Abstract
The current global pandemic outbreak of COVID-19, caused by the SARS-CoV-2, strikes an invincible damage to both daily life and the global economy. WHO guidelines for COVID-19 clinical management includes infection control and prevention, social distancing and supportive care using supplemental oxygen and mechanical ventilator support. Currently, evolving researches and clinical reports regarding infected patients with SARS-CoV-2 suggest a potential list of repurposed drugs that may produce appropriate pharmacological therapeutic efficacies in treating COVID-19 infected patients. In this study, we performed virtual screening and evaluated the obtained results of US-FDA approved small molecular database library (302 drug molecule) against two important different protein targets in COVID-19. Best compounds in molecular docking were used as a training set for generation of two different pharmacophores. The obtained pharmacophores were employed for virtual screening of ChEMBL database. The filtered compounds were clustered using Finger print model to obtain two compounds that will be subjected to molecular docking simulations against the two targets. Compounds complexes with SARS-CoV-2 main protease and S-protein were studied using molecular dynamics (MD) simulation. MD simulation studies suggest the potential inhibitory activity of ChEMBL398869 against SARS-CoV-2 main protease and restress the importance of Gln189 flexibility in inhibitors recognition through increasing S2 subsite plasticity.
Keywords: SARS-CoV-2, Virtual screening, Pharmacophore elucidation, ChEMBL database, Molecular dynamics (MD)
Graphical abstract
1. Introduction
The horrible global pandemic outbreak of COVID-19 (coronavirus disease 2019) swept the global community and the healthcare systems worldwide like a storm. Most if not all human beings were caught off guard without suitable defense mechanisms to cope or control such a pandemic. COVID-19, caused by that novel coronavirus species (SARS-CoV-2), has recently been identified and characterized (Zhou and Huang, 2020). Coronaviruses are huge family of viruses that named for their crown-like spikes that found on their surface. This family composed of four main sub-groupings of coronaviruses, known as alpha, beta, gamma, and delta (Andersen et al., 2020; Zhou et al., 2020b). SARS-CoV-2 belongs to the beta sub-group, and it considered to be the third member of a group of dangerous coronaviruses types including SARS-CoV and MERS-CoV that caused sever fatal symptoms to infected patients (Andersen et al., 2020; Wang et al., 2005; Zaki et al., 2012; Zhou et al., 2020a). It is believed that the causative virus of the current pandemic infected humans through zoonotic transmission (Saxena, 2020; Zimmermann and Curtis, 2020). Genetically, SARS-COV-2 is a positive-sense, single stranded RNA virus which encodes for two open reading frames (Ceraolo and Giorgi, 2020; Zhou and Huang, 2020). These open reading frames are designated as 1A and 1B. They code for some proteases called 3CLpro and PLpro. These proteases could latterly cleave the polypeptide into a) 16 non-structural proteins (Nsp) that are considered as essential viral enzymes which play major roles in replication and packaging of the virus within the host cell and b) 4 structural proteins that contribute to the outer structure of the virus (Clark et al., 2020; Corman et al., 2020; Zaki et al., 2012). In addition, this virus utilizes RNA-dependent RNA polymerase (RdRp) for replication and transcription of the viral genome. Like other Coronaviruses, the outer surface of the SARS-CoV-2 virus is made of Spike (S) protein, envelope (E) protein, membrane (M) protein and the Nucleocapsid (N) protein. The M and E proteins are responsible for virus morphogenesis and assembly. Also, they constitute the outer shell of the viral genetic material RNA. The RNA is also guarded by the Nucleocapsid protein. The Spike protein (S) is at the precedence of infection. It interacts with the ACE-2 receptor on the host cell surface so as to promote viral fusion to the cell membrane during initiation of viral infection. (DeDiego et al., 2011; Haines et al., 2020; Nieto-Torres et al., 2015)
At present, a lot of research efforts have been invested to develop vaccines around the world. Till now, there isn't any approved vaccine or specific effective drug molecule targeting the SARS-CoV-2. Hence, it remains a major challenge and a time race to prevent and treat that huge number of severely sick patients suffering from COVID-19 fatal attacks and to put an end for high COVID-19 mortality rate. Until we have a proper vaccine or specific therapeutic drug targeting SARS-CoV-2, “repurposed” drugs that have been approved by the FDA for other indications are used to treat COVID-19 patients.
Repurposing and repositioning approach of drugs (Khan et al., 2020) is a drug development technique that used to discover new applications of already existing approved drug molecules in various novel diseases. Nowadays, this process becomes a universal strategy mainly for novel rare diseases (Pushpakom et al., 2019). Many advantages for this strategy are observed including, fewer clinical trial steps and rapid results which will minimize the cost and time (Hodos et al., 2016). The advantages of drug repurposing discovery approach can also be used to discover potent lead molecules to overcome the COVID-19 pandemic. This strategy is based on virtual screening of database of compounds against major SARS-CoV-2 viral proteins that are gaining importance in anti-CoV drug design like the main protease and SARS-CoV-2 spike S protein aiming identification and repurposing of proper drug molecule for possible therapeutic effect against COVID-19.
Implementing this strategy, Wu, Canrong, et al. (Wu et al., 2020) virtually screened a library of ZINC drug database against main protease (3CLpro), Spike, RNA-dependent RNA polymerase (RdRp), and papain like protease (PLpro). Also, Peele, K. Abraham, et al. (Peele et al., 2020) make use of repurposing drug strategy by applying a virtual screening approach on library of antiviral databases including US-FDA approved drugs and plant-derived natural drugs against Coronavirus main protease (pdb id: 6LU7) protein. Structure-based virtual screening of FDA-approved drug bank database that resulted in 20 hits against 3CLpro was performed by Chatterjee, Shilpa, et al. (Chatterjee et al., 2020) Recently, Sen et al. virtually screened 10 important spices to identify potential inhibitors for both main protease and spike with the support of molecular dynamic simulations. (Sen et al., 2020). Finally, Huynh et al., explored mechanisms of clinically oriented SARS-CoV-2 inhibitors including naturally occurring rutin as main protease inhbitors.(Huynh et al., 2020a, b)
In view of the aforementioned findings, this study was developed to obtain appropriate drug molecules that could be repurposed to be used against the SARS-CoV-2 virus. A virtual screening of ChEMBL database against two targets of COVID-19 including the main protease (pdb id: 6LU7) and Spike S protein (pdb: 7C8V) which gain importance in anti-CoV drug design. This virtual screening will be based on two generated pharmacophore models based on training set generated from US-FDA approved antiviral database against COVID-19.
2. Materials and methods
2.1. Molecular docking simulations
Molecular operating environment (MOE) 2019.0102 (Diukendjieva et al., 2020) was used for Ligand/protein preparation, molecular docking, ligand-protein interaction, scoring, poses’ visualization and pharmacophore building. Docking processes were performed via MMFF94x protocol using docking placement: Triangular matcher, Rescoring: London dG, Forcefield and refinement: Affinity dG.
2.1.1. Preparation of target protein structures
Two SARS-CoV-2 viral proteins gaining importance in anti-CoV drug design through evolving researches (Chakraborti et al., 2020) were retrieved from Protein Data Bank (www.rcsb.org) including the main protease (pdb id: 6LU7) and SARS-CoV-2 spike S protein (pdb id: 7C8V).
Concerning, the main protease (pdb id: 6LU7) the protein structure was checked and repaired through automatic correction and fixation order of MOE to check for any errors in the atom's type and connection were done. A sufficient number of hydrogen atoms were added during protonation step. The binding site was determined by confining ligand N3. Then, the co-crystallized water molecules and bounded ligand N3 were removed. After docking completion, observation and filtration of results through scoring values and visualized poses was performed.
While in case of the spike S protein (pdb id: 7C8V), the protein structure was checked and repaired using automatic correction and fixation orders of MOE. Also, sufficient number of hydrogen atoms were added during protonation step. The binding pocket was determined and isolated using “site finder” operating process of the MOE and dummy atoms of the pocket were created. Then, the co-crystallized water molecules and bounded synthetic nanobody SR4 were removed. After docking completion, observation and filtration of results through scoring values and visualized poses was performed.
2.1.2. Preparation of tested drug molecules
The tested compounds were prepared on MOE in the form of 3D model library. After checking their structures and the formal charges on atoms by the 2D depiction, these compounds were subjected to energy minimization process. Then the partial charges were automatically calculated. Finally, the tested compounds libraries were saved in the form of an MDB file to be used in the docking calculations with target COVID-19 proteins.
2.1.3. Validation of docking
Calculating the root mean square deviation (RMSD) was used as a tool for the validation of the docking protocol for proteins having co-ligand in their active sites. The RMSD is predicted via redocking of the co-ligand on its target protein then superimposing the redocked co-ligand on its original co-crystallized bounded conformation. The RMSD of 3CLpro (6LU7) appeared in the acceptable range with value equal to 0.95 Å. The SARS-CoV-2 known spike receptor inhibitors (Nafamostat) was collected from literature and compared the selected hits with this known inhibitor. (Hoffmann et al., 2020)
2.2. Pharmacophore generation
Ligand-based pharmacophore models were generated from training set given from the molecular docking simulation process. The training set was prepared to generate bioactive conformers and give the suitable ionization states according to the given protocols in MOE 2019.0102. Queries of pharmacophore were generated according to pharmacophore elucidation protocol of MOE 2019.0102 with the default settings. The given results were clustered according the overlap and they were selected for further verification by fitting to a decoy training set. Consequently, Gunner-Henry (GH) score can be calculated through specification of total molecules (D), total actives (A), total hits (Ht) and true positives (Ha). Then, we can determine (GH-score) according to this equation (Lin et al., 2019). The chosen query within all given pharmacophore model should achieve the highest GH value which is not less than 0.7.
2.3. Virtual screening
2.3.1. Virtual screening by pharmacophore models
ChEMBL database was downloaded and prepared using MOE 2019.0102 by molecule database wash protocol to adjust the ionization state of compounds and rebuild the 3D structure. The pharmacophore searching of the prepared database was performed using first the pharmacophore of 3CLpro by the protocol of pharmacophore searching of MOE 2019.0102. The given hits were clustered by the choosing from the Hits drop down menu “Best per conformation”. During this process RMSD values were asked to be calculated. Consequently, the output database was subjected to searching with S protein pharmacophore with the same protocol of the previous step.
Four-point pharmacophore model was generated from the training set of all compounds used for pharmacophore generation by the protocol given from MOE 2019.0102. First, we calculate the field with the scores of four-point pharmacophore model by Compute, Fingerprint then calculate. This model can be saved from Compute, Model then Fingerprint. This model will be saved as a separate file with respect to that the score should be minimum and tanimoto score should be selected. The output database from the second step of pharmacophore searching would be subjected to this fingerprint model by using Compute, model then evaluate followed by selection of the file of the fingerprint model to predict the activity. The given result was used to rank the compounds and the best two results were selected for further investigation by molecular docking simulations.
2.4. Molecular dynamics
All atom molecular dynamics simulation was performed for the selected protein-ligand complexes using GROMACS 2020.3. (Abraham et al., 2015) The calculations were done on GPU enabled system using NVIDIA GeForce GTX 1050 Ti GPU. Charmm36 all-atom force field (Lee et al., 2016) was used to generate protein topology while ligands were parameterized using SwissParam server (Zoete et al., 2011). Complexes were built using ligand coordination obtained from docking studies then complexes were boxed in a dodecahedron box and solvated with TIP3P (Mark and Nilsson, 2001) explicit water molecules. Systems were neutralized by the addition of required number of Na+ or Cl− ions. Systems energy was minimized using steepest descent algorithm with a maximum force of 1000 kJ mol−1 nm−1. Equilibration using NVT and NPT ensembles for 100 ps each was done afterward then production run was done for 50 ns. Temperature was controlled at 300 K using the V-rescale algorithm (Bussi et al., 2007) while pressure was maintained using the Parrinello-Rahman barostat (Parrinello and Rahman, 1981) as required. The LINear Constraint Solver (LINCS) algorithm (Hess et al., 1997) was used for bond's length constraints and Particle mesh Ewald (PME) method (Darden et al., 1993) was used for long-range electrostatics calculations. For all simulations, 2 fs timestep was used. Van der Waals cut-off distance (rvdw) was set to 1.2 nm. Trajectories from the production run were used for analysis after correction of periodic boundary condition (PBC) using trajconv.
3. Results and discussion
3.1. Generation of pharmacophore of 3CL pro and spike S protein of SARS-CoV-2
3.1.1. Molecular docking on both 3CLpro and spike S protein to obtain active ligands
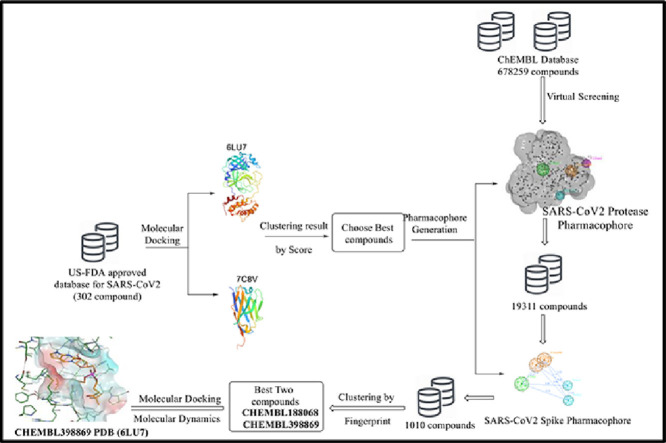
In this study, an immediately qualifying strategy is constructed to use a small molecule database library of compounds (Antiviral Library from Selleck, Catalog No.L7000) consisting of 302 drug molecules including existing powerful drugs from various virus treatments in addition to other pharmacologically active drugs that have been approved by the FDA for other indications after filtration and reformatting process. Molecular docking simulations were performed for this database on both 3CLpro and Spike S protein to obtain the best ligands from which we would generate two different pharmacophores as shown in Fig. 1 .
Fig. 1.
Protocol for the generation of pharmacophore of both main protease 3CLpro and spike s protein of SARS-CoV-2.
During this study, it was observed that N3 ligand had the ability to bind group of amino acid residues of 3CLpro receptor binding site via covalent linkage with Cys145 and non-covalent linkage through both hydrogen bonding that includes His163, His164, Gln189, Glu166, Thr190 and hydrophobic interactions including His41. Literature survey, showed in addition more important residues like Phe140, Leu141, Asn142, His164, Met165, His172, and Gln192 that interact with the ligand. Therefore, all of these amino acid residues can be considered as crucial interaction points of 3CLpro receptor binding site (Chatterjee et al., 2020). Molecular docking simulation illustrated that compounds of Table 1 showed binding poses with a group of the N3 binding amino acid residues with moderate acceptable binding scores ranging from -8.1703 to -6.6102. Verbascoside and orientin that were isolated from various plant origins like Cichorium intybus (chicory) (Barry, 1998) and Phyllostachys species (bamboo leaves) (Cho et al., 2008), respectively are reported as natural antiviral products (Kalita et al., 2012; Shawky et al., 2020). They showed proper binding poses that resemble N3 ligand binding amino acid residues. They fit the pocket by non-covalent interactions including hydrogen bonding and hydrophobic interactions. In addition, they display the highest binding scores against 3CLpro in this study with values equal to -8.1703 and -8.1212, respectively (Table 1).
Table 1.
Results of the assumed promising compounds docked against main protease 3CLpro.
| Compound name | Structure | Scoring | Binding residues |
|---|---|---|---|
| Verbascoside | -8.1703 | His163(a), Gly143(a), Gln189(b), Thr25(b) | |
| Orientin | -8.1212 | Thr190(a), Gln189(b) | |
| 1,2,3,4,6-o-Pentagalloylglucose | -7.6110 | Glu166(a), Met165(b) | |
| Rutin | -7.0150 | Leu141(a), Met165(b) | |
| Dehydroandrographolide | -6.9562 | His163(a), Glu166(a) | |
| Ginsenoside | -6.8529 | Glu166(a), Thr190(a) | |
| Epicatechin | -6.7489 | His164(a) | |
| Darunavir | -6.7299 | His163(a), Thr45(a) | |
| Remdesivir | -6.6101 | Gly143(a), Thr24(a), His41(b) |
(a) = Hydrogen bonding, (b) = Hydrophobic interactions
Regarding the molecular docking simulations against S protein, studying the obtained results revealed that the drug molecules Amentaflavone, Isochlorogenic acid, Scutellarin, Remdesivir, Letermovir, Hydroxychloroquine, Baicalin, Fosamprenavir and Doxorubicin exhibit different proper binding poses with a group of the Spike protein S amino acid residues with acceptable binding scores ranging from -6.4993 to -2.3514 as shown in Table 2 . The highest binding score records were observed for Amentaflavone and Isochlorogenic acid, followed by Scutellarin, Remdesivir and Letermovir that appeared with slight little binding values as shown in Table 2. A group of Spike protein amino acid residues appeared as common binding centers for the previously mentioned drug molecules that could be considered as promising targets for constructing common interaction points such as Arg403, Tyr505, Tyr453 and Glu406. The interactions between the previous structures and the amino acids residues were diverse between hydrogen bonding and hydrophobic interactions. Remdesivir, Amentaflavone and Scutellarin binding poses showed the best binding interaction with Spike protein amino acid residues including large number of residues involvement regardless of binding score values. Finally, it was observed that small and moderate sized molecules only were favored as suitable sized molecules by the MOE program for the Spike protein binding site which appear to be a narrow small pocket in contrast to 3CL-pro. Interestingly, this group of drug molecules are reported as potent antiviral scaffolds. Amentoflavone and Scutellarin are naturally extracted from Torreya nucifera and Scutellaria Baicalensis (Idrees et al., 2020). They were previously tested against SARS-CoV species and proved potent antiviral effect especially Scutellarin as reported. (Fuzimoto and Isidoro, 2020) Remdesivir also was reported as effectively inhibitor for the recently emerged novel coronavirus (2019-SARS-CoV-2) in vitro and for the previously explored SARS-CoV and MERS-CoV. Based on previously mentioned results and literature survey these promising molecules need to be furtherly studied against COVID-19.
Table 2.
Results of the assumed promising compounds docked against Spike protein.
| Compound name | Structure | Scoring | Binding residues |
|---|---|---|---|
| Amentoflavone | -6.4993 | Glu406(a), Tyr505(b), Leu455(b), Arg403(b) | |
| Isochlorogenic | -5.7526 | Arg403(a), Tyr453(a), Asn501(a) | |
| Scutellarin | -4.1546 | Arg403(a), Ser494(a), Tyr453(a), Tyr505(b) | |
| Remdesivir | -4.0641 | Glu406(a), Gln409(a), Lys417(a), Arg403(a), Tyr505(a), Tyr505(b) | |
| Letermovir | -3.7321 | Tyr495(a), Tyr505(b) | |
| Hydroxychloroquine | -3.3509 | Glu406(a), Arg403(a), Tyr505(b) | |
| Baicalin | -2.7901 | Arg403(a), Tyr505(a), Tyr505(b) | |
| Fosamprenavir | -2.7319 | Arg403(a), Tyr505(a) | |
| Doxorubicin | -2.3514 | Glu406(a), Arg403(a), Tyr505(a), Tyr505(b) |
(a) = Hydrogen bonding, (b) = Hydrophobic interaction
3.1.2. Pharmacophore generation
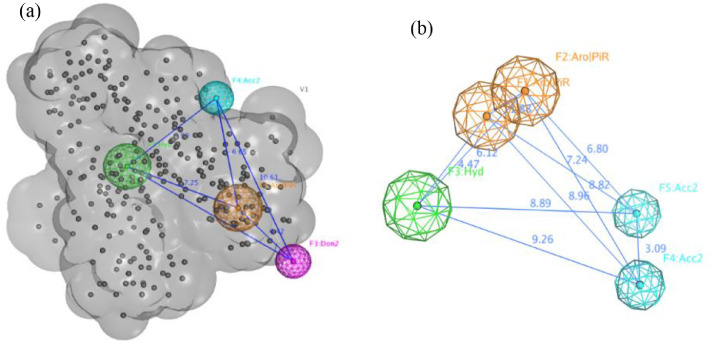
The given ligands from the aforementioned docking simulations were utilized as a training set for generation of both pharmacophores for both protease and spike protein of SARS-CoV-2. The generated pharmacophore for 3CLpro of SARS-CoV-2 was consisted of four features (HBA, HBD, Hyd. and Aro.) surrounded by the ligand volume (Fig. 2 ). On the other hand, the generated pharmacophore for S protein of SARS-CoV-2 illustrated 5 features as 2 of them are aromatic related to fused heterocyclic rings in addition to 2 HBA. and 1 Hyd. (Fig. 2). The given pharmacophores were validated by refitting of a decoy database, contacting all the active generated ligands in addition to inactive ligands to insure their filtration ability by calculation of Gunner-Henry score which is 0.762 for 3CLpro pharmacophore and 0.815 for Spike pharmacophore (Edgar et al., 2000; Guner and Bowen, 2013).
Fig. 2.
(a) Generated pharmacophore for 3CLpro of SARS-CoV-2 showing 4 features (HBA (Acc.), Hyd, Aro and HBD(Don.)) with the distance between them in Å supported by ligand volume as constrictor. (b) Generated pharmacophore for Spike S protein of SARS-CoV-2 showing 5 different features (2 Aro, Hyd, and 2 HBA.).
3.2. Virtual screening of ChEMBL database
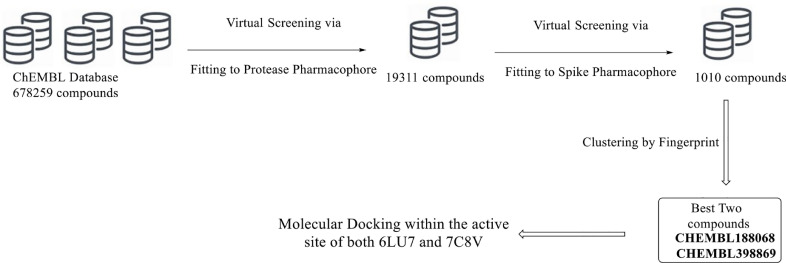
The protocol of virtual screening was performed to filter ChEMBLE database by the workflow shown in Fig. 3 . The prepared database was filtered by fitting to the generated pharmacophore of 3CLpro of SARS-CoV-2 which lead to a filtered database with 19311 compounds. This indicates that this pharmacophore has the power of filtration with about 2.8%. The output database from the first step is the subjected to another filtration process by the other pharmacophore of Spike protein. This process leads to a screened database with 1010 compound. The given RMSD value of these compounds due to fitting on the pharmacophore ranges from 0.4813 to 1.1123.
Fig. 3.
Virtual Screening protocol of ChEMBL database to obtain two compounds with potential activity against SARS-CoV-2.
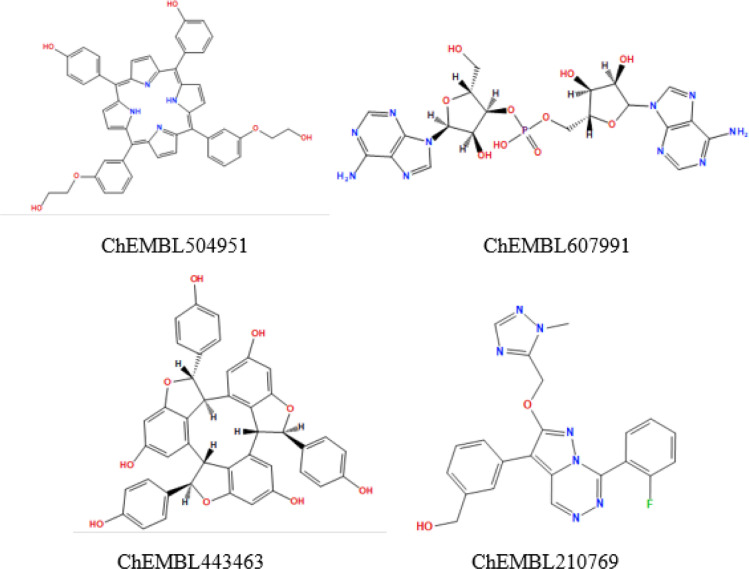
It is worthy to mention that the compounds in this database displayed four different types of chemical skeletons; nucleoside skeleton as ChEMBL607991, compounds with central 4 pyrrole rings, porphyrin skeleton, connected to each other as ChEMBL504951, compounds with central large ring system as ChEMBL443463 and compounds with fused heterocyclic ring with other aromatic and aliphatic substituents as ChEMBL210769 (Fig. 4 ). These compounds were finally clustered by a 4-point fingerprint model that was generated from the active ligands used in two pharmacophore generation encode the information for the features from them (Cereto-Massagué et al., 2015). This fingerprint model was used to predict the activity of the 1010 compounds then the best two compounds ChEMBL188068 and ChEMBL398869 (Table 3 ) were selected for the docking step for further analysis for the predicted activity.
Fig. 4.
Examples for 4 types of skeletons of the compounds from the database generated after filtration with the second pharmacophore.
Table 3.
Best compounds with the highest predictive activity according to fingerprint model.
| Compound name | Structure | Predicted activity according to fingerprint model | RMSD |
|---|---|---|---|
| ChEMBL188068 | 0.4343 | 1.0624 | |
| ChEMBL398869 | 0.4282 | 1.0838 |
3.3. Molecular docking of compounds ChEMBL188068 and ChEMBL398869
The best two ChEMBL compounds raised from the performed virtual screening against the previously prepared pharmacophores of SARS-CoV-2 main protease 3CLpro and the spike S protein were subjected to re-docking process on both targets and the given results were displayed in Table 4 .
Table 4.
Docking scores and featured interaction of the given compounds ChEMBL188068 and ChEMBL398869 when docked with both 3CLpro and S protein of SARS-CoV-2.
| Compound name | Structure | Scoring | Target protein | Binding residues |
|---|---|---|---|---|
| ChEMBL188068 | -5.9287 | Main protease 3CLpro | His163, Phe140 | |
| -8.3618 | S-protein | Tyr495 | ||
| ChEMBL398869 | -7.2007 | Main protease 3CLpro | His163, Leu141, His41, Gly143 | |
| -6.8146 | S-protein | Glu406, Tyr505 |
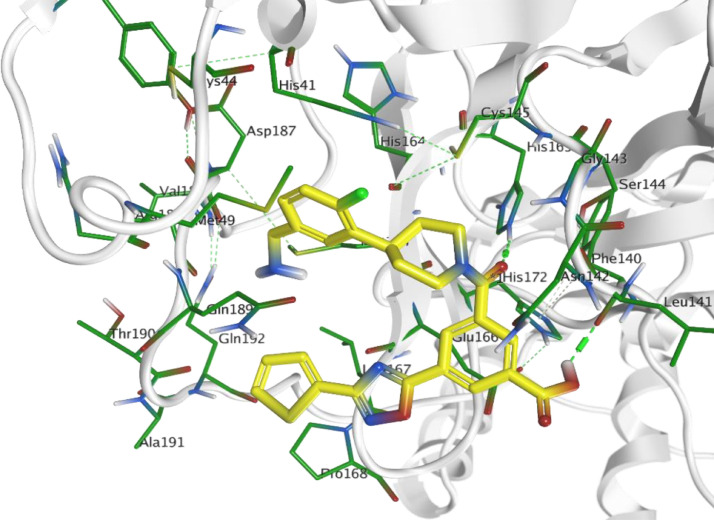
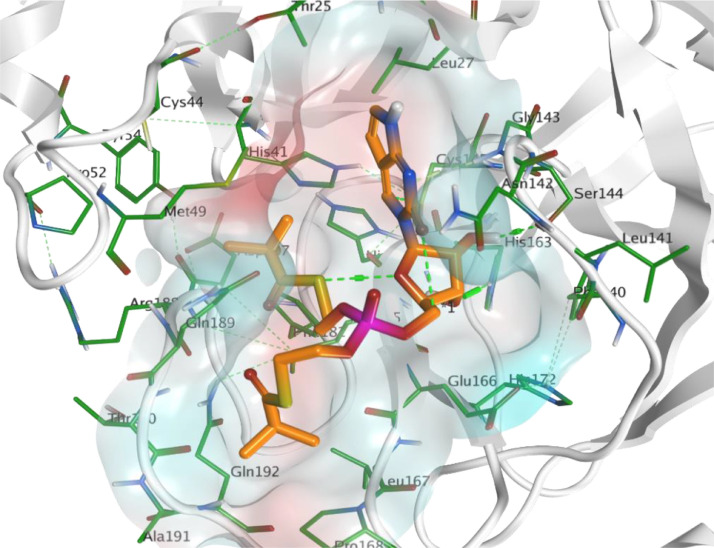
Observing the obtained results for the main protease 3CLpro target revealed that ChEMBL188068 compound succeeded to possess a binding score equal to -5.928706. It interacts within the binding pocket by H-bonding interaction with His163 and Phe140 residues. Additionally, p-fluorobenzylamine moiety was embedded in the hydrophobic region extended from Gln189 passing with Met165, His41 till Arg188 (Fig. 5 ). While, ChEMBL398869 compound showed better binding score equals to -7.200737. It possesses an observed interaction within the active site by H-bonding interaction with a group of target residues His163 and Leu141 by the hydroxyl groups of the 5-membered sugar. It was observed that there was an intramolecular interaction between S atom, in one of the extended aliphatic chains from phosphate group, and oxygen of ribose sugar. This was the mean reason for the extension of this aliphatic chain within the pocket of Gln189, Arg188, Asp187, His41 and Met165 (Fig. 6 ).
Fig. 5.
Docking pose of ChEMBL188068 (yellow) within the active site of 3CLpro (PDB: 6LU7).
Fig. 6.
Docking pose of ChEMBL398869 (orange) showing the hydrophobic extension of the aliphatic chain within the active site of 3CLpro which is surrounded by hydrophobic pocket (color index: hydrophilic: cyan and lipophilic: red) (PDB: 6LU7).
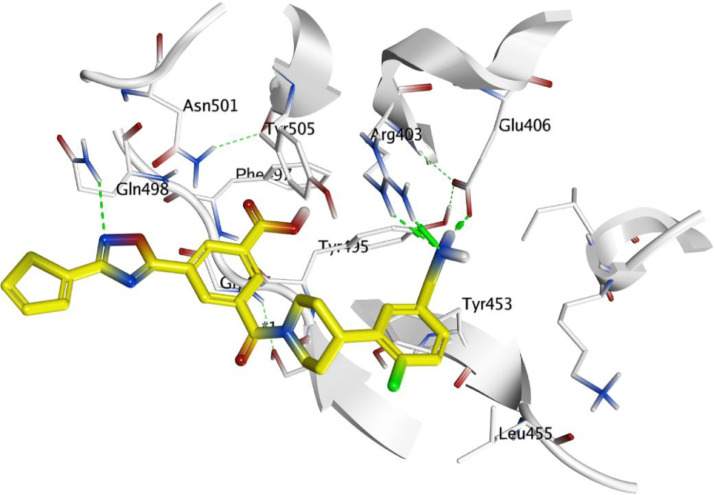
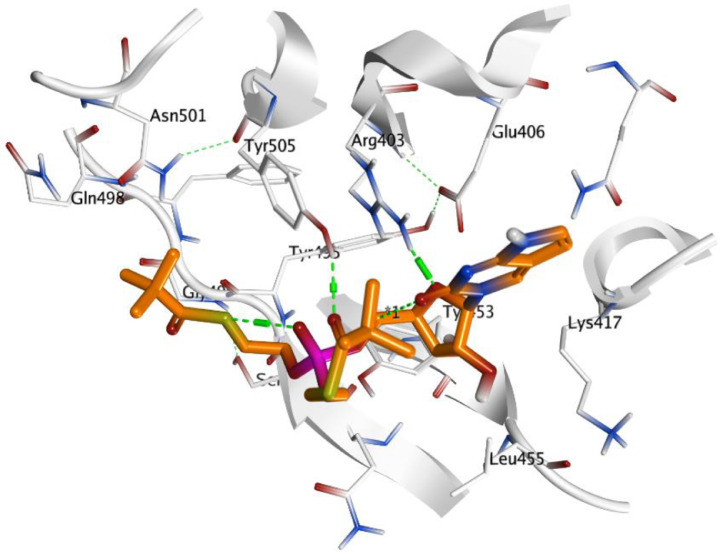
In Parallel, studying docking results for the Spike S protein target revealed moderate binding score results for both ChEMBL188068 and ChEMBL398869 compounds were -8.3618 and -6.8146, respectively. ChEMBL188068 could interact with Arg403 and Glu406 via hydrogen bond with the terminal amino of aminobenzyl moiety. The N atom of 1,2,4-oxadiazole ring can accept hydrogen bond from Gln498 as shown in Fig. 7 . While ChEMBL398869 displayed two hydrogen bonds with Arg403, Tyr505 residues through H-bonding interaction Fig. 8 .
Fig. 7.
Docking pose of ChEMBL188068 (carbon marked with yellow) within the active site of S protein (PDB: 7C8V).
Fig. 8.
Docking pose of ChEMBL398869 (carbon marked with orange) within the active site S protein (PDB: 7C8V).
3.4. Molecular dynamics simulations
To study the stability of the complexes generated by docking, molecular dynamics (MD) simulations were performed. Four complexes were studied which includes ChEMBL188068 docked in the active sites of main protease (complex 1) and S-protein (Complex 2). The other 2 complexes are for ChEMBL398869 in the active sites of main protease (complex 3) and in the active sites of S-protein (Complex 4). All complexes (Table 5 ) were initially boxed in dodecahedron boxes, solvated with TIP3P explicit water molecules and the system was neutralized. Minimization and equilibration with NVT and NPT ensembles were done before the production run. The four studied complexes were run for 50 ns and then the collected trajectories were analyzed. A summary of the molecular dynamics simulation results for the four complexes is shown in Table 5.
Table 5.
Summary of the molecular dynamics (MD) simulation results.
| Complex 1 | Complex 2 | Complex 3 | Complex 4 | |
|---|---|---|---|---|
| Ligand | ChEMBL188068 | ChEMBL188068 | ChEMBL398869 | ChEMBL398869 |
| Target | Main protease | S-protein | Main protease | S-protein |
| Average Heavy atoms RMSD (nm) | 1.19 | 1.27 | 0.49 | 2.60 |
| Average Number of H-Bond | 1.1 | 1.5 | 2.5 | 0.7 |
| Radius of gyration (Å) | 22.5363 | 18.2784 | 22.5389 | 18.26299 |
| Average RMSF (Å) | 1.35 | 1.05 | 1.42 | 1.06 |
| Binding Energy (KJ/mol) | 207.31 ± 20.4 | 120.01 ± 22.2 | 206.35 ± 15.43 | 82.29 ± 20.6 |
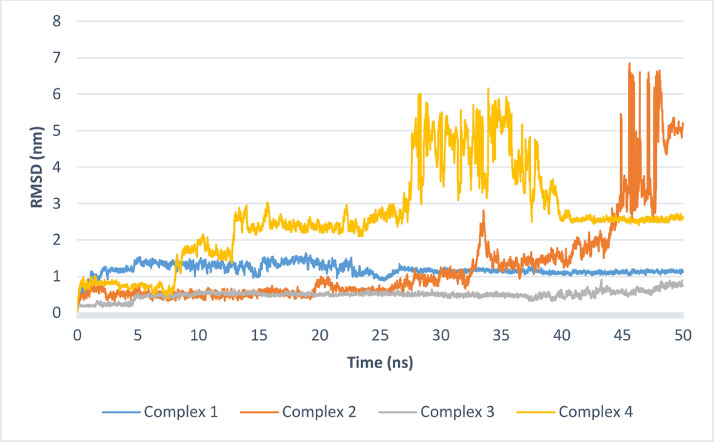
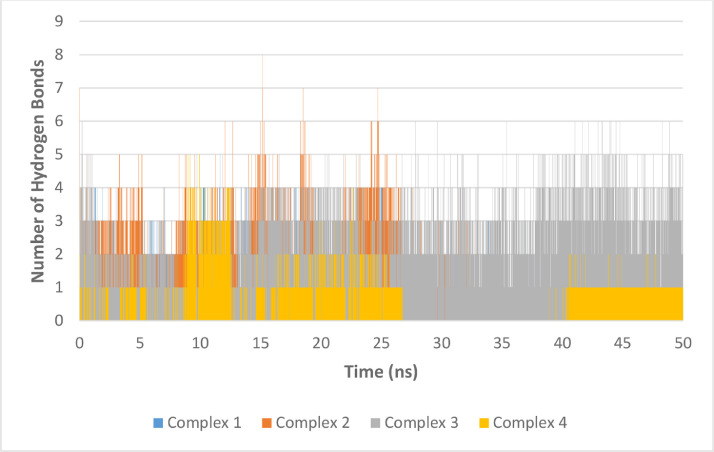
To investigate the stability of the studied complexes several factors are examined after extraction from the production run trajectories. A plot of the root mean square deviation (RMSD) during production run (Fig. 9 ) demonstrate the stability of the complex. Among the four complexes, complexes with the main protease (complexes 1 and 3) showed better stability and less RMSD variability compared to the other two complexes. Complex 3 was more stable with average RMSD of about half that of complex 1 (0.49 nm for complex 3 versus 1.19 nm for complex 3). Although these RMSD values are slightly high (especially with complex 1), The coefficient of variation in the RMSD values are 13.1 and 22.9 % which suggest low fluctuation. Complex 2 initially showed good stability but the RMSD started to increase after 30 ns. Also, the number of hydrogen bonds formed between the ligand and protein were investigated during the MD run and complex 3 showed the highest number of hydrogen bonds during the production run ( Fig. 10 ).
Fig. 9.
RMSD of ligand heavy atoms during the production run for the 4 complexes studied.
Fig. 10.
Number of hydrogen bonds formed between the ligand and targets in each complex during the production run.
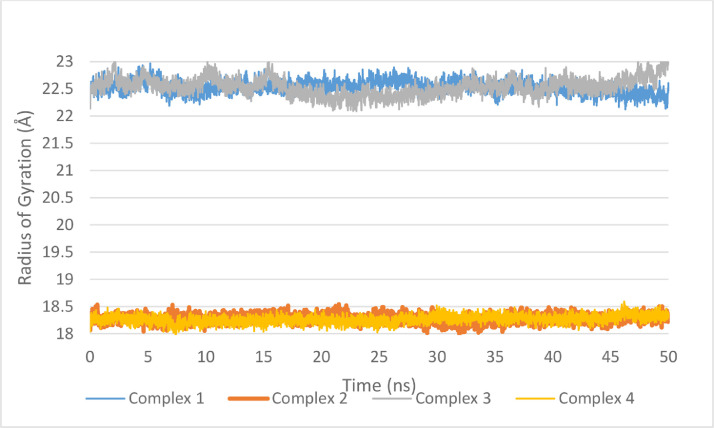
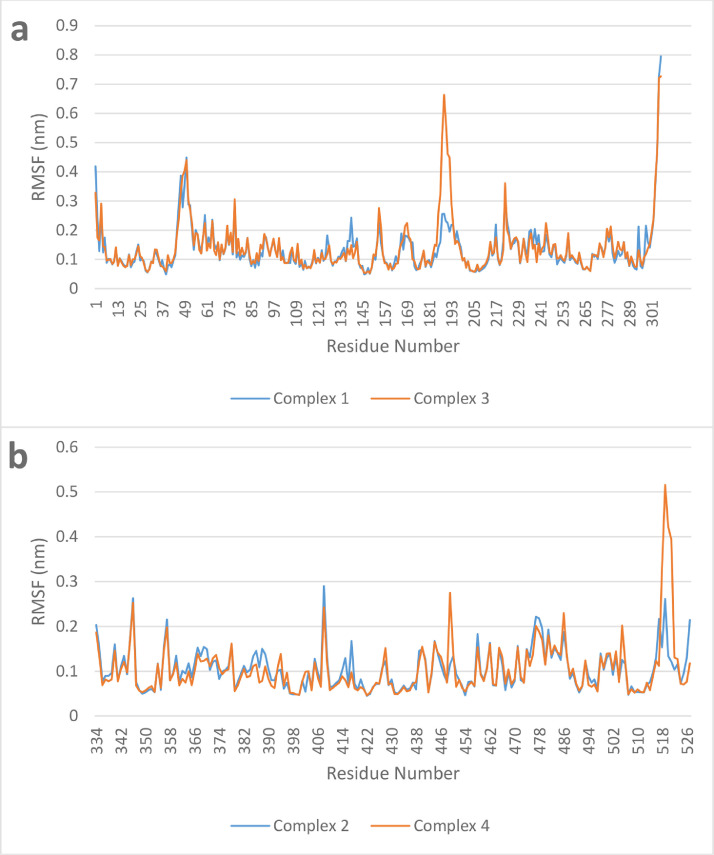
Compound ChEMBL398869 in the active site of main protease (Complex 3) also showed the best results with an average of 2.5 hydrogen bonds during the run which is 1 hydrogen bond higher than the next complex, complex 2. Furthermore, radius of gyration is a measure of structure compactness. Higher radius of gyration (Rg) indicates structure distortion after insertion of the ligand. All four complexes have shown stable Rg during the whole production run as can be seen in Fig. 11 . In addition, protein residue root mean square fluctuation (RMSF) can give an indication of the residues involved in binding and their move during the MD run to accommodate the ligand. For example, if we compare the RMSF of both compounds in the active site of main protease (Fig. 12 a), we can see that the RMSF is very similar except the loop formed around Gln189 (186-193). This indicate that the Gln189 has changed its structure to accommodate the ligand better. This agrees well with the results reported earlier indicating the plasticity of S2 subsite due to the conformational stability of Gln189.(Zhang et al., 2020a; Zhang et al., 2020b) Comparison of the RMSF of S-protein residues in complex 2 and 4 (Fig. 12b), on the other hand, does not show much change in the active site area. This structural change is a very important hallmark of main protease that could be very useful during the design of selective inhibitor against SARS-CoV-2 main protease.
Fig. 11.
Radius of gyration for each complex during the production run.
Fig. 12.
Root mean square fluctuation (RMSF) of each residue in the studied complexes. (a) SARS-Cov-2 main protease. (b) SARS-CoV-2 S-protein.
The results of the MD simulation suggest and support that both of our fished compound, ChEMBL398869, could be a promising SARS-CoV-2 main protease inhibitor and worth further study and biological testing.
4. Conclusion
In this study, virtual screening of ChEMBL database was performed using two generated pharmacophores for both targets 3CLpro and S protein of SARS-CoV2. These two pharmacophores were generated by a training set selected from docking results of US-FDA approved database for COVID-19 treatment. This process was followed by fingerprint model to obtain two compounds ChEMBL188068 and ChEMBL398869 that passes all these steps. Molecular dynamic simulations proved the stability of ChEMBL3988969 in the pocket of 3CLpro. It possesses significant H-bonds with His163 and Leu141. In addition to the extension of the side aliphatic chain of this compound within the pocket of Gln189, Arg188, Asp187, His41 and Met165. Molecular dynamic simulations of this compound within the main protease illustrated the importance of Gln189 flexibility in inhibitors recognition through increasing S2 subsite plasticity. According to this study, ChEMBL3988969 is considered as a novel candidate for screening as inhibitor against SARS-CoV2 main protease.
CRediT authorship contribution statement
Mohamed A. Said: Conceptualization, Methodology, Writing - original draft, Writing - review & editing. Amgad Albohy: Methodology, Software, Writing - original draft, Writing - review & editing. Mohamed A Abdelrahman: Methodology. Hany S. Ibrahim: Conceptualization, Methodology, Writing - original draft, Supervision, Writing - review & editing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ejps.2021.105744.
Appendix. Supplementary materials
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softw.X. 2015;1:19–25. [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry T. The feeding value of chicory (Cichorium intybus) for ruminant livestock. J. Agric. Sci. 1998;131:251–257. doi: 10.1017/S002185969800584X. [DOI] [Google Scholar]
- Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereto-Massagué A., Ojeda M.J., Valls C., Mulero M., Garcia-Vallvé S., Pujadas G. Molecular fingerprint similarity search in virtual screening. Methods. 2015;71:58–63. doi: 10.1016/j.ymeth.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Chakraborti, S., Bheemireddy, S., Srinivasan, N., 2020. Repurposing drugs against main protease of SARS-CoV-2: mechanism based insights supported by available laboratory and clinical data. http://doi.org/10.26434/chemrxiv.12057846.v2. [DOI] [PubMed]
- Chatterjee S., Maity A., Chowdhury S., Islam M.A., Muttinini R.K., Sen D. In silico analysis and identification of promising hits against 2019 novel coronavirus 3C-like main protease enzyme. J. Biomol. Struct. Dynam. 2020:1–14. doi: 10.1080/07391102.2020.1787228. [DOI] [PubMed] [Google Scholar]
- Cho S.-H., Choi Y.-J., Rho C.-W., Choi C.-Y., Kim D.-S., Cho S.-H. Reactive oxygen species and cytotoxicity of bamboo (phyllostachys pubescens) sap. Korean J. Food Preserv. 2008;15:105–110. [Google Scholar]
- Clark A., Jit M., Warren-Gash C., Guthrie B., Wang H.H., Mercer S.W., Sanderson C., McKee M., Troeger C., Ong K.L. Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: a modelling study. Lancet Glob. Health. 2020;8:e1003–e1017. doi: 10.1016/S2214-109X(20)30264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jiménez-Guardeño J.M., Regla-Nava J.A., Alvarez E., Oliveros J.C., Zhao J., Fett C., Perlman S., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diukendjieva A., Zaharieva M.M., Mori M., Alov P., Tsakovska I., Pencheva T., Najdenski H., Křen V., Felici C., Bufalieri F. Dual SMO/BRAF inhibition by Flavonolignans from Silybum marianum. Antioxidants. 2020;9:384. doi: 10.3390/antiox9050384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar S.J., Holliday J.D., Willett P. Effectiveness of retrieval in similarity searches of chemical databases: a review of performance measures. J. Mol. Graph. Modell. 2000;18:343–357. doi: 10.1016/S1093-3263(00)00061-9. [DOI] [PubMed] [Google Scholar]
- Fuzimoto A.D., Isidoro C. The antiviral and the coronavirus-host protein pathways inhibiting properties of herbs and natural compounds-additional weapons in the fight against the COVID-19 pandemic? J. Tradition. Complement. Med. 2020 doi: 10.1016/j.jtcme.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guner O.F., Bowen J.P. Pharmacophore modeling for ADME. Curr. Top. Med. Chem. 2013;13:1327–1342. doi: 10.2174/15680266113139990037. [DOI] [PubMed] [Google Scholar]
- Haines B.M., Shah R.C., Smidt J.M., Albright B.J., Cardenas T., Douglas M.R., Forrest C., Glebov V.Y., Gunderson M.A., Hamilton C.E. Observation of persistent species temperature separation in inertial confinement fusion mixtures. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-14412-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. https://doi.org/10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [Google Scholar]
- Hodos R.A., Kidd B.A., Khader S., Readhead B.P., Dudley J.T. Computational approaches to drug repurposing and pharmacology. Wiley interdisciplinary reviews. Syst. Biol. Med. 2016;8:186. doi: 10.1002/wsbm.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Schroeder S., Kleine-Weber H., Müller M.A., Drosten C., Pöhlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/aac.00754-20. e00754-00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T., Wang H., Luan B. In silico exploration of the molecular mechanism of clinically oriented drugs for possibly inhibiting SARS-CoV-2’s main protease. J. Phys. Chem. Lett. 2020;11:4413–4420. doi: 10.1021/acs.jpclett.0c00994. [DOI] [PubMed] [Google Scholar]
- Huynh T., Wang H., Luan B. Structure-based lead optimization of herbal medicine rutin for inhibiting SARS-CoV-2′s main protease. Phys. Chem. Chem. Phys. 2020;22:25335–25343. doi: 10.1039/D0CP03867A. [DOI] [PubMed] [Google Scholar]
- Idrees M., Khan S., Memon N.H., Zhang Z. Effect of the phytochemical agents against the SARS-CoV and some of them selected for application to COVID-19: a mini-review. Curr. Pharm. Biotechnol. 2020 doi: 10.2174/1389201021666200703201458. [DOI] [PubMed] [Google Scholar]
- Kalita S., Kumar G., Karthik L., Rao K.V.B. A review on medicinal properties of lantana camara linn. Res. J. Pharm. Technol. 2012;5:711–715. [Google Scholar]
- Khan R.J., Jha R.K., Amera G.M., Jain M., Singh E., Pathak A., Singh R.P., Muthukumaran J., Singh A.K. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J. Biomol. Struct. Dynam. 2020:1–14. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.-Y., Ho Y., Liu H.-L. Structure-based pharmacophore modeling to discover novel CCR5 inhibitors for HIV-1/cancers therapy. J. Biomed. Sci. Eng. 2019;12:10–30. http://10.4236/jbise.2019.121002 [Google Scholar]
- Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105:9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- Nieto-Torres J.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- Peele K.A., Potla Durthi C., Srihansa T., Krupanidhi S., Ayyagari V.S., Babu D.J., Indira M., Reddy A.R., Venkateswarulu T.C. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Inf. Med. Unlocked. 2020;19 doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., Norris A., Sanseau P., Cavalla D., Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. http://10.1038/nrd.2018.168 [DOI] [PubMed] [Google Scholar]
- Saxena A. Drug targets for COVID-19 therapeutics: ongoing global efforts. J. Biosci. 2020;45:87. doi: 10.1007/s12038-020-00067-w. http://10.1007/s12038-020-00067-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D., Debnath P., Debnath B., Bhaumik S., Debnath S. Identification of potential inhibitors of SARS-CoV-2 main protease and spike receptor from 10 important spices through structure-based virtual screening and molecular dynamic study. J. Biomol. Struct. Dynam. 2020:1–22. doi: 10.1080/07391102.2020.1819883. http://10.1080/07391102.2020.1819883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawky E., Nada A.A., Ibrahim R.S. Potential role of medicinal plants and their constituents in the mitigation of SARS-CoV-2: identifying related therapeutic targets using network pharmacology and molecular docking analyses. RSC Adv. 2020;10:27961–27983. doi: 10.1039/d0ra05126h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Yan M., Xu H., Liang W., Kan B., Zheng B., Chen H., Zheng H., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Liu Y.-F., Guo R.-T., Liu X.-N., Zhan L.-H., Zhou D.-H., Zhao A., Hai R., Yu D., Guan Y., Xu J. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect Dis. 2005;11:1860–1865. doi: 10.3201/eid1112.041293. http://10.3201/eid1112.041293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A., van BS, Bestebroer TM, Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. http://10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. http://10.1021/acs.jmedchem.9b01828 [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. http://10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Huang J. Current findings regarding natural components with potential anti-2019-nCoV activity. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.00589. http://10.3389/fcell.2020.00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Lou Y.X., Wang X., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. http://10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Curtis N. Coronavirus infections in children including COVID-19: an overview of the epidemiology, clinical features, diagnosis, treatment and prevention options in children. Pediatr. Infect Dis. J. 2020;39:355–368. doi: 10.1097/INF.0000000000002660. http://10.1097/INF.0000000000002660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V., Cuendet M.A., Grosdidier A., Michielin O. Swiss Param: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.