Mycobacteriophage phiT46-1 is a newly isolated Mycobacterium phage that was isolated by spontaneous release from Mycobacterium abscessus strain Taiwan-46 and infects M. abscessus strain BWH-C. Phage phiT46-1 is unrelated to previously described mycobacteriophages, has a 52,849-bp genome, and includes a polymorphic toxin-immunity cassette associated with type VII secretion systems.
ABSTRACT
Mycobacteriophage phiT46-1 is a newly isolated Mycobacterium phage that was isolated by spontaneous release from Mycobacterium abscessus strain Taiwan-46 and infects M. abscessus strain BWH-C. Phage phiT46-1 is unrelated to previously described mycobacteriophages, has a 52,849-bp genome, and includes a polymorphic toxin-immunity cassette associated with type VII secretion systems.
ANNOUNCEMENT
Mycobacterium abscessus is a nontuberculous mycobacterium (NTM) that is ubiquitous in the environment and is common in water and soil (1). M. abscessus is a common cause of pulmonary and disseminated infections in immunocompromised individuals, particularly those with cystic fibrosis (2, 3). Antibiotic treatment of NTM infections is challenging, with widespread multidrug resistance and nonresponsiveness to antibiotic therapy (4). Mycobacteriophages are viruses that infect mycobacterial hosts and, although many have been isolated on Mycobacterium smegmatis, few infect M. abscessus (5). Isolation and characterization of M. abscessus phages will advance our understanding of M. abscessus and their potential therapeutic utility (5).
Many strains of M. abscessus carry prophages (6–9) and are expected to spontaneously release phage particles. Phage phiT46-1 was isolated by plating a culture supernatant of M. abscessus Taiwan-46 (provided by Chidiebere Akusobi and Eric Rubin) onto a lawn of M. abscessus BWH-C on solid medium at 37°C, using standard methods (10). Phage were picked from infected areas, plaque purified, and amplified on BWH-C, and DNA was extracted by phenol-chloroform-isoamyl alcohol extraction (10). A sequencing library was prepared from genomic DNA using a NEBNext Ultra II FS kit with dual-indexed barcoding and was included as one of a pool of 48 phage genome libraries on an Illumina MiSeq system, yielding 931,342 paired-end 300-base reads and 2,400-fold coverage of the phiT46-1 genome. These reads were assembled using Newbler v2.9 with default settings, yielding a single 52,849-bp contig with a G+C content of 64%. The contig was evaluated for completeness and accuracy using Consed v29. Sequencing read alignments did not identify unique genome ends, and either there are multiple distinct termini or the contig is circularly permuted (11); for genome representation, it was linearized with coordinate 1 at the first codon of the small terminase subunit gene. Phage phiT46-1 is not closely related to other actinobacteriophages (nucleotide identities span <4% of the total genome length [12]) but shares several virion structural genes with cluster Q phages (13). phiT46-1 does not infect M. smegmatis.
The programs GeneMarkS v4.30 (14), Glimmer v3.02 (15), Phamerator Actino_prophage v5 (16), and DNA Master v5.23.5 (http://cobamide2.bio.pitt.edu) were used to identify 78 protein-coding genes in the phiT46-1 genome. All tools were run with default parameters unless otherwise specified. The genome has no tRNA genes, as indicated by ARAGORN v1.2.41 (17). Of the 78 predicted genes, 45% were assigned putative functions using BLAST (18) and HHpred (19, 20). The virion structure and assembly genes suggest that phiT46-1 has a siphoviral morphology (family Siphoviridae), and repressor and tyrosine integrase genes are consistent with its temperate nature. The predicted early lytic genes also include an HNH endonuclease, a phosphoadenosine phosphosulfate (PAPS) reductase, an oxidoreductase, WhiB, and a RecET-like recombination system.
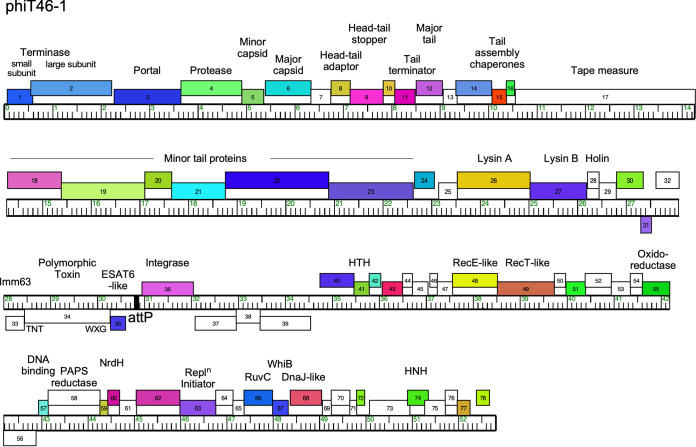
Interestingly, phiT46-1 contains a cassette coding for a polymorphic toxin (PT), an immunity (Imm63) protein, and an ESAT-6-like protein (Fig. 1). The 50-kDa PT has an N-terminal WXG-100 motif (21) and a C-terminal domain containing a tuberculosis necrotizing toxin (TNT) (22). The genomic location of the cassette, adjacent to the integration apparatus, suggests that these genes may be lysogenically expressed (23), likely with secretion via a type VII secretion pathway (24). The TNT motif is associated with escape of M. tuberculosis from phagosomes, and phiT46-1 is thus implicated in the survival of M. abscessus T46 in infected human cells.
FIG 1.
Genome organization of phage phiT46-1. The linearized viral genome of phage phiT46-1 is shown with genes represented by colored boxes either above or below the genome, reflecting rightward and leftward transcription, respectively; gene numbers are shown in each box. Genes are colored according to the “phamily” designations (16), with white boxes representing “orphams” with no close relatives in this data set. Phamily assignments were determined using Phamerator (16) and the Actino_prophage database (version 5). Predicted gene functions are indicated.
Data availability.
The phiT46-1 sequence is available in GenBank with accession no. MW353181, and sequencing reads are available with accession no. SRX9186031.
ACKNOWLEDGMENTS
This work was supported by funding from grant GM131729 from the National Institutes of Health and grant GT12053 from the Howard Hughes Medical Institute.
We thank Chidiebere Akusobi and Eric Rubin for bacterial strains.
REFERENCES
- 1.Dowdell K, Haig SJ, Caverly LJ, Shen Y, LiPuma JJ, Raskin L. 2019. Nontuberculous mycobacteria in drinking water systems: the challenges of characterization and risk mitigation. Curr Opin Biotechnol 57:127–136. doi: 10.1016/j.copbio.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee MR, Sheng WH, Hung CC, Yu CJ, Lee LN, Hsueh PR. 2015. Mycobacterium abscessus complex infections in humans. Emerg Infect Dis 21:1638–1646. doi: 10.3201/2109.141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fedrizzi T, Meehan CJ, Grottola A, Giacobazzi E, Fregni Serpini G, Tagliazucchi S, Fabio A, Bettua C, Bertorelli R, De Sanctis V, Rumpianesi F, Pecorari M, Jousson O, Tortoli E, Segata N. 2017. Genomic characterization of nontuberculous mycobacteria. Sci Rep 7:45258. doi: 10.1038/srep45258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 5.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25:730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann JL, Daffe M, Brosch R, Risler JL, Gaillard JL. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassi M, Drancourt M. 2014. Genome analysis reveals three genomospecies in Mycobacterium abscessus. BMC Genomics 15:359. doi: 10.1186/1471-2164-15-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sassi M, Gouret P, Chabrol O, Pontarotti P, Drancourt M. 2014. Mycobacteriophage-drived diversification of Mycobacterium abscessus. Biol Direct 9:19. doi: 10.1186/1745-6150-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glickman C, Kammlade SM, Hasan NA, Epperson LE, Davidson RM, Strong M. 2020. Characterization of integrated prophages within diverse species of clinical nontuberculous mycobacteria. Virol J 17:124. doi: 10.1186/s12985-020-01394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkis GJ, Hatfull GF. 1998. Mycobacteriophages. Methods Mol Biol 101:145–173. doi: 10.1385/0-89603-471-2:145. [DOI] [PubMed] [Google Scholar]
- 11.Russell DA 2018. Sequencing, assembling, and finishing complete bacteriophage genomes. Methods Mol Biol 1681:109–125. doi: 10.1007/978-1-4939-7343-9_9. [DOI] [PubMed] [Google Scholar]
- 12.Russell DA, Hatfull GF. 2017. PhagesDB: the actinobacteriophage database. Bioinformatics 33:784–786. doi: 10.1093/bioinformatics/btw711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris P, Marinelli LJ, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2008. Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination. J Bacteriol 190:2172–2182. doi: 10.1128/JB.01657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besemer J, Borodovsky M. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res 33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2011. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Remmert M, Biegert A, Hauser A, Soding J. 2011. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 20.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conrad WH, Osman MM, Shanahan JK, Chu F, Takaki KK, Cameron J, Hopkinson-Woolley D, Brosch R, Ramakrishnan L. 2017. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc Natl Acad Sci U S A 114:1371–1376. doi: 10.1073/pnas.1620133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J, Siroy A, Lokareddy RK, Speer A, Doornbos KS, Cingolani G, Niederweis M. 2015. The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD. Nat Struct Mol Biol 22:672–678. doi: 10.1038/nsmb.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dedrick RM, Jacobs-Sera D, Bustamante CA, Garlena RA, Mavrich TN, Pope WH, Reyes JC, Russell DA, Adair T, Alvey R, Bonilla JA, Bricker JS, Brown BR, Byrnes D, Cresawn SG, Davis WB, Dickson LA, Edgington NP, Findley AM, Golebiewska U, Grose JH, Hayes CF, Hughes LE, Hutchison KW, Isern S, Johnson AA, Kenna MA, Klyczek KK, Mageeney CM, Michael SF, Molloy SD, Montgomery MT, Neitzel J, Page ST, Pizzorno MC, Poxleitner MK, Rinehart CA, Robinson CJ, Rubin MR, Teyim JN, Vazquez E, Ware VC, Washington J, Hatfull GF. 2017. Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol 2:16251. doi: 10.1038/nmicrobiol.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. 2016. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol 14:677–691. doi: 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The phiT46-1 sequence is available in GenBank with accession no. MW353181, and sequencing reads are available with accession no. SRX9186031.