Pediatric acute myeloid leukemia (AML) is a rare, heterogeneous disease, characterized by recurrent cytogenetic and molecular aberrations.1,2 Genetic aberrations are the most important prognostic factor, besides early response to treatment.3,4 In pediatric AML, improvement of clinical outcome has reached a plateau, despite intensive chemotherapy and allogeneic hematopoietic stem cell transplantation (HSCT) in selected cases, leading to event-free survival rates of approximately 55-60% in the current era.1,2,5 Hence, identification of prognostic subgroups is important for risk-group stratification.1,5 Furthermore, novel therapeutic options should be explored for pediatric AML, with the objective of improving survival especially for patients with high-risk subtypes, such as those with the NUP98-NSD1 rearrangement (NUP98-KDM5A+).6,7
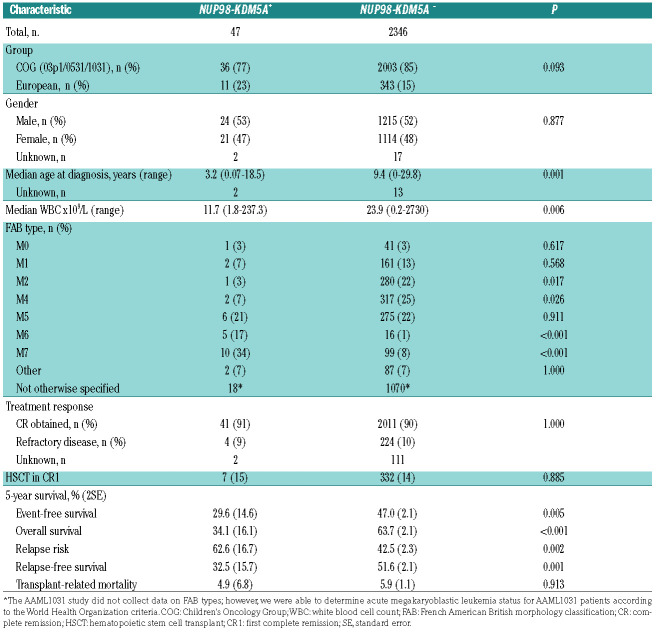
Here, we present the results of a collaborative study between the Children’s Oncology Group (COG) and European AML study groups, aimed to define the biological and clinical characteristics of AML patients carrying NUP98-KDM5A outside acute megakaryoblastic leukemia. The methodology and analysis are described in detail in the Online Supplementary Methods. In total 2,393 patients were included from various trials by the COG, AIEOP (Associazione Italiana di Ematologia e Oncologia Pediatrica), BFM (Berlin-Frankfurt-Münster) group, CPH (Czech Pediatric Hematology Working Group), DCOG (Dutch Childhood Oncology Group) and LAME (Leucémie Aiquë Myéloblastique Enfant). NUP98- KDM5A rearrangements were detected in 47/2,393 pediatric AML cases (2.0%) with similar prevalence in the COG and European cohorts (Table 1). The median age of the NUP98-KDM5A+ patients was significantly lower than that of patients without this fusion gene (3.2 vs. 9.4 years; P=0.001) (Table 1, Online Supplementary Figure S1A). The median white blood cell count of NUP98KDM5A+ patients was significantly lower than that of NUP98-KDM5A− patients (11.7x109/L vs. 23.9x109/L, P=0.006). Previously described as a recurrent rearrangement in acute megakaryoblastic leukemia, this study identified NUP98-KDM5A in all French-American-British (FAB) types of AML, with the rearrangement being present most frequently in M7 (34%), M5 (21%) and M6 (17%) (Online Supplementary Figure S1B).8 Interestingly, these characteristics were different from those previously described for NUP98-NSD1-rearranged pediatric AML (n=37), in which patients had a median age of 10.4 years (range, 1.2-19.4), a median white blood cell count of 181.2x109/L, and the rearrangement was most frequent in FAB types M4/M5 (51%).7,9
Table 1.
Clinical characteristics of pediatric patients with or without NUP98-KDM5A rearrangements.
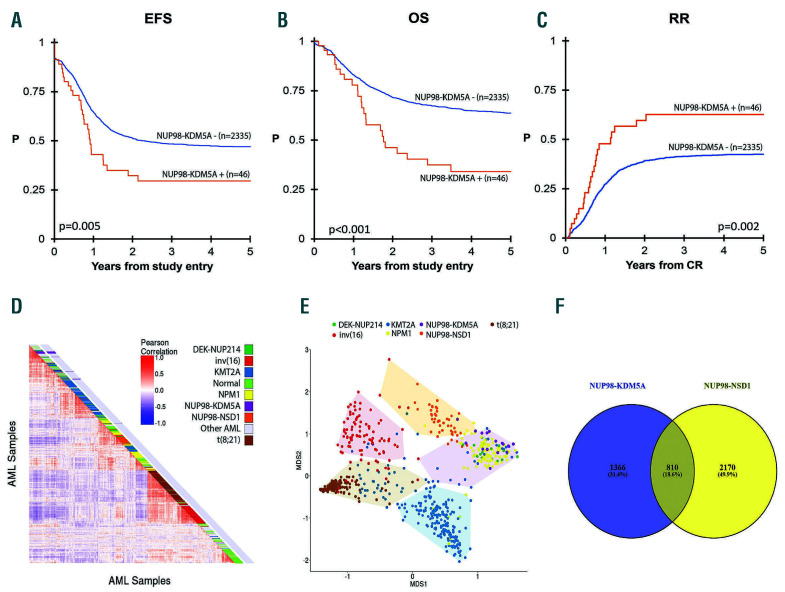
Figure 1.
Survival and gene expression of NUP98-KDM5A+ and NUP98-KDM5- acute myeloid leukemia. (A) Kaplan-Meier survival curve of event-free survival (EFS) of NUP98-KDM5A+ versus NUP98-KDM5– patients. (B) Kaplan-Meier survival curve of overall survival (OS) of NUP98-KDM5A+ versus NUP98-KDM5A– patients. (C) Relapse risk (RR) of NUP98-KDM5A+ versus NUP98-KDM5A– patients. (D) Unsupervised hierarchical clustering analysis by pairwise sample correlations (Pearson R). (E) HOX expression-based clustering using principal component analysis. The five distinct groups were determined using K-means clustering and depicted in convex hulls. (F) Venn diagram of differentially expressed genes in NUP98-KDM5A+ and NUP98-NSD1– cases as compared to other subtypes of acute myeloid leukemia (excluding those with NUP98 rearrangements).
In concordance with other studies, we found that NUP98-KDM5A+ patients lacked other common AML fusions.8,10 Mutations in genes that are recurrently mutated in other AML subtypes, such as RAS, WT1 and FLT3, occurred with very low frequency in NUP98-KDM5A+ cases, suggesting that the fusion itself may have a sufficient transforming effect (Online Supplementary Table S1).
There was not a significant difference in complete remission rates between NUP98-KDM5A+ and NUP98- KDM5A− patients (Table 1). Minimal residual disease (MRD) data were available for 31 NUP98-KDM5A+ patients; 17/31 (55%) NUP98-KDM5A+ patients were MRD+ (>10-3 blasts detected) at the end of induction, compared to 471/1740 NUP98-KDM5A− patients (27%, P<0.001). MRD status did not clearly influence outcome, as survival rates of NUP98-KDM5A+ patients were similar between MRD+ patients (4-year event-free survival of 27% ± 25.4% and overall survival of 29.9% ± 31.7%), and MRD– patients (4-year event-free survival of 36% ± 25.6% [P=0.30] and overall survival of 42.9% ± 26.5% [P=0.65]) (Online Supplementary Figure S2A and B). These survival rates were comparable to those of NUP98- KDM5A− but MRD+ patients (5-year event-free survival of 30.8% ± 3.4% and overall survival of 48.5% ± 3.7%), and significantly lower than survival rates of NUP98- KDM5A− patients who were MRD– (5-year event-free survival of 58.3% ± 2.2% [P<0.001] and overall survival of 73.3% ± 2.0% [P<0.001]). However, as the numbers of NUP98-KDM5A+ patients with available MRD data were low, definitive conclusions cannot be drawn.
Event-free and overall survival rates of NUP98- KDM5A+ patients were nearly superimposable with 5-year survival rates of 29.6% ± 14.6%, and 34.1% ± 16.1%, respectively. This illustrates that NUP98- KDM5A+ AML is more difficult to rescue than other AML subtypes. The relapse risk of NUP98-KDM5A+ patients was 62.6% ± 16.7%. These outcomes were significantly worse when compared to those of NUP98-KDM5A patients who had a 5-year event-free survival of 47.0% ± 2.1% (P=0.005), overall survival of 63.7% ± 2.1% (P≤0.001), and a relapse risk of 42.5 ± 2.3 (P=0.002) (Figure 1A-C). Previously described NUP98-NSD1+ cases had similar outcomes to NUP98-KDM5A+ patients cases, with a 5-year event-free survival rate of 22.2% ± 6.9% and an overall survival rate of 45.7% ± 8.7%.7,9 Based on multivariable analysis, NUP98-KDM5A was an independent risk factor for poor overall survival and relapse, with hazard ratios of 1.77 for overall survival (P=0.009) and 1.62 for relapse risk (P=0.049), but not for event-free survival (hazard ratio 1.40, P=0.08) (Online Supplementary Table S2). Seven NUP98-KDM5A+ patients underwent HSCT in first complete remission. Among these patients, five relapsed and four subsequently died.
Gene expression analysis using RNA-sequencing data from 1,035 patients, including 16 with NUP98-KDM5A+ and 49 with NUP98-NSD1+, revealed 2,252 differentially expressed genes between NUP98-KDM5A+ and NUP98- KDM5A− cases, of which 1,542 were upregulated genes (Online Supplementary Table S3). However, hierarchical clustering did not cluster the majority of NUP98- KDM5A+ cases together, but scattered them among samples with normal and other karyotypes (Figure 1D).
The top upregulated genes included many HOXB genes (Online Supplementary Table S3). The upregulation of both HOXA and HOXB genes has been previously reported in NUP98-rearranged cases, and represents a common pathway to leukemia development, as it is also shared with NPM1-mutated AML, t(8;16)-rearranged AML, and others. 7,11 HOXA/B-gene-based principle coordinate analysis, excluding patients with rare or unknown driving aberrations, revealed several subgroups (Figure 1E). NUP98-KDM5A clustered together with NPM1-mutated cases and DEK-NUP214, while NUP98-NSD1 clustered separately. The difference between NUP98-KDM5A and NUP98-NSD1 was further underlined by comparing gene expression profiles with other AML cases. This revealed 2,176 differentially expressed genes in NUP98-KDM5A+ cases and 2,980 differentially expressed genes in NUP98- NSD1+ cases (Figure 1F, Online Supplementary Tables S3 and S4). Among these differentially expressed genes, 810 were shared between the two groups: 68 genes were upregulated in both groups, 48 were downregulated and 694 had opposing expression profiles (Online Supplementary Table S5).
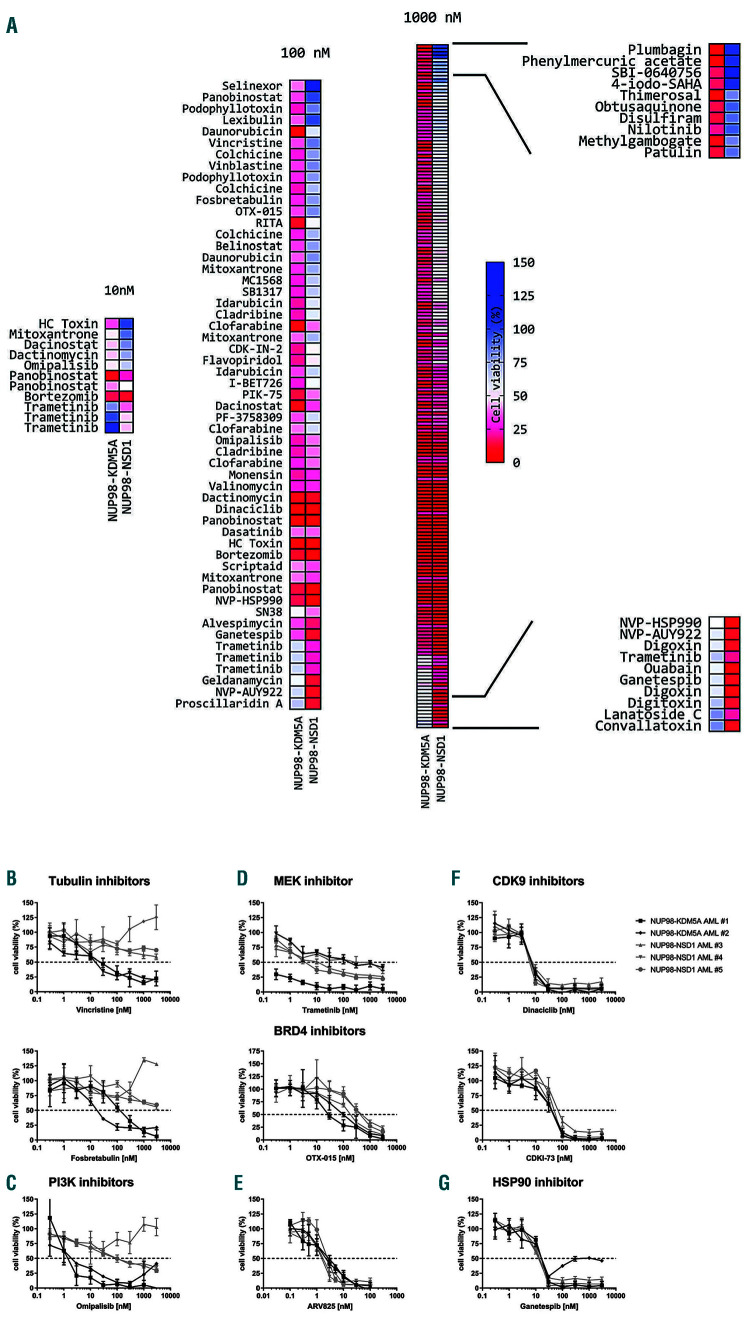
Figure 2.
Top hits of the drug screening and validation of the most promising candidate drugs on primary NUP98-KDM5A+ and NUP98-NSD1- acute myeloid leukemia samples. (A) Heatmaps of the most effective hits from the drug library screens at 10 nM, 100 nM and 1,000 nM on primary samples from a case of NUP98-KDM5A+ acute myeloid leukemia (AML) and a case of NUP98-NSD1+ AML, ranked by difference in cell viability. Drugs occurring multiple times in the heatmap indicate that the drug was present in multiple screened drug libraries. (B-G) Dose-response curves of the selected candidate drugs on primary NUP98-KDM5A+ (n=2) and NUP98-NSD1+ (n=3) AML samples to the tubulin inhibitors vincristine (top) and fosbretabulin (bottom) (B), the PI3K inhibitor omipalisib (C), the MEK inhibitor trametinib (D), the BRD4 inhibitors OTX-015 (top) and ARV825 (bottom) (E), the CDK9 inhibitors dinaciclib (top) and CDKI-73 (bottom) (F), and the HSP90 inhibitor ganetespib (G). Data are based on a 4-day MTT assay and normalized to values in controls treated with dimethylsulfoxide.
Gene set enrichment analysis of the differentially expressed genes revealed upregulation of targets of E2F and FLT3 in both NUP98-rearranged subgroups (Online Supplementary Tables S6-S9). TP53 and HDAC targets were downregulated in both rearrangements. Interestingly, STAT5, NF1 and NOTCH1 pathways and targets were upregulated in NUP98-KDM5A cases and downregulated in NUP98-NSD1 cases, whereas the MYC pathway was upregulated in NUP98-NSD1 and downregulated in NUP98-KDM5A cases. Connectivity mapping using the 90th percentile absolute log foldchange of the differentially expressed genes indicated different potential targets for treatment of NUP98-KDM5A when compared to NUP98-NSD1 cases (Online Supplementary Figure S3). Both NUP98-KDM5A and NUP98-NSD1 cases had negative median tau scores for histone deacetylase inhibitors. Furthermore, NUP98- KDM5A cases had a median tau score of -21.4 for microtubule inhibitors, indicating that these inhibitors could reverse the gene expression signature of the NUP98- rearranged patients.
In search of treatment options, high-throughput screening of more than 4,000 compounds (Online Supplementary Table S10) was performed on primary samples from a patient with NUP98-KDM5A+ and a patient with NUP98-NSD1+ AML, and revealed an overall resistance profile to different drugs (Online Supplementary Figure S4). At a drug concentration of 1,000 nM, 146 unique compounds were identified that inhibited cell viability by >70% in one or both samples. At 100 nM, 41 compounds were found to inhibit cell viability by >60%, and at 10 nM eight drugs inhibited cell viability by >50% (Figure 2A). Etoposide and cytarabine, chemotherapeutics used in standard AML treatment, were not identified as top hits at the doses of 10 nM and 100 nM. A total of nine drugs, comprising six drug target classes, namely microtubule, PI3K, MEK, HSP90, CDK9 and BRD4 inhibitors, were selected for further validation on NUP98-KDM5A+ (n=2) and NUP98-NSD1+ (n=3) primary AML samples (Online Supplementary Table S11), as well as the CHRF-288-11 cell line harboring a NUP98-KDM5A fusion (Online Supplementary Figure S5). These validation experiments confirmed the effects observed in the highthroughput drug screening (Online Supplementary Figure S2B-G and S6).
In concordance with the connectivity map analysis, tubulin inhibitors such as vincristine and fosbretabulin, decreased cell viability in vitro in NUP98-KDM5A+ cases, while showing limited cell toxicity in NUP98-NSD1+ cases (Online Supplementary Figure S7, Online Supplementary Table S12). In concordance with our pathway analysis, which showed upregulation of the STAT5 pathway in NUP98-KDM5A+ AML as compared to other types of AML and downregulation in NUP98-NSD1+ AML, the PI3K inhibitor omipalisib decreased cell viability in NUP98-KDM5A+ cases with a half maximal inhibitory concentration (IC50) of 1.6 nM - 2.2 nM), whereas it had little effect on NUP98-NSD1+ cases (IC50= 95.4 nM - 3000 nM) (Figure 2C). Trametinib, a MEK inhibitor with a manageable safety profile in pediatric patients, produced variable responses in both tested NUP98 fusions, but in all cases had an IC50 value of less than ~300 nM (Figure 2D)12. Both NUP98-fusion types responded to drugs targeting BRD4, CDK9 and HSP90, highlighting that the fusions do not solely have distinctive features but also common leukemia hallmarks (Figure 2E-G). Although these compounds produce promising in vitrro responses, implementation in pediatric AML first awaits testing in phase I/II trials in adults. Furthermore, in vivo drug testing is required as in vitro response does not always imply in vivo response.13
Overall, these data show the value of including screening for NUP98-KDM5A rearrangements as part of the standard survey in children with AML irrespectively of their AML FAB subtype. Although HSCT in first complete remission did not seem to prevent relapse in these cases, HSCT is currently the most effective post-remission therapy for preventing relapse. Therefore, we suggest that NUP98-KDM5A+ AML deserves stratification into the high-risk group, and that HSCT in first complete remission should be considered. We showed that NUP98-KDM5A+ and NUP98-NSD1+ cases of AML have different clinical and biological characteristics, and may benefit from different types of targeted treatment.
Supplementary Material
Acknowledgments
The authors would like to thank Sandra Mimoso Pinhanços, Bianca Koopmans, Luke Jones, Susan Arentsen-Peters and Patricia Garrido Castro for their assistance in the in vitro drug screens and validation.
References
- 1.Zwaan CM, Kolb EA, Reinhardt D, et al. Collaborative efforts driving progress in pediatric acute myeloid leukemia. J Clin Oncol. 2015; 33(27):2949-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia. Hematol Oncol Clin North Am. 2010;24(1):35-63. [DOI] [PubMed] [Google Scholar]
- 3.Balgobind BV, Hollink IH, Arentsen-Peters ST, et al. Integrative analysis of type-I and type-II aberrations underscores the genetic heterogeneity of pediatric acute myeloid leukemia. Haematologica. 2011;96(10):1478-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coenen EA, Raimondi SC, Harbott J, et al. Prognostic significance of additional cytogenetic aberrations in 733 de novo pediatric 11q23/MLL-rearranged AML patients: results of an international study. Blood. 2011;117(26):7102-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Rooij JD, Zwaan CM, van den Heuvel-Eibrink M. Pediatric AML: from biology to clinical management. J Clin Med. 2015;4(1):127-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niktoreh N, Walter C, Zimmermann M, et al. Mutated WT1, FLT3- ITD, and NUP98-NSD1 fusion in various combinations define a poor prognostic group in pediatric acute myeloid leukemia. J Oncol. 2019;2019:1609128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118(13):3645-3656. [DOI] [PubMed] [Google Scholar]
- 8.de Rooij JD, Hollink IH, Arentsen-Peters ST, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia. 2013;27(12):2280-2288. [DOI] [PubMed] [Google Scholar]
- 9.Bolouri H, Farrar JE, Triche T, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Struski S, Lagarde S, Bories P, et al. NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia. 2017;31(3):565-572. [DOI] [PubMed] [Google Scholar]
- 11.Bisio V, Zampini M, Tregnago C, et al. NUP98-fusion transcripts characterize different biological entities within acute myeloid leukemia: a report from the AIEOP-AML group. Leukemia. 2017;31(4):974-977. [DOI] [PubMed] [Google Scholar]
- 12.McCowage GB, Mueller S, Pratilas CA, et al. Trametinib in pediatric patients with neurofibromatosis type 1 (NF-1)–associated plexiform neurofibroma: A phase I/IIa study. J Clin Oncol. 2018;36(15_suppl): 10504-10504. [Google Scholar]
- 13.Aplenc R, Meshinchi S, Sung L, et al. The addition of bortezomib to standard chemotherapy for pediatric acute myeloid leukemia has increased toxicity without therapeutic benefit: a report from the Children's Oncology Group. Blood. 2016;128(22):899. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.