Abstract
Our core timekeeping mechanism, the circadian clock, plays a vital role in immunity. Although the mechanics of circadian control over the immune response is generally explained by transcriptional activation or repression derived from this clock's transcription-translation negative-feedback loop, research suggests that some regulation occurs beyond transcriptional activity. We comprehensively profiled the transcriptome and proteome of murine bone marrow-derived macrophages and found that only 15% of the circadian proteome had corresponding oscillating mRNA, suggesting post-transcriptional regulation influences macrophage clock regulatory output to a greater extent than any other tissue previously profiled. This regulation may be explained by the robust temporal enrichment we identified for proteins involved in degradation and translation. Extensive post-transcriptional temporal-gating of metabolic pathways was also observed and further corresponded with daily variations in ATP production, mitochondrial morphology, and phagocytosis. The disruption of this circadian post-transcriptional metabolic regulation impaired immune functionality. Our results demonstrate that cell-intrinsic post-transcriptional regulation is a primary driver of circadian output in macrophages and that this regulation, particularly of metabolic pathways, plays an important role in determining their response to immune stimuli.
Much of life on earth is exposed to the regular day/night cycle of the planet and has evolved circadian rhythms to anticipate these predictable changes (Eckel-Mahan and Sassone-Corsi 2013). Circadian rhythms are generated by a molecular oscillator, or clock, which adjusts innumerable cellular functions to regulate behaviors, encompassing everything from luminescence in fungi to sleep in humans (Dunlap 1999; Patke et al. 2020). In concordance with this, many facets of immunity are controlled by circadian rhythms, and the dysregulation of circadian rhythms can be detrimental to the immune response (Scheiermann et al. 2018). Chronic circadian disruption is associated with an increased risk for metabolic, cardiovascular, and neurodegenerative diseases, all sharing underlying inflammatory components, leading to the hypothesis that dysregulation of immunity, for example, chronic inflammation, is a critical link between disease and circadian rhythms (Evans and Davidson 2013; Scheiermann et al. 2018). Further, studies with myeloid-specific clock knockouts have demonstrated that the cell-intrinsic clock in myeloid immune cells plays a critical role in dictating this regulation/dysregulation, independent of systemic cues (Nguyen et al. 2013; Sato et al. 2014; Xu et al. 2014; Curtis et al. 2015; Huo et al. 2017; Kiessling et al. 2017; Early et al. 2018).
The canonical view of physiological circadian regulation on immunity is via a transcription-translation-based negative-feedback loop, where the heterodimeric transcription factor positive-arm complex regulates the expression of both the negative arm as well as a host of other genes not involved in the feedback loop, termed clock-controlled genes (ccgs) (Hurley et al. 2016; Takahashi 2017). Clock genes in an interlocking auxiliary loop (e.g., Rev-erbα) also regulate ccgs and have been shown to play an important role in macrophages’ circadian functions (Gibbs et al. 2012; Lam et al. 2013; Sato et al. 2014). Utilizing many different organisms and tissue types, transcriptome profiling studies have found that large proportions of transcripts oscillate circadianly and are highly tissue-specific (Hughes et al. 2009; Abruzzi et al. 2011; Zhang et al. 2014; Mure et al. 2018). However, recent evidence suggests the importance of circadian post-transcriptional and post-translational mechanisms, resulting in circadianly oscillating proteins not derived from oscillating transcripts and vice versa (Reddy et al. 2006; Kojima et al. 2011; Chiang et al. 2014; Hurley et al. 2014, 2018; Mauvoisin et al. 2014; Robles et al. 2014; Wang et al. 2017a, 2018; Green 2018; Mauvoisin and Gachon 2020). Despite this mounting evidence, there is a lack of studies utilizing adequate sampling depth, resolution, replication, and time course length to generate highly powered data sets probing the regulatory relationships between the circadian transcriptome and proteome, and there is little information on how this post-transcriptional regulation impacts the immune response (Hughes et al. 2017).
To provide such a data set, we analyzed the global transcriptome and proteome of murine bone marrow-derived macrophages (BMDMs) every 2 h, over 48 h, in triplicate using RNA-seq and tandem mass tag mass spectrometry (TMT-MS), respectively. Macrophages were selected for this investigation as previous work has shown that macrophages possess robust circadian clocks and demonstrate circadian control of their key immune functions, such as phagocytosis, cytokine release, differentiation, and migration in/out of various tissue compartments (Bourin et al. 2002; Hayashi et al. 2007; Keller et al. 2009; Gibbs et al. 2012; Silver et al. 2012, 2018; Sato et al. 2014; Wang et al. 2016; Kiessling et al. 2017; Pick et al. 2019). Macrophages play a vital role in the initiation, sustainment, and resolution of both acute and chronic inflammation, and it has been suggested that the disruption of these circadian rhythms in immunity and inflammation may play a critical part in the negative impact of clock disruption on human health (Bechtold et al. 2010; Arjona et al. 2012; Cermakian et al. 2013; Evans and Davidson 2013; Curtis et al. 2014; Geiger et al. 2015; Labrecque and Cermakian 2015; Scheiermann et al. 2018). Finally, as macrophages live for months at a time, circulating in the bloodstream before long in-tissue residencies, the influence of their circadian clocks would be of more consequence than shorter-lived immune cells (van Furth and Cohn 1968). However, much is left to be desired in the study of macrophage rhythms as most prior studies have used transcriptional and ex/in vivo methods to determine macrophage output pathways, complicating what rhythms are generated by the presence of systemic circadian cues versus what is intrinsically controlled by macrophage clocks. Here, we report and characterize a broad array of cellular functions that are circadianly regulated by the macrophage-intrinsic clock.
Results
Analysis of the macrophage circadian transcriptome and proteome demonstrates extensive circadian regulation
To determine if the substantial, physiologically relevant, circadian post-transcriptional regulation noted in other tissue types was conserved in immune cells, bone-marrow derived macrophages were selected for in vitro interrogation as they are derived from primary cells, making their clock output more relevant to in vivo conditions and are terminally differentiated, limiting potential confounding effects of the cell cycle. BMDMs were derived from bone marrow progenitor cells extracted from Per2::Luc C57BL/6J mice and differentiated with recombinant M-CSF (Yoo et al. 2004). Flow cytometry confirmed that >99% of cells displayed the macrophage-specific markers ITGAM (also known as CD11b) and ADGRE1 (also known as F4/80), demonstrating complete differentiation into macrophages (Supplemental Fig. S1A,B; Zhang et al. 2008). We extracted total RNA and protein from these serum-shock-synchronized BMDMs every 2 h over 48 h in triplicate, totaling six independent time courses (Fig. 1A; Supplemental Fig. S1C). Luminescence traces confirmed that our protocol resulted in reliable, ∼24-h PER2 oscillations, and sampling for all time courses began at 16 h post-serum shock to avoid artifactual gene expression that occurs immediately following serum shock (Supplemental Fig. S1D; Balsalobre et al. 1998).
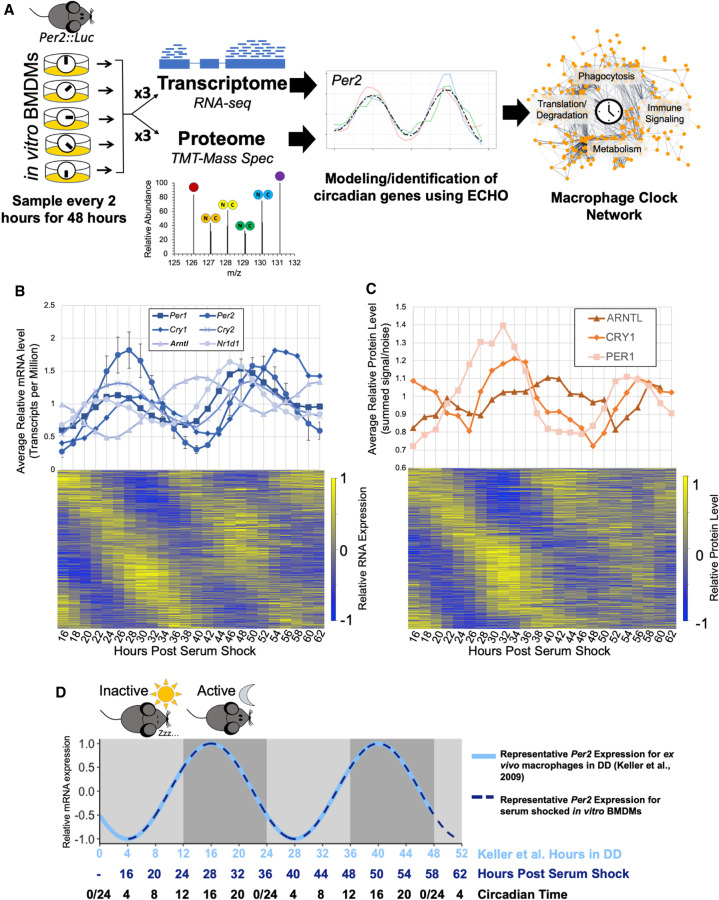
Figure 1.
Multi-omics profiling details extensive circadian regulation of the macrophage transcriptome and proteome. (A) A schematic of the analysis of macrophage circadian regulation. Bone marrow derived macrophages (BMDMs) from Per2::Luc mice were synchronized in vitro via serum shock and sampled in triplicate for global proteome and transcriptome profiling with 10-plex tandem mass tag mass spectrometry and RNA-seq, respectively. Circadianly oscillating genes were identified using the ECHO program to determine the macrophage circadian protein–protein interaction network. (B) All detected clock gene transcripts oscillated circadianly, and error bars show the standard deviation for Per2 as a representative gene for the variation between triplicate values. Heat map in bottom panel shows relative expression for all identified circadian transcripts. (C) All detected clock proteins exhibited circadian abundances with delay from their respective transcripts. Points shown are an average of the summed signal/noise of peptides for the identified protein for up to three replicates depending on the detection at the given time point. Heat map in bottom panel shows relative expression for all identified circadian proteins. (D) A schematic summarizing how circadian time (CT) for our in vitro synchronized experimentation was inferred from comparison of Per2 mRNA oscillation timing we observed to the oscillation of Per2 mRNA reported in macrophages extracted from light-entrained mice transferred to constant dark conditions (Keller et al. 2009). Shading represents the relative inactive period (light gray) and active periods (dark gray) during the Dark:Dark (DD) experiment by Keller et al. Because mice are nocturnal, CT0/24 is the onset of the inactive period and CT12 is the onset of the active period.
Quantitative RNA-sequencing was performed to analyze the transcriptome over time. In short, samples were processed so as to quantify both coding and noncoding RNA expression in transcripts per million (TPM) at each time point (Supplemental Table S1; Supplemental Fig. S2A). Quantitative proteomic analysis was completed using tandem mass tag mass spectrometry. Protein samples were isobarically mass-tagged after digestion and analyzed utilizing eight multiplex groups. Across all multiplexes, 162,920 unique peptides were identified in at least one time point, comprising a total of 10,162 distinct proteins detected, with 6290 proteins detected in every time point (Supplemental Table S2). The transcriptome and proteome data were preprocessed and analyzed using the previously published tools LIMBR and ECHO, designed for circadian time course analysis (Supplemental Fig. S2B,C; Supplemental Data S3, S4; De los Santos et al. 2017, 2020; Crowell et al. 2019). After LIMBR preprocessing, a total of 6050 proteins and 36,451 RNAs qualified for further analysis. These transcripts and proteins were categorized as oscillating/not oscillating using the ECHO program in the free-run mode (see Methods; De los Santos et al. 2020).
Once we identified our oscillating proteins and transcripts, we examined both data sets for clock gene levels. We found that each identified positive- and negative-arm clock gene known to oscillate in a circadian manner did so at both the RNA (Fig. 1B) and protein levels (Fig. 1C). Clock gene transcripts including Arntl (also known as Bmal1), Clock, Cry1, Cry2, Nr1d1, Per1, and Per2 were identified as circadian, with low variation between replicates (Fig. 1B, top panel; Supplemental Fig. S2C). Due to known low abundance levels and our stringent detection cutoffs, no protein products of clock genes were classified as detected in the proteome. However, manual examination of pre-LIMBR curated average levels of clock gene proteins at each time point indicated oscillations of both positive- (ARNTL) and negative-arm (PER1, CRY1) clock proteins (Fig. 1C, top panel).
Based on the ECHO-modeled periods for known clock gene transcripts, genes were considered to be circadianly oscillating if the Benjamini–Hochberg adjusted P-value was <0.05, the amplitude change coefficient was between ±0.15, and the period between 20 and 28 h. This classified 5790 transcripts as circadian, 15.8% of the detected macrophage transcriptome (Fig. 1B). A similar proportion, 15.3%, of coding transcripts were circadian (n = 3160) within the coding-only detected transcriptome (n = 20,699). When analyzing the distributions of modeled parameters, we noted a bias toward a shorter period length (between 20 and 22 h) and that most oscillations decreased in amplitude (damped) over time (Supplemental Fig. S2D; De los Santos et al. 2017, 2020). For ECHO-modeled protein periods, we broadened the period range of what we considered circadian to 18–30 h to accommodate higher levels of technical noise attributable to proteomics-based techniques (Hurley et al. 2018). Analysis of the proteome identified 1778 proteins as circadian, meaning 29% of the detected proteome is under circadian control (Fig. 1C). There was little bias in period length detected and more widespread damping over time as compared to the transcriptome (Supplemental Fig. S2D). In both cases, the majority of genes in the circadian RNA and protein sets had P-values far lower than 0.01, indicating statistically accurate modeling of these genes by ECHO (Supplemental Fig. S2D, lower panel).
Post-transcriptional and post-translational controls in the late phase contribute to circadian output
In an exhaustive study, Mure et al. concluded that there was broad circadian gating on mammalian transcripts but did not investigate the impact of these transcriptional waves on protein levels and, by extension, cell physiology (Mure et al. 2018). We therefore probed our data sets for circadianly gated timing of the peak phases of RNA and protein and found that macrophage circadian RNA and protein levels both peak in two anti-phase “waves” when visualized on a 24-h scale (Fig. 2A). While our experiment was done in vitro, circadian time (CT) that is comparable to murine macrophage 24-h timing in vivo was inferred by comparing Per2 mRNA oscillations observed in our time course to previous work done by Keller et al. with macrophages extracted from light-entrained mice (Keller et al. 2009). To orient our in vitro results to what could be expected in vivo, we determined that our time course beginning at a trough of Per2 mRNA expression at post-serum shock hour 16 is equivalent to the trough in Per2 expression occurring at CT4 in murine macrophages (Fig. 1D). Thus, we observed one smaller wave of circadian genes generally peaking during the “early” time points, corresponding to early circadian time (CT3–15, circadian RNAs n = 2724, circadian proteins n = 807), and one larger wave with peaks around the “later” time points/circadian time phase (CT15–3, circadian RNAs n = 3066, circadian proteins n = 947) (Fig. 2A). When we subset our RNA data to only include circadian RNAs that encoded proteins (n = 2801), the RNA waves became more evident, occurring ∼2 h before the corresponding protein wave (CT1–13 circadian RNAs n = 1415, CT13–1 circadian RNAs n = 1386) (Fig. 2A).
Figure 2.
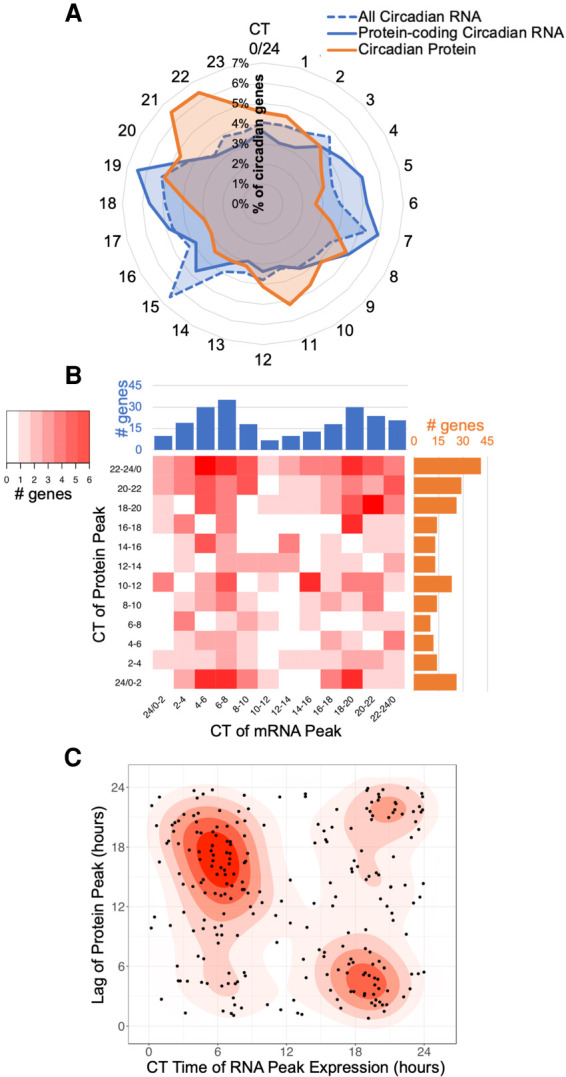
Comparison of circadian transcripts and proteins reveals a significant role for post-transcriptional and post-translational regulation. (A) The percentage of total oscillating transcripts (blue) or proteins (orange) peaking at a given time was plotted on a radial histogram by circadian time of peak, binned in 1-h windows. (B) A heat map comparing the difference in the peak phases of corresponding oscillating transcripts (blue) and proteins (orange) binned into 2-h intervals over circadian time for genes circadian at both the mRNA and protein level. (C) Density graph displaying the lag time to protein creation from the peak time of corresponding oscillating transcript levels for genes circadian at both the mRNA and protein level.
The fixed delay between RNA and protein peaks suggested a correlation between the temporally related RNA and protein waves; that is, mRNAs are transcribed and shortly after translated into proteins, as would be suggested by the canonical circadian regulation framework. To determine if these general temporal waves aligned with specific protein/RNA relationships, we subset our data sets to examine only genes that were detected and rhythmic in both the transcriptome and proteome. This identified 250 genes that were circadian at both the mRNA and protein level (Supplemental Fig. S3A). We compared the peak time of each mRNA with the peak time of its corresponding protein and found a low correlation between the RNA and protein pairs. While the majority of oscillating proteins with oscillating mRNA peaked during the late wave of translation, the peak phase of their corresponding mRNAs was evenly distributed between the two mRNA waves (Fig. 2B). The dominance of this late wave peak for protein abundance appeared to be specific to proteins with associated oscillating mRNA, as the peak level timings of proteins that did not have oscillating mRNA were more evenly distributed (Supplemental Fig. S3B,C). When the lag times of the peak protein levels were plotted against the timing of peak mRNA levels, it was apparent that mRNAs peaking in the early phase had a longer phase delay to translation as compared to mRNAs peaking in the late phase (Fig. 2C). Transcripts that peaked near CT6 took 15–18 h to reach their respective protein peak, whereas transcripts peaking around CT18 only took 3–6 h to reach their protein peak. This suggested that translation of transcripts occurs more readily during the late phase, implying temporal differences in post-transcriptional regulation.
We then expanded our analysis to investigate the overall relationship between oscillating RNAs and proteins. We found that, while our transcriptome largely encompassed our proteome (n = 5833 total genes), there was little overlap between circadian transcripts and circadian proteins (Supplemental Fig. S3A,D). Sixty-eight percent of circadian transcripts did not result in a circadian protein, and 86% of circadian proteins did not derive from a circadian transcript. As this discrepancy was higher than has been previously reported, we wanted to rule out that this lack of overlap was due to the more lenient period cutoffs used for proteins to be considered circadian (Hurley et al. 2018). We therefore evaluated the overlap using the more or less lenient period cutoffs for both data sets (Supplemental Fig. S3E,F) and found that the ratios remained skewed toward minority overlap, indicating strong evidence for post-transcriptional regulation in these BMDMs.
Post-transcriptional and post-translational mechanisms are abundant in the late phase
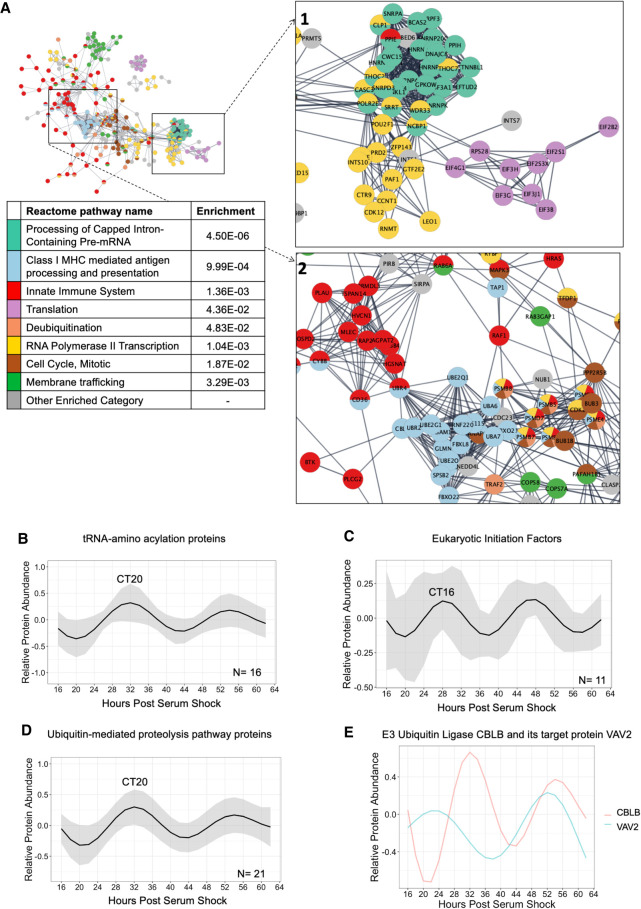
The evidence of post-transcriptional circadian control described above led us to examine clock-timed biological processes that could contribute to this regulation. Past studies have shown that translation may be the key process underlying circadian post-transcriptional regulation, and gene ontological enrichment analysis to examine the 250 genes rhythmic at both the mRNA and protein levels found that they were enriched for processes involved in translation (Bonferroni P-value = 2.72 × 10−4, fold enrichment = 5.18) (Supplemental Table S3; Ashburner et al. 2000; Hurley et al. 2014, 2018; Caster et al. 2016; Fabregat et al. 2018; Mi et al. 2019). Therefore, we examined the circadian proteome to determine if a significant proportion of the clock output has functionality in translation. First, we split the proteins into two groups evenly with time based on the temporal “waves” observed in Figure 2A. Proteins that peaked in abundance between CT3 and CT15 were considered “early wave” proteins and proteins that peak between CT15 and CT3 were considered “late wave” proteins. Both categories were assessed for enrichments for Reactome categories using Amigo/Panther enrichment analysis tools, as well as interrogated to determine protein–protein interactions (Ashburner et al. 2000; Fabregat et al. 2018; Mi et al. 2019). Proteins belonging to significantly enriched (Bonferroni adjusted P-value < 0.05) Reactome categories were color-coded according to their category and graphed by their protein–protein interactions with STRINGdb (Fig. 3A; Supplemental Data S1, S2; Szklarczyk et al. 2015). We further utilized Phase Set Enrichment Analysis (PSEA) with curated murine Gene Ontology gene sets to investigate protein enrichment by time of day (Supplemental Fig. S4A; Zhang et al. 2016; Bares and Ge 2019).
Figure 3.
Functional pathway enrichments indicate late phase post-transcriptional/-translational regulation. (A) A global view of the STRINGdb network of the proteins in enriched Reactome categories that peak in the late wave (CT15–3), with insets focusing on key interactions, including (1) Translation, RNA Polymerase II Transcription, and Processing of Capped Intron-Containing Pre-mRNA, and (2) Class I MHC mediated antigen processing and presentation, and Innate Immune System. Interactions shown as strings are filtered to highest confidence interactions (confidence > 0.9). Colors denote independent Reactome categories. Gray coloring represents genes from enriched Reactome categories that did not represent a large proportion of the protein–protein interactome or described redundant categories. (B) An average of the modeled fits of all 16 circadian proteins in the tRNA-amino acelyation reactome category. Shading indicates ±1 standard deviation of models at each time point. The circadian time of the first peak is labeled. (C) An average of the modeled fits of all 11 circadian proteins identified as elogation initiation factors. (D) An average of the modeled fits of all 21 circadian proteins in the ubiquitin-proteasome category, as defined by KEGG. Shading indicates ±1 standard deviation of models at each time point. (E) Modeled fits for the E3 ubiquitin ligase CBLB and a protein it targets for degradation, VAV2.
Proteins peaking in the late wave (CT15–3) were enriched in translational categories (“Processing of Capped Intron-Containing Pre-mRNA,” “RNA polymerase II Transcription,” and “Translation”), and the proteins involved in these processes were highly clustered (Fig. 3A, inset 1). Using PSEA, we additionally identified that proteins peaking in the late phase were involved in tRNA aminoacylation, the ligation of amino acids to tRNA to be used in translation (Supplemental Fig. S4A). We then modeled the levels of 16 different rhythmic enzymes catalyzing the formation of tRNA and calculated the average relative level of tRNA enzymes over the day. We found that tRNA-synthesizing enzymes generally peaked during the late phase (CT20) (Fig. 3B; Supplemental Fig. S4B). We also identified several circadian eukaryotic initiation factors, which are another important contributor to the translational machinery (Fig. 3C; Supplemental Fig. S4C). While circadian eukaryotic initiation factors exhibited more variation in their phases and shorter periods, an average of all oscillations demonstrates a peak in the late phase (CT16). These peaks in translation-associated proteins possibly underlie the wave of circadian proteins that arose rapidly from circadian RNA at CT18, as well as circadian proteins that arose from noncircadian RNA from CT18–22 (see Fig. 2; Supplemental Fig. S3).
However, the late wave peak in translational regulation did not explain the peak of circadian proteins near CT10 without associated circadian RNAs. Previous models suggested that a peak in ubiquitin ligases antiphase to protein peak phases would suggest that these proteins are going through timed degradation (Lück et al. 2014). We noted that the late phase was also enriched for ubiquitin ligases and proteasomal subunits (Fig. 3A, inset 2). Moreover, PSEA identified an enrichment for ubiquitin-protein ligases in the late phase (Supplemental Fig. S4A). We modeled and calculated the average level of rhythmic proteins involved in protein ubiquitination/degradation over the day. We found that the average peak of protein degrading enzymes was in the late phase (Fig. 3D; Supplemental Fig. S4D). This suggests that ubiquitin-targeted proteolysis may explain the peak of circadian proteins near CT10 without associated circadian RNAs. To probe this further, we identified a ubiquitin-protein ligase and its targeted protein to examine their temporal relationship (Fig. 3E; Supplemental Fig. S4E). The E3 ubiquitin-protein ligase CBLB is known to be particularly important for immune cell function due to its ability to modulate immune signaling by specifically targeting protein tyrosine kinases (PTKs) for degradation and other proteins important to this PTK signaling network, such as VAV2 (Lutz-Nicoladoni et al. 2015). VAV2 is a guanine exchange factor with regulatory roles in phagocytosis, adhesion, and migration via PTK-based toll-like receptor signaling (Miletic et al. 2007). As would be expected with a targeted-degradation regulatory relationship, we found that VAV2 had higher abundance when CBLB had lower abundance, and vice versa (Fig. 3E; Supplemental Fig. S4E).
Post-transcriptional circadian regulation in the early phase is enriched in immunometabolic regulatory pathways
With the demonstration of post-transcriptional regulation specific to the late wave, we next investigated the enrichment of the proteins in the early wave (CT3–15). Highly enriched categories included the “Citric Acid Cycle and Respiratory Electron Transport” and “Metabolism of Vitamins and Cofactors” (Fig. 4A). PSEA identified several central energy-producing pathways and mitochondrial cellular components in the early wave (Supplemental Fig. S4A). While both the CT15–3 and CT3–15 waves were enriched for “Innate Immunity,” the proteins involved in the early wave (CT3–15) appeared to share more interactions with proteins involved in receptor tyrosine kinase signaling and metabolism (Fig. 4A, inset). These data characterize the early wave as a time for energy production via the regulation of central metabolic pathways.
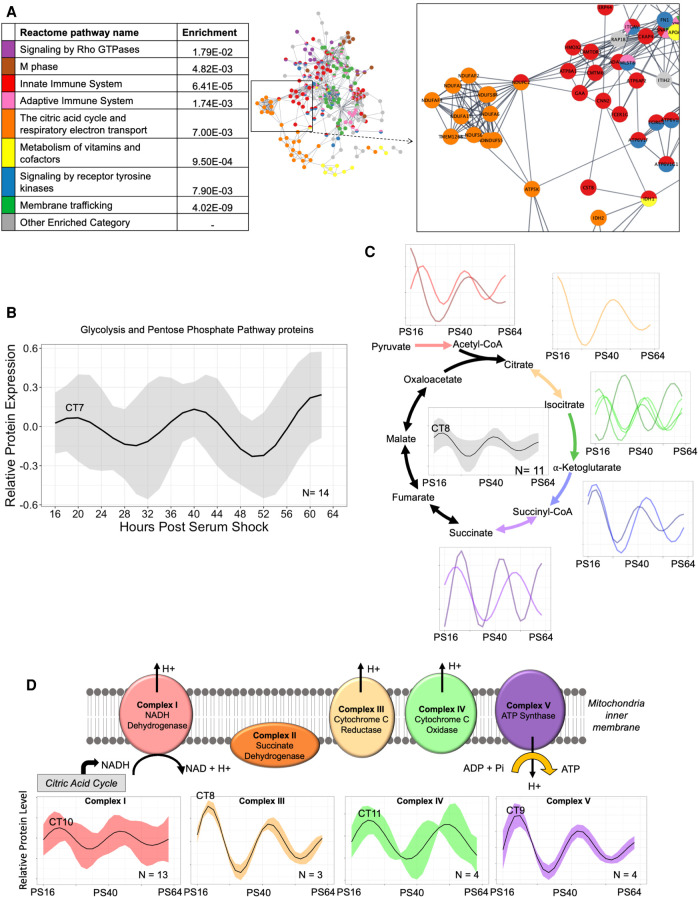
Figure 4.
Circadian regulation of central metabolic pathways coordinates mitochondrial ATP synthesis in the early phase. (A) A global view of the STRINGdb network of the proteins in enriched Reactome categories that peak in the early wave (CT3–15), with an inset focusing on key interactions, including the citric acid cycle and respiratory electron transport. Interactions shown as strings are filtered to highest confidence interactions (confidence > 0.9). Colors denote independent Reactome categories. Gray coloring represents genes from enriched Reactome categories that did not represent a significant proportion of the protein–protein interactome or described redundant categories. (B) An average of the oscillations for circadian proteins in the glycolysis and pentose phosphate pathways (PPPs). The average relative protein abundance model of all circadian proteins identified in each phase is represented, with the shaded region representing the standard deviation between models at each time point. The circadian time of the first peak is labeled. (C) A schematic of the subunits involved in the TCA cycle which had component proteins identified as rhythmic. An average curve and shaded standard deviation of the overall TCA cycle (center) as well as individual modeled fits for all circadian proteins in the identified corresponding enzyme complex are shown next to each step. The circadian time of the first peak is labeled. (D) A schematic of the subunits involved in the ETC which had component proteins identified as rhythmic. Average curve and shaded standard deviation of the modeled fits for all circadian proteins in the identified enzyme complex are shown below. The circadian time of the first peak is labeled.
The timing of the macrophage metabolic state is of interest as metabolic programming is increasingly recognized as a critical factor in determining macrophage immune responses, such as characterizing polarization within the M1 pro-inflammatory/M2 anti-inflammatory spectrum (Blagih and Jones 2012; Tang and Mauro 2017; Van den Bossche et al. 2017). To determine if there is circadian regulation of these metabolic pathways in macrophages, we analyzed which proteins circadianly oscillated in the protein complexes involved in the production of ATP (Fig. 4B–D). To ultimately produce ATP and other energy mediators (e.g., NADH), glucose first must undergo glycolysis. Parallel to glycolysis, the pentose phosphate pathway (PPP) utilizes glucose to produce NADPH and other metabolites that can feed back into glycolysis. Evaluation of circadian proteins in the glycolysis pathway and the PPP revealed 14 proteins, 11 functioning in glycolysis and three in the PPP, with the average oscillation peaking in the early phase. In contrast, phosphofructokinases (PFKL, PFKM, PFKP), the mediators of the rate-limiting first step of glycolysis, peak during the late phase (Supplemental Fig. S5A; Bonora et al. 2012). Previous work in Neurospora crassa demonstrated circadianly regulated proteins in the PPP peak in abundance antiphase to circadianly regulated glycolysis proteins (Hurley et al. 2018). We found evidence of a similar arrangement in macrophages, where the PPP proteins were circadianly regulated, peaking in the late wave, antiphase to glycolysis rate-limiting enzymes (Supplemental Fig. S5B). This suggests that energy production via glycolysis versus PPP are primed to occur at different times of the circadian day.
The product of glycolysis, pyruvate, can be further metabolized in the mitochondria via the tricarboxylic acid (TCA) cycle to generate an abundance of NADH. Eleven protein subunits in five protein complexes in the TCA cycle were circadianly regulated, and all but one peaked during the early phase (Fig. 4C; Supplemental Fig. S5C). Importantly, this included the enzyme catalyzing the initial entry of metabolites derived from glycolysis into the TCA cycle, as well as the majority of subunits belonging to protein complexes that facilitate irreversible reactions. Thus, this targeted circadian regulation is likely to propel the TCA cycle in the forward, NADH-producing direction during the early phase. Moreover, in the electron transport chain (ETC), which utilizes the NADH produced by the TCA cycle to generate large amounts of ATP, 24 subunits comprising parts of four ETC complexes oscillated with a circadian period in our data set (Fig. 4D; Supplemental Fig. S5D). The average time of proteins reaching their peak levels in the ETC was also in the early phase, slightly delayed from that seen for the TCA cycle. As M2-like, anti-inflammatory macrophages favor ATP production via stable oxidative phosphorylation, our findings suggest that naïve macrophages resemble an anti-inflammatory immunometabolic state in early phase (Van den Bossche et al. 2017). To also examine if this circadian regulation of central metabolic pathways is largely derived from post-transcriptional/-translational mechanisms, we analyzed which circadian proteins have corresponding circadian mRNA and found that only four—one in glycolysis, one in pentose phosphate, and two in the electron transport chain categories—were circadian at the mRNA level. Thus, the majority of circadian regulation of metabolic enzymes occurs at the post-transcriptional level in macrophages.
Post-transcriptional circadian regulation influences ATP production and mitochondrial morphology
Due to the significant circadian post-transcriptional regulation in the TCA cycle and ETC, we hypothesized that there would be a resulting oscillation in the respiratory rate of mitochondria, that is, the macrophage respiratory rate. To analyze daily variations in macrophage respiratory rates, we utilized a Seahorse assay to measure oxygen consumption rate (OCR) at both the trough and peak of PER2 abundance over one complete cycle, as determined by luminescence tracings (Supplemental Fig. S6A). For each time point, a basal respiratory rate was measured (Fig. 5A). As predicted by our analysis of the proteome, where we noted a peak in oxidative phosphorylation protein abundance in the early phase, basal respiratory rates were significantly higher during the early phase at CT4 compared to CT16 (Fig. 5A). OCR was measured after the addition of oligomycin, an ATP synthase-inhibitor, to provide a measure of ATP production. CT4 exhibited a higher capacity for mitochondrial ATP production as compared to CT16 (Fig. 5B). Fluoro-carbonyl cyanide phenylhydrazone (FCCP) was then added to assess the maximal respiration of the mitochondria. We found that maximal respiration decreased as a function of time as opposed to an oscillatory manner (Supplemental Fig. S6B). Finally, Rotenone/Antimycin A was added to inhibit mitochondrial function and measure spare respiratory capacity. The spare respiratory capacity decreased with time similar to what was observed for maximal capacity (Supplemental Fig. S6C). As a whole, this suggests that the circadian clock generates daily variations in basal rate metabolism and ATP-linked oxygen consumption in macrophages, similar to what was also observed in fibroblasts (Schmitt et al. 2018).
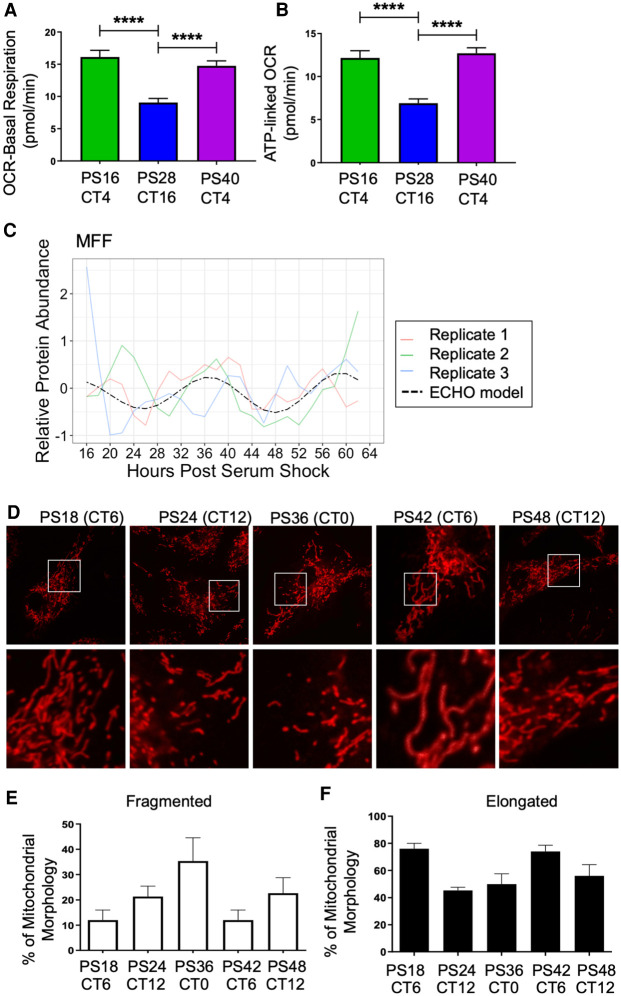
Figure 5.
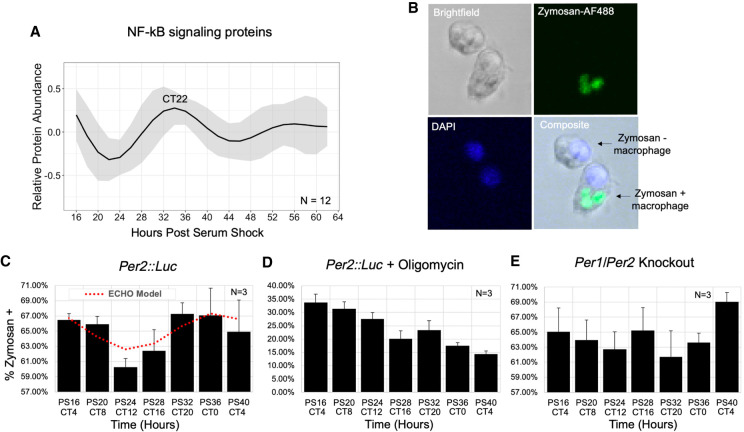
Circadian regulation of metabolism and mitochondrial morphology leads to a peak in mitochondrial-linked oxygen consumption rate in the early phase. Seahorse-derived measurements at different time points for the rate of (A) basal respiration and (B) ATP-linked OCR, in picomoles/min for 50,000 cells/well and 30 total wells per time point. (****) P ≤ 0.0001. (C) The oscillation of MFF protein in the proteomics time course. Across circadian time in synchronized BMDMs, mitochondria were stained using MitoTracker Red CMXRos (D), and the amount of fragmented (E) and elongated (F) mitochondria were quantified by confocal microscopy by measuring the length of over 50 mitochondrial particles per cell for 25 cells per sample in triplicate independent time courses. Mitochondria with intermediate lengths were considered “tubular” and therefore neither elongated nor fragmented.
Mitochondria regulate energy production through the coordination of fission (fragmentation of mitochondria) and fusion (elongation of mitochondria). Fused mitochondrial morphology supports higher levels of ATP generation via oxidative phosphorylation (Angajala et al. 2018). A key protein controlling fission is dynamin 1-like protein (DNM1L), also known as DRP1, a GTPase which is recruited to the mitochondrial outer membrane by Mitochondrial fission factor (MFF) (Smirnova et al. 2001; Otera et al. 2010; Wang et al. 2017b). In our data set, MFF protein oscillated in a circadian manner, but its transcript did not (Fig. 5C). When we surveyed additional proteins involved in the fission/fusion pathway, we found that FIS1 and DNM1L were also rhythmic (Supplemental Fig. S6D,E; Supplemental Table S4). We postulated that the oscillation of MFF would lead to an oscillation in mitochondrial morphology that would also coincide with the observed variations in basal/ATP-linked oxygen consumption. To investigate this, mitochondrial morphology was assessed over time by imaging macrophages stained with MitoTracker Red (Fig. 5D). Indeed, an increase in the percentage of fragmented, that is, fissioned, mitochondria was observed in the late phase at CT0/24 (Fig. 5E), coinciding with the peak in MFF protein abundance (Fig. 5C). The highest percentage of elongated, that is, fused, mitochondria occurred at CT6, coinciding with the time of day with the highest basal and ATP-linked oxygen consumption (Fig. 5F).
Circadian post-transcriptional regulation of metabolism impacts the phagocytic immune response
Mitochondrial function is highly influential to immune responses (Mills and O'Neill 2016). We thus proposed that circadian regulation of mitochondrial dynamics and ATP production would have distinct consequences on a macrophage's response to immunological stimuli. One of the characteristic functions of macrophages in their role as mediators of the immune response and inflammation is their phagocytic activity. Phagocytosis involves complex networking of surface receptors, signaling cascades, and cytoskeletal modifications, making phagocytosis an energy-demanding process requiring interconnection with cellular metabolism. In addition to metabolic regulation, PSEA for circadian proteins identified in the late wave the ontological category “Positive Regulation of I-Kappa B Kinase NF-kB Cascade,” a category that highlights proteins involved in the signaling pathway that leads to the phosphorylation and degradation of I-Kappa B, enabling a robust inflammatory response via NF-kB signaling (Fig. 6A; Supplemental Fig. S4A; Liou 2002).
Figure 6.
Macrophage phagocytosis of zymosan shows metabolism-related circadian variation in vitro. (A) An average of the modeled fits of all 12 circadian proteins in the NF-kB signaling pathway. The circadian time of the first peak is labeled. (B) Representative confocal microscopy images demonstrating identification of macrophages positive and negative for phagocytosis of zymosan-AlexaFluor488. Cells were also DAPI-stained to aid in cell counting. The levels of phagocytosis are reported as a percent of cells with one or more particles of zymosan in (C) Per2::Luc, (D) Per2::Luc co-incubated with oligomycin, and (E) Per1/Per2 knockout BMDMs at a 4-h resolution for 24 h (N = 3). Trace of significant circadian oscillation as determined by ECHO (BH adj P-value = 0.0026) is plotted on the Per2::Luc graph in a dotted red line. Error bars represent standard error of the mean (SEM) for triplicate samples. All parameters for ECHO and JTK analyses, including noncircadian oscillatory fits for D and E, are reported in Supplemental Table S5.
To determine if macrophage phagocytic activity was circadianly timed in our in vitro model, we measured the phagocytosis of AF488-conjugated zymosan, a yeast-derived bioparticle, as zymosan had previously been shown to be phagocytized with daily variation by ex vivo murine peritoneal macrophages and is also known to exert inflammatory signaling primarily via TLR2/Dectin-1 receptors, which have cooperating signaling cascades converging on NF-kB signaling (Fig. 6A,B; Gantner et al. 2003; Dennehy and Brown 2007; Hayashi et al. 2007). Luminescence tracking further examined that the addition of zymosan and/or oligomycin was not radically altering clock gene expression for the duration of our experiment or ablating PER2 rhythmicity, though long-term, immune challenge at CT4 decreased the amplitude of the oscillation, whereas immune challenge at CT16 did not. (Supplemental Fig. S7A–C). Consistent with previous studies on circadian phagocytosis, BMDMs derived from Per2::Luc mice were observed to phagocytize zymosan in a temporally dependent manner, with the lowest phagocytosis rate at CT12 and the highest at CT0 (Fig. 6C), with an 11.4% increase in efficiency at the peak versus the trough (Hayashi et al. 2007; Oliva-Ramírez et al. 2014).
Both ECHO and JTK analysis confirmed the significance of this variation's fit to a 24-h oscillatory pattern (Supplemental Table S5; Hughes et al. 2010; De los Santos et al. 2017, 2020). To determine if circadian metabolic regulation was the driving force in the temporal organization of phagocytosis, we added the ATP synthase inhibitor oligomycin A to the phagocytosis assay at each time point and measured phagocytosis 30 min later (Fig. 6D; Jastroch et al. 2010). We noted an expected decrease in phagocytosis, which was exacerbated over time, yet with no evidence of oscillatory daily variation in phagocytosis in these cells (Fig. 6D; Supplemental Table S5). To eliminate the possibility that the phagocytosis variation was dictated independently of clock function, we analyzed phagocytosis rates from BMDMs derived from Per1/Per2 double-knockout mice, which are arrhythmic in all tissues (Bae et al. 2001; Zheng et al. 2001), and found that phagocytosis was not oscillatory in Per1/Per2 knockout BMDMs (Fig. 6E; Supplemental Table S5). Modeling the levels of the signaling proteins involved in the I-KappaB Kinase/NF-kB cascade demonstrated that these proteins peaked at the same time as increased phagocytosis activity (Fig. 6A,C; Supplemental Fig. S7D), suggesting that circadian signaling pathway regulation in crosstalk with circadian metabolic regulation could also play a role in the generation of the temporal variations observed.
Discussion
Our comprehensive profiling of the macrophage transcriptome and proteome over circadian time revealed extensive physiologically relevant circadian post-transcriptional regulation (Fig. 1; Supplemental Fig. S3) via two distinct yet discordant waves of circadian transcripts and proteins occurring antiphase to each other (Fig. 2). Our high-powered study, optimized to identify as many oscillating genes as possible, observed a marked lack of overlap between circadian mRNA and protein as compared to previous studies, establishing that post-transcriptional/-translational mechanisms play a particularly significant role in macrophages. These findings refute the canonical view that transcriptional activation or repression from clock genes is the dominant driver of circadian output, as has been described in other tissue types where the magnitude of such discrepancies between circadian mRNAs/proteins is smaller (Reddy et al. 2006; Kojima et al. 2011; Chiang et al. 2014; Hurley et al. 2014, 2018; Mauvoisin et al. 2014; Robles et al. 2014; Wang et al. 2017a, 2018; Green 2018; Mauvoisin and Gachon 2020). This discordance may extend to all mammalian tissue types, as previous studies have not been able to achieve the power of our extended, densely sampled study (Reddy et al. 2006; Kojima et al. 2011; Mauvoisin et al. 2014; Robles et al. 2014; Green 2018; Wang et al. 2018; Mauvoisin and Gachon 2020). It remains possible, however, that macrophages are unique in the extent of their circadian post-transcriptional regulation. This regulation may be necessary due to the macrophages’ need to respond quickly to a wide variety of potential immunological stimuli (Abbas et al. 2018). Our previous work utilizing an identical experimental design and similar analysis in the fungal model N. crassa did not indicate a high prevalence of post-transcriptional regulation, supporting that our findings represent a novel lack of correlation between rhythmic mRNA and protein (Hurley et al. 2018). However, future work deeply evaluating both proteomes and transcriptomes from other immune cells and tissues is needed to determine if this is the case.
Post-transcriptional regulation in mammalian tissues has been suggested to primarily derive from systemic cues, such as hormones, nutrients, and nerve signals (Koike et al. 2012; Kojima et al. 2012; Menet et al. 2012; Partch et al. 2014). However, our data showed that extensive post-transcriptional regulation occurred in macrophages devoid of systemic cues in vitro, contradicting this hypothesis and demonstrating that the macrophages’ endogenous clock network alone can produce oscillations beyond the canonical mechanism of transcriptional regulation. In contrast to solid tissues, where the stochasticity between millions of cells can be buffered against by systemic regulation producing an overall circadian output from the solid tissue, macrophages are largely independent in their surveillance and initiation of early stages of inflammation (Nagoshi et al. 2004; Forger and Peskin 2005; Lande-Diner et al. 2015). Our work suggests that macrophages’ unique roles in the body may have driven them to rely heavily upon their intrinsic clock to regulate output by driving post-transcriptional oscillations in protein abundance, suggesting a novel mechanism for circadian regulation. Enrichment analysis revealed that protein degradation, in addition to translational mechanisms also reported in other tissues, potentially plays a role in macrophage post-transcriptional circadian regulation, suggesting that control of protein homeostasis is a source of this intrinsic post-transcriptional regulation (Figs. 2, 3; Lück et al. 2014; Caster et al. 2016; Hurley et al. 2018). Although enrichments for proteins with functions in various pathways were identified, it is beyond the scope of our high-throughput methods to definitively determine how, or if, these pathways would ultimately be influenced. Ribosome and degradation profiling would be needed to shed additional light on the impact and origins of protein abundance oscillations, as this cannot be deciphered by our steady-state measurements. Additionally, future work and low-throughput validation of gene oscillations are warranted to determine how our in vitro findings relate to macrophages in vivo with the additional influence of systemic cues. There are several possibilities to explain how proteins without rhythmic mRNAs could be targeted for rhythmic synthesis or degradation by clock-controlled mechanisms. For example, regulation could occur via the control of selective initiation/elongation factors, splicing complexes, transport across the nuclear membrane or towards ribosomes/proteasomes, or via stabilizing/destabilizing interactions with clock gene proteins either directly or indirectly through the temporal formation of protein complexes (Wong et al. 2020).
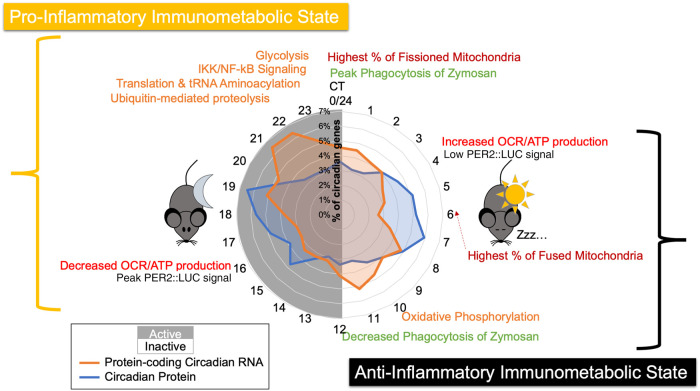
Our deeply sampled data set proved to be a valuable resource to investigate the impact of circadian regulation on macrophage physiology, demonstrating that the circadian clock generates distinct immunometabolic states at opposing times of day, with the inactive phase primed for ATP synthesis via oxidative phosphorylation and the active phase poised for glycolysis (Fig. 7). These antiphase immunometabolic states were defined by their different enrichments of circadian proteins (Figs. 3, 4) and corresponding daily variations in both metabolic rate and mitochondrial morphology (Fig. 5). Although protein abundance does not necessarily equate to increased activity or efficiency of a metabolic pathway, previous work in Neurospora provides evidence that circadian regulation of enzyme abundance serves to control metabolic pathways such as xylitol synthesis (Hurley et al. 2018). In macrophages, it is known that the lipopolysaccharide-induced metabolic reprogramming to a pro-inflammatory state is coordinated by the transcription factor HIF1A, which up-regulates expression of glycolytic enzymes (Tannahill et al. 2013). Therefore, it is feasible that clock regulation of metabolic enzyme abundance could prime macrophage metabolic programming to be pro- or anti-inflammatory. Indeed, our data suggest that this regulation is meaningful in generating metabolic consequences. Our Seahorse assay confirmed the existence of a daily variation in basal and ATP-linked cellular respiration, and our examination of mitochondrial morphology further supports the temporal organization of high and low metabolic states corresponding with post-transcriptional regulation of oxidative phosphorylation proteins (Peek et al. 2013; Langston et al. 2017; Schmitt et al. 2018).
Figure 7.
Summary model of the circadian regulation of distinct immunometabolic states. A summary schematic displaying key findings, spatially organized by circadian time of day, with a radial histogram in the center to indicate when circadian proteins and circadian mRNAs peak in abundance. Orange text represents enriched categories identified in the proteome, red represents the occurrence of mitochondrial morphology/functional observations, green represents phagocytosis assay observations, and black text shows the trough and peak of PER2 levels as determined by PER2::LUC luminescence signal. For reference, Post Shock 16, our first time course time point, is equivalent to CT4, as described in Figure 1D.
In concordance with our finding that clock output temporally primes macrophage immunometabolic states, zymosan phagocytosis demonstrated 24-h variation consistent with rhythmic regulation. This is similar to observations made in previous studies, yet differences in the presumed phases of optimal phagocytic activity were seen, likely due to differences in the macrophage niches, pathogens, and experimental designs used (Hayashi et al. 2007; Tognini et al. 2017; Geiger et al. 2019; Kitchen et al. 2020). We further found that both a clock knockout strain and mitochondrial disruptor ablated this rhythm (Fig. 6). This suggests that the distinct immunometabolic states generated by the circadian clock impact immunological responses, yet our approach could not definitively isolate metabolic regulation from other possible regulatory mechanisms underlying circadian influence on phagocytosis. Our data as a whole support that post-transcriptional circadian regulation metabolically primed naïve macrophages to resemble an anti-inflammatory phenotype (Fig. 7), which characteristically has robust ATP production via mitochondrial oxidative phosphorylation. At the opposite time of day, the immunometabolic state resembled a pro-inflammatory programming, as shunting oxidative phosphorylation in favor of glycolysis is a hallmark of pro-inflammatory macrophages (Van den Bossche et al. 2017; Carroll et al. 2019). A circadianly timed proclivity for inflammatory responses was consistent with our findings that zymosan is more readily phagocytized at the end of the active phase (Fig. 6C), which is consistent with the majority of studies in both humans and nocturnal animals, showing that immune challenges are generally less effectively neutralized when exposed during the inactive phase or under circumstances of circadian disruption when macrophages are not primed for appropriate inflammatory responses (Geiger et al. 2015; Nagy and Haschemi 2015; Van den Bossche et al. 2017). However, exact timing of the immunometabolic phases coordinated by the circadian clock requires future examination to determine the relationship between observed peaks in protein abundance and related metabolic/immunological activity. As circadian control of metabolic pathways has been shown to be conserved in a multitude of tissue types and organisms, our study serves as a foundation to generate hypotheses about the diverse functional impacts of metabolic regulation in a multitude of biological and disease contexts.
In addition to the protective roles of inflammation in pathogen defense, inflammation also plays an important part in the pathology of diseases associated with chronic circadian disruption, leading many to hypothesize that the dysregulation of immunity plays a major role in the link between chronic circadian disruption and associated diseases (Evans and Davidson 2013; Musiek and Holtzman 2016; Shostak 2017). Our work provides further evidence for this link, as disruption to the intricate metabolic and clock regulatory networks we identified in macrophages could result in a pathological dysregulation of immunity, potentially fueling chronic inflammation that contributes to disease. Our data sets and analyses provide a robust foundation on which we can build to understand the role of circadian clocks in macrophages. This could ultimately inform novel strategies for prevention and treatment of diseases associated with chronic circadian disruption and inflammation.
Methods
BMDM differentiation and synchronization
Bone marrow was collected from tibias/fibulas of 3- to 6-mo-old male Per2::Luc or Per1/Per2 knockout mice (C57BL/6J) and differentiated with DMEM supplemented with M-CSF and 10% FBS. After 7 d, differentiated macrophages were synchronized by a 24-h starve in serum-free media followed by 2-h serum shock with Leibovitz supplemented with 50% FBS. Cells were assayed via LumiCycle32 (Actimetrics) in Leibovitz media containing Luciferin and 10% FBS. See Supplemental Methods for further details.
RNA-sequencing
Every 2 h over 48 h starting at 16 h post-serum shock for three independent time courses, RNA was extracted using standard Qiagen RNeasy protocols for analysis via RNA-seq. Paired-end reads were sequenced with an Illumina HiSeq and mapped to the Mus musculus GRCm38.87 reference genome using the DRAGEN pipeline. Hit counts were quantified with HTSeq and normalized to transcripts per million. See Supplemental Methods for further details.
Proteomics
Every 2 h over 48 h starting at 16 h post-serum shock for three independent time courses, protein was extracted with 8 M Urea buffer for analysis via multiplex tandem mass tag mass spectrometry. A total of eight 10-plexes were run on an Orbitrap Lumos spectrometer, each containing nine isobarically tagged randomized samples and one pooled sample shared between all multiplexes for comparison. Peptides were identified by their MS2 spectra and mapped to UniProt reference Mus musculus proteome UP000000589. See Supplemental Methods for further details.
Omics data processing and detection of rhythms
RNA-seq and proteomics data were further processed to remove batch effects using the LIMBR pipeline with default parameters and removing peptides/transcripts with <70% detection across all 72 time points. Any missing values for remaining peptides were then imputed by LIMBR (Crowell et al. 2019). Batch effect-adjusted data were then analyzed for rhythms with ECHO (v3.0) in free-run mode with smoothing and normalization options (see Supplemental Data S3, S4; De los Santos et al. 2020). Genes were considered circadianly oscillating if BH-adjusted P-value < 0.05, and forcing coefficient between 0.15 and −0.15, as recommended in De los Santos et al. (2020), as well as if the period was between 20 and 28 for RNA and 18 and 30 for proteins, based on evaluating the known clock gene periods as modeled by our ECHO analysis method. To compare the circadian times at which transcripts/proteins peak, CT phase times were adjusted by period length. To compare genes between data sets, UniProt numbers were used to look up current MGI primary gene names. Duplicate circadian proteins due to database differences or multiple isoforms present were manually curated by choosing a representative protein based on which had more peptides quantified.
Reactome enrichment and STRINGdb analysis
The list of circadian proteins generated with ECHO was bisected into two groups based on circadian adjusted time of peak: those peaking between CT3–15 and those peaking between CT15–3. Each list was analyzed with PantherDB's PANTHER overrepresentation test using Reactome version 65 (2019-03-02) against the full Mus musculus database. Categories were considered enriched if the Bonferroni corrected P-value was <0.05. For STRINGdb visualization and analysis, both lists were subset to only include proteins that belong to at least one enriched category. STRINGdb networks were then generated and colored by Reactome category using the stringApp for Cytoscape and selecting to visualize only high-confidence interactions (>0.9) (Doncheva et al. 2019).
Mitochondrial dynamics
For Seahorse analysis, 50,000 murine macrophages were synchronized via serum shock as described previously in three 12-h staggered groups in a 96-well plate, then simultaneously assayed at postshock time points 16, 28, and 40 with a Seahorse XF instrument according to the manufacturer's protocol. For mitochondrial morphology analysis, macrophages were synchronized in glass-bottom dishes, and 50 nM MitoTracker Red CMXRos was added to media 30 min prior to each time point. Cells were imaged via confocal microscope, and the mitochondrial length of more than 50 mitochondrial particles per cell was measured with ImageJ in over 25 cells per experimental condition, out of three independent experiments. Mitochondria were then divided into two different categories based on length, as mitochondria of <1 µm (fragmented) and >3 µm (elongated), as described by Park et al. (2013). See Supplemental Methods for further details.
Zymosan phagocytosis
Macrophages from Per2::Luc or Per1/Per2 knockout mice were synchronized utilizing two groups staggered by 12 h. FBS-opsonized zymosan (AF488 labeled) was added (1:100 cell:zymosan) to cell culture media for triplicate time points every 4 h over 24 h, with or without 1.5 µM oligomycin. Cells were washed, formalin-fixed, and DAPI-stained, then imaged with confocal fluorescence microscope. Using ImageJ, the number of macrophages with positive zymosan-AF488 signal was counted to calculate the percentage of zymosan-positive macrophages out of at least 100 total macrophages observed per sample. See Supplemental Methods for further details.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE157878. The mass spectrometry proteomics data generated in this study have been submitted to ProteomeXchange (http://www.proteomexchange.org) under accession number PXD022260.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Dr. Kristen Bennett and Hannah De los Santos of the Rensselaer Institute for Data Exploration and Applications for their help with ECHO analyses, Gretchen Clark at Rensselaer for her help with macrophage work, and other members of the Hurley lab for their valuable discussion and feedback. We also thank Sergey Pryshchep of the Rensselaer Microscopy Core for his technical expertise and the BioResearch Core for animal husbandry. Finally, we thank the Dunlap/Loros labs at Dartmouth for generously sharing Per1/Per2 knockout mice. This work was supported by the National Institutes of Health, National Institute of General Medical Sciences (T32GM067545 to E.J.C. and GM128687 to J.M.H.); Rensselaer startup funds (to J.M.H.); Consejo Nacional de Ciencia y Tecnología (CVU440823 to M.P.C-S); the Science Foundation Ireland Career Development Award (17/CDA/4688 to A.M.C.); and the Irish Research Council Laureate Award (IRCLA/2017/110 to A.M.C.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.263814.120.
References
- Abbas A, Lichtman A, Pillai, S. 2018. Innate immunity. In Cellular and molecular immunology, 9th ed (ed. Merritt J), pp. 82–90. Elsevier, Philadelphia. [Google Scholar]
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. 2011. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev 25: 2374–2386. 10.1101/gad.178079.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angajala A, Lim S, Phillips JB, Kim J-H, Yates C, You Z, Tan M. 2018. Diverse roles of mitochondria in immune responses: novel insights into immuno-metabolism. Front Immunol 9: 1605 10.3389/fimmu.2018.01605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjona A, Silver AC, Walker WE, Fikrig E. 2012. Immunity's fourth dimension: approaching the circadian–immune connection. Trends Immunol 33: 607–612. 10.1016/j.it.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. 2001. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30: 525–536. 10.1016/S0896-6273(01)00302-6 [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–937. 10.1016/S0092-8674(00)81199-X [DOI] [PubMed] [Google Scholar]
- Bares V, Ge X. 2019. gskb: Gene Set data for pathway analysis in mouse. R package version 1.18.0 10.18129/B9.bioc.gskb [DOI]
- Bechtold DA, Gibbs JE, Loudon ASI. 2010. Circadian dysfunction in disease. Trends Pharmacol Sci 31: 191–198. 10.1016/j.tips.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Blagih J, Jones RG. 2012. Polarizing macrophages through reprogramming of glucose metabolism. Cell Metab 15: 793–795. 10.1016/j.cmet.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Bonora M, Patergnani S, Rimessi A, De Marchi E, Suski JM, Bononi A, Giorgi C, Marchi S, Missiroli S, Poletti F, et al. 2012. ATP synthesis and storage. Purinergic Signal 8: 343–357. 10.1007/s11302-012-9305-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourin P, Ledain AF, Beau J, Mille D, Lévi F. 2002. In-vitro circadian rhythm of murine bone marrow progenitor production. Chronobiol Int 19: 57–67. 10.1081/CBI-120002677 [DOI] [PubMed] [Google Scholar]
- Carroll RG, Timmons GA, Cervantes-Silva MP, Kennedy OD, Curtis AM. 2019. Immunometabolism around the clock. Trends Mol Med 25: 612–625. 10.1016/j.molmed.2019.04.013 [DOI] [PubMed] [Google Scholar]
- Caster SZ, Castillo K, Sachs MS, Bell-Pedersen D. 2016. Circadian clock regulation of mRNA translation through eukaryotic elongation factor eEF-2. Proc Natl Acad Sci 113: 9605–9610. 10.1073/pnas.1525268113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian N, Lange T, Golombek D, Sarkar D, Nakao A, Shibata S, Mazzoccoli G. 2013. Crosstalk between the circadian clock circuitry and the immune system. Chronobiol Int 30: 870–888. 10.3109/07420528.2013.782315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C-K, Mehta N, Patel A, Zhang P, Ning Z, Mayne J, Sun WYL, Cheng H-YM, Figeys D, Kramer A. 2014. The proteomic landscape of the suprachiasmatic nucleus clock reveals large-scale coordination of key biological processes. PLoS Genet 10: e1004695 10.1371/journal.pgen.1004695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell AM, Greene CS, Loros JJ, Dunlap JC, Wren J. 2019. Learning and Imputation for Mass-spec Bias Reduction (LIMBR). Bioinformatics 35: 1518–1526. 10.1093/bioinformatics/bty828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis AM, Bellet MM, Sassone-Corsi P, O'Neill LAJ. 2014. Circadian clock proteins and immunity. Immunity 40: 178–186. 10.1016/j.immuni.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Curtis AM, Fagundes CT, Yang G, Palsson-McDermott EM, Wochal P, McGettrick AF, Foley NH, Early JO, Chen L, Zhang H, et al. 2015. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci 112: 7231–7236. 10.1073/pnas.1501327112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De los Santos H, Collins EJ, Hurley JM, Bennett KP. 2017. Circadian rhythms in Neurospora exhibit biologically relevant driven and damped harmonic oscillations. In Proceedings of the Eighth ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, pp. 455–463. Boston 10.1145/3107411.3107420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De los Santos H, Collins EJ, Mann C, Sagan AW, Jankowski MS, Bennett KP, Hurley JM. 2020. ECHO: an application for detection and analysis of oscillators identifies metabolic regulation on genome-wide circadian output. Bioinformatics 36: 773–781. 10.1093/bioinformatics/btz617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy KM, Brown GD. 2007. The role of the β-glucan receptor Dectin-1 in control of fungal infection. J Leukoc Biol 82: 253–258. 10.1189/jlb.1206753 [DOI] [PubMed] [Google Scholar]
- Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. 2019. Cytoscape stringApp: network analysis and visualization of proteomics data. J Proteome Res 18: 623–632. 10.1021/acs.jproteome.8b00702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. 1999. Molecular bases for circadian clocks. Cell 96: 271–290. 10.1016/S0092-8674(00)80566-8 [DOI] [PubMed] [Google Scholar]
- Early JO, Menon D, Wyse CA, Cervantes-Silva MP, Zaslona Z, Carroll RG, Palsson-McDermott EM, Angiari S, Ryan DG, Corcoran SE, et al. 2018. Circadian clock protein BMAL1 regulates IL-1β in macrophages via NRF2. Proc Natl Acad Sci 115: E8460–E8468. 10.1073/pnas.1800431115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K, Sassone-Corsi P. 2013. Metabolism and the circadian clock converge. Physiol Rev 93: 107–135. 10.1152/physrev.00016.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Davidson AJ. 2013. Health consequences of circadian disruption in humans and animal models. Prog Mol Biol Transl Sci 119: 283–323. 10.1016/B978-0-12-396971-2.00010-5 [DOI] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. 2018. The Reactome pathway Knowledgebase. Nucleic Acids Res 46: D649–D655. 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger DB, Peskin CS. 2005. Stochastic simulation of the mammalian circadian clock. Proc Natl Acad Sci 102: 321–324. 10.1073/pnas.0408465102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. 2003. Collaborative induction of inflammatory responses by Dectin-1 and toll-like receptor 2. J Exp Med 197: 1107–1117. 10.1084/jem.20021787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger SS, Fagundes CT, Siegel RM. 2015. Chrono-immunology: progress and challenges in understanding links between the circadian and immune systems. Immunology 146: 349–358. 10.1111/imm.12525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger SS, Curtis AM, O'Neill LAJ, Siegel RM. 2019. Daily variation in macrophage phagocytosis is clock-independent and dispensable for cytokine production. Immunology 157: 122–136. 10.1111/imm.13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, et al. 2012. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci 109: 582–587. 10.1073/pnas.1106750109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB. 2018. Circadian posttranscriptional regulatory mechanisms in mammals. Cold Spring Harb Perspect Biol 10: a030692 10.1101/cshperspect.a030692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Shimba S, Tezuka M. 2007. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull 30: 621–626. 10.1248/bpb.30.621 [DOI] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB, Barsh GS, 2009. Harmonics of circadian gene transcription in mammals. PLoS Genet 5: e1000442 10.1371/journal.pgen.1000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. 2010. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25: 372–380. 10.1177/0748730410379711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Abruzzi KC, Allada R, Anafi R, Arpat AB, Asher G, Baldi P, de Bekker C, Bell-Pedersen D, Blau J, et al. 2017. Guidelines for genome-scale analysis of biological rhythms. J Biol Rhythms 32: 380–393. 10.1177/0748730417728663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo M, Huang Y, Qu D, Zhang H, Wong WT, Chawla A, Huang Y, Tian XY. 2017. Myeloid Bmal1 deletion increases monocyte recruitment and worsens atherosclerosis. FASEB J 31: 1097–1106. 10.1096/fj.201601030R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Dasgupta A, Emerson JM, Zhou X, Ringelberg CS, Knabe N, Lipzen AM, Lindquist EA, Daum CG, Barry KW, et al. 2014. Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proc Natl Acad Sci 111: 16995–17002. 10.1073/pnas.1418963111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Loros JJ, Dunlap JC. 2016. Circadian oscillators: around the transcription–translation feedback loop and on to output. Trends Biochem Sci 41: 834–846. 10.1016/j.tibs.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Jankowski MS, De los Santos H, Crowell AM, Fordyce SB, Zucker JD, Kumar N, Purvine SO, Robinson EW, Shukla A, et al. 2018. Circadian proteomic analysis uncovers mechanisms of post-transcriptional regulation in metabolic pathways. Cell Syst 7: 613–626.e5. 10.1016/j.cels.2018.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastroch M, Divakaruni AS, Mookerjee S, Treberg JR, Brand MD. 2010. Mitochondrial proton and electron leaks. Essays Biochem 47: 53–67. 10.1042/bse0470053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk H-D, Kramer A, Maier B. 2009. A circadian clock in macrophages controls inflammatory immune responses. Proc Nat Acad Sci 106: 21407–21412. 10.1073/pnas.0906361106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling S, Dubeau-Laramée G, Ohm H, Labrecque N, Olivier M, Cermakian N. 2017. The circadian clock in immune cells controls the magnitude of Leishmania parasite infection. Sci Rep 7: 10892 10.1038/s41598-017-11297-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen GB, Cunningham PS, Poolman TM, Iqbal M, Maidstone R, Baxter M, Bagnall J, Begley N, Saer B, Hussell T, et al. 2020. The clock gene Bmal1 inhibits macrophage motility, phagocytosis, and impairs defense against pneumonia. Proc Natl Acad Sci 117: 1543–1551. 10.1073/pnas.1915932117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo S-H, Huang H-C, Kumar V, Lee C, Kim T-K, Takahashi JS. 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354. 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Shingle DL, Green CB. 2011. Post-transcriptional control of circadian rhythms. J Cell Sci 124: 311–320. 10.1242/jcs.065771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Sher-Chen EL, Green CB. 2012. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev 26: 2724–2736. 10.1101/gad.208306.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrecque N, Cermakian N. 2015. Circadian clocks in the immune system. J Biol Rhythms 30: 277–290. 10.1177/0748730415577723 [DOI] [PubMed] [Google Scholar]
- Lam MTYY, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. 2013. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498: 511–515. 10.1038/nature12209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L, Stewart-Ornstein J, Weitz CJ, Lahav G, Lippincott-Schwartz J. 2015. Single-cell analysis of circadian dynamics in tissue explants. Mol Biol Cell 26: 3940–3945. 10.1091/mbc.E15-06-0403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston PK, Shibata M, Horng T. 2017. Metabolism supports macrophage activation. Front Immunol 8: 61 10.3389/fimmu.2017.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou HC. 2002. Regulation of the immune system by NF-κB and IκB. J Biochem Mol Biol 35: 537–546. 10.5483/bmbrep.2002.35.6.537 [DOI] [PubMed] [Google Scholar]
- Lück S, Thurley K, Thaben PF, Westermark PO. 2014. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep 9: 741–751. 10.1016/j.celrep.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Lutz-Nicoladoni C, Wolf D, Sopper S. 2015. Modulation of immune cell functions by the E3 ligase CBL-b. Front Oncol 5 10.3389/fonc.2015.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvoisin D, Gachon F. 2020. Proteomics in circadian biology. J Mol Biol 432: 3565–3577. 10.1016/j.jmb.2019.12.004 [DOI] [PubMed] [Google Scholar]
- Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, Waridel P, Quadroni M, Gachon F, Naef F. 2014. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci 111: 167–172. 10.1073/pnas.1314066111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. 2012. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1: e00011 10.7554/eLife.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47: D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletic AV, Graham DB, Montgrain V, Fujikawa K, Kloeppel T, Brim K, Weaver B, Schreiber R, Xavier R, Swat W. 2007. Vav proteins control MyD88-dependent oxidative burst. Blood 109: 3360–3368. 10.1182/blood-2006-07-033662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, O'Neill LA. 2016. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur J Immunol 46: 13–21. 10.1002/eji.201445427 [DOI] [PubMed] [Google Scholar]
- Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, et al. 2018. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359: eaao0318 10.1126/science.aao0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek ES, Holtzman DM. 2016. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science 354: 1004–1008. 10.1126/science.aah4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U, 2004. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119: 693–705. 10.1016/j.cell.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Nagy C, Haschemi A. 2015. Time and demand are two critical dimensions of immunometabolism: the process of macrophage activation and the pentose phosphate pathway. Front Immunol 6: 164 10.3389/fimmu.2015.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. 2013. Circadian gene Bmal1 regulates diurnal oscillations of Ly6Chi inflammatory monocytes. Science 341: 1483–1488. 10.1126/science.1240636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva-Ramírez J, Moreno-Altamirano MMB, Pineda-Olvera B, Cauich-Sánchez P, Sánchez-García FJ. 2014. Crosstalk between circadian rhythmicity, mitochondrial dynamics and macrophage bactericidal activity. Immunology 143: 490–497. 10.1111/imm.12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158. 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Choi H, Min J-S, Park S-J, Kim J-H, Park H-J, Kim B, Chae J-I, Yim M, Lee D-S. 2013. Mitochondrial dynamics modulate the expression of pro-inflammatory mediators in microglial cells. J Neurochem 127: 221–232. 10.1111/jnc.12361 [DOI] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24: 90–99. 10.1016/j.tcb.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patke A, Young MW, Axelrod S. 2020. Molecular mechanisms and physiological importance of circadian rhythms. Nat RevMol Cell Biol 21: 67–84. 10.1038/s41580-019-0179-2 [DOI] [PubMed] [Google Scholar]
- Peek CB, Affinati AH, Ramsey KM, Kuo H-Y, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, et al. 2013. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342: 1243417 10.1126/science.1243417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick R, He W, Chen C-S, Scheiermann C. 2019. Time-of-day-dependent trafficking and function of leukocyte subsets. Trends Immunol 40: 524–537. 10.1016/j.it.2019.03.010 [DOI] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GKY, Chesham J, Odell M, Lilley KS, et al. 2006. Circadian orchestration of the hepatic proteome. Curr Biol 16: 1107–1115. 10.1016/j.cub.2006.04.026 [DOI] [PubMed] [Google Scholar]
- Robles MS, Cox J, Mann M. 2014. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet 10: e1004047 10.1371/journal.pgen.1004047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Sakurai T, Ogasawara J, Takahashi M, Izawa T, Imaizumi K, Taniguchi N, Ohno H, Kizaki T. 2014. A circadian clock gene, Rev-erbα, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. J Immunol 192: 407–417. 10.4049/jimmunol.1301982 [DOI] [PubMed] [Google Scholar]
- Scheiermann C, Gibbs J, Ince L, Loudon A, 2018. Clocking in to immunity. Nat Rev Immunol 18: 423–437. 10.1038/s41577-018-0008-4 [DOI] [PubMed] [Google Scholar]
- Schmitt K, Grimm A, Dallmann R, Oettinghaus B, Restelli LM, Witzig M, Ishihara N, Mihara K, Ripperger JA, Albrecht U, et al. 2018. Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab 27: 657–666.e5. 10.1016/j.cmet.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Shostak A. 2017. Circadian clock, cell division, and cancer: from molecules to organism. Int J Mol Sci 18: 873 10.3390/ijms18040873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver AC, Arjona A, Walker WE, Fikrig E. 2012. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity 36: 251–261. 10.1016/j.immuni.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver AC, Buckley SM, Hughes ME, Hastings AK, Nitabach MN, Fikrig E. 2018. Daily oscillations in expression and responsiveness of toll-like receptors in splenic immune cells. Heliyon 4: e00579 10.1016/j.heliyon.2018.e00579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland D-L, van der Bliek AM, Pollard TD. 2001. Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell 12: 2245–2256. 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS. 2017. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18: 164–179. 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C-Y, Mauro C. 2017. Similarities in the metabolic reprogramming of immune system and endothelium. Front Immunol 8: 837 10.3389/fimmu.2017.00837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. 2013. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496: 238–242. 10.1038/nature11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini P, Thaiss CA, Elinav E, Sassone-Corsi P. 2017. Circadian coordination of antimicrobial responses. Cell HostMicrobe 22: 185–192. 10.1016/j.chom.2017.07.007 [DOI] [PubMed] [Google Scholar]
- Van den Bossche J, O'Neill LA, Menon D. 2017. Macrophage immunometabolism: where are we (going)? Trends Immunol 38: 395–406. 10.1016/j.it.2017.03.001 [DOI] [PubMed] [Google Scholar]
- van Furth R, Cohn ZA. 1968. The origin and kinetics of mononuclear phagocytes. J Exp Med 128: 415–435. 10.1084/jem.128.3.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pati P, Xu Y, Chen F, Stepp DW, Huo Y, Rudic RD, Fulton DJR, Cermakian N. 2016. Endotoxin disrupts circadian rhythms in macrophages via reactive oxygen species. PLoS One 11: e0155075 10.1371/journal.pone.0155075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mauvoisin D, Martin E, Atger F, Galindo AN, Dayon L, Sizzano F, Palini A, Kussmann M, Waridel P, et al. 2017a. Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab 25: 102–117. 10.1016/j.cmet.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Subramanian M, Yurdagul A, Barbosa-Lorenzi VC, Cai B, de Juan-Sanz J, Ryan TA, Nomura M, Maxfield FR, Tabas I. 2017b. Mitochondrial fission promotes the continued clearance of apoptotic cells by macrophages. Cell 171: 331–345.e22. 10.1016/j.cell.2017.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Song L, Liu M, Ge R, Zhou Q, Liu W, Li R, Qie J, Zhen B, Wang Y, et al. 2018. A proteomics landscape of circadian clock in mouse liver. Nat Commun 9: 1553 10.1038/s41467-018-03898-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DCS, Seinkmane E, Stangherlin A, Zeng A, Rzechorzek NM, Beale AD, Day J, Reed M, Chew SP, Putker M, et al. 2020. CRYPTOCHROME suppresses the circadian proteome and promotes protein homeostasis. bioRxiv 10.1101/2020.05.16.099556 [DOI]
- Xu H, Li H, Woo S-L, Kim S-M, Shende VR, Neuendorff N, Guo X, Guo T, Qi T, Pei Y, et al. 2014. Myeloid cell-specific disruption of Period1 and Period2 exacerbates diet-induced inflammation and insulin resistance. J Biol Chem 289: 16374–16388. 10.1074/jbc.M113.539601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong H-K, Oh WJ, Yoo OJ, et al. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci 101: 5339–5346. 10.1073/pnas.0308709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Goncalves R, Mosser DM. 2008. The isolation and characterization of murine macrophages. Curr Protoc Immunol 83: 14.1.1–14.1.14. 10.1002/0471142735.im1401s83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci 111: 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Podtelezhnikov AA, Hogenesch JB, Anafi RC. 2016. Discovering biology in periodic data through Phase Set Enrichment Analysis (PSEA). J Biol Rhythms 31: 244–257. 10.1177/0748730416631895 [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, et al. 2001. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105: 683–694. 10.1016/S0092-8674(01)00380-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.