Abstract
COVID-19 pandemic caused by SARS-CoV-2 infection continue to cause the morbidity and mortality in many countries. Limitations of the gold standard qRT-PCR for diagnosis of this infection includes need for expensive equipment, specialized molecular laboratory, and experienced staff. Currently, CRISPR-based diagnostic method was approved by the U.S. FDA for rapid detection. Several studies developed SARS-CoV-2 detection based on CRISPR-Cas12a platform; however, the validations with RNA extracted from clinical specimens were limited. Therefore, this study evaluated the clinical performance of previously described CRISPR-Cas12a based diagnostic assays for SARS-CoV-2. According to the results, the CRISPR-Cas12a assays on N1 and S genes provided diagnostic accuracy (≥ 95 %) comparable to the qRT-PCR results. The assays with E, N2 and S genes yielded acceptable sensitivity of detection (≥ 95 %) whereas N1 and S genes provided outstanding specificity of detection (100 %). Preferably, multiple target genes should be detected by using CRISPR-Cas12a to ensure the most effective SARS-CoV-2 detection. Therefore, the N1 and S genes would be attractive target genes for SARS-CoV-2 detection based on CRISPR-Cas12a.
Keywords: SARS-CoV-2, CRISPR-Cas12a, Clinical specimen, Performance
COVID-19 (Coronavirus disease 2019) pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The first emergence of SARS-CoV-2 was in Wuhan, China in December 2019 which caused pneumonia in patients and then spread rapidly to multiple countries (Zhou et al., 2020). Recently, SARS-CoV-2 infection has continued to increase the mortality rate around the world. Epidemiological studies revealed that the virus is easily spread by droplets in human-to-human transmission (Shereen et al., 2020). Consequently, rapid detection is crucial to monitor and limit the spread of infections.
The WHO recommends a viral detection protocol based on quantitative reverse transcription-polymerase chain reaction (qRT-PCR) from nasopharyngeal (NP) or throat swab that is collected in a viral transport medium (VTM). Extracted viral RNA from specimens are reverse transcribed and amplified within specific genes for detection of SARS-CoV-2 based on qRT-PCR (Corman et al., 2020; Radbel et al., 2020). However, only 4 commercial reagents have been approved for SARS-CoV-2 diagnosis by the Centers for Disease Control and Prevention (CDC) (Esbin et al., 2020). Meanwhile, the limitations of qRT-PCR include the need for expensive instruments, a specialized molecular laboratory, appropriately trained staff, and approximately 4 h turnaround time for detection. Therefore, several studies have attempted to develop rapid nucleic acid diagnostic tests for SARS-CoV-2 such as Recombinase polymerase amplification (RPA), Loop-mediated isothermal amplification (LAMP) and CRISPR-based diagnostic test (Kilic et al., 2020). Interestingly, the CRISPR-based diagnostic method was recently approved by the U.S. Food and Drug Administration (FDA) (Joung et al., 2020)
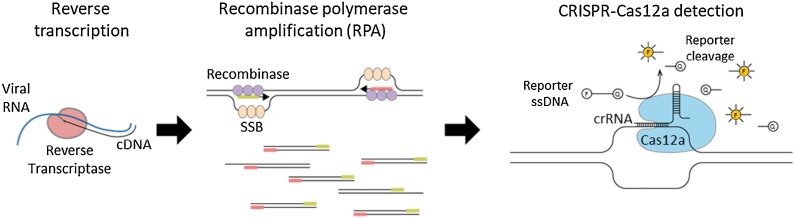
The CRISPR-based diagnostic method for SARS-CoV-2 detection consists of DNA Endonuclease Targeted CRISPR Trans Reporter (DETECTR) (Broughton et al., 2020), SHERLOCK Testing in One Pot for COVID-19 (STOPCovid) (Joung et al., 2020), specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) (Patchsung et al., 2020) based on CRISPR-Cas12a, CRISPR-Cas12b and CRISPR-Cas13, respectively. However, only Cas12a is commercially available to allow researchers to perform the CRISPR-based diagnosis. The Cas12a recognizes and cleaves double stranded DNA target that complement with the CRISPR RNA (crRNA) called “cis-cleavage activity”. After that, Cas12a randomly cleaves single stranded DNA probes conjugated with fluorescence-quencher called “trans-cleavage activity” (Broughton et al., 2020) (Fig. 1 ). Several studies were developed on the CRISPR-Cas12a platform for rapid and sensitive detection of SARS-CoV-2. However, the validations of the CRISPR-Cas12a assay with RNA extracted from clinical specimens were limited. Therefore, this study aims to perform clinical sample evaluation of CRISPR-Cas12a based COVID-19 diagnostic assays.
Fig. 1.
Schematic representation of SARS-CoV-2 detection based on CRISPR-Cas12a. The process includes reverse transcription, isothermal amplification by RPA and specific target detection based on CRISPR-Cas12a.
Nasopharyngeal and/or throat swab samples were collected from COVID-19 suspected patients (N = 107) in VTM (MP Biomedicals, USA) media obtained from the Institute for Urban Disease Control and Prevention (IUDC), Thailand. The viral RNA was extracted from 200 μL of specimens using a magLEAD 12gC instrument with a magLEAD Consumable Kit (Precision System Science, Japan) following the manufacturer’s instructions. SARS-CoV-2 detection was confirmed by Allplex™ 2019-nCoV Assay (Seegene, Korea) (Farfour et al., 2020). The qRT-PCR (cut-off at Ct ≤ 38) was interpreted as positive for SARS-CoV-2. The left-over extracted RNA (N = 107) was reverse transcribed into cDNA by using RevertAid RT Reverse Transcription Kit (Thermo Scientific™, USA) with random hexamers according to the standard protocol. The cDNA samples were tested under code with different CRISPR-Cas12a based assays.
Targeted genes amplification was performed by TwistAmp® Basic recombinase polymerase amplification (RPA) Kit (TwistDx, United Kingdom) with specific primer sets depending on different assays (Table 1 ). RPA is an isothermal amplification method in which (1) the recombinase binds with primers complementary to DNA template, then (2) single-stranded DNA-binding protein (SSB) displaces the double stranded DNA template and (3) the strand-displacing polymerase exponentially amplify the target gene products (Piepenburg et al., 2006) (Fig. 1). Briefly, lyophilized RPA was resuspended by rehydration buffer and then mixed with 480 nM forward primer and 480 nM reverse primer. In the last step, 14 mM of MgOAC and 1 μL of cDNA template were added to the reaction mixture. Targeted genes of SARS-CoV-2 were amplified by incubating at 39 °C for 30 min, then the reaction was inactivated at 75 °C for 5 min. The CRISPR-Cas12a detection method consists of 1X NEBuffer 2.0 (New England Biolabs, USA), 30 nM crRNA, 330 nM EnGen® LbaCas12a endonuclease (New England Biolabs, USA), 200 nM 5′ 6-FAM / 3′ BHQ-1®, Dual Labeled Fluorescent Probe and DEPC-treated water in a total reaction volume of 15 μL. The amplified products were added in a CRISPR-Cas12a reaction and then incubated at 39 °C for 15 min. The fluorescence was detected under BluPAD Dual LED Blue/White Light Transilluminator (BIO-HELIX, Taiwan). The fluorescence results were separately read under code and interpreted by 3 technicians
Table 1.
Sequences of primers and crRNAs for detection of SARS-CoV-2 based on different CRISPR-Cas12a assays.
| No | Gene | Primer name | Sequence (5′-3′) | Reference |
|---|---|---|---|---|
| 1 | N1 gene | RT-AIOD-CRISPR F primer | AGGCAGCAGTAGGGGAACTTCTCCTGCTAGAAT | Ding et al. (2020) |
| RT-AIOD-CRISPR R primer | TTGGCCTTTACCAGACATTTTGCTCTCAAGCTG | |||
| crRNA1 | UAAUUUCUACUAAGUGUAGAUCAUCACCGCCAUUGCCAGCC | |||
| crRNA2 | UAAUUUCUACUAAGUGUAGAUUUGCUGCUGCUUGACAGAUU | |||
| 2 | N2 gene | 2019-nCoV_N2-F_RPA | ACAAGGAACTGATTACAAACATTGGCCGCAAA | Broughton et al. (2020) |
| 2019-nCoV_N2-R_RPA | TTCCATGCCAATGCGCGACATTCCGAAGAA | |||
| N-gene gRNA | UAAUUUCUACUAAGUGUAGAUCCCCCAGCGCUUCAGCGUUC | |||
| 3 | E gene | E-Sarbeco_F1_RPA | GAAGAGACAGGTACGTTAATAGTTAATAGCGT | Broughton et al. (2020) |
| E-Sarbeco-R2_RPA | ACGTTAACAATATTGCAGCAGTACGCACACA | |||
| E-gene gRNA | UAAUUUCUACUAAGUGUAGAUGUGGUAUUCUUGCUAGUUAC | |||
| 4 | S gene | SARS2_spike1-F | CCACTGAGAAGTCTAACATAATAAGAGGCTG | Mayuramart et al. (2020) |
| SARS2_spike1-R | AATAAACTCTGAACTCACTTTCCATCCAACT | |||
| SARS2_spike2-F | AATCTATCAGGCCGGTAGCACACCTTGTAAT | |||
| SARS2_spike2-F | TCCACAAACAGTTGCTGGTGCATGTAGAAGTT | |||
| SARS2-S1 | UAAUUUCUACUAAGUGUAGAUGAUUCGAAGACCCAGUCCCU | |||
| SARS2-S2 | UAAUUUCUACUAAGUGUAGAUCAAUCAUAUGGUUUCCAACC |
Note: Underline sequence represents spacer sequence.
The results obtained from CRISPR-Cas12a assays were compared with the gold standard qRT-PCR (RdRp, E and N genes). Clinical performance of the assay in terms of sensitivity, specificity, positive predictive values (PPV), negative predictive values (NPV), and diagnostic accuracy was calculated by a diagnostic test evaluation calculator (https://www.medcalc.org/calc/diagnostic_test.php) based on the formula and calculations as shown in Supplementary Table 1.
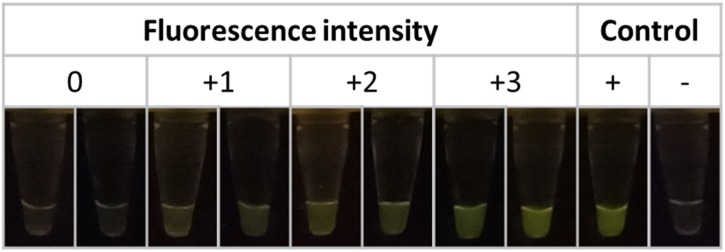
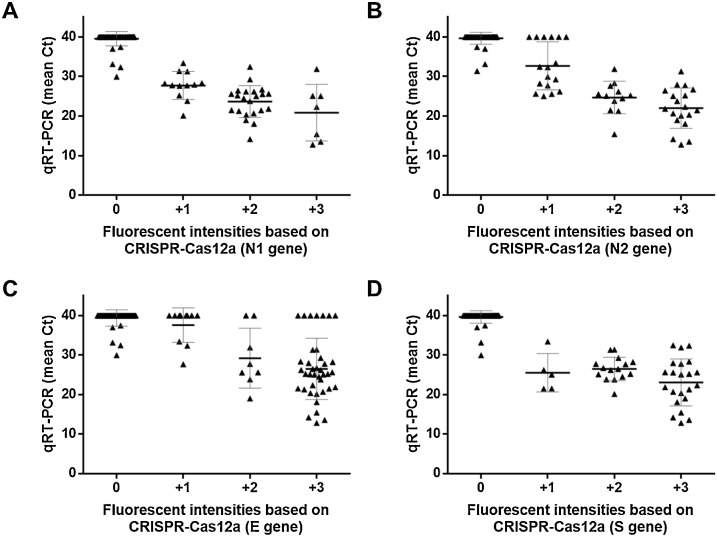
RPA primers and crRNAs specific to target genes including E (envelope), S (spike) and N (nucleoprotein) of SARS-CoV-2 were validated with clinical specimens obtained from the same cohort. Interpretations of the CRISPR-Cas12a assays were performed based on fluorescent intensities as negative (0) and positive (+1, +2 and +3) (Fig. 2 ). The qRT-PCR results showed that 44 and 63 samples were detected as positive and negative for SARS-CoV-2, respectively. The correlations between fluorescence score based on different CRISPR-Cas12a assays and the mean Ct obtained from qRT-PCR are shown in Fig. 3 . In general, the samples negative for SARS-CoV-2 yielded Ct higher than 40 from qRT-PCR and no fluorescent signal from CRISPR-Cas12a assays. On the other hand, the samples positive with higher titers of SARS-CoV-2 yielded lower Ct values from qRT-PCR and higher fluorescent signals based on N1 and S genes of CRISPR-Cas12a assays (Fig. 3A and D). There were 6 false-positive samples (scored as +1) obtained from the N2 gene of CRISPR-Cas12a assay, but these samples were negative (Ct > 38) based on qRT-PCR (Fig. 3B). In addition, 17 false-positive samples (scored as +1 (N = 8), +2 (N = 2) and +3 (N = 7)) obtained from E gene of CRISPR-Cas12a assay yielded a negative result (Ct > 38) based on qRT-PCR (Fig. 3C). A few false-negative results (scored as 0) were also obtained from all the CRISPR-Cas12a assays, while these samples yielded Ct values lower than 38 based on qRT-PCR (Fig. 3).
Fig. 2.
Representative results of fluorescence intensities based on CRISPR-Cas12a assays. The results scored as 0, +1, +2 and +3 depending on fluorescent intensities under BluPAD Dual LED Blue/White Light Transilluminator.
Fig. 3.
The correlations between the mean Ct obtained from qRT-PCR and fluorescent score based on different CRISPR-Cas12a assays for A. N1 gene, B. N2 gene, C. E gene and D. S gene of SARS-CoV-2. The bold line represents the mean Ct, the grey lines represents S.D. and each dot represents each sample.
The results yielded from different CRISPR-Cas12a based assays were compared with qRT-PCR and evaluated for their clinical performance. In general, 4 assays based on CRISPR-Cas12a tested in this study yielded sensitivities range from 93.2 % to 97.7 % whereas the specificity range from 84.1 % to 100 %. The diagnostic accuracy (89.7 %–98.1 %) of CRISPR-Cas12a based assays were comparable to qRT-PCR (Table 2 & Supplementary Table 1). The detection of the N2 gene yielded moderate performance with 95.5 % sensitivity, 90.5 % specificity and 92.5 % diagnostic accuracy. The results revealed that detection of the E gene (Broughton et al., 2020) of SARS-CoV-2 based on CRISPR-Cas12a provides the highest sensitivity (97.7 %). On the other hand, S (Mayuramart et al., 2020) and N1 (Ding et al., 2020) genes yielded the highest specificity (100 %). Interestingly, S (Mayuramart et al., 2020) and N1 (Ding et al., 2020) genes represented acceptable diagnostic accuracy as high as 98.1 % and 97.2 %, respectively (Table 2).
Table 2.
Clinical performance of SARR-CoV-2 detection based on different CRISPR-Cas12a assays compared with gold standard qRT-PCR.
| No | Gene | TPa | FPb | TNc | FNd | PPVe | NPVf | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | N1 | 41 | 0 | 63 | 3 | 100 % | 95.5 % | 93.2 % | 100 % | 97.2 % |
| 2 | N2 | 42 | 6 | 57 | 2 | 87.5% | 96.6 % | 95.5 % | 90.5 % | 92.5 % |
| 3 | E | 43 | 10 | 53 | 1 | 81.1% | 98.2 % | 97.7 % | 84.1 % | 89.7 % |
| 4 | S | 42 | 0 | 63 | 2 | 100 % | 96.2 % | 95.5 % | 100 % | 98.1 % |
The details of calculation were summarized in Supplementary Table 1.
True Positive (TP).
False Positive (FP).
True Negative (TN).
False Negative (FN).
Positive predictive value (PPV).
Negative predictive value (NPV).
In the current pandemic, there is no vaccine available to prevent against SARS-CoV-2 infection. Therefore, rapid and accurate viral detections could decrease the spread of infections. Several studies have attempted to develop CRISPR-Cas12a based detection methods suitable for point-of-care (POC) diagnosis for SARS-CoV-2. Broughton et al. developed the CRISPR-Cas12a assay to detect E and N genes of SARS-CoV-2 by using a pair of primers and a crRNA for each gene (Broughton et al., 2020). Meanwhile, Ding et al. established the method based on a pair of primers and 2 regions of crRNAs within the N gene (Ding et al., 2020). Mayuramart et al. reported the assay based on 2 pairs of primers and 2 regions of crRNAs for the S gene (Mayuramart et al., 2020). The current study evaluated the performance of these primers and crRNAs for SARS-CoV-2 detection based on CRISPR-Cas12a assays with RNA extracted from the same clinical specimens. Although the conditions of RPA, Cas12a reaction, fluorescent detection and signal scoring system used in our study were slightly different from the optimal conditions described in previous reports (Broughton et al., 2020; Ding et al., 2020; Mayuramart et al., 2020), all the assays were performed under the same conditions and tested against samples from the same cohort to ensure appropriate evaluation of the assays.
According to our study (Table 2 & Supplementary Table 1), the CRISPR-Cas12a assays based on N1 (Ding et al., 2020) and S (Mayuramart et al., 2020) genes yielded diagnostic accuracy (≥ 95 %) comparable to the qRT-PCR results). The CRISPR-Cas12a assays within E, N2 (Broughton et al., 2020) and S (Mayuramart et al., 2020) genes provided suitable sensitivity of detection (≥ 95 %) while N1 (Ding et al., 2020) and S (Mayuramart et al., 2020) genes provided excellent specificity of detection (100 %). Ideally, multiple target genes should be detected by using CRISPR-Cas12a to ensure the most effective SARS-CoV-2 detection. Therefore, the N1 and S genes might be attractive target genes for SARS-CoV-2 detection based on CRISPR-Cas12a.
Ethical statement
The study protocol was approved by the Institutional Review Board, Faculty of Medicine, Chulalongkorn University (IRB No. 302/63).
Authors contributions
PN performed the experiments and drafted the manuscript. OM assisted in results interpretation. SR and JP assisted in sample processing. NC and SS coordinated the project. YP provided the clinical samples. SP designed the study and revised the manuscript. All the authors have read and approved the accuracy and integrity of the manuscript.
CRediT authorship contribution statement
Pattaraporn Nimsamer: Validation, Formal analysis, Investigation, Writing - original draft. Oraphan Mayuramart: Validation, Investigation, Visualization. Somruthai Rattanaburi: Investigation. Naphat Chantaravisoot: Visualization. Suthat Saengchoowong: Project administration. Jiratchaya Puenpa: Resources. Yong Poovorawan: Resources. Sunchai Payungporn: Conceptualization, Methodology, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank the staff of Institute for Urban Disease Control and Prevention (IUDC), Thailand and the Center of Excellence in Clinical Virology, Faculty of Medicine, Chulalongkorn University for assistance with specimen collections. This work was supported by the Ratchada Pisek Sompoch Fund, Faculty of Medicine, Chulalongkorn University [RA(P0)002/63]; National Research Council of Thailand (NRCT); Thailand Research Fund (TRF) [RSA6180035]; National Science and Technology Development Agency (NSTDA) [P-17-51377]; and the Innovation Fund to fight against COVID-19 [Taejai].
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2021.114092.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020:1–5. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V., Bleicker T., Brünink S., Drosten C., Zambon M., Organization W.H. World Health Organization; Geneva: 2020. Diagnostic Detection of Wuhan Coronavirus 2019 by Real-time RT-PCR. January 13. [Google Scholar]
- Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020 doi: 10.1261/rna.076232.120. 076120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfour E., Lesprit P., Visseaux B., Pascreau T., Jolly E., Houhou N., Mazaux L., Asso-Bonnet M., Vasse M., group, S.-C.-F.H.s The Allplex 2019-nCoV (Seegene) assay: which performances are for SARS-CoV-2 infection diagnosis? Eur. J. Clin. Microbiol. Infect. Dis. 2020:1–4. doi: 10.1007/s10096-020-03930-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N.-G., Woolley A.E., Segel M., Barretto R.P., Ranu A., Macrae R.K., Faure G. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 2020;383:1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic T., Weissleder R., Lee H. Molecular and immunological diagnostic tests of COVID-19–current status and challenges. Iscience. 2020 doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayuramart O., Nimsamer P., Rattanaburi S., Chantaravisoot N., Khongnomnan K., Chansaenroj J., Puenpa J., Suntronwong N., Vichaiwattana P., Poovorawan Y., Payungporn S. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Exp. Biol. Med. 2020 doi: 10.1177/1535370220963793. 0, 1535370220963793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020:1–10. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radbel J., Jagpal S., Roy J., Brooks A., Tischfield J., Sheldon M., Bixby C., Witt D., Gennaro M.L., Horton D.B. Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is comparable in clinical samples preserved in saline or viral transport medium. J. Mol. Diagn. 2020;22:871–875. doi: 10.1016/j.jmoldx.2020.04.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020 doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. BioRxiv; 2020. Discovery of a Novel Coronavirus Associated With the Recent Pneumonia Outbreak in Humans and Its Potential Bat Origin. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.