ABSTRACT
Circadian clock operates autonomously in each cell and drives the approximately 24-h rhythm in individual tissues and organs. It is known that the evening complex (EC) components and GI are required for ambient temperature perception and thermomorphogenesis in higher plants. Our previous study found that PRR9 and 7 are required for the lengthened period of the circadian rhythm in roots, and they are responsible for the temperature overcompensation in shoots. However, the molecular mechanism of the circadian clock, especially in different tissues, in response to temperature oscillations remains largely unknown. Here, we studied the transcript levels of EC genes and GI of the prr7 prr9 mutant shoots and roots in response to 22°C or 28°C, respectively. The results showed that PRR9, 7 in roots inhibited the transcripts accumulation of ELF3, ELF4, and LUX at 28°C. In addition, loss-of-function of both PRR9 and 7 caused an increase in GI expression at 22°C, but warm temperature of 28°C limited the negative effect of PRR9, 7 on GI in roots. Our findings proposed a temperature-dependent molecular basis for root-specific circadian clock and indicated the critical role for PRR9, 7 in negatively regulating ELF3, ELF4, LUX, and GI in the circadian gating of thermoresponse.
KEYWORDS: Circadian clock, PRRs, evening complex, warm temperature, root
Circadian clock, as a time-keeping mechanism, drives a large variety of physiological rhythms to synchronize with cyclic changes of environmental zeitgebers, such as light and temperature.1 The five members of Pseudo-Response Regulators (PRRs) family are expressed sequentially from morning to evening during a 24-h day, and function as negative regulators of morning-phased CCA1, LHY, and RVE8 in the transcriptional feedback loops of the circadian clock.2 The long periodicity in the prr7 prr9 mutant continues to lengthen as the ambient temperature rises to 30°C, demonstrating a temperature overcompensation phenotype.3,4 In addition, temperature compensation in the prr7 prr9 mutants is related to the increased expression of CCA1 and LHY at warm temperature. CCA1 and LHY mainly act as transcription repressors in the circadian negative feedback loops, and their targets include PRR5, PRR1/TOC1, and ELF3-ELF4-LUX evening complexes (EC) genes. EC components perceive the changes in ambient temperature, and warm temperature inhibits EC gene expression; and as a result, ELF3 regulates thermoresponsive growth and flowering.5–7 Whereas the relationship between PRR9, 7 and ELF3-ELF4-LUX complex in thermoresponse and temperature compensation of the circadian clock remains largely unknown.
Circadian rhythms run autonomously in individual organ, apical tissue, and single cell of plants.8–10 Period length in the roots is much longer than that in the shoot. The core oscillators in the shoot apex and vasculature, like in the central clock of mammals, play a dominant role in the perception of photosynthetic signals and orchestrates the entire circadian network in whole plant.9,11–13 In a recent study, we found that the detached hairy roots of soybean perceive the intensity of red and blue light, and respond to discontinuous light or temperature pulses applied within the 24 h, which cause the phase shift of circadian rhythm.14 This finding implied that the root clock independently responds to external zeitgebers. We also found protein–protein interactions (PPI) between core oscillators present an approximately 24-h rhythms in both shoots and roots, and the period length of PPI rhythm in roots is longer than that in the shoots.15 What is more, mutation of PRR9 and PRR7 caused lengthened period in roots to be insignificant, indicating that PRR9, 7 is required for autonomous root clock. However, at 28°C warm temperature, prr7 prr9 mutant has shown overcompensation only in the shoots, but not in the roots.15 In summary, we speculate that there are more circadian components involved in the molecular mechanism for temperature signal feeding into root clock.
Here, to explore the molecular architecture of the circadian clock in the temperature responses, we investigated the transcriptional feedback loops mediated by shoot- or root-specific expression of PRR9 and PRR7. Shoots and roots of prr7 prr9 mutant and wild-type plant were isolated and transferred to continuous light, 22°C or 28°C, respectively. Samples were collected during the second free-running cycle, and then transcripts enrichment of ELF3, ELF4, and LUX were analyzed by qRT-PCR. All the three EC complex genes showed higher expression in roots, at both 22°C and 28°C, than that in shoots (Figure 1). The result suggested that, like PRR9 and 7, the EC complex has a specific effect on the circadian clock in the roots. In addition, at ZT24-48 of the constant light and temperature conditions, the peaks of the EC gene expression in the prr7 prr9 mutant moved to the daytime, especially the peaks in the roots at 28°C appeared near ZT27. We speculated that the long-period phenotype of the prr7 prr9 double mutant under the free-running conditions is a cause of the peaking shift of the circadian expression pattern.
Figure 1.
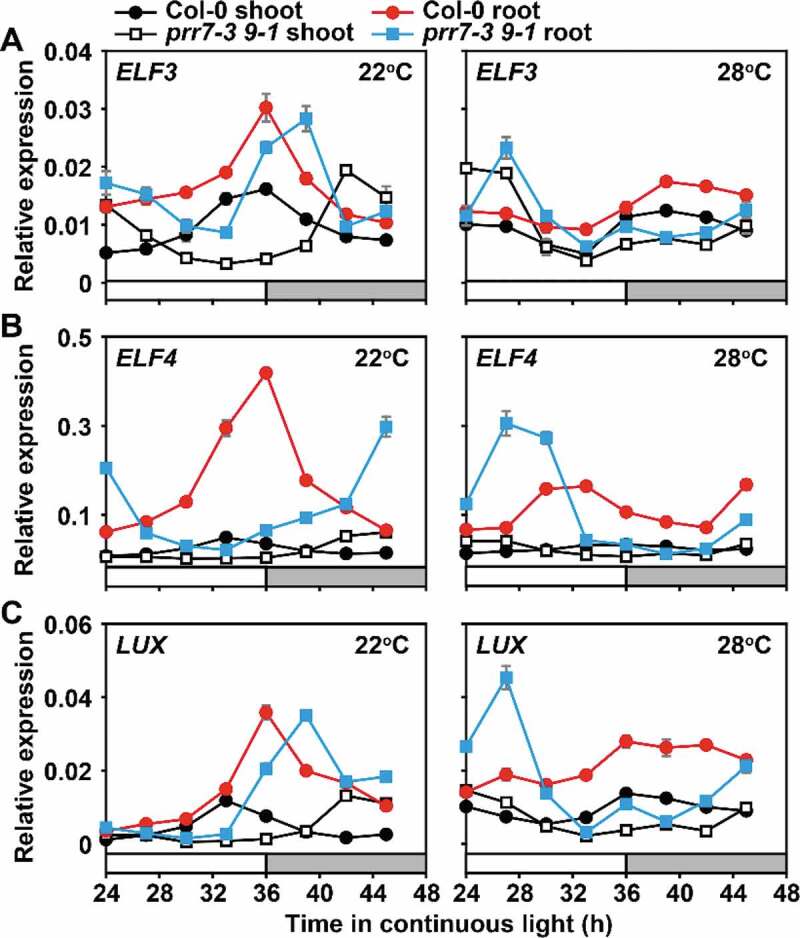
PRR9, 7 in roots inhibit EC complex gene expression under warm temperature. Transcript accumulation of ELF3 (a), ELF4 (b), and LUX (c) were examined in prr7 prr9 mutant and wild-type plants. The seedlings were grown under 12:12 LD cycles at 22°C for 14 days. After the shoots and roots were separated, they were transferred to 22°C or 28°C, respectively, under constant light conditions. Samples were harvested every 3 h over a 24-h cycle for RNA extraction and qRT-PCR analysis. The reference gene IPP2 was used in the quantitative expression analysis. The data are presented as the mean ± SEM of three technical replicates from one of three independent biological experiments; all experiments yielded congruent results. White or gray bars represent subjective day or subjective night, respectively
Further analysis found that at 22°C, the peak expression of ELF3, ELF4, and LUX in the prr7 prr9 mutants delayed significantly under free-running conditions, but the overall accumulation or amplitude of the EC-related gene mRNA in the mutant and wild-type were similar in the 24-h day, both in shoots (black circle vs. empty square) or roots (red circle vs. blue square) (Figure 1). Under the 28°C condition, the peak expression of ELF3 in the shoots of the prr7 prr9 mutant increased slightly, but the overall expression levels of ELF4 and LUX were similar to those of the wild type in the 24-h day. However, in roots, the peak expression of ELF3, ELF4, LUX in the prr7 prr9 was higher than that of the wild type (Blue square vs. red circle). This result indicated that PRR9, 7 in roots inhibited the ELF3, ELF4, LUX expression at warm temperature. Recently, the expression of PRR9, 7, 5, and TOC1 were analyzed at 22°C or 28°C separately,15 and the results showed that PRR9 accumulation at 28°C was more than that at 22°C observed only in root tissues. Collectively, we provide the hypothesis that the expression of PRR9 increases in roots at warm temperature, thus inhibiting the accumulation of EC genes.
Evening-phased GIGANTEA (GI) modulate the TOC1 and PRR5 accumulation in the evening by stabilizing ZEITLUPE (ZTL) protein,16 but the GI–ZTL interaction becomes extremely weak at warm temperature.17 The GI mRNA slightly increased as the temperature increased from 17°C to 27°C.18 Compared to 22°C, the gi mutant has a shorter period than the wild type at both 12°C and 27°C, indicating that GI accounts for temperature compensation. In order to determine the morning oscillators' regulation of GI in the interlocked loops, we examined GI transcript accumulation in the prr7 prr9 mutant (Figure 2). Compared with 22°C, the amplitude of GI rhythmic expression in wild-type and mutant shoots decreased at 28°C; however, there was no significant difference in overall transcript accumulation in shoots and roots in the 24-h day. But it is worth noting that at 22°C, the increase in GI expression in the prr7 prr9 mutant was greater than that at 28°C during the subjective night. This result indicated that the inhibitory effect of PRR9, 7 on GI was weakened at warm temperature. According to the RNA-seq data that has been studied, GI expression is up-regulated in the prr975 mutant.19 ChIP-seq data also indicate that PRR7 may bind to the GI promoter region.20 Therefore, we speculated that PRR7 potentially directly inhibited the expression of GI. In addition, PRR9 and PRR7 may also indirectly regulate GI expression through CCA1/LHY and other circadian components. To verify the above hypothesis, the transcription activation experiment in transient transfected protoplasts and EMSA can be used to further analyze the mechanism of PRR9 and PRR7 in regulating the circadian responses to the temperature. In addition, Salomé et al. used the whole Arabidopsis seedlings to analyze the rhythmic expression of GI during ZT72-96 at 12°C, 22°C, and 30°C.4 In this study, we used the detached shoots and roots to analyze the tissue specificity of the rhythmic expression during ZT24-48 at 22°C or 28°C. Comparing the results of the two studies, it is consistent that compared with 22°C, GI expression in the prr7 prr9 mutant was down-regulated at 28°C or 30°C, and the GI amplitude was reduced in both wild-type and the mutant. The difference is that at 30°C, the expression of GI increases in the wild-type whole plant; at 28°C, the GI peak expression decreases in the wild-type shoots, the trough value increases based on the rhythmic expression trace, and there were no significant changes in the roots during the 24-h day.
Figure 2.
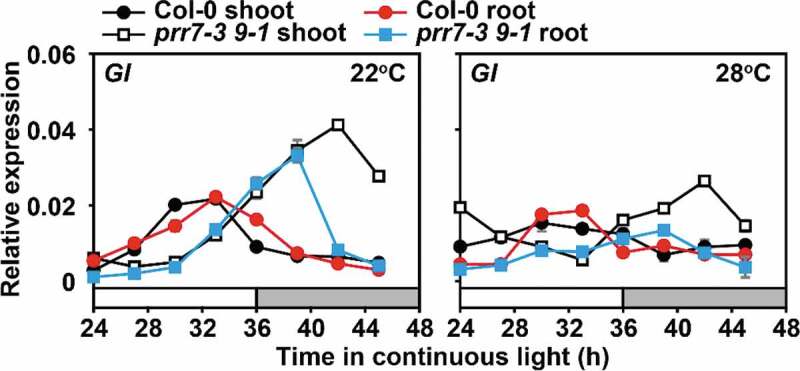
The negative regulation of PRR9, 7 in roots on GI is weakened at warm temperature. Transcript levels of GI were analyzed at 22°C and 28°C, respectively. Arabidopsis seedlings of the prr7 prr9 mutant and wild-type plants were grown under 12:12 LD cycles at 22°C for 14 days. After the shoots and roots were separated, they were transferred to 22°C or 28°C, respectively, under constant light conditions. Samples were harvested every 3 h over a 24-h cycle for RNA extraction and qRT-PCR analysis. The reference gene IPP2 was used in the quantitative expression analysis. The data are presented as the mean ± SEM of three technical replicates from one of three independent biological experiments; all experiments yielded congruent results. White or gray bars represent subjective day or subjective night, respectively
It is known that the products of the photosynthesis from shoots and leaves,8 and the EC complex component ELF4 protein21 regulate the root clock through transportation, thus maintaining the synchronization of the circadian rhythms between the organs. But this mechanism is not necessary for the self-sustained circadian rhythms in the roots. When the plant was intact, the circadian rhythm of the root was longer than that of the shoots; but when the shoot and root were separated, the circadian rhythm of the root was still longer than that of the shoots,9,15,22 which indicated that the root clock independently drives the circadian rhythm and own a lengthened period not entirely dependent on the signal transmission of vasculature and leaves. In addition, the plant root system, like the shoots on the ground, also experiences daily and seasonal changes of environmental temperature. In this study, in transcriptional feedback loops of root-specific clock, PRR9 and PRR7 inhibited the expression of evening-phased genes, ELF3, ELF4, LUX, and GI, at a warm temperature of 28°C. The proposed molecular model may function to modulate the self-sustained circadian rhythm and the temperature compensation in roots. In addition, warm temperature affects the inhibitory effect of PRR9, 7 on GI expression, forming a regulatory pathway of the circadian clock in anticipation of the thermocycles. Yet the genetic interactions between PRR9, 7 and GI in the temperature compensation and thermomorphogenesis need to be further determined.
Funding Statement
This work was supported by grants from National Natural Science Foundation of China to X.X. (U1904202, 31570285) and Q.X. (31670285), the Natural Science Foundation of Hebei (17966304D) and the Hebei Hundred Talents Program (E2016100018) to Q.X
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Authors contribution
X.X., Q.X. conceived the project and wrote the article. L.Y. performed most of the experiments, L.Y., Y.H., S.L. performed the reference gene analysis. L.Y., Q.X., and X.X. analyzed the data.
References
- 1.Greenham K, McClung CR.. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet. 2015;16:1–4. doi: 10.1038/nrg3976. [DOI] [PubMed] [Google Scholar]
- 2.Hsu PY, Harmer SL.. Wheels within wheels: the plant circadian system. Trends Plant Sci. 2014;19:240–249. doi: 10.1016/j.tplants.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salomé PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salomé PA, Weigel D, McClung CR. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7 and PRR9 in temperature compensation. Plant Cell. 2010;22:3650–3661. doi: 10.1105/tpc.110.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. Phytochromes function as thermosensors in Arabidopsis. Science. 2016;354:886–889. [DOI] [PubMed] [Google Scholar]
- 6.Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, Yamashino T. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in arabidopsis thaliana. Plant Cell Physiol. 2014;55:958–976. doi: 10.1093/pcp/pcu030. [DOI] [PubMed] [Google Scholar]
- 7.Song YH, Kubota A, Kwon MS, Covington MF, Lee N, Taagen ER, Laboy Cintron D, Hwang DY, Akiyama R, Hodge SK, et al. Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nat Plants. 2018;4:824–835. doi: 10.1038/s41477-018-0253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, Nimmo HG. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science. 2008;322:1832–1835. doi: 10.1126/science.1161403. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi N, Hirata Y, Aihara K, Mas P. A hierarchical multi-oscillator network orchestrates the arabidopsis circadian system. Cell. 2015;163:148–159. doi: 10.1016/j.cell.2015.08.062. [DOI] [PubMed] [Google Scholar]
- 10.Thain SC, Hall A, Millar AJ. Functional independence of circadian clocks that regulate plant gene expression. Curr Biol. 2000;10:951–956. doi: 10.1016/S0960-9822(00)00630-8. [DOI] [PubMed] [Google Scholar]
- 11.Endo M, Shimizu H, Nohales MA, Araki T, Kay SA. Tissue-specific clocks in arabidopsis show asymmetric coupling. Nature. 2014;515:419–422. doi: 10.1038/nature13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marti MC, Webb AA. Plant science: leaf veins share the time of day. Nature. 2014;515:352–353. doi: 10.1038/nature13936. [DOI] [PubMed] [Google Scholar]
- 13.McClung CR. A fibre-optic pipeline lets the root circadian clock see the light. Plant Cell Environ. 2018;41:1739–1741. doi: 10.1111/pce.13343. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Yuan L, Su T, Wang Q, Gao Y, Zhang S, Jia Q, Yu G, Fu Y, Cheng Q, et al. Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ. 2020;43:637–648. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Wang L, Yuan L, Song Y, Sun J, Jia Q, Xie Q, Xu X. Molecular investigation of organ-autonomous expression of Arabidopsis circadian oscillators. Plant Cell Environ. 2020;43:1501–1512. [DOI] [PubMed] [Google Scholar]
- 16.Kim W-Y, Fujiwara S, Suh -S-S, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 17.Kim TS, Wang L, Kim YJ, Somers DE. Compensatory mutations in GI and ZTL may modulate temperature compensation in the circadian clock. Plant Physiol. 2020;182:1130–1141. doi: 10.1104/pp.19.01120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould PD, Locke JCW, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. 2006;18:1177–1187. doi: 10.1105/tpc.105.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- 20.Liu T, Carlsson J, Takeuchi T, Newton L, Farré EM. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013;76:101–114. [DOI] [PubMed] [Google Scholar]
- 21.Chen WW, Takahashi N, Hirata Y, Ronald J, Porco S, Davis SJ, Nusinow DA, Kay SA, Mas P. A mobile ELF4 delivers circadian temperature information from shoots to roots. Nat Plants. 2020;6:416–426. doi: 10.1038/s41477-020-0634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bordage S, Sullivan S, Laird J, Millar AJ, Nimmo HG. Organ specificity in the plant circadian system is explained by different light inputs to the shoot and root clocks. New Phytol. 2016;212:136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]


