Abstract
TSA-seq mapping suggests that gene distance to nuclear speckles is more deterministic and predictive of gene expression levels than gene radial positioning. Gene expression correlates inversely with distance to nuclear speckles, with chromosome regions of unusually high expression located at the apex of chromosome loops protruding from the nuclear periphery into the interior. Genomic distances to the nearest lamina-associated domain are larger for loop apexes mapping closest to nuclear speckles, suggesting the possibility of conservation of speckle-associated regions. To facilitate comparison of genome organization by TSA-seq, we reduced required cell numbers 10- to 20-fold for TSA-seq by deliberately saturating protein-labeling while preserving distance mapping by the still unsaturated DNA-labeling. Only ∼10% of the genome shows statistically significant shifts in relative nuclear speckle distances in pair-wise comparisons between human cell lines (H1, HFF, HCT116, K562); however, these moderate shifts in nuclear speckle distances tightly correlate with changes in cell type–specific gene expression. Similarly, half of heat shock-induced gene loci already preposition very close to nuclear speckles, with the remaining positioned near or at intermediate distance (HSPH1) to nuclear speckles but shifting even closer with transcriptional induction. Speckle association together with chromatin decondensation correlates with expression amplification upon HSPH1 activation. Our results demonstrate a largely “hardwired” genome organization with specific genes moving small mean distances relative to speckles during cell differentiation or a physiological transition, suggesting an important role of nuclear speckles in gene expression regulation.
The mammalian genome is organized nonrandomly in the nucleus (Ferrai et al. 2010; Meldi and Brickner 2011; Bickmore and van Steensel 2013). A radial distribution of genome organization with a gradient of gene expression increasing from nuclear periphery to interior has been demonstrated in multiple tissue types across a wide range of metazoans (Takizawa et al. 2008b; Bickmore 2013). However, this radial positioning and its correlation with gene expression are highly stochastic. Within a cell population, a given allele can vary widely in its radial position; similarly, different genes with similar transcriptional activity can vary widely in their mean radial positions (Takizawa et al. 2008b). Thus, the functional significance of radial gene positioning remains unclear (Takizawa et al. 2008b).
Subsequent measurement of DNA ligation interaction frequencies using Hi-C suggested the genome could be divided into two major compartments, “B” and “A” (Lieberman-Aiden et al. 2009), corresponding closely to lamina-associated domains (LADs) and inter-LADs (iLADs) (Kind et al. 2015; van Steensel and Belmont 2017), as well as late-replicating versus early-replicating chromosome regions, respectively (Ryba et al. 2010). These discoveries instead suggested a binary division of the genome into peripherally localized heterochromatin and interiorly localized euchromatin. More recently, higher resolution Hi-C further subdivided the genome into two A subcompartments and three primary B subcompartments (Rao et al. 2014), whereas newer genomic methods such as SPRITE (Quinodoz et al. 2018) and GAM (Beagrie et al. 2017) suggested multiway interactions occurring more frequently in the nuclear interior at nucleoli and nuclear speckles (Quinodoz et al. 2018). These newer results thus pointed to additional nuclear bodies/compartments shaping nuclear genome architecture, consistent with previous microscopy analysis of chromosome positioning relative to nuclear bodies (Shopland et al. 2003; Wang et al. 2004; Pederson 2011; Politz et al. 2013; van Steensel and Belmont 2017; Jagannathan et al. 2018).
In particular, nuclear speckles, previously defined as inter-chromatin granule clusters by electron microscopy (Fakan and Puvion 1980), have alternatively been proposed to act as storage sites for factors primarily related to RNA processing (Spector and Lamond 2011) or as gene expression hubs for a subset of active genes (Hall et al. 2006; Chen and Belmont 2019). These two functions are not necessarily mutually exclusive. Coupling between transcription, RNA processing, and RNA export is now appreciated (Galganski et al. 2017), and nuclear speckles are known to be enriched in factors related to transcriptional pause release, RNA capping and splicing, polyadenylation and cleavage factors, and RNA export (Saitoh et al. 2004; Galganski et al. 2017; Dopie et al. 2020) and have been proposed to be sites of post-transcriptional splicing as well as events related to facilitation of nuclear export (Xing et al. 1995; Smith et al. 1999; Johnson et al. 2000; Dias et al. 2010; Girard et al. 2012; Carvalho et al. 2017; Wang et al. 2018).
Indeed, the Lawrence laboratory demonstrated the preferential localization near nuclear speckles of approximately half of about 25 highly expressed genes (Hall et al. 2006). Several described speckle-associated genes express at high levels in specific cell types, including collagen 1A1 (COL1A1) in fibroblasts (Smith et al. 1999), cardiac myosin heavy chain locus (MYH6/MYH7) in myotubes (Smith et al. 1999), mono-allelically expressed GFAP in astrocytes (Takizawa et al. 2008a), and beta-globin (HBB) in erythroblasts (Brown et al. 2006). Two of these genes localize away from nuclear speckles in cell types in which they are inactive: MYH6/MYH7 localizes near nucleoli in fibroblasts (Xing et al. 1995), while HBB localizes near the nuclear periphery in pre-erythrocytes (Brown et al. 2006) and fibroblasts (Bian et al. 2013). Thus, it was natural to expect significant numbers of genes showing large movements relative to nuclear speckles. This expectation was further increased by live-cell imaging of directed movements after heat shock of HSPA1A plasmid transgene arrays to nuclear speckles (Khanna et al. 2014). Moreover, both HSP70 BAC transgenes containing HSPA1A/HSPA1B/HSPA1L or only HSPA1B and endogenous gene loci showed amplification of heat shock-induced HSP70 gene (HSPA1A/HSPA1B/HSPA1L) gene expression when in contact with nuclear speckles (Kim et al. 2020).
These microscopy results provided limited support for the idea of nuclear speckles serving as a gene expression hub due to the small number of gene loci examined. Moreover, they did not determine the relative closeness to nuclear speckles of this group of genes compared to the distance to nuclear speckles of all other genes.
To determine the true prevalence of gene association with nuclear speckles, we developed a novel method to map spatial chromosomal distances relative to a specific nuclear compartment/body (Chen et al. 2018). Based on tyramide signal amplification (TSA) (Bobrow et al. 1989; Raap et al. 1995), which amplifies immunostaining through generation of tyramide free radicals by horseradish peroxidase (HRP), TSA-seq exploits the exponential decay gradient of tyramide labeling from a point source to convert sequencing reads into a “cytological ruler,” reporting on mean chromosomal distances from immunostained nuclear compartments. TSA-seq measures cytological distances on a micron distance scale and thus is fundamentally different from molecular proximity methods such as ChIP-seq or DamID, which measure molecular distances on a nanometer distance scale (Chen et al. 2018).
Applying TSA-seq to K562 cells revealed a nuclear lamina to speckle axis correlating with a gradient in gene expression levels and DNA replication timing. Hi-C A1 and A2 active subcompartment regions formed the apexes of chromatin loops protruding into the nuclear interior from the nuclear periphery, with A1 apexes localizing closer to nuclear speckles than A2 apexes (Chen et al. 2018). Distances in DNA base pairs to the nearest LAD were larger for A1 than A2 loop apexes, suggesting that these A1 loop apexes may have evolved to be positioned larger distances from LAD boundaries and closest to nuclear speckles, predicting also that these speckle-associated regions might be largely conserved across cell types.
To test this “hardwiring” model of genome organization relative to nuclear speckles, here, we applied a more sensitive, “super-saturation” TSA-seq method to compare relative nuclear speckle distances genome-wide in four human cell lines and changes within one cell line after heat shock.
Results
Greater than 10-fold increased sensitivity using “super-saturation” TSA-seq
Core histones showed very low TSA- labeling by western blotting, presumably due to inaccessibility of their tyrosine residues. In contrast, tyramide free radicals labeled DNA uniformly across the genome (Chen et al. 2018). To avoid bias toward chromosomal regions with high nonhistone protein content, TSA-seq was based on tyramide-labeling of DNA, even though the low efficiency of TSA DNA labeling translated into a requirement for ∼100 million to 1 billion cells per replicate, depending on the target and desired spatial resolution (Chen et al. 2018), hindering routine application of TSA-seq.
An obvious solution to increase TSA labeling would be to increase the tyramide concentration and/or TSA reaction time. However, TSA staining of nuclear speckles with increasing tyramide-biotin concentrations and reaction times appeared to increase nonspecific staining, with the biotin signal spreading throughout the nucleus and even cytoplasm. We hypothesized that increased tyramide free radical production caused a progressive saturation of protein tyrosines spreading outward from nuclear speckles, with the weaker DNA tyramide labeling, masked by this stronger protein labeling, still far from saturation and still specific for the staining target.
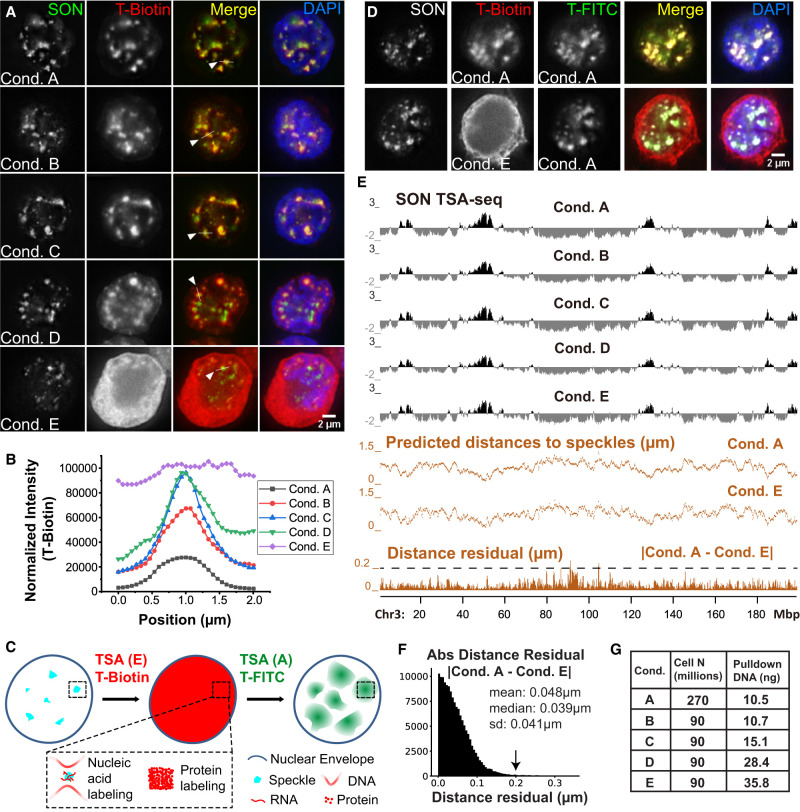
To test this hypothesis, we compared anti-SON nuclear speckle TSA labeling in K562 cells using progressively higher tyramide-biotin concentrations and longer reaction times (Conditions A–E) (Fig. 1A; Supplemental Fig. S1A), using an affinity-purified, antipeptide polyclonal antibody highly specific for nuclear speckles, as demonstrated previously by comparison with other anti-SON and speckle antibodies using both light microscopy and TSA-seq mapping (Chen et al. 2018). Condition A corresponds to our original, “TSA-seq 1.0” conditions (Chen et al. 2018). TSA labeling increased from Conditions A to C but plateaued, with increases in background staining, from Conditions C to E (Fig. 1A,B). Condition E produced strong tyramide-biotin labeling in the cytoplasm, with no nuclear speckle staining appearing above the nucleoplasmic background staining. We then “blocked” protein tyrosine groups using high concentrations of tyramide-biotin (Condition E) and performed a second TSA using tyramide-FITC (Condition A) (Fig. 1C). Consistent with our hypothesis of saturation of protein TSA labeling, while Condition E TSA labeling produced diffuse biotin-labeling throughout the cell, a second TSA labeling (Condition A) revealed nuclear speckle-specific labeling in K562 (Fig. 1D, bottom) and HFFc6 cells (Supplemental Fig. S1B, bottom).
Figure 1.
TSA-seq 2.0 enables 10- to 20-fold increase in sensitivity but preserves distance mapping capability. (A) TSA Conditions A–E (K562 cells) show varying nuclear speckle specificity. SON immunostaining of speckles (green), streptavidin tyramide-biotin staining (red), merged channels, plus DNA (DAPI, blue). (B) Tyramide-biotin intensities along line profiles spanning nuclear speckles in A for Conditions A–E. (C) Schema predicting results from following Condition E tyramide-biotin TSA staining with Condition A tyramide-FITC TSA staining, assuming Condition E saturates protein but not DNA tyramide-labeling. (D) Experimental results for schema in C. Top row: Control showing two consecutive rounds of Condition A (nonsaturating) TSA-labeling using tyramide-biotin and then tyramide-FITC. Bottom row: Same as top row but using Condition E for first TSA-labeling. SON immunostaining (gray), tyramide-biotin (red), tyramide-FITC (green), merged channels, plus DAPI (blue). (E) SON TSA-seq mapping results for Conditions A–E showing TSA-seq enrichment scores (black tracks), estimated speckle distances (Conditions A and E, middle orange tracks), and residuals (absolute magnitude) between Conditions A and E distances (bottom, orange track). (F) Histogram of distance residuals in E: number (y-axis), residual value (x-axis). (G) Cell numbers used and pulldown DNA yields for TSA Conditions A–E.
Average tyramide-biotin labeling increased from ∼1 biotin /200 kb using Condition A to ∼1 biotin/7.5 kb using Condition E (Supplemental Fig. S1C), predicting ∼1 biotin/0.94 kb over the ∼eightfold-enriched speckle TSA-seq peaks. Sonicating DNA to ∼100−600 bp ensures few DNA fragments with multiple biotins, maintaining linearity between TSA labeling and pulldown read number.
Speckle TSA-seq maps generated using Conditions A–E were both qualitatively and quantitatively similar (Fig. 1E; Supplemental Figs. S2A, S3C). Converting TSA-seq signals to predicted nuclear speckle distances using exponential fitting of immuno-FISH data (Supplemental Tables S1, S2; Methods) demonstrated that distance residuals between Conditions A and E were nearly all smaller (<0.05 µm mean and median) than the microscopy diffraction limit of ∼0.25 µm (Fig. 1E,F; Supplemental Fig. S2). Thus, using Condition E, we increased DNA pulldown by 10- to 20-fold relative to TSA-seq 1.0 (Condition A) (Fig. 1G; Supplemental Figs. S3D, S4C), reducing required cell numbers to ∼10−15 million (to obtain ∼5 ng pulldown DNA), without significant quantitative changes in nuclear speckle distance estimations.
Speckle-associated domains remain largely conserved, but small, statistically significant changes highly correlate with gene expression changes
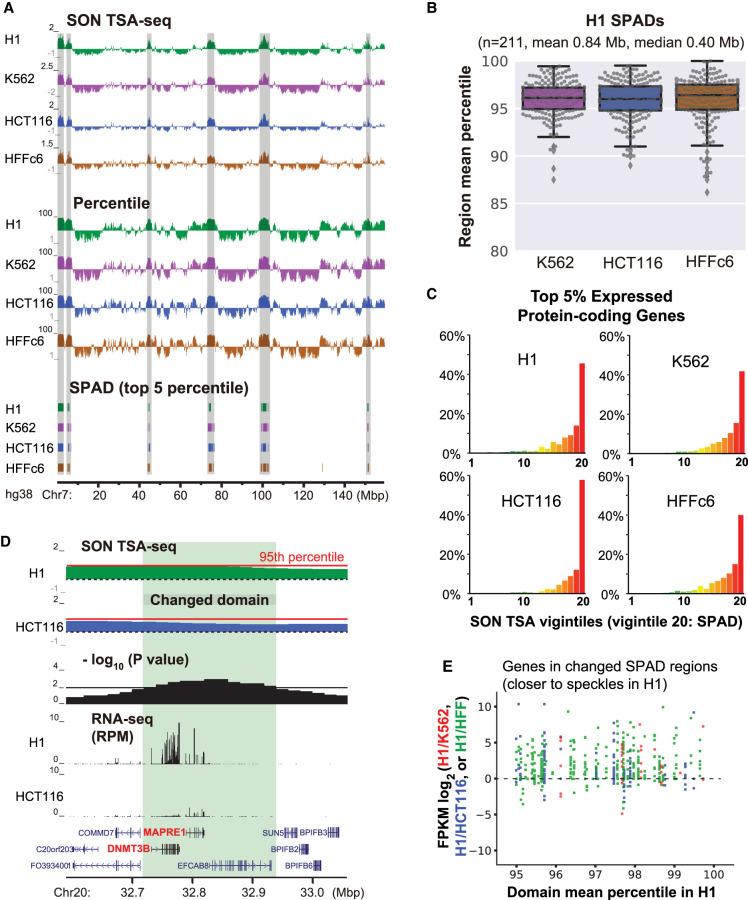
Previously, we defined chromosome regions with the top 5% nuclear speckle TSA-seq scores as SPeckle-Associated Domains (SPADs), as they were near-deterministically positioned near nuclear speckles. Specifically, several chromosome regions with speckle TSA-seq scores in this range examined by FISH showed near 100% of alleles localizing within 0.5 μm of nuclear speckles (mean estimated distances of <0.32 μm) (Chen et al. 2018). To test whether SPADs are conserved among different cell types, we applied SON TSA-seq to four cell lines (Figs. 1A, 2A; Supplemental Table S3; Supplemental Figs. S3A, S4A), each with two biological replicates: K562 erythroleukemia cells, HCT116 colon carcinoma cells, HFFc6 human foreskin fibroblast cells immortalized by hTERT expression, and H1 human embryonal stem cells (hESCs).
Figure 2.
SPADs remain close to nuclear speckles and show high gene expression in all cell lines, yet small position shifts even closer to nuclear speckles correlate with increased gene expression. (A) SON TSA-seq enrichment score (top) and SON TSA-seq signal as a genome-wide percentile score (middle) tracks (20-kb bins), and segmented SPeckle Associated Domains (SPADs) (bottom) in four human cell lines (Chr 7). SPADs were defined as contiguous bins, each with >95th percentile score (Methods). (B) TSA-seq score percentile distributions of H1 SPADs in other three cell lines. Box plots (with data points as dots) show median (inside line), 25th (box bottom) and 75th (box top) percentiles, 75th percentile to highest value within 1.5-fold of box height (top whisker), 25th percentile to lowest value within 1.5-fold of box height (bottom whisker), and outliers (diamonds). (C) Distributions of percentages (y-axis) of top 5% expressed protein-coding genes versus SON TSA-seq vigintiles (division of percentile scores, 1−100, into 20 bins of 5% size each; x-axis) in four cell lines. (D) Top to bottom (light green highlight shows domain repositioned relative to speckles): comparison of H1 and HCT116 smoothed SON TSA-seq enrichment score tracks (black dashed lines: zero value; red solid lines indicate 95th percentiles of TSA-seq enrichment scores), −log10 P-values per bin (significance of change in rescaled SON TSA-seq scores), RNA-seq RPM values, and gene annotation (GENCODE). (E) Expression changes of protein-coding genes in SPADs (in H1) closer to speckles in H1: scatterplots show log2-fold changes in FPKM ratios (y-axis) of H1/K562, H1/HCT116, or H1/HFF versus H1 TSA-seq score percentiles (x-axis).
Overlapping of SPADs across all four cell lines revealed that 56.4% (129.3 Mbp) are classified as SPADs (>95th percentile) in all four cell lines, 12.5% (28.6 Mbp) in three cell lines, 13.0% in two cell lines (29.7 Mbp), and 18.1% (41.5 Mbp) in one cell line, out of 229.1 Mbp total (Supplemental Fig. S5F). In the SON TSA-seq score percentile, most SPADs remain at the >90th percentile and 100% of SPADs remain at the >80th percentile in all four cell lines (Fig. 2A,B; Supplemental Fig. S5A–C). Thus, SPADs are largely conserved, particularly considering that regions in the top 10% of TSA-seq scores show an ∼5-percentile variation between biological replicates (Supplemental Fig. S5D,E).
SPADs identified in each cell line were enriched in the top 5% of expressed genes (Fig. 2C) and had the highest FPKM values in all four cell lines (Supplemental Fig. S5G,I), suggesting that SPADs were correlated with genomic domains with constitutively high, overall gene expression. Also, SPADs were enriched in housekeeping genes (Supplemental Fig. S5H).
We then asked if statistically significant changes in the speckle proximity of SPADs correlated with changes in gene expression using pair-wise comparisons between cell lines. Starting with all genomic regions defined as SPADs in either cell line, we identified SPAD subregions 100 kb or larger that showed significant changes in TSA-seq scores. These “changed” subregions were SPADs in both lines (>95th genomic percentile), which shifted closer to speckles in one line, or else were non-SPADs in one line but changed to SPADs in the other. Changed SPAD regions closer to speckles in H1 compared with HFFc6 (HCT116 or K562) accounted for 13.4% (5.2% or 1.8%) of H1 SPADs, with changes ranging from 1.3 to 13.2 (1.6 to 7.2, or 2.7 to 10.0) and mean changes of 4.4 (4.1 or 5.0) in TSA-seq genomic percentile.
These small relative decreases in SPAD speckle distances in H1 cells were accompanied by a pronounced bias toward increased gene expression (Fig. 2D,E).
Moderate distance changes relative to nuclear speckles correlate with genome-wide changes in gene expression and histone modifications
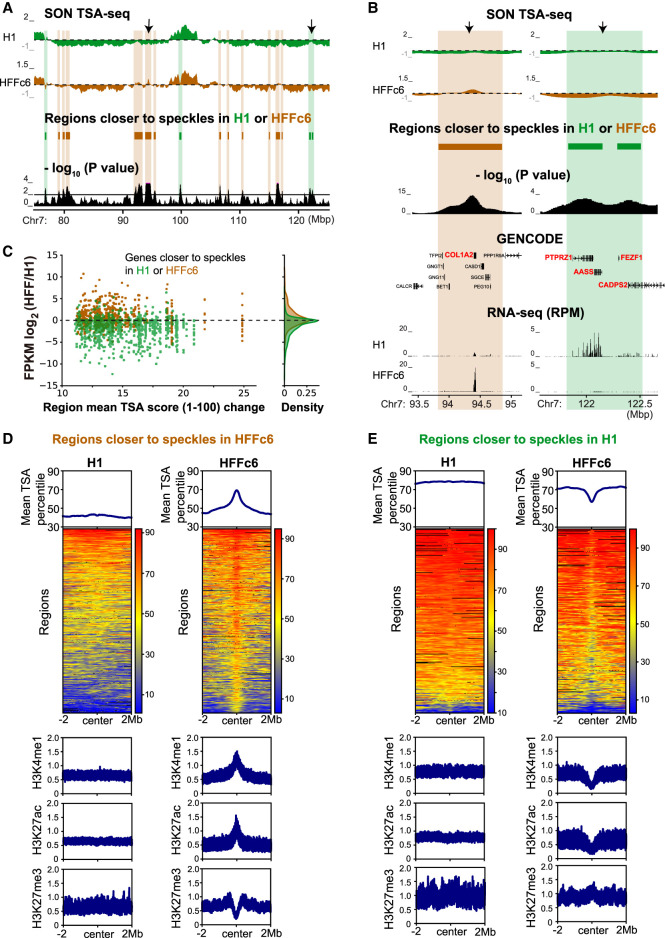
We next extended our statistical analysis to the entire genome. Comparing H1 and HFFc6, we identified 494 domains positioned closer to speckles in HFFc6 (408 kb average size) and 367 domains (396 kb average size) closer to speckles in H1 (e.g., Fig. 3A, highlighted). Using the same threshold, we detected only two changed domains between biological replicates, a false discovery rate of 0.23%. Similarly, we identified changed regions comparing H1 with HCT116 or K562 (Supplemental Fig. S6A,D).
Figure 3.
Chromosome regions changing position relative to speckles in H1 versus HFFc6 cells show changes in gene expression and histone modifications. (A) Top to bottom (domains shifting position highlighted): H1 (green) and HFFc6 (brown) smoothed SON TSA-seq enrichment scores (dashed lines: zero value), domains repositioned relative to speckles, and −log10 P-values per bin (significance of change in rescaled SON TSA-seq scores). (B,C) Strong bias toward increased gene expression in domains shifting closer to nuclear speckles: (B) zoomed view of two regions (arrows in A) plus gene annotation (GENCODE) and RNA-seq RPM values; (C) scatterplots show log2-fold changes in FPKM ratios (y-axis) between HFFc6 and H1 versus absolute values of changes in mean scaled TSA-seq scores (x-axis). Green (brown) dots are genes closer to speckles in H1 (HFFc6). Kernel density plots show gene density (right). (D,E) TSA-seq percentiles and histone modifications fold-enrichment for regions significantly closer to speckles in HFFc6 (D) or H1 (E) ±2 Mbps from each region center. Top: TSA-seq percentiles averaged over all regions; middle: heat maps with color-coded TSA-seq score percentiles; bottom: histone modification fold-enrichment for H3K4me1, H3K27Ac, or H3K27me3 averaged over all regions.
No changed domains showed large shifts in SON TSA-seq scores. Normalizing TSA-seq scores on a scale from 1 to 100 (Supplemental Methods), statistically significant changes ranged only from ∼10–26. We used our distance calibration in K562 cells to convert the SON TSA-seq score into an estimated mean distance to nuclear speckles. Estimated mean distances to speckles in K562 cells (Supplemental Tables S1, S2; Methods) linearly correlated with scaled TSA-seq scores (1−100) (Supplemental Fig. S7A). Domains that significantly shifted in relative nuclear speckle distance comparing H1 and K562 cells showed predicted mean distance shifts from 0.113 to 0.375 µm (Supplemental Fig. S7B).
Despite these modest position shifts, again we saw correlated changes in gene expression, similar to what we had seen for changed SPADs. Domains closer to nuclear speckles in one cell line typically show one or more genes with increased gene expression level in this cell line (e.g., Fig. 3B; Supplemental Fig. S6B,E), including many genes with tissue-specific expression, such as collagen genes in HFF fibroblasts (Fig. 3B; Supplemental Fig. S9). Genome-wide analysis confirmed this expression bias (Fig. 3C; Supplemental Figs. S6C,F, S7C). Comparing H1 versus HFFc6 cells, 628 genes show log2-fold changes ≥1, 379 genes ≥2, and 248 genes ≥3 in domains closer to speckles in H1; 91%, 94%, and 96% of these genes, respectively, show higher expression in H1. Of 307 genes with log2-fold changes ≥1, 161 genes ≥2, and 94 genes ≥3 in domains closer to speckles in HFFc6, 66%; 77%, and 83% of these genes, respectively, show higher expression in HFF.
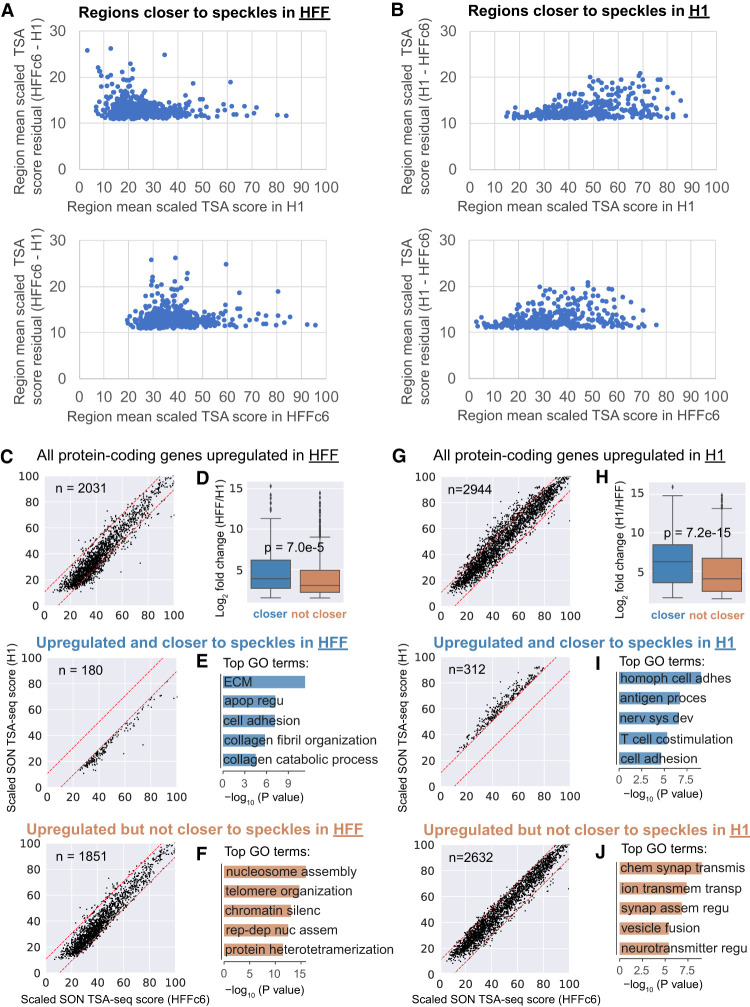
Examining changed domains in H1 versus HFFc6 revealed an interesting asymmetry in the pattern of nuclear speckle distance shifts (Fig. 3D,E). Regions shifting closer to speckles in HFFc6 showed flat TSA-seq score profiles in H1 cells changing to local peaks in HFFc6 (Fig. 3D, top); these peaks were ∼600 kb in full width at half-maximum (FWHM) and associated with increased active and decreased repressive histone modifications (Fig. 3D, bottom). Conversely, regions shifting further from nuclear speckles in HFFc6 showed flat TSA-seq score profiles in H1 changing to local valleys in HFFc6 (Fig. 3E, top); these valleys were ∼400 kb FWHM and associated with decreased active and increased repressive histone marks (Fig. 3E, bottom). Domains moving toward (away from) speckles in HFFc6 cells generally showed low (high) SON TSA-seq scores in H1 versus intermediate (intermediate) scores in HFFc6 (Fig. 4A,B).
Figure 4.
Shifts in domain positioning relative to speckles are small and correlate with changes in expression of cell type–specific genes. (A,B) Scatterplots show change in scaled SON TSA-seq scores (1−100 range) averaged over each domain (y-axis) with a significant score change for domains closer to speckles in HFFc6 (A) or H1 (B) as a function of scaled TSA-seq scores averaged over each domain (x-axis) in H1 (top) or HFFc6 (bottom). (C,G) Scatter plots show scaled SON TSA-seq scores (1−100 range) in H1 hESCs (y-axis) versus HFFc6 (x-axis) for protein-coding genes (dots) that are up-regulated in HFF (C) or in H1 (G): top panels: all up-regulated genes; middle panels: up-regulated genes in regions closer to nuclear speckles in HFFc6 (C) or in H1 (G); bottom panels: up-regulated genes in regions that are not closer to nuclear speckles. Red dashed lines represent thresholds for statistically significant changes in scaled HFFc6 versus H1 SON TSA-seq scores. (D,H) Comparison of log2-fold changes in gene expression for protein-coding genes significantly up-regulated in HFF (D, HFF/ H1) or in H1 (H, H1/HFF) for: up-regulated genes in regions closer to nuclear speckles in HFFc6 (D, left, blue) or in H1 (H, left, blue) versus up-regulated genes in regions that are not closer to nuclear speckles in HFFc6 (D, right, orange) or in H1 (H, right, orange) (Welch's t-test). Box plots show median (inside line), 25th (box bottom), and 75th (box top) percentiles, 75th percentile to highest value within 1.5-fold of box height (top whisker), and 25th percentile to lowest value within 1.5-fold of box height (bottom whisker). (E,F,I,J) Top Gene Ontology (GO) terms for protein-coding genes up-regulated in HFF (E,F) or in H1 (I,J) for up-regulated genes in regions closer to nuclear speckles in HFFc6 (E) or in H1 (I) versus up-regulated genes in regions not closer to nuclear speckles in HFFc6 (F) or in H1 (J). Abbreviated GO terms: (E) ECM: extracellular matrix organization; apop regu: negative regulation of apoptotic process; (F) chromatin silenc: chromatin silencing at rDNA; rep-dep nuc assem: DNA replication-dependent nucleosome assembly; (I) homoph cell adhes: homophilic cell adhesion via plasma membrane adhesion molecules; antigen proces: antigen processing and presentation of peptide or polysaccharide antigen via MHC class II; nerv sys dev: nervous system development; (J) chem synap transmis: chemical synaptic transmission; ion transmem transp: ion transmembrane transport; synap assem regu: positive regulation of synapse assembly; neurotransmitter regu: calcium ion-regulated exocytosis of neurotransmitter.
Genes with significantly higher expression in regions that position closer to nuclear speckles are enriched in cell type–specific functions
We next examined changes in gene expression in these changed chromosome domains. Approximately 10% of genes showing increased gene expression in one cell line versus another were in changed domains closer to nuclear speckles (Fig. 4C,G). Differentially expressed (DE) genes in domains closer to speckles showed a moderate shift in their distribution toward higher fold-changes in expression, although there was extensive overlap in expression changes with DE genes in domains that did not move closer (Fig. 4D,H).
Instead, Gene Ontology (GO) analysis suggests DE genes in regions closer to speckles in one cell line are enriched in genes with cell type–specific functions, including extracellular matrix organization and collagen-related functions for regions closer to speckles in HFFc6 fibroblasts (Fig. 4E) and homophilic cell adhesion related to stem cell colony formation and maintenance (Pieters and van Roy 2014) in H1 hESCs (Fig. 4I), as compared to different GO terms for genes which changed expression in domains that did not move relative to nuclear speckles (Fig. 4F,J). “Cell adhesion” function was enriched in DE genes that were closer to speckles in either H1 or HFFc6 cells (Fig. 4E,I).
Genome-wide analysis of changes in genome organization relative to nuclear speckles after heat-shock reveals partial “hardwiring” of heat shock protein gene loci
To further investigate the possible “hardwiring” and dynamics of genome organization relative to nuclear speckles, we applied TSA-seq mapping to K562 cells undergoing heat shock. As previously reported (Spector and Lamond 2011), nuclear speckles round up and enlarge after heat shock (Supplemental Fig. S10A). Live-cell imaging recently has revealed that at least some of this speckle enlargement after heat shock occurs through directed movement of small speckles toward larger nuclear speckles, followed by speckle fusion (Kim et al. 2019). Despite these significant changes in nuclear speckle size and shape, speckle movement, and significant changes in gene expression, including thousands of down-regulated genes 30 min after heat shock (42°C) (Vihervaara et al. 2017), we found nearly no significant genome-wide changes in SON TSA-seq scores after 30 min to 2 h of heat shock (Supplemental Fig. S10B). TSA-seq score differences between control and heat shocked samples were largely comparable to differences between biological replicates (Supplemental Fig. S10C).
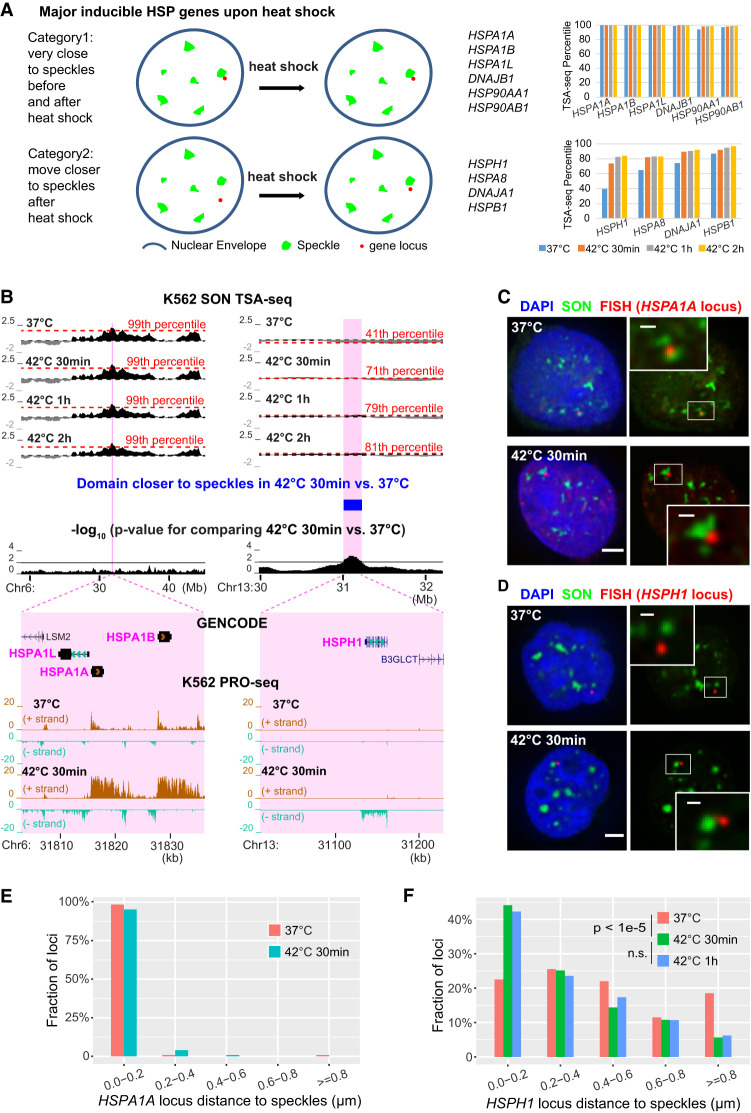
These unchanged chromosome regions included four of eight chromosome loci—HSPA1A/HSPA1B/HSPA1L, DNAJB1, HSP90AA1, and HSP90AB1 (Fig. 5A,B; Supplemental Fig. S11)—containing major induced heat shock protein (HSP) genes (based on PRO-seq [Vihervaara et al. 2017]). Each of these four heat shock gene loci score near the 100th genomic percentile in speckle proximity. This prepositioning so close to speckles before heat shock is striking given our recent demonstration of increased (decreased) nascent HSPA1A transcript production within several minutes after nuclear speckle association (disassociation) of a HSPA1B BAC transgene and a several-fold increased gene expression of the endogenous HSPA1A/HSPA1B/HSPA1L locus in human HAP1 cells (Kim et al. 2020).
Figure 5.
Approximately half of inducible heat shock protein (HSP) gene loci are prepositioned very close to nuclear speckles in SPADs while remaining gene loci shift closer to speckles after heat shock. (A) Schema (left) and TSA-seq percentiles at 0 (blue), 30 min (orange), 1 h (gray), and 2 h (gold) after heat shock (right) for genes prepositioned near speckles (top) versus those moving closer after heat shock (bottom). (B) TSA-seq and PRO-seq profiles of HSPA1A (left) and HSPH1 (right) loci in K562 cells after heat shock. Top to bottom: smoothed SON TSA-seq enrichment scores for control or heat shock times (red dashed lines: labeled TSA-seq score percentiles; black dashed lines: zero value), significantly changed domains comparing 37°C versus 42°C 30 min (blue rectangular bar, right), −log10 P-values for comparison of rescaled SON TSA-seq scores between 37°C and 30 min heat shock (20-kb bins), and zoomed view of the highlighted two regions (magenta) plus gene annotation (GENCODE) and K562 PRO-seq tracks showing normalized read count (BPM, 50-bp bins). (C,D) 3D immuno-FISH for HSPA1A (red, C) or HSPH1 (red, D) plus SON immunostaining (green) and merged channels with DAPI (blue, left; scale bars: 2 μm) in K562 cells at 37°C (top) or at 42°C for 30 min (bottom). Insets: 3× enlargement of the white-boxed image area (right, scale bars: 0.5 μm). (E,F) Nuclear speckle distance distributions for HSPA1A (E) or HSPH1 (F) FISH signals in control versus after heat shock. HSPA1A: n = 119 (37°C) or 124 (42°C 30 mins). HSPH1: n = 200 (37°C) or 195 (42°C 30 min) or 225 (42°C 1 h), combination of two biological replicates; χ2 test of homogeneity, P = 3.793 × 10−6 between 37°C and 42°C 30 min, P = 0.9341 (n.s.) between 42°C 30 min and 42°C 1 h.
We did identify 13 chromosome regions ∼100–680 kbp in size, together comprising 3.36 Mbp, positioned closer to speckles after 30 min heat shock. Four of these 13 regions contained the remaining four of eight heat shock chromosome loci containing major induced HSP genes in K562 cells identified by PRO-seq (Vihervaara et al. 2017): HSPH1, HSPA8, DNAJA1, HSPB1 (Fig. 5A,B; Supplemental Fig. S11). Their position shifts closer to nuclear speckles with heat shock were in a similar modest range observed for regions that changed speckle position in pair-wise comparisons of cell lines (Fig. 3C; Supplemental Figs. S6C,F, S8): 5–30 changes in SON TSA-seq genomic percentiles (30, HSPH1 locus; 17, HSPA8 locus; 15, DNAJA1 locus; 5, HSPB1 locus) and 11–13 in scaled (1−100) score changes (13, HSPH1 locus; 12, HSPA8 locus; 12, DNAJA1 locus; 11, HSPB1 locus), averaged over the regions. In contrast, only six regions shifted to positions further from nuclear speckles.
HSPA1A/HSPA1B/HSPA1L deterministic positioning near nuclear speckles; HSPH1 movement toward nuclear speckles, and large-scale chromatin decondensation after heat shock associated with increased nascent transcripts
We next applied 3D immuno-FISH to investigate the nuclear positioning of HSP genes relative to nuclear speckles, choosing the two loci showing the smallest and largest distances prior to heat shock. As predicted by TSA-seq, DNA FISH showed near-deterministic positioning of the HSPA1A/HSPA1B/HSPA1L locus near speckles in K562 cells: ∼90%–100% alleles localized within 0.2 µm from speckles before and after heat shock (0.024 µm and 0.040 µm mean, 0 µm and 0 µm median distances before and after 30 min heat shock, respectively) (Fig. 5C,E; Supplemental Fig. S12A). Similar deterministic positioning near speckles was seen in four additional human cell lines before and after heat shock (WI38 and IMR-90 normal fibroblasts, hTERT-immortalized Tig3 fibroblasts, and HCT116 cells) (Supplemental Fig. S12B–E).
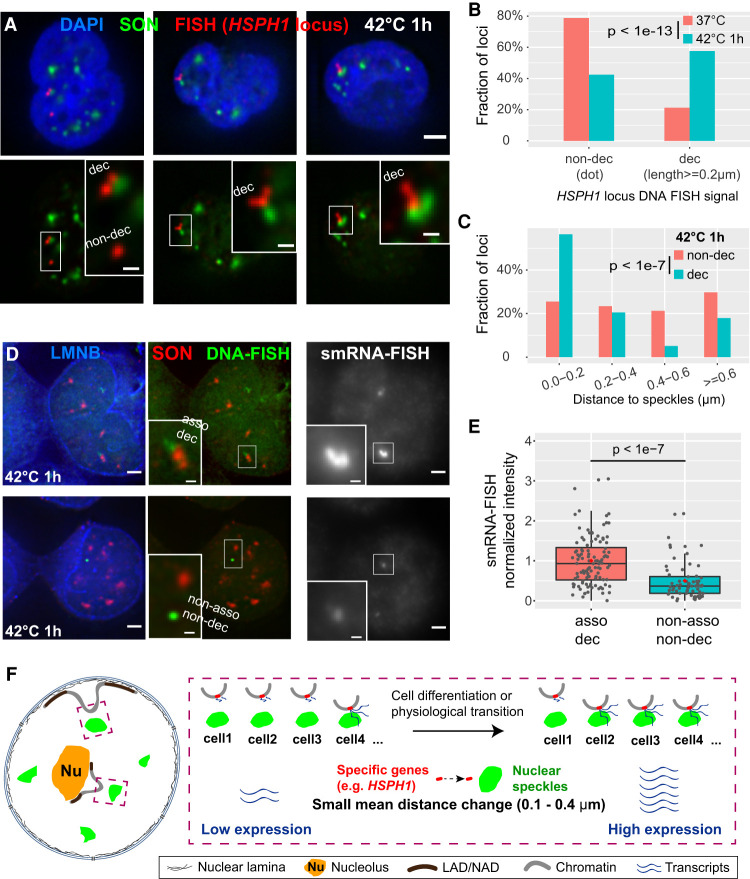
In contrast, the HSPH1 locus showed a significant shift in the nuclear speckle distance distribution 30 and 60 min after heat shock (Fig. 5D,F), with an ∼twofold increase in alleles localized within 0.2 μm from nuclear speckles (∼20% to ∼40%) together with an ∼fourfold reduction in alleles localized 0.8 μm or further from nuclear speckles (∼20% to ∼5%) (Fig. 5F).
In addition to the HSPH1 shift toward nuclear speckles, a fraction of HSPH1 alleles showed a significantly decondensed DNA FISH signal (“dec”) (Fig. 6A,B), which increased from ∼20% to ∼60% after heat shock. This decondensed FISH signal was typically observed for HSPH1 alleles associated with nuclear speckles after heat shock (Fig. 6A,B), appearing as elongated, fiber-like FISH signals, frequently with one region of the FISH signal touching a nuclear speckle (Fig. 6A). In contrast, round, diffraction-limited FISH signals (“non-dec”) represented ∼80% of HSPH1 alleles before heat shock. The speckle distance distribution of decondensed versus non-decondensed alleles was shifted toward lower distances with an increase from ∼25% to ∼55% for alleles localized within 0.2 μm from nuclear speckles (Fig. 6C).
Figure 6.
Both distance shift toward speckles and large-scale chromatin decondensation of HSPH1 locus after heat shock correlate with gene expression amplification in K562 cells. (A) 3D immuno-FISH for HSPH1 locus (red) plus SON immunostaining (green) and merged channels with DAPI (blue, top; scale bar: 2 μm) in K562 cells after heat shock (42°C 1 h). Types of FISH signals: left: decondensed (upper, elongated) and non-decondensed signals (lower, dot); middle and right: decondensed signals. Insets: 3× enlargement of region in white box (bottom, scale bars: 0.5 μm). (B) Increase in decondensed HSPH1 loci fraction after heat shock: n = 198 (37°C) or 224 (42°C 1 h), combination of two biological replicates; χ2 test of homogeneity, P = 3.045 × 10−14. (C) Speckle distance distributions for different cases of HSPH1 locus FISH signals in K562 cells after 1 h heat shock (42°C): n = 141 (non-decondensed, dot) or 200 (decondensed, ≥0.2 μm in length), combination of two biological replicates; χ2 test of homogeneity, P = 2.74 × 10−8 between non-decondensed and decondensed. (D) Combined DNA- and single-molecule RNA (smRNA)-immuno-FISH for HSPH1 locus showing DNA-FISH signal (green), SON immunostaining (red), Lamin B1/B2 (LMNB) immunostaining signal (blue), and smRNA-FISH (gray) in K562 cells after 1 h heat shock (scale bars: 2 μm). Decondensed chromatin structure (elongated DNA-FISH signal) with speckle association (top) and non-decondensed (dot DNA-FISH signal) not associated with speckles (bottom) and their corresponding smRNA-FISH signal (right). Insets: 3× enlargement of white-boxed area (scale bar: 0.5 μm). Single optical section is shown containing center (in z) of DNA FISH signal; RNA-FISH images correspond to projected sum of 20 optical sections. (E) Normalized smRNA-FISH signals corresponding to different chromatin states based on DNA immuno-FISH. Combination of two biological replicates: asso dec: distance to speckles <0.4 µm and signal length ≥0.4 µm, n = 108, mean = 1.00, median = 0.93; non-asso non-dec: distance ≥0.6 µm and dot signal, n = 68, mean = 0.50, median = 0.37. Box plots (data points: gray dots, means: red dots) show median (inside line), 25th (box bottom) and 75th (box top) percentiles, 75th percentile to highest value within 1.5-fold of box height (top whisker), 25th percentile to lowest value within 1.5-fold of box height (bottom whisker). Welch's t-test: P = 1.179 × 10−8. (F) Cartoon model: small mean distance changes relative to nuclear speckles predicted by TSA-seq may reflect changes in the distribution of distances such that an increased fraction of alleles shows close association with nuclear speckles with an accompanying amplification of gene expression.
Single-molecule RNA FISH (smRNA FISH) was followed by DNA FISH to correlate differential HSPH1 expression with both HSPH1 nuclear speckle distance and chromatin decondensation. We defined two opposite subclasses of alleles: “speckle-associated with decondensation” (distance to speckles < 0.4 µm and signal length ≥0.4 µm) or “non-speckle-associated without decondensation” (distance to speckles ≥0.6 µm and dot signal). Our rationale was that non-decondensed, speckle-associated alleles or non-speckle associated, decondensed alleles might represent intermediate, transient states. We used a smRNA FISH probe set spanning the 5′ UTR (two oligos) and first intron (28 oligos) of the HSPH1 gene to measure nascent transcript levels (Supplemental Table S4). Speckle-associated, decondensed HSPH1 alleles showed an ∼2.0-fold mean (2.5-fold median) increased nascent transcript smRNA FISH signal relative to non-speckle-associated, non-decondensed alleles (Fig. 6D,E), similar to increased nascent transcript signals of HSPA1A/HSPA1B/HSPA1L and several flanking genes, observed after nuclear speckle association (Kim et al. 2020).
Discussion
Our results suggest a conserved genome organization relative to nuclear speckles. Approximately 84% of the genome showed no significant position change relative to nuclear speckles in comparisons of hESCs with all three differentiated cell types—fibroblasts (HFFc6), eythroleukemia (K562), and colon carcinoma (HCT116)—and ∼90% unchanged in single, pair-wise comparisons with hESCs. This contrasts sharply with the ∼50% of changes in the LAD versus inter-LAD organization encountered in these same cell type comparisons of DamID mapping (K562 and HCT116 [van Schaik et al. 2020]; H1 and HFF: T van Schaik and B van Steensel, unpubl. [https://data.4dnucleome.org]) and ∼30% of changes in mouse cell type comparisons (Meuleman et al. 2013). (This binary LAD/inter-LAD division may underestimate statistically significant “changed” DamID signals.)
Moreover, the small percentage of the genome that did change position showed only modest estimated mean distance changes relative to speckles. However, these modest position changes of chromatin domains ∼100 kbp–5.8 Mbp in size closer (further) to nuclear speckles highly correlated with increased (decreased) gene expression and active (repressive) histone marks (Fig. 6F). Preliminary work and analysis suggest that these nuclear speckle distance shifts inversely correlate with nuclear lamina distance shifts, suggesting movement of these changed chromatin domains closer to speckles rather than movement of speckles closer to the changed domains.
This largely fixed genome positioning relative to nuclear speckles suggests that the genome may have evolved to position some genes near nuclear speckles to facilitate their induced expression, as supported by our analysis of heat shock-inducible gene loci: 4/8 of the major heat shock-induced loci are already positioned very close (top 1%–2% of genome) to nuclear speckles before heat shock in five human cell lines examined. The remaining four loci were among the 13 genomic loci (∼0.1% of genome) that moved closer to nuclear speckles within 30 min of heat shock in K562 cells. Nuclear speckle association of HSPH1 correlated with increased nascent transcript levels, as observed for several genes at the HSPA1A locus (Kim et al. 2020).
Notably, live-cell imaging recently revealed that HSPA1B transgenes not already located near nuclear speckles required an additional several minutes to move to nuclear speckles and reach maximum nascent transcript production (Kim et al. 2020). Thus, this “prewiring” of the endogenous HSPA1A/HSPA1B/HSPA1L locus very close to nuclear speckles likely ensures robust heat shock activation across all alleles in all cells. We anticipate a similar effect of close speckle proximity on the robust activation of the other three heat shock gene loci, and possibly other stress-induced genes, that are prewired near nuclear speckles. More generally, our observed positioning of SPADs, defined in one cell type, close to nuclear speckles in other cell lines suggests that this prewiring concept may apply to a much larger set of genes.
One still puzzling aspect of our results is that this bias in differential gene expression is not only seen for regions that gain or lose very close proximity to nuclear speckles but also for regions that shift closer to nuclear speckles but without reaching very close proximity. We estimated that these modest changes in TSA-seq scores corresponded to actual estimated mean distance shifts of only 0.113–0.375 μm (Supplemental Figs. S7, S8). We speculate that the functional significance of these intermediate distance shifts relative to nuclear speckles may instead be related to shifts in their distance relative to other, still unknown nuclear compartments, for instance, RNA Pol II clusters/condensates (Cisse et al. 2013; Boehning et al. 2018; Cho et al. 2018; Guo et al. 2019), which themselves are positioned nonrandomly relative to nuclear speckles.
Although APEX-based proximity labeling methods have been reported to label proteins/RNAs (Gao et al. 2018; Myers et al. 2018; Fazal et al. 2019), and, more recently, chromatin (Kurihara et al. 2020; Tran et al. 2021), TSA-seq is able to map genomic regions through unbiased DNA labeling, without the need to genetically modify the cells to express APEX, and is capable of reporting mean chromosomal distances from immunostained nuclear compartments by exploiting the exponential decay gradient of tyramide labeling. Using DNA, rather than chromatin, labeling and pulldown also avoids nonlinearities that would be introduced by the supersaturation of protein tyrosines described here. With the development of our more sensitive TSA-seq 2.0 method, we are now better positioned to explore how the genome is organized relative to other nuclear compartments, in addition to nuclear speckles. This includes both known compartments, including the nuclear lamina, nuclear pores, PML bodies, Cajal bodies, and pericentric heterochromatin, as well as currently less-well-characterized compartments identified by interesting antibody staining patterns. TSA-seq is particularly well-suited to explore the genome spatial relationship to liquid–liquid phase-separated condensates that might otherwise be difficult to map by methods such as DamID, ChIP-seq, or CUT&RUN. Future extensions of TSA-seq to multiple nuclear compartments in many cell types and states should greatly increase our understanding of the role of nuclear compartmentalization in nuclear genome organization and function.
Methods
Cell culture
K562 cells were obtained from the ATCC and cultured following the ENCODE Consortium protocol (http://genome.ucsc.edu/ENCODE/protocols/cell/human/K562_protocol.pdf). H1-ESC (WA01), HCT116, and HFF-hTert-clone 6 cells were obtained through the 4D Nucleome Consortium and cultured according to 4DN SOPs (https://www.4dnucleome.org/cell-lines.html). A detailed description for reagents and culturing procedures is provided in Supplemental Methods.
TSA staining, TSA-seq, and data processing
TSA staining and TSA-seq procedures were modified from our previous publication (Chen et al. 2018). Detailed experimental procedures, quality control methods, and TSA-seq data processing pipeline are provided in Supplemental Methods.
Microscopy
3D optical sections were acquired with 0.2-μm z-steps using DeltaVision SoftWoRx software (GE Healthcare) and either an Applied Precision Personal DeltaVision microscope system with a 60× oil objective (NA 1.4) and a CoolSNAP HQ2 charge-coupled device camera or a DeltaVision OMX microscope system with a 100× (NA 1.4) objective lens and an Evolve 512 Delta EMCCD Camera operated in wide-field mode. Image deconvolution and registration also were done using SoftWoRx. Image intensity line profiles were measured with FIJI software (ImageJ, NIH) using the Plot Profile function and normalized by exposure time and the % transmitted exciting light. Normalized intensity plots were generated using OriginPro 2018 software (OriginLab).
TSA distance prediction and comparison between different staining conditions
To convert TSA-seq scores into mean speckle distances, we used our previously published FISH data (Chen et al. 2018) and applied a new “hybrid” method for distance calibration, as described in Supplemental Methods. To compare these speckle distances estimated from TSA-seq data produced using two different TSA staining conditions, we generated histograms of their distance residuals over each 20-kb bin using a 0.005-µm histogram binning interval.
SPAD calling and cell type comparisons
We ranked smoothed TSA-seq enrichment scores over all 20-kb bins genome-wide from largest to smallest excluding hg38 unmapped regions, dividing these ranked scores into 100 equal sized groups, defined as percentiles 1 to 100 (lowest to highest scores). Adjacent 20-kb bins ranked in the 96th−100th percentile were merged to segment these regions as SPADs.
To compare SPADs called in one cell line with TSA-seq score percentiles in other cell lines, we calculated the mean TSA-seq score percentile over each SPAD region in each of the other cell lines and plotted these values in a box plot, with dots showing all values. We used Intervene (Khan and Mathelier 2017) (version 0.6.4) to generate a four-way Venn diagram using 20-kb binned BED files of the four cell lines.
Identification of genomic domains that show different nuclear positions relative to speckles in different cell lines
To identify genomic regions that change nuclear position relative to speckles in cell line pair-wise comparisons, we adapted a previously published method (Peric-Hupkes et al. 2010) that compares the variability of scores in two cell lines with the variability observed in biological replicates. The null hypothesis tested is whether a region is statistically different from the values of the biological replicates of the two different cell lines. The detailed statistical method is provided in Supplemental Methods.
Correlating TSA-seq data with other genomic features
Methods for RNA-seq, CUT&RUN-seq, and PRO-seq data processing, and integrative analysis with TSA-seq data are described in Supplemental Methods (Supplemental Tables S5–S8). CUT& RUN-seq data were generated by the Steven Henikoff laboratory (Seattle, WA) using an automated CUT&RUN platform (Janssens et al. 2018).
Estimating nuclear speckle mean distance changes between H1 versus K562 using TSA-seq
We first correlated the scaled TSA-seq score in each 20-kb genomic bin in K562 cells with estimated mean nuclear speckle distances (Methods, TSA distance prediction), generating a score-distance dictionary. Using mean scaled TSA-seq scores for each changed domain in K562 or H1 cells, we then converted these into distances based on this score-distance dictionary for K562 cells. We then calculated the changes in distance predicted in K562 cells for shifts in scaled TSA-seq scores corresponding to the shift observed comparing scaled scores in H1 versus K562.
FISH
3D immuno-FISH and single-molecule RNA FISH are modified from our previous publications (Chen et al. 2018; Tasan et al. 2018; Kim et al. 2020). Experimental details and the newly developed sequential RNA- and DNA- FISH procedure and methods for FISH data analysis are provided in Supplemental Methods.
Software availability
TSA-seq normalization software is available at https://github.com/zocean/Norma/tree/master/TSA-seq_2.0 (Supplemental Codes C1). Codes for all genomic data analyses are available at https://github.com/lgchang27/TSA-Seq-2020v2 (Supplemental Codes C2).
Data access
The TSA-seq data generated in this study have been submitted to the 4DN Data Portal (https://data.4dnucleome.org/) under accession numbers 4DNEXUUZ3QJY, 4DNEXZ8L4YL6, 4DNEX2JUPDUP, 4DNEXY7SLH9Y, 4DNEXUTOBNQO, 4DNEX9APWCUE, 4DNEX6U8TS3Y, 4DNEXI7XUWFK, 4DNEXJYIQ1LL, 4DNEXIRQ3TRW, 4DNEX1969D8U, 4DNEX82F2EAZ, 4DNEXD6PVVIG, 4DNEX8XT4LOP, 4DNEXYD1WGJD, 4DNEXVGZUN76, 4DNEXCAJWM6W, 4DNEXI5ZVEAW, 4DNEXTBEE5EZ, 4DNEXDL3SDJS, 4DNEXFTPDSNA, 4DNEXYYVG4NT, 4DNEX4ANWWVQ, 4DNEX5CWKK1V, 4DNEXPTYCQQ1, 4DNEXXL427TJ, 4DNEXCLGBHDE, 4DNEX2PH2EGA, 4DNEXWI62POK, 4DNEXHBXW5R2, 4DNEXPZYYA7S, 4DNEXK1AOFGT, 4DNEX4DZEZ7G, 4DNEXNS4ZCCO, 4DNEX4G8UEFD, 4DNEXNBF8QSE, 4DNEXT2NZVA6, 4DNEX67FKCUC, 4DNEX8553UTO, 4DNEXA1SMWU3, 4DNEXHGVQ3G6, and 4DNEX4I7I7AT (summarized in Supplemental Table S9).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the UIUC Biotechnology Center for guidance with preparation of sequencing libraries and quality control. We thank Drs. K.V. Prasanth, William Brieher, Lisa Stubbs, and Huimin Zhao (UIUC, Urbana, IL) for helpful suggestions. We thank Derek Janssens (Henikoff lab, Fred Hutchinson Cancer Research Center, Seattle, WA) for generating and sharing CUT&RUN-seq data. We thank Belmont lab members for sharing reagents and providing suggestions. We thank members of the 4D-Nucleome Consortium and the Belmont NOFIC U54 Center for helpful suggestions and feedback. This work was supported by National Institutes of Health grants R01 GM58460 (A.S.B.) and U54 DK107965 (A.S.B. and J.M.).
Author contributions: L.Z. designed and performed all experiments, collected data, and conducted microscopy image analysis with A.S.B.’s guidance. L.Z. and Y.Z. analyzed genomic data with guidance from A.S.B. and J.M. Y.Z. developed the TSA-seq normalization software and updated this software to its new version with guidance from J.M. and A.S.B. Y.C. developed and standardized TSA-seq 1.0 protocols and advised on the development of TSA-seq 2.0. O.G. contributed to the “hybrid” distance-calibration mapping approach. Y.W. processed raw CUT&RUN-seq data. L.Z. and A.S.B. wrote the manuscript with critical suggestions from other co-authors. A.S.B. supervised the overall study.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.266239.120.
References
- Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. 2017. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543: 519–524. 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Khanna N, Alvikas J, Belmont AS. 2013. β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J Cell Biol 203: 767–783. 10.1083/jcb.201305027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA. 2013. The spatial organization of the human genome. Annu Rev Genomics Hum Genet 14: 67–84. 10.1146/annurev-genom-091212-153515 [DOI] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. 2013. Genome architecture: domain organization of interphase chromosomes. Cell 152: 1270–1284. 10.1016/j.cell.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Bobrow MN, Harris TD, Shaughnessy KJ, Litt GJ. 1989. Catalyzed reporter deposition, a novel method of signal amplification application to immunoassays. J Immunol Methods 125: 279–285. 10.1016/0022-1759(89)90104-X [DOI] [PubMed] [Google Scholar]
- Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, et al. 2018. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol 25: 833–840. 10.1038/s41594-018-0112-y [DOI] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. 2006. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol 172: 177–187. 10.1083/jcb.200507073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T, Martins S, Rino J, Marinho S, Carmo-Fonseca M. 2017. Pharmacological inhibition of the spliceosome subunit SF3b triggers exon junction complex-independent nonsense-mediated decay. J Cell Sci 130: 1519–1531. 10.1242/jcs.202200 [DOI] [PubMed] [Google Scholar]
- Chen Y, Belmont AS. 2019. Genome organization around nuclear speckles. Curr Opin Genet Dev 55: 91–99. 10.1016/j.gde.2019.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adam SA, Goldman R, van Steensel B, Ma J, Belmont AS. 2018. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol 217: 4025–4048. 10.1083/jcb.201807108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, Cisse II. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361: 412–415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341: 664–667. 10.1126/science.1239053 [DOI] [PubMed] [Google Scholar]
- Dias AP, Dufu K, Lei H, Reed R. 2010. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun 1: 97 10.1038/ncomms1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopie J, Sweredoski MJ, Moradian A, Belmont AS. 2020. Tyramide signal amplification mass spectrometry (TSA-MS) ratio identifies nuclear speckle proteins. J Cell Biol 219: e201910207 10.1083/jcb.201910207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Puvion E. 1980. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int Rev Cytol 65: 255–299. 10.1016/s0074-7696(08)61962-2 [DOI] [PubMed] [Google Scholar]
- Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, Ting AY. 2019. Atlas of subcellular RNA localization revealed by APEX-seq. Cell 178: 473–490.e26. 10.1016/j.cell.2019.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, de Castro IJ, Lavitas L, Chotalia M, Pombo A. 2010. Gene positioning. Cold Spring Harb Perspect Biol 2: a000588 10.1101/cshperspect.a000588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galganski L, Urbanek MO, Krzyzosiak WJ. 2017. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res 45: 10350–10368. 10.1093/nar/gkx759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XD, Tu LC, Mir A, Rodriguez T, Ding Y, Leszyk J, Dekker J, Shaffer SA, Zhu LJ, Wolfe SA, et al. 2018. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9–APEX2. Nat Methods 15: 433–436. 10.1038/s41592-018-0006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Will CL, Peng J, Makarov EM, Kastner B, Lemm I, Urlaub H, Hartmuth K, Lührmann R. 2012. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun 3: 994 10.1038/ncomms1998 [DOI] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, et al. 2019. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572: 543–548. 10.1038/s41586-019-1464-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB. 2006. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288A: 664–675. 10.1002/ar.a.20336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Cummings R, Yamashita YM. 2018. A conserved function for pericentromeric satellite DNA. eLife 7: e34122 10.7554/eLife.34122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens DH, Wu SJ, Sarthy JF, Meers MP, Myers CH, Olson JM, Ahmad K, Henikoff S. 2018. Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. Epigenetics Chromatin 11: 74 10.1186/s13072-018-0243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Primorac D, McKinstry M, McNeil J, Rowe D, Lawrence JB. 2000. Tracking Col1a1 RNA in osteogenesis imperfecta: splice-defective transcripts initiate transport from the gene but are retained within the Sc35 domain. J Cell Biol 150: 417–432. 10.1083/jcb.150.3.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Mathelier A. 2017. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics 18: 287 10.1186/s12859-017-1708-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N, Hu Y, Belmont AS. 2014. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol 24: 1138–1144. 10.1016/j.cub.2014.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Han KY, Khanna N, Ha T, Belmont AS. 2019. Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J Cell Sci 132: jcs226563 10.1242/jcs.226563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Venkata NC, Hernandez Gonzalez GA, Khanna N, Belmont AS. 2020. Gene expression amplification by nuclear speckle association. J Cell Biol 219: e201904046 10.1083/jcb.201904046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. 2015. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163: 134–147. 10.1016/j.cell.2015.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara M, Kato K, Sanbo C, Shigenobu S, Ohkawa Y, Fuchigami T, Miyanari Y. 2020. Genomic profiling by ALaP-seq reveals transcriptional regulation by PML bodies through DNMT3A exclusion. Mol Cell 78: 493–505.e8. 10.1016/j.molcel.2020.04.004 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldi L, Brickner JH. 2011. Compartmentalization of the nucleus. Trends Cell Biol 21: 701–708. 10.1016/j.tcb.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. 2013. Constitutive nuclear lamina–genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23: 270–280. 10.1101/gr.141028.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SA, Wright J, Peckner R, Kalish BT, Zhang F, Carr SA. 2018. Discovery of proteins associated with a predefined genomic locus via dCas9–APEX-mediated proximity labeling. Nat Methods 15: 437–439. 10.1038/s41592-018-0007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. 2011. The nucleolus. Cold Spring Harb Perspect Biol 3: a000638 10.1101/cshperspect.a000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters T, van Roy F. 2014. Role of cell–cell adhesion complexes in embryonic stem cell biology. J Cell Sci 127: 2603–2613. 10.1242/jcs.146720 [DOI] [PubMed] [Google Scholar]
- Politz JC, Scalzo D, Groudine M. 2013. Something silent this way forms: the functional organization of the repressive nuclear compartment. Annu Rev Cell Dev Biol 29: 241–270. 10.1146/annurev-cellbio-101512-122317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174: 744–757.e24. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raap AK, van de Corput MPC, Vervenne RAM, van Gijlswijk RPM, Tanke HJ, Wiegant J. 1995. Ultra-sensitive FISH using peroxidase-mediated deposition of biotin- or fluorochrome tyramides. Hum Mol Genet 4: 529–534. 10.1093/hmg/4.4.529 [DOI] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770. 10.1101/gr.099655.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. 2004. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell 15: 3876–3890. 10.1091/mbc.e04-03-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. 2003. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol 162: 981–990. 10.1083/jcb.200303131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. 1999. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol 144: 617–629. 10.1083/jcb.144.4.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3: a000646 10.1101/cshperspect.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. 2008a. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev 22: 489–498. 10.1101/gad.1634608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T. 2008b. The meaning of gene positioning. Cell 135: 9–13. 10.1016/j.cell.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasan I, Sustackova G, Zhang L, Kim J, Sivaguru M, HamediRad M, Wang Y, Genova J, Ma J, Belmont AS, et al. 2018. CRISPR/Cas9-mediated knock-in of an optimized TetO repeat for live cell imaging of endogenous loci. Nucleic Acids Res 46: e100 10.1093/nar/gky501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran JR, Paulson DI, Moresco JJ, Adam SA, Yates JR, Goldman RD, Zheng Y. 2021. An APEX2 proximity ligation method for mapping interactions with the nuclear lamina. J Cell Biol 220: e202002129. 10.1083/jcb.202002129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik T, Vos M, Peric-Hupkes D, Hn Celie P, van Steensel B. 2020. Cell cycle dynamics of lamina-associated DNA. EMBO Rep 21: e50636 10.15252/embr.202050636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. 2017. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169: 780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A, Mahat DB, Guertin MJ, Chu T, Danko CG, Lis JT, Sistonen L. 2017. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun 8: 255 10.1038/s41467-017-00151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, Sheer D. 2004. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J Cell Biol 164: 515–526. 10.1083/jcb.200305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang L, Wang J, Chen S, Shi M, Cheng H. 2018. Intronless mRNAs transit through nuclear speckles to gain export competence. J Cell Biol 217: 3912–3929. 10.1083/jcb.201801184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT Jr., McNeil JA, Lawrence J. 1995. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol 131: 1635–1647. 10.1083/jcb.131.6.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.