse neurons (Prabakaran et al. 2014), and harbor deleterious mutations from cancer and other inherited diseases (Erady et al. 2021). Despite these examples, the vast majority of nORFs have no known function, and their exclusion from canonical genome annotations means that many of the studies that could uncover roles for nORFs do not even consider them.
In this study, we have investigated the potential functional importance of a curated set of nORFs genome-wide. We begin at a broad scale, investigating the heritability associated with nORF regions for several human traits and diseases. We then narrow our focus from nORF regions to specific classes of nORF variants (e.g., nORF stop-gained variants) to evaluate potential signals of negative selection, which would be indicative of functional importance. Finally, we move to specific genetic variants, such as those known to cause disease, to investigate whether their pathogenicity can be explained by their effect on nORFs. In particular, we highlight disease mutations that appear benign to canonical proteins but highly deleterious to nORFs, the clearest potential examples of nORF functional importance and hence warranting reinterpretation of some variants of benign or unknown significance.
Results
Data overview
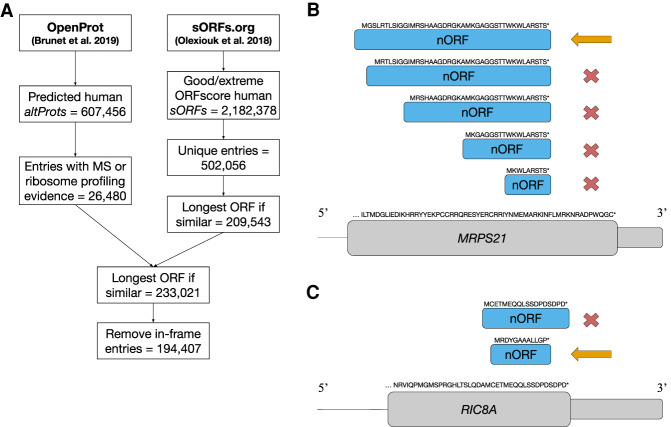
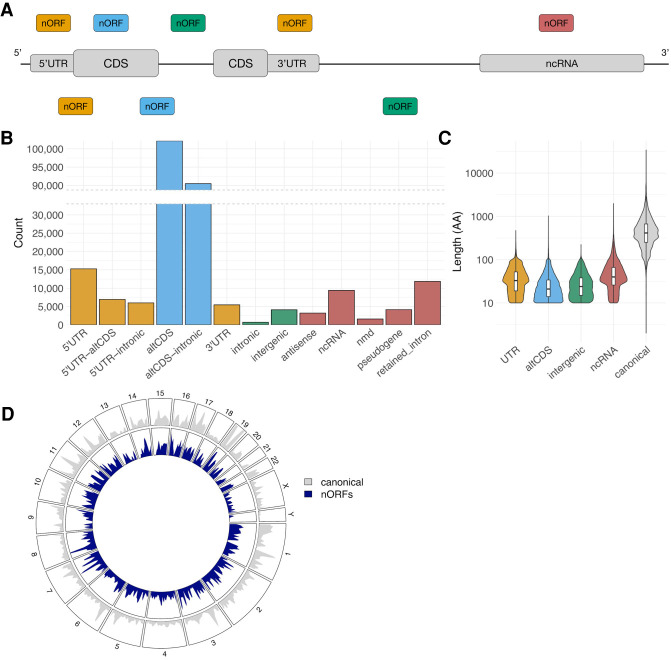
The nORFs data set contains 194,407 ORFs curated from OpenProt (Brunet et al. 2019) and sORFs.org (Fig. 1A; Olexiouk et al. 2018), which we have made publicly available on the nORFs.org platform (Fig. 2). The curation steps (Fig. 1A) involved selecting unique ORFs with translation evidence from MS or ribosome profiling experiments that are distinct from each other (Fig. 1B) and from canonical proteins (Fig. 1C). These nORFs were annotated with respect to canonical transcripts and CDS, and they are found in diverse locations in the genomes such as overlapping canonical CDSs in alternate frames (altCDSs), in UTRs, in noncoding RNAs (ncRNAs), and in intronic/intergenic regions (Fig. 3A). From the 194,407 nORFs, we found that 98,577 (50.7%) fully overlap canonical CDSs, 31,361 (16.1%) overlap CDSs and intron regions, 28,067 (14.4%) overlap 5′ UTRs, 5509 (2.8%) overlap 3′ UTRs, 19,909 (10.2%) overlap ncRNAs, and 4836 (2.5%) fully map to intronic or intergenic regions (Fig. 3B). The length distribution of nORFs for each major annotation category falls mostly below 100 amino acids, with mean lengths of 39.8 aa, 27.6 aa, 29.9 aa, and 54.4 aa for UTR, altCDS, intergenic, and ncRNA nORFs respectively, much smaller, as reported (Erady et al. 2021), than the mean canonical protein length of 557.3 aa (Fig. 3C). They are found spread throughout all 22 autosomes, in both sex chromosomes, and on mitochondrial DNA, similar to canonical CDS (Fig. 3D).
Figure 1.
Flow chart for the curation of the nORFs data set. (A) Steps illustrating the workflow to curate nORFs entries. From OpenProt, all predicted human altProts were filtered to entries with MS or ribosome profiling evidence (26,480). From sORFs.org, all human sORFs with an ORFscore of good or extreme were filtered to unique entries (502,056) and then summarized to the longest ORF at sites with multiple ORFs. Entries were then merged, the longest ORF was selected at multiple ORF sites, and in-frame entries were removed, leaving a total of 194,407 nORFs in the final data set. (B) An example of selecting the longest ORF for five small ORFs (smORFs) in an alternative frame of the final coding exon of the MRPS21 gene. In cases in which the ORFs share the same end site and differ only by their start site, we retain the longest ORF, indicated by the orange arrow, and remove the shorter ORFs, indicated by the red cross. (C) An example of removing in-frame entries in which two smORFs overlap the CDS of the RIC8A gene. The ORF in the same frame as the RIC8A CDS is removed from the data set as indicated by the red cross, whereas the second ORF in a different frame is retained in the data set, indicated by the orange arrow.
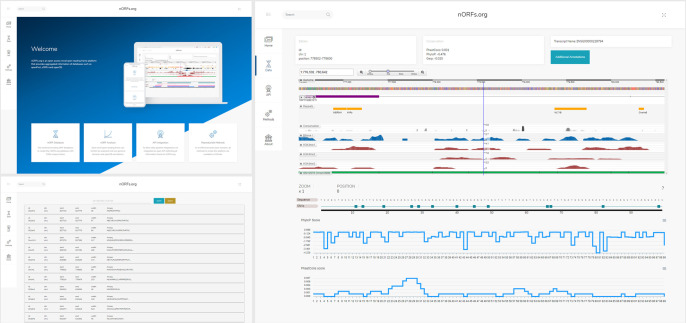
Figure 2.
Overview of platform. The nORFs.org platform contains six individual pages (three shown above) that introduce the platform, methods, and nORF entries. The nORF detail page (right) is divided into three sections: The meta data section includes the unique identifier, genomic position, and experimental sources; the (biodalliance) genome browser displays genes, repeats, conservation, and epigenetic information such as DNase I binding sites, and histone modifications (H3K4me1-me3); and the protein sequence section can be used for biostatistical pipelines to display variants, topology, and alternative splicing.
Figure 3.
nORF genomic annotations. (A) Schematic of common nORF locations with respect to a typical protein coding gene and a ncRNA. (B) Number of nORFs per genomic annotation. (C) Distribution of amino acid length of major categories of nORF annotations and canonical UniProtKB/Swiss-Prot proteins (The UniProt Consortium 2019). (D) Distribution of nORFs and canonical CDS from GENCODE throughout human chromosomes. Canonical and nORF scales are not proportionate.
We compared this nORF data set with previously published uORF data set (McGillivray et al. 2018). We note that the sources of uORF entries from McGillivray et al. (2018) (Fritsch et al. 2012; Lee et al. 2012; Gao et al. 2015) are three of the ribosome profiling experiments also used as input for the sORFs.org data set. By comparing the 188,802 “likely active” uORFs from McGillivray et al. (2018), with the 194,407 nORFs from this work, we find that there are 15,082 entries that are identical or highly similar (share stop codon but differ in start codon) between data sets. The majority of these shared entries fall, as expected, under the nORFs classified as 5′ UTR (7333) or 5′ UTR-altCDS (3681). The entries in the nORFs data set not found in the uORF data set can be attributed to the broader set of experiments used as input from sORFs.org and OpenProt and to the broader focus of any all unannotated ORFs compared with the specific uORF focus of McGillivray et al. (2018). As the 188,802 “likely active” uORFs from McGillivray et al. (2018) would have been found in sORFs.org data set, those not found in the nORFs data set would have been filtered out at one of data curation steps performed (e.g., good/extreme ORFscore, longest ORF if similar, removing in-frame entries) (Fig. 1A).
Heritability
We investigated the heritability of nORF regions in the genome to assess their importance to human traits and disease. To achieve this, we applied stratified LD score regression (S-LDSC) (Finucane et al. 2015; Gazal et al. 2017) with the baseline-LF model (Gazal et al. 2018) developed to assess both common (minor allele frequency [MAF] ≥ 5%) and low-frequency (0.5%≤ MAF <5%) heritability in complex traits. As applied by Gazal et al. (2018), we used a UK10K (The UK10K Consortium 2015) LD reference panel, analyzing 40 heritable, complex UK Biobank (Bycroft et al. 2018) traits restricted to 409,000 individuals with UK ancestry.
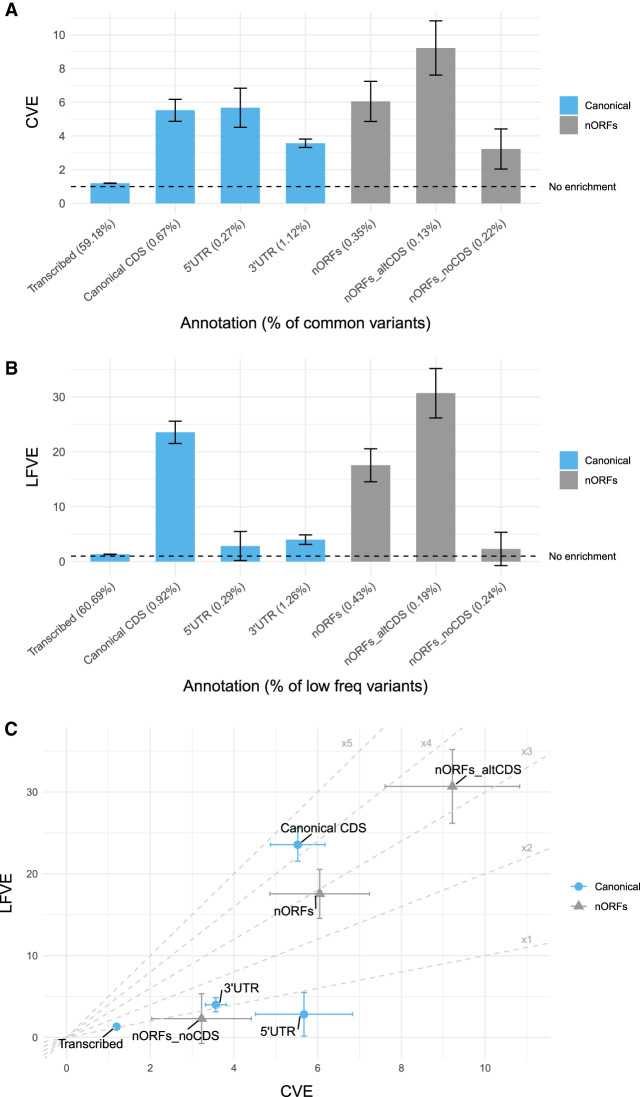
With this, we analyzed all baseline-LF model annotations and custom annotations (see Methods). For 67 baseline-LF annotations and for our seven custom annotations, we calculated heritability enrichments in each of the 40 UK Biobank traits. For each annotation, common variant enrichment (CVE) and low-frequency variant enrichment (LFVE) were meta-analyzed across 27 independent traits (Supplemental Tables S1, S2). To interpret heritability enrichments of nORFs, we focus on four custom annotations from canonical genes—transcribed regions, CDS, 5′ UTR, and 3′ UTR—and three custom annotations from nORFs—all nORFs, nORF regions overlapping canonical CDS (norfs_altCDS), and nORF regions not overlapping canonical CDS (nORFs_noCDS). Results from common variant heritability show that nORFs have a similar CVE (6.0 ± 1.2, P = 8 × 10−4) to canonical CDS (5.5 ± 0.7, P = 7 × 10−7) and that this CVE in nORFs is concentrated in the subset that overlaps canonical CDS (9.2 ± 1.6, P = 2 × 10−4) rather than those that do not (3.2 ± 1.2, NS) (Fig. 4A). In low-frequency variant heritability, we found that again nORFs have a comparable enrichment (17.6 ± 3.0, P = 1 × 10−8) to canonical CDS (23.6 ± 2.0, P = 1 × 10−31) but that the difference between LFVE in nORFs overlapping CDS (30.7 ± 4.5, P = 7 × 10−11) and nORFs not overlapping CDS (2.3 ± 3.0, NS) was more pronounced (Fig. 4B).
Figure 4.
Meta-analysis of heritability partitioned across 27 UK Biobank traits for nORF regions. Heritability enrichment was compared for canonical gene annotation from GENCODE versus nORF annotation for (A) common variation enrichment (CVE), defined as the proportion of common variant heritability explained by the annotation divided by the proportion of common variants in the annotation; (B) low-frequency variant enrichment (LFVE), defined similarly to CVE; and (C) the LFVE/CVE ratio. Higher enrichments suggest more functional importance in the studied traits and diseases.
Higher ratios of LFVE/CVE have been associated with CDSs, theorized to be owing to natural selection keeping trait relevant variation at lower frequencies (Gazal et al. 2018). Here we found that canonical CDSs showed the highest LFVE/CVE ratio (4.3×), with all nORFs (2.9×) and nORFs overlapping CDSs (3.3×) showing high ratios but nORFs not overlapping CDSs showing a ratio below one (0.7×), more comparable to that of 5′ UTRs (0.5×) (Fig. 4C). These results suggest that nORFs outside of coding regions may have less functional importance, but nORFs in the altCDS and canonical CDS show a possible additive effect on heritability. From the results, however, we cannot distinguish between heritability coming from canonical CDS versus nORFs. We attempt to disentangle the potential functional importance of nORFs from canonical CDS in the following analyses by examining specific nORF variant classes.
Mutability-adjusted proportion of singletons
To examine the potential functional importance of nORFs separately from canonical CDS, we drew on variant frequencies from the Genome Aggregation Database (gnomAD) data sets, made up of 125,748 exome sequences and 15,708 genome sequences (Karczewski et al. 2020). Specifically, we used the mutability-adjusted proportion of singletons (MAPS) score, which measures selection against classes of variants in a population (Lek et al. 2016; Karczewski et al. 2020). This measure is based on the principle that damaging classes of variants are kept at lower frequencies by natural selection. It compares the number of observed singletons for a particular variant class against the number of expected singletons under neutral selection, with a higher MAPS score being indicative of stronger selection against that variant class.
Variant bins for MAPS analysis were created using Ensembl's variant effect predictor (VEP) (McLaren et al. 2016) to annotate the gnomAD exomes and genomes variants in the context of both nORFs and canonical genes. Selection patterns for nORFs are unclear when considering nORF annotations in isolation, likely owing to consequences in canonical frames confounding the results (Supplemental Fig. S1). We therefore stratified our analysis by canonical consequence to examine the selection on nORF variants independently of their effect on canonical genes. We focus on seven annotations in canonical frames and five in nORFs, for a total of 35 variant bins that vary substantially in bin size (Supplemental Fig. S2). For each bin, the MAPS score was calculated for the exomes (Supplemental Table S3) and genomes (Supplemental Table S4) data set.
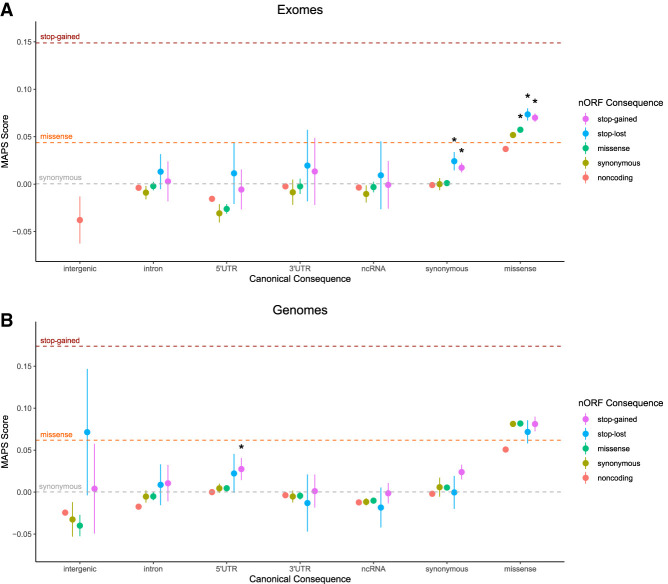
We observe that across most canonical consequences, variants annotated as stop-lost or stop-gained in nORFs show higher MAPS scores than the remainder of the canonical consequences, suggesting additional selective pressure on these variants (Fig. 5). Several of the larger, and therefore better powered, bins showed significant differences in MAPS scores. For instance, for all exome data set variants that are annotated as synonymous in canonical proteins, those that have stop-lost or stop-gained effects in nORFs show significantly higher MAPS scores than variants that fall outside of nORFs (both permuted P < 1 × 10−4) or are that are synonymous in nORFs (both permuted P < 1 × 10−4) (Fig. 5A). Similarly, when considering all canonical missense variants, those that have missense, stop-lost, or stop-gained effects in nORFs, all show significantly higher MAPS scores than variants that fall outside of nORFs (all permuted P < 1 × 10−4) or are that are synonymous in nORFs (all permuted P < 1 × 10−4) (Fig. 5A). From the genomes, four of the five significant bins from the exomes analysis were also significantly different from variants falling outside of nORFs but not from synonymous nORF variants (Fig. 5B; Supplemental Table S4). We also observed in the genomes data set that 5′ UTR variants from canonical proteins showed significantly higher MAPS scores if they caused a stop-gained effect in a nORF rather than a falling outside of nORFs (permuted P = 1 × 10−4) or synonymous in nORFs (permuted P = 7 × 10−4) (Fig. 5B). Overall, these results indicate selective pressure against deleterious nORF variants, suggesting that many of these variants may not be benign like current annotations would suggest.
Figure 5.
nORF stop-lost and stop-gained variants show signals of negative selection. The mutability-adjusted proportion of singletons (MAPS) was calculated for 35 variant bins of SNVs from gnomAD exomes (A) and genomes (B). Higher values indicate an enrichment of lower-frequency variants, suggesting negative selection. The canonical annotation of the bin is indicated along the x-axis, whereas the nORF annotation is indicated by color. Noncoding refers to variants falling outside of nORF regions. Dotted lines correspond to results from bins of only canonical annotations previously reported (Karczewski et al. 2020). (*) Permuted P adj < 0.05 versus noncoding bin and <0.05 versus synonymous bin with the same canonical consequence.
Disease mutations in nORF contexts
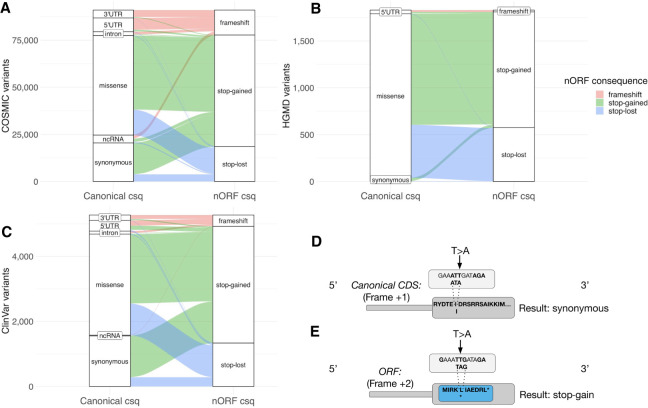
Considering that stop-lost and stop-gained variants in nORFs show signals of negative selection, we investigated potential disease-causing variants that could be owing to these mutation types. We first examined somatic cancer mutations from the Catalogue Of Somatic Mutations In Cancer (COSMIC) database (Tate et al. 2019). We annotated the 6.2 million coding and 19.7 million noncoding somatic variants using VEP in the context of nORFs and then canonical annotations. Although COSMIC variant sets are expected to be dominated by passenger mutations, their functional interpretation is key to identifying the cancer-causing genes and variants. We highlight 109,000 potential frameshift, stop-gained, or stop-lost variants in nORFs that have a less severe consequence in canonical genes (Fig. 6A; Supplemental Table S5).
Figure 6.
Reinterpreting COSMIC, HGMD, and ClinVar mutations in the context of nORFs. The canonical consequence and nORF consequence of (A) 109,000 somatic cancer mutations from COSMIC, (B) 1852 disease mutations from HGMD, and (C) 5269 disease mutations from ClinVar. Bins with 10 or fewer variants are not shown. These mutations would likely be interpreted as benign or missense in canonical genes but may have more severe consequences in nORFs. (D) A theoretical example of a disease variant that results in a synonymous mutation in canonical CDS but a stop-gain mutation in a nORF from an alternative reading frame (E).
We then performed a similar analysis to annotate known human disease variants present in the Human Gene Mutation Database (HGMD) (Stenson et al. 2017) and ClinVar (Landrum et al. 2018) databases. We identified 1852 variants from HGMD and 5269 variants from ClinVar that are frameshift, stop-gained, or stop-lost variants in nORFs but have less severe consequences in canonical genes (Fig. 6B,C; Supplemental Table S6).
To create a short list of disease mutations most likely to have a nORF-related cause, we further prioritized the COSMIC, HGMD, and ClinVar disease mutations. Specifically, we identified the top 20 cancer-associated genes with mutations with benign consequences in CDS but with deleterious consequences in the nORFs (Supplemental Table S7), 34 HGMD variants classified as disease causing (Supplemental Table S8), and 14 ClinVar variants classified as pathogenic or likely pathogenic (Supplemental Table S9) that have benign consequences in canonical annotations but stop-loss or stop-gain consequences in nORFs. We show an example in which a theoretical synonymous disease variant has a stop-gained effect on a nORF overlapping canonical CDS (Fig. 6D,E), which would normally be missed as a potential mechanism of pathogenicity.
Discussion
Following the advent of proteogenomics, ribosome profiling, and massively parallel sequencing studies, a key observation was that the entire genome has the potential to encode transcriptional and translational products. It was observed that noncanonical transcription and translation are not bound by classical motifs for transcriptional start or stop sites, polyadenylation, AUG start codons, single CDS per transcript, or numerous other signatures associated with the conventional gene definitions. Beyond the lack of conventional signatures to identify them, there is no consensus on how nORFs should be classified, with research groups often focusing on specific types or sizes of nORFs. We have undertaken a systematic analysis to collate and reclassify these nORFs into an accessible data set available to the wider community. This data set was created with the goal of facilitating investigations into nORF signatures for transcription, translation, regulation, and function. In this study, we curated and annotated 194,407 nORFs with translation evidence from MS or ribosome profiling and assessed their functional significance using global genomic properties. We found signals of functional importance for nORFs from heritability of common and low-frequency variants, negative selection against classes of nORF variants, and disease mutations potentially explained by nORFs consequences.
Our observations demonstrate that nORFs show large heritability enrichments characteristic of CDSs in both common and low-frequency variation. We also show that these enrichments are vastly different when dividing nORFs into those that overlap canonical CDSs and those that do not (Fig. 4). The nORF regions that do not overlap CDSs show modest heritability enrichments, largely similar to other noncoding regions such as UTRs, which are not indicative of functional importance on the level of canonical CDSs. In contrast, nORF regions overlapping canonical CDSs show large heritability enrichments, higher than only canonical CDSs. This suggests possible functional importance in nORFs and CDSs showing an additive effect on heritability. An alternative explanation for these enrichments is that the nORFs regions investigated are not causally adding heritability but instead have been identified in a subset of highly enriched canonical CDSs owing to confounding factors such as gene expression, gene identity, or sequence composition. Future investigations might attempt to control for these factors by using within-gene control sites (i.e., canonical CDSs not overlapped by nORFs from only those genes with an overlapping nORF).
When considering both the canonical and nORF consequences of variants, MAPS scores reveal selection acting to keep nORF stop-lost and stop-gained variants at lower frequencies than other variants with the same canonical consequence (Fig. 5). This signal was significant in exonic regions from the exomes data set and in 5′ UTRs from the genomes data set, possibly owing to each being better powered in these respective areas. We also note that where there is signal of selection, the magnitude of that selection for stop-gained variants in nORFs appears notably smaller than that of stop-gained variants in canonical frames. This gap can be attributed to several possible reasons. First, of the 194,407 nORFs in our data set, there are surely both false-positive detections of translation and detected translation of nORFs that do not create functional products, which would dilute the selection signal. Second, nORF products that are functional may have more specialized, cell-specific, or context-specific functions than do canonical proteins. This could mean that the selection pressure against a “true” set of stop-gained variants from functional nORFs could be weaker than the pressure acting on canonical genes and lead to a lower expected MAPS score. Nevertheless, these results suggest that nORF encoded protein products may have functional importance, motivating further analysis of disease mutations in which they may be relevant.
Investigation of this showed that numerous variants in disease mutation databases could potentially have nORF-related mechanisms of pathogenicity such as stop-lost, stop-gained, or frameshift mutations. We identified candidate HGMD disease mutations and ClinVar pathogenic/likely-pathogenic mutations with benign effects in canonical genes for which we believe nORF consequences should be considered as possible mechanisms of pathogenicity, similar to uORF-perturbing variants recently found to be disease causing (Whiffin et al. 2020). These examples highlight the potential impact of annotating disease mutations for their nORF consequence.
Although this study has added valuable insights into noncanonical translation products, it does have limitations. First, some entries gathered in our data set may be false-positive detections of translation or be translation events of proteins with no function. This may dilute signals of functional importance and should be kept in mind when using the data set. The difference in entry count is clearly weighted toward sORFs (12% vs. 88%); however, the difference in sequence context is not quite as pronounced (22% vs. 78%) owing to the length distribution of OpenProt being substantially higher. We believe that both databases add substantial value to the nORFs data set with the advantages of the sORFs database being that it focuses on Ribo-seq (OpenProt is primarily MS) and that it does not use several of the constraints of OpenProt, which only considers ORFs above 30 codons and limits ORFs to ATG start codons and canonical transcripts. Although we acknowledge that there may be concerns as to the sORF scoring methods, we have filtered out >90% of the 2.1 million sORFs.org entries to the 209,000 with the filtering methods described in the paper (ORFscore with unique genomic mappings, longest ORF at shared stop sites, removing in-frame entries), which we believe substantially reduces false positives. Despite these measures, one could certainly hypothesize that the filtering strategies of OpenProt make it more likely to contain functional nORFs. Our investigations of this possibility have shown that perhaps the opposite is true; however, it is difficult to make conclusive statements because of lack of statistical power when analyzing the smaller OpenProt data set by itself. In an early analysis of averaged heritability partitioned across 11 UK Biobank traits, split by OpenProt and sORFs, we found that sORFs show more heritability than OpenProt entries; however, the confidence intervals here are mostly overlapping (Supplemental Fig. S4). We also reran our MAPS analysis using only OpenProt entries and found that they did not show particularly strong selection (Supplemental Fig. S5); however, this analysis is limited by lack of power as shown by the large confidence intervals. Based on these results, we find it unlikely there are strong functional signals from OpenProt being contaminated by entries from sORFs.org. In addition, the calculation of heritability and MAPS scores based on the source of the database suffers from statistical noise, making it difficult to draw meaningful conclusions, compared with the better-powered joint analyses.
Second, it is by no means a comprehensive catalog of translation products in the human genomes; more nORFs are sure to be found as more investigations of translation products are performed. Third, heritability enrichment estimates for nORF regions do not directly estimate the contribution of nORFs but of any causal heritability signals in the region investigated. This is particularly relevant for the nORF regions that overlap canonical CDSs where the relative contribution of these factors cannot reliably be distinguished. Despite this caveat, the increase in heritability enrichment is an interesting finding that suggests a possible contribution of nORFs to the heritability of traits and disease. Last, for disease mutations potentially explained by nORFs, we caution that individual causal mechanisms cannot be confidently determined without weighing original translation evidence associated with the nORF, other possible mechanisms (e.g., regulatory), and potential follow-up functional analysis. Nevertheless, these disease mutation examples show the potential of annotating variants for their consequence in nORFs to explain their pathogenicity.
In this work, we developed a consistent, comprehensive, and accessible nORF resource that will aid future investigations into noncanonical translation products for the community. We have used this data set to make insights into the potential functional impacts of nORFs in the human genome. We have shown heritability enrichments associated with nORFs, particularly those overlapping canonical CDS. We then showed selective pressure acting on potentially deleterious nORF variants, suggesting their potential functional importance. Finally, we annotated disease mutations with nORF consequences, showing a potential to uncover plausible mechanisms and to generate hypothesis of their pathogenicity. In future investigations, this technique may be a valuable addition for discerning pathogenic mechanisms for rare disease diagnosis or in common disease phenotypes. If these investigations are successful, nORFs could be a set of new potential drug targets for disease treatment.
Methods
Selection of sources for evidence of nORFs
Three existing databases with entries that qualify as nORFs were considered for inclusion in the nORFs data set: OpenProt (Brunet et al. 2019), sORFs.org (Olexiouk et al. 2018), and SmProt (Hao et al. 2018). SmProt was not used because of inconsistencies in data (e.g., incorrect genomic coordinate annotations) and lack of details in their methods to reanalyze the data, specifically in regards to their MS evidence (Olexiouk et al. 2018). In contrast, OpenProt and sORFs.org have shown commitment to providing consistent, verifiable, and maintained data and were therefore used as the main sources for the nORFs data set.
OpenProt (Release 1.3) predicts all possible ORFs with an ATG start codon and a minimum length of 30 codons that map to an Ensembl (Zerbino et al. 2018) or RefSeq (O'Leary et al. 2016) transcript. They identified 607,456 altORFs that are neither canonical ORFs nor an isoform of those ORFs but are in noncoding regions or an alternate frame to canonical CDSs. Although OpenProt maps to both Ensembl and RefSeq transcripts, we focus exclusively on the Ensembl annotations for compatibility with the sORFs.org data set and other downstream analyses. From the altORFs mapped to Ensembl transcripts, we consider the 26,480 altORFs with translation evidence from MS (21,708), ribosome profiling (5059), or both (398).
The sORFs.org database (downloaded April 30, 2019) uses notably different inclusion criteria, annotating “sORFs” with translation evidence from 43 human ribosome profiling experiments and then adding MS evidence found in publicly available data sets. The sORFs are defined as ORFs between 10 and 100 codons using any of four start codons—“ATG,” “CTG,” “TTG,” or “GTG”—and are not restricted to known transcripts.
Curation of nORFs
The curation steps we performed to create a nORF data set are detailed in Figure 1. The final data set that we created (1) contains only nORFs with translation evidence from either MS or ribosome profiling, (2) contains no duplicate or highly similar entries, and (3) contains only ORFs clearly distinct from currently annotated canonical proteins.
We used 607,456 predicted altORFs from OpenProt and filtered to the 26,480 entries with MS or ribosome profiling evidence of translation. From over 2.1 million sORFs.org entries with “good” or “extreme” ORFscore (Bazzini et al. 2014), 502,056 entries with unique genomic mappings were extracted (Fig. 1A). The next step involved processing similar entries in the sORFs.org data set that shared the same stop site and amino acid sequences up to differing start sites. A characteristic example is shown in Figure 1B, where in an alternative frame of the final coding exon of the MRPS21 gene, sORFs.org provides evidence for five smORFs sharing the same end site and differing only by their start site. This is common in the sORFs.org data set because of the ambiguity in ribosome profiling experiments to identify the correct translation start site, unless specifically using methods that search for them (e.g., ribosome profiling with antibiotics used to trap newly initiated ribosomes at start codons) (Olexiouk et al. 2018; Weaver et al. 2019). Although ideally the correct start site(s) would be identified through experiments, these data are not currently available. For consistency and simplicity, we have selected the longest ORF in these cases, which may not always represent the true translated ORF but will always encompass all ORFs identified at these sites. We emphasize this ambiguity in the correct start site as an important limitation to be kept in mind when using the data set. In all, the selection of the longest ORF at ambiguous start sites further reduced extracted sORFs.org entries to 209,543.
Next, the OpenProt and sORFs.org data sets were merged, 1028 redundant entries between the data sets were removed, and 1976 cases of ambiguous start sites between the two data sets were resolved by again taking the longest ORF, resulting in a merged total of 233,021 entries. The small number of overlapping or similar entries between the two data sets can be partly attributed to different inclusion criteria for ORFs between the databases (i.e., ORF length, start codon, transcript requirement) and the main source of entries (sORFs from ribosome profiling and OpenProt predominantly from MS).
Finally, we separated all entries that were in-frame with canonical CDSs, as the translation evidence from these entries cannot be unambiguously resolved as to whether they are from a canonical protein product or an independent nORF embedded within a canonical protein. We identified 38,614 such entries and removed them, leaving a total of 194,407 entries in the final nORFs data set. An example case is shown in Figure 1C, where two smORFs overlap the CDS of the RIC8A gene. One of these ORFs is in the same frame as the RIC8A CDS and was therefore filtered out, whereas the second ORF is in a different frame and retained in the data set. Following this final curation step, all entries in the nORF data set that overlap canonical CDS are in a different frame from and do not share amino acid sequence with that CDS.
Annotation of nORFs
We annotated each nORF with reference to human GENCODE (v30) gene annotations (Frankish et al. 2019). The annotation categories included nORFs mapping to UTRs or CDS of protein coding transcripts, ncRNAs, or intergenic regions. When multiple annotations were possible, owing to multiple transcripts in a region, annotations were prioritized by first selecting full overlaps with protein coding transcripts, particularly those that overlap canonical CDSs in an alternative reading frame (altCDSs), followed by full overlaps with ncRNA transcripts, then by partial transcript overlaps, and finally intronic or intergenic regions. Our detailed prioritization summary is shown in Supplemental Figure S3.
When using GENCODE 34 (latest version), our pipeline identifies 194,291 rather than 194,407 nORFs, meaning that between releases 30 and 34, 116 nORFs became part of canonical CDS as newly identified genes or as part of new coding transcripts of existing genes. We find it encouraging that some nORFs are becoming canonical CDS, and plan to regularly update our GENCODE reference in future iterations of the nORFs database.
Database and web platform
To reduce the threshold of accessibility, databases need to be accessible with minimal requirements of tools or prior knowledge. We therefore built an online platform with representational state transfer (REST) application programming interface (API) functionality. This online platform acts as an entry and lookup point for individual entries, whereas the REST API is feature compatible with existing bioinformatics pipelines. We made the curated and annotated GRCh38 raw data set available in BED and GTF formats, as well as a downloadable nORFs.org UCSC track. Considering reproducible research guidelines, we used git as a versioning tool and uploaded the repository to GitHub under an MIT license (https://github.com/PrabakaranGroup/nORFs.org).
S-LDSC heritability analysis
As applied previously (Gazal et al. 2018), we obtained summary statistics for 40 heritable, complex UK Biobank (Bycroft et al. 2018) traits (downloaded from https://data.broadinstitute.org/alkesgroup/UKBB/UKBB_409K/) that were restricted to 409,000 individuals with UK ancestry. We then generated an LD reference panel for UK ancestry to match the summary statistics with 3567 UK10K (The UK10K Consortium 2015) whole-genome sequencing (WGS) samples from the ALSPAC and TWINSUK cohorts.
With these inputs, we analyzed a total of 177 genomic annotations, each corresponding to a defined set of variants, for their heritability enrichment. Of the 177, 163 are together known as the previously described baseline-LF model (Gazal et al. 2018). We added to the analysis 14 custom annotations, from seven functional annotations doubled for common variants and low-frequency variants. Of these seven, three custom annotations were nORF related: one for all nORFs and two in which nORFs were split at the variant level to those regions that overlap canonical CDS (norfs_altCDS) and those that do not (nORFs_noCDS). The remaining four were canonical annotations from GENCODE: transcribed regions, CDS, 5′ UTRs, and 3′ UTRs. It should be noted that similar annotations appear to be already present in the baseline-LF model, but they were generated from a different reference set than our nORFs (Gusev et al. 2014) and their “coding” annotation contains UTRs, which ours does not.
For the baseline-LD functional annotations and our custom annotations, we calculated CVE and LFVE for each of the 40 UK Biobank traits. CVE is the proportion of common heritability (h2C) divided by the proportion of common single-nucleotide polymorphisms (SNPs) in the annotation, whereas LFVE is proportion of low-frequency heritability (h2LF) divided by the proportion of low-frequency SNPs in the annotation:
Meta-analysis of results was conducted using random-effects meta-analyses in the rmeta package on 27 independent traits (Gazal et al. 2018), indicated in Supplemental Table S1. All standard errors were computed using a block jackknife procedure (Bulik-Sullivan et al. 2015).
MAPS analysis
We calculated MAPS with gnomAD genomes and exomes by using publicly available code at GitHub (https://github.com/macarthur-lab/gnomad_lof). We modified the code to include variant bins based on both nORF consequences and canonical consequences, rather than only canonical consequences. We selected five nORF consequences of interest—missense, synonymous, stop-lost, stop-gained, and noncoding (intergenic + upstream gene + downstream gene)—and seven canonical consequences of interest—missense, synonymous, ncRNA, 5′ UTR, 3′ UTR, intronic, and intergenic. For each of these 35 (5 × 7) bins, MAPS calibrated expected variant frequencies to account for one surrounding base of context and CpG methylation, two factors known to influence the mutability of base pairs (Lek et al. 2016). The transformation between variant frequencies and the expected proportion of singletons was regressed against the observed proportion of synonymous variants in canonical proteins. As the MAPS score given to variant classes is a relative metric, this means that synonymous variants in canonical proteins were set as zero, and higher scores reflected more negative selection. We reported MAPS scores for bins with at least 100 variants in the gnomAD exomes or genomes data set, respectively.
P-values were calculated using a bootstrapping approach as applied previously (Whiffin et al. 2020). For a given bin with n variants, n variants were randomly sampled with replacement and used to calculate MAPS for two bins of interest: bin A and bin B. This was repeated over 10,000 permutations with the P-value being the proportion of permutations in which MAPS of bin B was less than MAPS of bin A.
Variant annotation
Variant annotation was performed using version 96 of VEP (McLaren et al. 2016) to investigate the consequences of variants in the context of canonical frames and nORFs. Variant sets were obtained for annotation as VCFs. These included gnomAD genomes and exomes (release 2.1.1) (Karczewski et al. 2020), HGMD (pro release 2019.2) (Stenson et al. 2017), ClinVar (release 2019 0708) (Landrum et al. 2018), and COSMIC coding and noncoding mutations (v89) (Tate et al. 2019). Each set of variants was annotated for their most severe consequence as defined by VEP with respect to (1) canonical gene annotations, corresponding to GENCODE 30 in GRCh38 or GENCODE 30 lifted over to GRCh37, and (2) nORF annotations provided as a custom GTF in the appropriate genome assembly.
When examining possible disease mutations that could be explained by nORF consequences, we first filtered variants from the disease mutations databases (COSMIC, HGMD, and ClinVar) to remove those with strongly deleterious annotations in canonical proteins (i.e., essential splice, frameshift, stop-gained, stop-lost, start-lost). We then further filtered these variant sets to those with possible pathogenic consequences in nORFs (stop-lost, stop-gained, and frameshift).
Software availability
The code used to curate, annotate, and analyze the nORFs data set is publicly available at GitHub (https://github.com/PrabakaranGroup/nORF-data-prep) and uploaded as Supplemental Code Files 1 and 2. To share the nORFs data set, we have also created https://norfs.org, an open source platform for the nORFs data set with implementation available at GitHub (https://github.com/PrabakaranGroup/nORFs.org) and uploaded as Supplemental Code File 3. A UCSC track download is also provided on the nORFs.org API page (https://norfs.org/api).
Competing interest statement
Cambridge Enterprise Limited, Indian Institute of Science Education and Research, and International Centre for Genetic Engineering and Biotechnology have filed patent applications related to the work described here. The title of the patent application is “Treatment of Diseases Associated with Variant Novel Open Reading Frames.” The U.S. Provisional Application was filed on December 9, 2020, application no. 63/123,454.
Supplementary Material
Acknowledgments
We thank RosettaHuB (https://rosettahub.com) for helping us to build applications using Amazon Web Services. S.P. is funded by the Cambridge-DBT lectureship; C.E. is funded by the Dr. Manmohan Singh scholarship; R.K. is funded by Biotechnology and Biological Sciences Research Council (BBSRC) Fellowship; N.M. is funded by Government of India and Trinity Barlow fellowship. We thank the anonymous reviewers and the editor for their critical and thoughtful comments.
Author contributions: M.D.C.N. did the nORF classification with help from C.E. and N.M.; R.K. built the database and web platform; M.D.C.N. did the MAPS, S-LDSC, and variant analyses; M.D.C.N. and S.P. interpreted the data and wrote the manuscript with assistance from R.K. and C.E.; M.H., D.N.C., and M.M. curated and provided the HGMD data set; S.P. designed and supervised the project.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.263202.120.
References
- Albuquerque JP, Tobias-Santos V, Rodrigues AC, Mury FB, da Fonseca RN. 2015. Small ORFs: a new class of essential genes for development. Genet Mol Biol 38: 278–283. 10.1590/S1415-475738320150009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SJ, Rothnagel JA. 2014. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet 15: 193–204. 10.1038/nrg3520 [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al. 2014. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 33: 981–993. 10.1002/embj.201488411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet MA, Levesque SA, Hunting DJ, Cohen AA, Roucou X. 2018. Recognition of the polycistronic nature of human genes is critical to understanding the genotype–phenotype relationship. Genome Res 28: 609–624. 10.1101/gr.230938.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet MA, Brunelle M, Lucier J-F, Delcourt V, Levesque M, Grenier F, Samandi S, Leblanc S, Aguilar J-D, Dufour P, et al. 2019. OpenProt: a more comprehensive guide to explore eukaryotic coding potential and proteomes. Nucleic Acids Res 47: D403–D410. 10.1093/nar/gky936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL, Neale BM. 2015. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47: 291–295. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J, et al. 2018. The UK Biobank resource with deep phenotyping and genomic data. Nature 562: 203–209. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Brunner A-D, Cogan JZ, Nuñez JK, Fields AP, Adamson B, Itzhak DN, Li JY, Mann M, Leonetti MD, et al. 2020. Pervasive functional translation of noncanonical human open reading frames. Science 367: 1140–1146. 10.1126/science.aay0262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erady C, Boxall A, Puntambekar S, Jagannathan NS, Chauhan R, Chong D, Meena N, Kulkarni A, Kasabe B, Prathivadi Bhayankaram K, et al. 2021. Pan-cancer analysis of transcripts encoding novel open reading frames (nORFs) and their potential biological functions. npj Genomic Medicine (in press). [DOI] [PMC free article] [PubMed]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, et al. 2015. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47: 1228–1235. 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J Gen Virol 93: 1385–1409. 10.1099/vir.0.042499-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch C, Herrmann A, Nothnagel M, Szafranski K, Huse K, Schumann F, Schreiber S, Platzer M, Krawczak M, Hampe J, et al. 2012. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res 22: 2208–2218. 10.1101/gr.139568.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian S-B. 2015. Quantitative profiling of initiating ribosomes in vivo. Nat Methods 12: 147–153. 10.1038/nmeth.3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazal S, Finucane HK, Furlotte NA, Loh P-R, Palamara PF, Liu X, Schoech A, Bulik-Sullivan B, Neale BM, Gusev A, et al. 2017. Linkage disequilibrium–dependent architecture of human complex traits shows action of negative selection. Nat Genet 49: 1421–1427. 10.1038/ng.3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazal S, Loh P-R, Finucane HK, Ganna A, Schoech A, Sunyaev S, Price AL. 2018. Functional architecture of low-frequency variants highlights strength of negative selection across coding and noncoding annotations. Nat Genet 50: 1600–1607. 10.1038/s41588-018-0231-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Lee SH, Trynka G, Finucane H, Vilhjálmsson BJ, Xu H, Zang C, Ripke S, Bulik-Sullivan B, Stahl E, et al. 2014. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet 95: 535–552. 10.1016/j.ajhg.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Zhang L, Niu Y, Cai T, Luo J, He S, Zhang B, Zhang D, Qin Y, Yang F, et al. 2018. SmProt: a database of small proteins encoded by annotated coding and noncoding RNA loci. Brief Bioinform 19: 636–643. 10.1093/bib/bbx005 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Brown CM, Chisnall MAW, Waterhouse PM, Macknight RC. 2016. The emerging world of small ORFs. Trends Plant Sci 21: 317–328. 10.1016/j.tplants.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Hu H, He L, Li L, Chen L. 2016. Apelin/APJ system as a therapeutic target in diabetes and its complications. Mol Genet Metab 119: 20–27. 10.1016/j.ymgme.2016.07.012 [DOI] [PubMed]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. 2018. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46: D1062–D1067. 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang S-X, Shen B, Qian S-B. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci 109: E2424–E2432. 10.1073/pnas.1207846109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ward CC, Jungreis I, Slavoff SA, Schwaid AG, Neveu J, Budnik BA, Kellis M, Saghatelian A. 2014. Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. J Proteome Res 13: 1757–1765. 10.1021/pr401280w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi PP. 2017. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541: 228–232. 10.1038/nature21034 [DOI] [PubMed] [Google Scholar]
- McGillivray P, Ault R, Pawashe M, Kitchen R, Balasubramanian S, Gerstein M. 2018. A comprehensive catalog of predicted functional upstream open reading frames in humans. Nucleic Acids Res 46: 3326–3338. 10.1093/nar/gky188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. 2016. The Ensembl variant effect predictor. Genome Biol 17: 122 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miravet-Verde S, Ferrar T, Espadas-García G, Mazzolini R, Gharrab A, Sabido E, Serrano L, Lluch-Senar M. 2019. Unraveling the hidden universe of small proteins in bacterial genomes. Mol Syst Biol 15: e8290.. 10.15252/msb.20188290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olexiouk V, Van Criekinge W, Menschaert G. 2018. An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res 46: D497–D502. 10.1093/nar/gkx1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S, Menschaert G, Payre F. 2017. In search of lost small peptides. Annu Rev Cell Dev Biol 33: 391–416. 10.1146/annurev-cellbio-100616-060516 [DOI] [PubMed] [Google Scholar]
- Prabakaran S, Hemberg M, Chauhan R, Winter D, Tweedie-Cullen RY, Dittrich C, Hong E, Gunawardena J, Steen H, Kreiman G, et al. 2014. Quantitative profiling of peptides from RNAs classified as noncoding. Nat Commun 5: 5429 10.1038/ncomms6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueyo JI, Magny EG, Sampson CJ, Amin U, Evans IR, Bishop SA, Couso JP. 2016. Hemotin, a regulator of phagocytosis encoded by a small ORF and conserved across metazoans. PLoS Biol 14: e1002395 10.1371/journal.pbio.1002395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A, Couso JP. 2015. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat Chem Biol 11: 909–916. 10.1038/nchembio.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN. 2017. The human gene mutation database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet 136: 665–677. 10.1007/s00439-017-1779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. 2019. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 47: D941–D947. 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UK10K Consortium. 2015. The UK10K project identifies rare variants in health and disease. Nature 526: 82–90. 10.1038/nature14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47: D506–D515. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver J, Mohammad F, Buskirk AR, Storz G. 2019. Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10: e02819-18 10.1128/mBio.02819-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiffin N, Karczewski KJ, Zhang X, Chothani S, Smith MJ, Evans DG, Roberts AM, Quaife NM, Schafer S, Rackham O, et al. 2020. Characterising the loss-of-function impact of 5′ untranslated region variants in 15,708 individuals. Nat Commun 11: 2523 10.1038/s41467-019-10717-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. 2018. Ensembl 2018. Nucleic Acids Res 46: D754–D761. 10.1093/nar/gkx1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.