Abstract
Cell fusion is essential for the development of multicellular organisms, and plays a key role in the formation of various cell types and tissues. Recent findings have highlighted the varied protein machinery that drives plasma-membrane merger in different systems, which is characterized by diverse structural and functional elements. We highlight the discovery and activities of several key sets of fusion proteins that together offer an evolving perspective on cell membrane fusion. We also emphasize recent discoveries in vertebrate myoblast fusion in skeletal muscle, which is composed of numerous multinucleated myofibers formed by the fusion of progenitor cells during development.
Cell Fusion and Its Importance in Physiology
The fusion of cellular membranes is a highly specialized biological process that is indispensable for muscle development, the sperm/egg fertilization event during sexual reproduction, and the formation of multinucleated osteoclasts, giant cells of the macrophage lineage, and syncytiotrophoblasts in the placenta. In each of these mammalian cell-fusion systems, many molecules are involved in fusion to varying degrees, although the magnitude to which they participate directly in the membrane fusion reaction has not always been clear. In recent years, however, the identification of various proteins that play key roles in driving membrane fusion provides an essential foundation for mechanistic investigation. A complex picture has emerged in which a diverse set of fusion machineries are active across different systems and organisms. However, much remains unknown about the mechanisms of the factors identified, and it is likely that many membrane-active proteins that drive fusion remain undiscovered.
We emphasize an evolving perspective on the diverse mechanisms harnessed by various systems to achieve fusion of plasma membranes. Instead of recounting details of all known fusion factors, we focus on representative classes of fusion-driving proteins that reveal emerging principles of fusion biology. We then concentrate our discussion on recent discoveries in the field of myoblast fusion, with an emphasis on membrane coalescence as a distinct step in the muscle progenitor differentiation program, further highlighting how the study of fusion-driving proteins has the potential to impact muscle biology. Skeletal muscle is composed of numerous myofibers, and each acquires multiple nuclei to properly develop and orchestrate locomotion and metabolism. Mononucleated muscle progenitors fuse together during development to form multinucleated fibers, and this cellular fusion process occurs throughout the lifetime of the muscle to allow regeneration and adaptations to exercise [1,2]. Thus, cell fusion is a central event that controls the health and maintenance of skeletal muscle, and regulated plasma membrane coalescence is crucial for these processes.
Membrane Fusion in Different Systems: Similarities and Divergence
Similar to the better-studied intracellular and viral fusion processes, cell–cell fusion events are driven by specialized proteins that remodel the phospholipid bilayer. Before membrane rearrangements, the broad consensus is that membranes are brought to within 10 nm of one another by surrogate cell-adhesion machineries. However, the energy barrier to bring them close enough (<10 nm) for the initial fusion connections to begin to form is extremely high owing to hydration repulsion. To overcome this barrier, which should involve surface dehydration, specialized proteins (i.e., fusogens) are required to initiate the biophysical fusion pathway. The most widely accepted pathway for fusion is the stalk–pore model, which involves formation of a hemifusion stalk intermediate, where the proximal monolayers of the fusing cells coalesce (hemifusion) (Figure 1). According to the canonical view, fusion then progresses by expansion of the stalk, thus causing the two distal uncoalesced monolayers to bend and form a bilayer (diaphragm). Disruption of this diaphragm leads to the formation of a fusion pore, which is then stabilized and expanded to culminate in syncytium formation (e.g., a multinucleated cytoplasm) [3]. A central paradigm of the stalk–pore model is that the cell membranes remain intact and are not leaky. Although alternative models propose that fusion may be leaky, because pores are formed outside the stalk intermediate and can be incorporated into the complex to culminate in a fusion pore leading to syncytial formation [4], there is less experimental evidence for these alternative models [5]. Therefore, we discuss how proteins drive cell–cell fusion from the perspective of the stalk–pore model. Independent of the biophysical model, it is clear that a major barrier to fusion is bringing membranes close enough for membrane rearrangements, and this might be achieved by diverse mechanisms in cell–cell fusion.
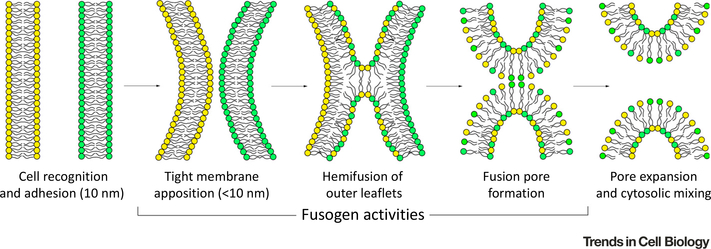
Figure 1. Essential Steps of the Hemifusion Stalk/Fusion-Pore Model of Cell Membrane Fusion.
A series of distinct membrane events must occur for two cells to fuse and merge their cytoplasmic contents. First, two cells must recognize and adhere to one another, followed by close membrane adhesion (to within 10 nm). The outer-membrane leaflets must then fuse, which results in mixing of lipids and the formation of an unstable membrane stalk intermediate. The formation and expansion of a fusion pore within the hemifused membrane then completes the reaction.
Although the general biophysical pathway for membrane coalescence may be conserved, the identification of fusion-activating proteins in different systems has revealed a surprising diversity. Indeed, these discoveries indicate that fusogens (loosely defined as proteins that mediate membrane remodeling leading to membrane merger) are structurally diverse and distinct, both in their biochemical functions and in the ancillary cellular machinery involved in their activities. For a comprehensive inventory of all putative fusogens, we refer readers to excellent recent reviews on the topic [6,7]. We focus on three representative types of cell–cell fusion proteins and their respective mechanisms: fusion family (FF) proteins (Caenorhabditis elegans development), fusion-associated small-transmembrane (FAST) proteins (nonenveloped viruses), and myomaker/myomerger (vertebrate myogenesis) (Figure 2).
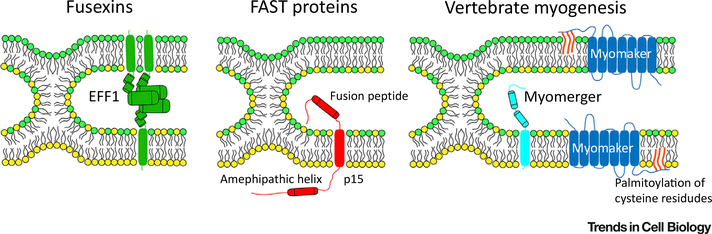
Figure 2. Diversity of Protein Machineries Driving Cell–Cell Fusion.
Representative fusogenic proteins from three classes are shown at the stage of the unstable hemifusion-stalk intermediate. EFF1, representing the fusexin family, acts in a trans-trimerization form to drive membrane fusion. The transmembrane domains perform a zippering-like action to bring opposing membranes together and drive pore formation. Fusion-associated small transmembrane (FAST) proteins act unilaterally to drive fusion of reovirus-infected cells. FAST proteins harness endogenous cellular machinery to promote cell adhesion. An extracellular fusion peptide inserts into the trans membrane, and an intracellular amphipathic helix is postulated to drive fusion pore formation. Myomaker and myomerger act independently at the membrane to drive a bipartite myoblast fusion mechanism. Myomaker is required for hemifusion to occur, and must be palmitoylated in its C-terminal region to function. Myomerger mediates pore formation through the activity of one or both extracellular helices.
FF Proteins
The FF proteins are members of a superfamily referred to as fusexins, a structurally homologous class that includes factors involved in sexual reproduction (e.g., the Arabidopsis gamete fusogen HAP2/GCS1) [8–10], enveloped virus entry (e.g., class II viral fusogen E1) [11,12], and in somatic cell fusion (including the FF proteins). The evolutionary and mechanistic implications of a shared fusion family in such diverse settings of fusion are currently unknown, but are at the forefront of current research in the field [7]. The two primary FF proteins are EFF-1 and AFF-1, closely related membrane glycoproteins identified in C. elegans, which during development drive the fusion of specific sets of epithelial cells within the reproductive tract, epidermis, and pharynx [13,14]. FF proteins are required on both of the fusing cells, and form trimers between the two membranes in trans [11]. A conformational change of the trimer follows, which bends the two opposing membranes into closer proximity. Final membrane merger is postulated to occur through a zippering action of the transmembrane domains acting in trans, similar in mechanism to the trans-oligomerization and hairpin formation of SNARE-mediated vesicle fusion. Remarkably, FF proteins can fuse heterologous cells and can act interchangeably in trans with one another and with other members of the fusexin family such as HAP2/GCS1 [8], indicating a high degree of conserved functionality within the class.
FAST Proteins
The FAST proteins are a group of viral fusogens encoded by the nonenveloped fusogenic reoviruses. FAST proteins are unique among nonenveloped viral fusogens in that they are responsible for cell–cell, but not virus–cell, membrane fusion [15,16]. They are characterized by their small size (<200 residues) and are distinct from other viral fusogens both in structure and mechanism. All FAST proteins possess a single transmembrane domain separating a minimal N-terminal ectodomain from a C-terminal endodomain of variable size. Unlike the complexes involved in virus–cell entry, FAST proteins rely on endogenous cellular adhesion factors and actin cytoskeletal rearrangements to accomplish the initial adhesion and membrane apposition necessary for fusion [17]. However, unlike the fusexins, FAST proteins are modular fusogens that are only required on one cell surface to drive membrane fusion [18], consistent with their supposed role in enhancing viral spread [19]. To achieve cell fusion, FAST proteins function by affecting both the cis and trans bilayers, and this is consistent with their unilateral requirement.
Myomaker/Myomerger
Myomaker and myomerger are essential for muscle formation in vertebrates [20–23]. Myomerger (discovered by three independent groups; also called myomixer or minion) exhibits some structural similarities to FAST proteins, whereas myomaker bears no resemblance to any known fusogens. Myomaker/myomerger expression and activity are highly regulated to the time and place of myoblast fusion during development, and also in adult muscle for regeneration, and as muscle adapts to exercise. Recent evidence supports a stepwise model in which myomaker is necessary for hemifusion whereas myomerger mediates pore formation [24]. These two proteins are discussed in further detail in the following section.
Comparison of these three categories of fusion proteins shows that, despite the convergent membrane events shared by all cell fusions, the identification of fusogenic proteins has uncovered a remarkable divergence of structure and function among the factors driving syncytialization. One explanation for these divergent fusogens is the context in which membrane fusion takes place. For example, the small size, unilateral activity, and high efficiency of FAST proteins are consistent with their goal to propagate the virus with little concern for effects on the cell. Viral evolution would select for proteins that act quickly in an unregulated manner. Endogenous cell fusion, however, leads to the development of a highly ordered cellular superstructure, complete with nuclear positioning, cytoskeletal arrangements, and membrane integrity, and therefore requires more sophisticated regulation. Intuitively, such a process likely requires (i) tight regulation of fusogenic activity to the time and place of membrane fusion, and (ii) close coordination with other cellular machinery. Studies on EFF-1 and myomaker/myomerger are consistent with the concept that developmental cell fusions are highly regulated. EFF-1 requires targeted delivery to the plasma membrane for proper function [25], and although myomaker localizes to intracellular vesicles, its activity can be blocked with an antibody that binds on the plasma membrane [26]. Another way cells could regulate fusion is to regulate different points of the pathway by different proteins, which was recently revealed for the myomaker/myomerger system.
Given that the above proteins drive fusion through unique structures and biochemical activities, one question regards the suitability of the term ‘fusogen’ to describe these diverse proteins. A fusogen typically refers to a protein that is necessary and sufficient to induce membrane merger directly at the site of fusion. Related to this, the ability to fuse membranes that do not normally fuse has been used as an experimental benchmark of fusogen status. Although several important factors have been identified that meet these standards, it is possible that this understanding of fusogens needs to be reevaluated given the divergence of mechanisms that is becoming increasingly apparent. Recent discoveries show that (i) not all fusion reactions take place in an identical fashion, and (ii) proteins can act independently or in combination at different steps of the reaction. Thus, assessing putative drivers of fusion by their similarities to a classical understanding of fusion may limit our ability to recognize proteins, whether currently known or unknown, that act through novel mechanisms or combinations to drive membrane merger. Another possibility is that a less-specialized protein machinery is harnessed for membrane remodeling at the time and place of cell fusion. Put simply, the ‘fusogens’ identified tomorrow may look nothing like the fusogens of today and yesterday. Exemplifying this trend is the identification of myomaker, a protein with no structural or mechanistic similarities to known fusogens, but that unequivocally plays a central role in myoblast fusion.
Myoblast Fusion
Recent years have seen exciting progress in the field of myoblast fusion. Insights from multiple model organisms have identified many molecules that are involved in the regulation and direct execution of fusion.
Myoblast Fusion in the Fly
Drosophila melanogaster has classically been the dominant experimental system for developmental myoblast fusion. Careful studies of body-wall muscles in Drosophila embryos and larvae have given rise to a model wherein mononucleated founder cells (FCs) fuse with multiple fusion-competent myoblasts (FCMs) to form the syncytial myofiber [27]. A large number of factors have been identified as playing cell-specific roles in the steps of FC-to-FCM fusion [28]. Immunoglobulin superfamily proteins Kirre/Duf and Rst/Irre-C are expressed in FCs and act redundantly to drive migration and association with FCMs [29,30], which themselves must express the nephrin homologs Sticks-and-stones (Sns) and Hibris (Hbs) [31,32]. Sns interacts with Kirre/Duf in trans to recognize and adhere to FCs and nascent myotubes. Many additional factors involved in larval myoblast fusion have been identified, and these have been covered in elegant detail elsewhere [28,33].
Recent developments in the field have specifically highlighted the role that actin cytoskeletal rearrangements play in mediating the asymmetrical fusion reaction between FC and FCM [34,35]. The discovery of actin-enriched cytoskeletal structures at sites of FC–FCM fusion has spurred much interest in the role that actin-mediated mechanical forces might play in driving membrane fusion [34]. Transmission electron microscopy has revealed projecting podosome-like membrane protrusions originating from the FCM that form an interface with the FC membrane [34]. This has led to a model in which several actin cytoskeletal regulators (notably WASP, Arp3/ArpC1, Rac1/Rac2, and related pathways [28]) mediate asymmetric actin structural rearrangements in the form of FCM ‘invasion’ [34] and FC ‘resistance’ [35]. These opposing mechanical forces have been postulated to drive close membrane apposition beyond the distance that cell adhesion molecules alone can mediate, thereby advancing cell fusion [28]. Although insights into these actin polymerization events and their regulatory pathways can no doubt inform our understanding of cell fusion, they do not fully account for the membrane events necessary to fuse two cells because fusogenic proteins are also required [36]. Mechanistically, it has been proposed that the relationship between the actin cytoskeleton and fusogens is that actin-propelled invasive protrusions promote membrane juxtaposition and fusogen engagement [36–38]. The evidence for this model is partly based on a heterologous fusion system in Drosophila cells using EFF-1 as the fusogen [36]. In this case, EFF-1 (a C. elegans fusogen) and Sns (an actin-modifying protein) were found to accumulate in the finger-like projections. Moreover, protrusions have been visualized by electron microscopy (EM) [36,39], and the actin cytoskeleton is required for EFF-1 localization [38]. However, there are also indications that there is more to understand before confirming the model. First, the presence of podosome-like structures has not been consistently observed in fusing myoblasts, including postembryonic Drosophila flight muscles [40], which could indicate their transient nature or that they are not universally required. Second, evidence is lacking that the opposing membranes within the podosome-like structure are closer than membranes outside the podosome, which would seem to be a central principle for the proposed actin cytoskeleton–fusogen engagement model. More broadly, it remains to be demonstrated that any proteins identified in Drosophila operate directly at the site of fusing FCM/FC membranes. Thus, studies on myoblast fusion in Drosophila have yielded detailed understanding about the general pathways that induce myoblast fusion, but there remain open questions regarding the fusogens that catalyze the biochemical events required for membrane coalescence.
Vertebrate Myoblast Fusion
The situation is somewhat reversed in the field of mammalian myoblast fusion, where, despite only modest understanding of the more general machinery involved in fusion, two proteins have been shown to be directly involved at the level of the plasma membrane. Myomaker, a skeletal muscle-specific seven-pass transmembrane protein, was the first such protein to be discovered. In the absence of myomaker, myoblasts do not form syncytia in culture or during mouse development or regeneration [20]. Ectopic expression of myomaker in fibroblasts induces the competency to fuse to muscle cells [20,41]; however, myomaker+ fibroblasts do not fuse to one another, indicating that myomaker alone is not sufficient for fusion. The discovery of myomerger, a second muscle-specific fusion protein, resolved the issue of sufficiency [21–23]. When coexpressed, myomaker and myomerger drive fibroblast fusion, indicating that the combination of the two proteins can perform the steps necessary for fusion. Further studies have revealed that myomaker and myomerger are conserved in vertebrates and function to drive muscle formation in zebrafish and chick [42–44]. In humans, hypomorphic mutations of myomaker have been identified as the underlying cause of the rare congenital myopathy, Carey–Fineman–Ziter syndrome [45,46]. Currently, no homologs for myomaker or myomerger have been identified in Drosophila.
Recent lines of investigation have revealed that myomaker and myomerger act independently to drive steps of the fusion pathway, where myomaker controls hemifusion competence and myomerger drives pore formation [24]. Lipid-mixing assays, in which membrane lipid dyes allow precise tracking of lipid exchange between cells, demonstrated that myomaker−/− myoblasts lose their ability to undergo hemifusion, whereas myomerger−/− myoblasts retain this ability. Furthermore, each protein was found to perform its activity in the absence of the other. Expression of myomerger can drive completion of the fusion reaction in fibroblasts initiated by the viral fusogen hemagglutinin, which in controlled conditions leads only to hemifusion. Because myomaker is not expressed in fibroblasts, these data indicate that myomerger does not require myomaker for activity. Conversely, myomaker-expressing myoblasts that lack myomerger could be rescued by membrane stress-inducing reagents. Together, these experiments support a model in which myomaker and myomerger have independent effects directly on myoblast membranes through which fusion is achieved.
This model of myomaker/myomerger activity thus provides a novel pathway by which fusion can be achieved via the delegation of specific steps to independently active proteins. It is noteworthy that the fusion reaction has been divided in muscle cells, but the reasons for this are not obvious. One possibility is that a divided fusion reaction provides more regulatory control that may help to maintain fidelity between partners in a system that has a lifelong reliance on fusion, such as what takes place in skeletal muscle. This pathway presents an exciting opportunity to study the myoblast fusion mechanism at the level of the membrane. Although a multitude of questions remain with respect to myoblast fusion (both in Drosophila and in mammals), we highlight several key unknowns emerging from the current myomaker/myomerger model.
Mammalian Myoblast Fusion: Emerging Areas of Investigation
What Is the General Mammalian Myoblast Fusion Pathway?
It is obvious that myomaker/myomerger do not operate in isolation, but instead cooperate as key players in an intricate but largely uncharacterized myoblast fusion sequence. Other factors previously identified may operate at different stages of the process, from signaling to cell adhesion to membrane remodeling (Figure 3A). For example, the need for actin cytoskeletal remodeling in mouse myoblast fusion is well established [39,47,48], and myomaker/myomerger-mediated fusion can be inhibited when actin polymerization is blocked [20]. Phosphatidylserine (PS) exposure is associated with myoblast fusion, as well as the associated BAI1 and stabilin 2 membrane PS receptors [49,50]. Annexins, cadherins, GRAF1/ferlin proteins, and the Rac1/ELMO/Bai3/Dock1 pathway have also been implicated [51–54]. Intriguing recent studies have identified the transforming growth factor (TGF)-β pathway as a brake signal on myoblast fusion, highlighting the fact that negative regulation of syncytialization is a crucial component to consider [55,56]. However, despite the many molecules implicated in fusion, any interactions between these factors and the myomaker/myomerger mechanisms remain unknown. In addition, mutants of many of these identified factors have been found to have relatively mild (e.g., BAI1/stabilin 2 [49]) or nonexistent (e.g., cadherins [57]) phenotypes in vivo. These discrepancies could be explained by the enhanced redundancy of function that is often seen in murine models, and also highlight the limitations of studying fusion exclusively in cultured cell lines, which does not reflect the full complexity of the tissue when modeling recognition, adhesion, and membrane apposition. A further difficulty is in separating myoblast fusion defects from upstream deficits in differentiation. To this end, adoption of heterologous fusion systems (e.g., induced fusion of normally nonfusing cells or viral pseudotyping) or fusion of synthetic membranes should be utilized.
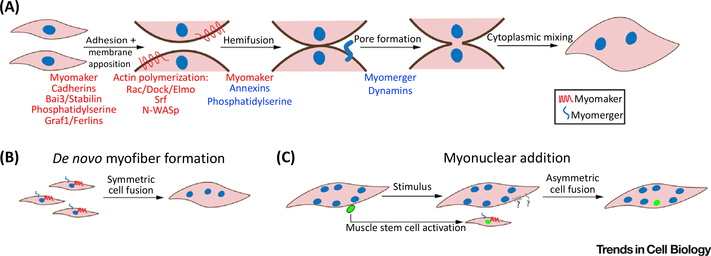
Figure 3. Mechanisms of Vertebrate Myoblast Fusion.
(A) Myoblast fusion involves a series of membrane modification steps. Key protein and lipid regulators are listed beneath each step. Knowledge of the precise stage at which a given factor regulates fusion is frequently absent, therefore factors are color-coded according to the strength of evidence that they act at a particular step (blue, high confidence; red, further clarification is needed). Highlighted are the bilateral requirement for myomaker and the unilateral requirement for myomerger. (B) Symmetric fusion between cells in similar states (myocytes to myocytes) occurs to form myofibers de novo during early development and adult regeneration. Myomaker and myomerger are shown bilaterally to highlight their expected coexpression in fusogenic myocytes. (C) Myonuclear addition to existing myofibers can also occur during postnatal development after myofiber number has been established, and after a stimulus (such as exercise or injury) in the adult. In this case, muscle stem cells become activated, and undergo an asymmetric fusion event (fusion between cells in different states – myocytes and myofibers). Whether fusion between myocytes and myofibers progresses through a unilateral or a bilateral mechanism, with respect to the requirement of myomaker or myomerger on each of the fusing cells, is not fully understood. Although myomaker/myomerger are required on the myocyte, it is not known whether they are active on the myofiber. Note that, in adult regenerative contexts, myocyte-to-myocyte fusion can also occur between this activated muscle stem-cell pool to generate de novo myofibers (not shown).
What is the Molecular Function of Myomaker?
The molecular basis of the activity of myomaker is an obvious area for future research. As a protein with no known homologs or structural similarities to known fusogens, deciphering its activity promises to provide novel insights into how membrane fusion can be accomplished. Hemifusion connections and lipid-mixing require trans engagement of apposing membranes to pull those membranes close to one another (<10 nm). In myoblasts, hemifusion competence requires bilateral expression of myomaker, and palmitoylation of its intracellular C terminus is necessary for activity [26]. However, the current model of myomaker membrane topology, based on live immunostaining of tagged constructs, indicates that myomaker is highly embedded in the membrane and lacks extensive extracellular domains that could tether two membranes [58], which is how EFF-1 and other classical fusogens function. Additional structural studies could yield greater clarification of the orientation of myomaker. On the basis of currently available data, it seems probable that myomaker acts in one of two ways: (i) through modification of the cis membrane to achieve fusion competency, or (ii) by recruitment of proteins or protein complexes, which do not need to be muscle-specific and may also possess fusion-independent activities, but when acted upon by myomaker can bring about fusion competency.
How Exactly Does Myomerger Drive Fusion Pore Formation?
Myomerger acts at the plasma membrane via its ectodomain to drive pore formation. This immediately raises the question of how a protein can act unilaterally and extracellularly to create a fusion pore. Currently, no known fusogens exert their activity in this exact manner, and investigation of myomerger activity could therefore uncover new mechanisms by which fusion is accomplished. Moreover, the myomerger ectodomain contains two closely positioned α-helices of different amphipathic character. This type of structure is unique among membrane-active proteins, and it will be important to understand how these extracellular helices cooperate to drive a fusion pore. Among other fusion proteins, FAST proteins bear comparison to myomerger. Although no outright homology exists between myomerger and FAST proteins, they nevertheless bear some similarities. In general, FAST proteins contain a small extracellular fusion peptide and an intracellular amphipathic helix which is thought to mediate pore formation [59–61], and these elements each bear some structural or functional resemblance to the membrane-proximal amphipathic helix of myomerger. It is clear, however, that the proteins are functionally distinct because myomerger alone is insufficient for fusion. Further studies will be necessary to elucidate the overlap in structure and function between myomerger and FAST proteins.
What Are the Roles of Myomaker and Myomerger on Differentiated Myofibers?
Another central question concerns the symmetry of myomaker/myomerger activity. Experiments performed with cultured C2C12 myoblasts have repeatedly demonstrated a bilateral requirement for myomaker and a unilateral requirement for myomerger, meaning that both fusing cells must express myomaker, but only one needs to express myomerger [20,21]. This is consistent with a model in which myomaker induction of hemifusion involves membrane modifications on both cells, whereas the role of myomerger in pore formation must somehow be mediated from one side alone. The requirements for these proteins in the fusing compartments of muscle are less clear in vivo, however. Within a native tissue context, myoblast fusion can take place in one of two scenarios: (i) fusion of multiple progenitors to form multinucleated myofibers de novo, or (ii) fusion of myoblasts to existing myofibers, occurring notably during postnatal growth and adult muscle hypertrophy or regeneration (Figure 3B,C) [62,63]. It is clear from both global and conditional knockout models that mononuclear muscle progenitors require myomaker for fusion of either type [20,63]. However, in the case of myoblast-to-myofiber fusion, it is not clear whether myomaker is also required on the myofiber membrane. Preliminary evidence in the synergist ablation model of muscle hypertrophy suggests that resident myonuclei in the myofiber do not actively upregulate myomaker [62], although this requires further study. It is interesting to speculate about whether there may be a differential requirement for myomaker on myoblast versus myofiber membranes, perhaps owing to some fusion ‘competency’ or readiness that the myofiber retains after formation, or due to the presence of other unknown factors. Furthermore, although myomerger knockout mice have established its necessity for progenitor fusion, the question of myomerger expression and function (if any) on myofibers remains to be pursued.
Concluding Remarks
The juxtaposition of myoblast fusion factors with FAST proteins and FF proteins/fusexins provides a helpful framework in which to consider ongoing investigations towards understanding fusion mechanisms in diverse systems. In particular, insights from myoblast fusion can inform other fields where the machinery is less well defined. Current knowledge in the fly versus the mouse underscores the difference between fusion-assisting molecules and true membrane-active drivers of fusion. Indeed, Drosophila fusogens have yet to be discovered, and may be different from the vertebrate myoblast fusogens because fusogens are typically more tissue- and species-specific, in contrast to the actin cytoskeleton that is generally required for various fusion events. Second, the bipartite (at least) mechanisms of myomaker/myomerger illustrate that not all systems possess a single self-sufficient fusogen. Sufficiency tests for fusion may in fact set a bar that precludes us from discovering more complex multifactor machineries, especially machineries that include factors and proteins that are conserved between different cell types. In short, there appears to be no ‘one-size-fits-all’ approach for studying factors involved in cell–cell fusion, highlighting the need for creativity and context-specific experimental strategies to further decipher mechanisms of cell–cell fusion in muscle cells (see Outstanding Questions). In addition, we expect that a greater understanding of the biochemical properties of fusing membranes will be a key component of future discoveries.
Outstanding Questions.
Many separate proteins and signaling mechanisms have been implicated in promoting vertebrate myoblast fusion. What are the relationships between these pathways, and how do they interact with the central myomaker/myomerger mechanism?
What is the molecular function of myomaker with respect to myoblast plasma membranes?
How does the extracellular domain of myomerger accomplish fusion pore formation?
What are the mechanistic differences between fusion of myoblasts to one another versus fusion between myoblasts and a myofiber?
What factors directly mediate membrane remodeling in Drosophila?
Highlights.
Cell–cell fusion in different systems is accomplished by specific fusogenic proteins that are notable for their structural and functional diversity.
Vertebrate myoblast fusion is driven by two independent skeletal muscle-specific proteins, myomaker and myomerger, whose expression are tightly regulated to the time of membrane fusion.
Myomaker and myomerger function at distinct membrane remodeling steps, creating a divided fusion reaction in myoblasts.
Acknowledgments
We thank Dilani Gamage of the laboratory of D.P.M. for expertise and helpful discussions. We apologize to our colleagues for the omission of many seminal studies concerning cell fusion owing to space constraints. This work was supported by grants to D.P.M. from the National Institutes of Health (NIH) National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS, grant R01AR068286), NIH National Institute on Aging (NIA, grant R01AG059605), and the Pew Charitable Trusts. M.J.P. was supported by the NIH National Heart, Lung, and Blood Institute (NHLBI, grant T32HL007752).
References
- 1.Yin H et al. (2013) Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chal J and Pourquie O (2017) Making muscle: skeletal myogenesis in vivo and in vitro. Development 144, 2104–2122 [DOI] [PubMed] [Google Scholar]
- 3.Chernomordik LV et al. (2006) Membranes of the world unite! J. Cell Biol. 175, 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katsov K et al. (2006) Field theoretic study of bilayer membrane fusion. II. Mechanism of a stalk-hole complex. Biophys. J. 90, 915–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haldar S et al. (2018) Lipid-dependence of target membrane stability during influenza viral fusion. J. Cell Sci. 132, jcs218321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brukman NG et al. (2019) How cells fuse. J. Cell Biol. 218, 1436–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez JM and Podbilewicz B (2017) The hallmarks of cell–cell fusion. Development 144, 4481–4495 [DOI] [PubMed] [Google Scholar]
- 8.Valansi C et al. (2017) Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J. Cell. Biol. 216, 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fedry J et al. (2017) The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell 168, 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinello JF et al. (2017) Structure–function studies link class II viral fusogens with the ancestral gamete fusion protein HAP2. Curr. Biol. 27, 651–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez-Vargas J et al. (2014) Structural basis of eukaryotic cell–cell fusion. Cell 157, 407–419 [DOI] [PubMed] [Google Scholar]
- 12.Kielian M and Rey FA (2006) Virus membranefusion proteins: more than one way to make a hairpin. Nat Rev Microbiol 4, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podbilewicz B et al. (2006) The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev Cell 11, 471–481 [DOI] [PubMed] [Google Scholar]
- 14.Sapir A et al. (2007) AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev Cell 12, 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciechonska M and Duncan R (2014) Reovirus FAST proteins: virus-encoded cellular fusogens. Trends Microbiol 22, 715–724 [DOI] [PubMed] [Google Scholar]
- 16.Duncan R (2019) Fusogenic reoviruses and their fusion-associated small transmembrane (FAST) proteins. Annu. Rev. Virol. 6, 341–363 [DOI] [PubMed] [Google Scholar]
- 17.Salsman J et al. (2008) A virus-encoded cell–cell fusion machine dependent on surrogate adhesins. PLoS Pathog 4, e1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clancy EK and Duncan R (2009) Reovirus FAST protein transmembrane domains function in a modular, primary sequence-independent manner to mediate cell–cell membrane fusion. J Virol 83, 2941–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan R et al. (1996) Avian reovirus-induced syncytium formation is independent of infectious progeny virus production and enhances the rate, but is not essential, for virus-induced cytopathology and virus egress. Virology 224, 453–464 [DOI] [PubMed] [Google Scholar]
- 20.Millay DP et al. (2013) Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499, 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn ME et al. (2017) Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat Commun 8, 15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi P et al. (2017) Control of muscle formation by the fusogenic micropeptide myomixer. Science 356, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q et al. (2017) The microprotein minion controls cell fusion and muscle formation. Nat Commun 8, 15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leikina E et al. (2018) Myomaker and myomerger work independently to control distinct steps of membrane remodeling during myoblast fusion. Dev Cell 46, 767–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smurova K and Podbilewicz B (2017) Endocytosis regulates membrane localization and function of the fusogen EFF-1. Small GTPases 8, 177–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamage DG et al. (2017) Insights into the localization and function of myomaker during myoblast fusion. J Biol Chem 292, 17272–17289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rushton E et al. (1995) Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development 121, 1979–1988 [DOI] [PubMed] [Google Scholar]
- 28.Lee DM and Chen EH (2019) Drosophila myoblast fusion: invasion and resistance for the ultimate union. Annu. Rev. Genet. Published online July 5, 2019. 10.1146/annurev-genet-120116-024603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strunkelnberg M et al. (2001) rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development 128, 4229–4239 [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Gomez M et al. (2000) Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell 102, 189–198 [DOI] [PubMed] [Google Scholar]
- 31.Bour BA et al. (2000) Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev 14, 1498–1511 [PMC free article] [PubMed] [Google Scholar]
- 32.Shelton C et al. (2009) The immunoglobulin superfamily member Hbs functions redundantly with Sns in interactions between founder and fusion-competent myoblasts. Development 136, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abmayr SM and Pavlath GK (2012) Myoblast fusion: lessons from flies and mice. Development 139, 641–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sens KL et al. (2010) An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J Cell Biol 191, 1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JH et al. (2015) Mechanical tension drives cell membrane fusion. Dev Cell 32, 561–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shilagardi K et al. (2013) Actin-propelled invasive membrane protrusions promote fusogenic protein engagement during cell–cell fusion. Science 340, 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JH et al. (2015) Mechanisms of myoblast fusion during muscle development. Curr Opin Genet Dev 32, 162–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y et al. (2017) Spectraplakin induces positive feedback between fusogens and the actin cytoskeleton to promote cell–cell fusion. Dev Cell 41, 107–120.e4 [DOI] [PubMed] [Google Scholar]
- 39.Randrianarison-Huetz V et al. (2018) Srf controls satellite cell fusion through the maintenance of actin architecture. J Cell Biol 217, 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhanyasi N et al. (2015) Surface apposition and multiple cell contacts promote myoblast fusion in Drosophila flight muscles. J Cell Biol 211, 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitani Y et al. (2017) In vivo myomaker-mediated heterologous fusion and nuclear reprogramming. Faseb j 31, 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landemaine A et al. (2014) Myomaker mediates fusion of fast myocytes in zebrafish embryos. Biochem Biophys Res Commun 451, 480–484 [DOI] [PubMed] [Google Scholar]
- 43.Shi J et al. (2017) Requirement of the fusogenic micropeptide myomixer for muscle formation in zebrafish. Proc Natl Acad Sci U S A 114, 11950–11955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo W et al. (2015) Myomaker, regulated by MYOD, MYOG and miR-140–3p, promotes chicken myoblast fusion. Int J Mol Sci 16, 26186–26201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Gioia SA et al. (2017) A defect in myoblast fusion underlies Carey–Fineman–Ziter syndrome. Nat Commun 8, 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedberg-Oldfors C et al. (2018) Carey–Fineman–Ziter syndrome with mutations in the myomaker gene and muscle fiber hypertrophy. Neurol Genet 4, e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruenbaum-Cohen Y et al. (2012) The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc Natl Acad Sci U S A 109, 11211–11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin NY et al. (2014) Dynamin and endocytosis are required for the fusion of osteoclasts and myoblasts. J Cell Biol 207, 73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hochreiter-Hufford AE et al. (2013) Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 497, 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SY et al. (2016) Stabilin-2 modulates the efficiency of myoblast fusion during myogenic differentiation and muscle regeneration. Nat Commun 7, 10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leikina E et al. (2013) Extracellular annexins and dynamin are important for sequential steps in myoblast fusion. J Cell Biol 200, 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamoud N et al. (2014) G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc Natl Acad Sci U S A 111, 3745–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krauss RS et al. (2017) Keep your friends close: cell– cell contact and skeletal myogenesis. Cold Spring Harb. Perspect. Biol. 9, a029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lenhart KC et al. (2014) GRAF1 promotes ferlindependent myoblast fusion. Dev Biol 393, 298–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Girardi F et al. (2019) TGFb signaling curbs cell fusion and muscle regeneration. bioRxiv, p. 557009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sieiro D et al. (2019) Auto-inhibition of myoblast fusion by cyclic receptor signalling. bioRxiv. Published online February 19, 2019. 10.1101/553420 [DOI] [Google Scholar]
- 57.Goel AJ et al. (2017) Niche cadherins control the quiescence-to-activation transition in muscle stem cells. Cell Rep 21, 2236–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Millay DP et al. (2016) Structure–function analysis of myomaker domains required for myoblast fusion. Proc Natl Acad Sci U S A 113, 2116–21121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Read J et al. (2015) Reovirus FAST proteins drive pore formation and syncytiogenesis using a novel helix-loop-helix fusion-inducing lipid packing sensor. PLoS Pathog 11, e1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barry C et al. (2010) Features of a spatially constrained cystine loop in the p10 FAST protein ectodomain define a new class of viral fusion peptides. J Biol Chem 285, 16424–16433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Top D et al. (2012) Cell–cell membrane fusion induced by p15 fusion-associated small transmembrane (FAST) protein requires a novel fusion peptide motif containing a myristoylated polyproline type II helix. J Biol Chem 287, 3403–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goh Q and Millay DP (2017) Requirement of myomaker-mediated stem cell fusion for skeletal muscle hypertrophy. eLife 6, e2000728186492 [Google Scholar]
- 63.Millay DP et al. (2014) Myomaker is essential for muscle regeneration. Genes Dev 28, 1641–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]