ABSTRACT
Long non-coding RNA (lncRNA) X inactive specific transcript (XIST) has been identified as an oncogenic lncRNA in a series of human cancers, including esophageal squamous cell carcinoma (ESCC). In this study, we aimed to further explore the underlying mechanism of XIST on ESCC progression. qRT-PCR assay was used to determine the levels of XIST and miR-129-5p. Western blot analysis was performed to assess cyclin D1 (CCND1) expression. Bioinformatic analysis was performed using starBase v2.0 software. Dual-luciferase reporter and RNA immunoprecipitation assays were employed to confirm the interaction between XIST and miR-129-5p or miR-129-5p and CCND1. Cell cycle progression and apoptosis were measured by flow cytometric analysis, and cell migration and invasion were detected by transwell assay. Mouse studies were used to observe the effect of XIST silencing on tumor growth in vivo. Our results indicated that XIST was upregulated and miR-129-5p was downregulated in ESCC. XIST silencing or miR-129-5p overexpression repressed cell cycle progression, proliferation, migration, invasion, and promoted the apoptosis in ESCC cells. Moreover, XIST directly interacted with miR-129-5p and repressed miR-129-5p expression. MiR-129-5p mediated the regulatory effect of XIST on ESCC cell progression in vitro, and XIST promoted CCND1 expression by sponging miR-129-5p. Additionally, XIST silencing inhibited tumor growth in vivo. Our findings suggested that XIST silencing repressed the progression of ESCC at least partly through regulating the miR-129-5p/CCND1 axis. Targeting XIST might be a potential therapeutic strategy for ESCC treatment.
KEYWORDS: ESCC, X inactive specific transcript (XIST), miR-129-5p, cyclin D1 (CCND1)
1. Introduction
Cancer of the esophagus is one of the most common human cancers, leading to over 400,000 deaths worldwide annually [1]. Esophageal squamous cell carcinoma (ESCC) accounts for about 90% of esophageal cancers and has a high prevalence in Asia and Africa [2]. The etiology of ESCC is complex and tightly population-dependent [2]. Despite advanced diagnosis and treatment methods, the overall 5-year survival rate of ESCC patients is less than 30% [3]. Therefore, it is very urgent to identify novel biomarkers and therapeutic targets for ESCC treatment.
Long non-coding RNAs (lncRNAs) are a heterogeneous group of RNAs with more than 200 nucleotides in length [4]. The dysregulation of lncRNAs plays a crucial role in human diseases, including cancers [5]. MicroRNAs (miRNAs) regulate gene expression by binding to the 3ʹ-untranslated region (3ʹ-UTR) of target mRNAs [6]. Recently, the competing endogenous RNAs (ceRNAs) hypothesis has proposed that lncRNAs regulate gene expression through sponging miRNAs, highlighting the importance of such interactions in human cancers [7,8].
LncRNA X inactive specific transcript (XIST) has been identified as a tumor promoter in a series of human cancers, such as retinoblastoma [9], cervical cancer [10], and colorectal cancer [11]. Moreover, XIST was reported to accelerate the malignant progression of ESCC by regulating the miR-101/EZH2 axis [12]. Nevertheless, the molecular mechanisms of XIST on ESCC progression are still largely unclear. In this study, we aimed to validate the detailed effect and molecular mechanisms of XIST on ESCC progression. The result of our research suggested that XIST regulated ESCC progression at least partly through sponging miR-129-5p and upregulating CCND1 expression.
2. Materials and methods
2.1. Tissues collection
Forty-two pairs of ESCC tissues and corresponding adjacent noncancerous tissues were collected from those patients who underwent the primary resection surgery at the Affiliated Cancer Hospital of Zhengzhou University. According to TNM staging, these patients were divided into two groups: stage III–IV group (n = 27) and stage I–II group (n = 15). No conventional therapy was performed before surgery. All samples were stored at −80°C until RNA extraction. Written informed consent was obtained from all patients, and the study was approved by the Ethics Committee of the Affiliated Cancer Hospital of Zhengzhou University.
2.2. Cell culture and transfection
Human esophageal epithelial cell line (Het-1A) and four ESCC cell lines (EC9706, KYSE150, KYSE450 and Eca-109) were purchased from Cell Bank of Chinese Academy of Sciences (Shanghai, China) and maintained in DMEM medium (Gibco, Grand Island, NY, USA) supplemented with 10% heated inactivated fetal bovine serum (FBS, Gibco), 1% streptomycin/penicillin (Gibco) at 37°C under 5% CO2 and saturated moisture.
To explore the effect of XIST on ESCC, EC9706 and Eca-109 cells at 60–70% confluence were transfected with 50 nM of silencer select predesigned small interfering RNAs (si-XIST-1/-2, Applied Biosystems, Foster City, CA, USA) or siRNA negative control (si-NC, Applied Biosystems), 20 ng of XIST overexpression plasmid (Vector-XIST, Applied Biosystems) or its negative control (Vector, Applied Biosystems) using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the instructions of manufacturers. To study the role of miR-129-5p, cells were transfected with 20 nM of miR-129-5p mimics (Applied Biosystems) or mimics negative control (miR-NC mimics, Applied Biosystems), 20 nM of miR-129-5p inhibitor (anti-miR-129-5p, Applied Biosystems) or inhibitor negative control (anti-miR-NC, Applied Biosystems). Transfected cells were performed further analysis 48 h after transfection.
2.3. RNA preparation, reverse transcription and quantitative real-time PCR (qRT-PCR)
Total RNA from tissues and cells was extracted using the reagent ISOGEN (Nippon Gene, Toyama, Japan) according to the instructions of manufacturers. For XIST and CCND1 mRNA detection, cDNA was synthesized by using a High-Capacity Reverse Transcription Kit (Applied Biosystems) and then subjected to qRT-PCR using the SYBR Green Master Mix (Applied Biosystems) on the LightCycler 480 Real-Time PCR System (Roche Life Science, Indianapolis, IN, USA). The expression levels of XIST and CCND1 mRNA were calculated by the 2−ΔΔCt method with GAPDH as the internal control. For miR-129-5p detection, RNA was reverse-transcribed using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems) and qRT-PCR was performed using the TaqMan MicroRNA Assay kit (Applied Biosystems) with U6 as a housekeeping gene for normalization. The following primers (5’-3’) were used: XIST-Forward: TCAGCCCATCAGTCCAAGATC and XIST-Reverse: CCTAGTTCAGGCCTGCTTTTCAT, CCND1-Forward: TCCTACTACCGCCTCACA and CCND1-Reverse: ACCTCCTCCTCCTCCTCT, miR-129-5p-Forward: ACCCAGTGCGATTTGTCA and miR-129-5p-Reverse: ACTGTACTGGAAGATGGACC, GAPDH-Forward: TCGACAGTCAGCCGCATCTTCTTT and GAPDH-Reverse: ACCAAATCCGTTGACTCCGACCTT, U6-Forward: GCTTCGGCAGCACATATACTAAAAT and U6-Reverse: CGCTTCACGAATTTGCGTGTCAT.
2.4. Flow cytometric analysis of cell cycle progression and apoptosis
Cell cycle analysis was performed using Cycletest™ Plus DNA Reagent kit (BD Biosciences, Bedford, MA, USA) according to the protocols of manufacturers. In brief, transfected cells (1 × 105) were labeled with 10 µl of PI in the presence of RNase for 30 min in the dark, and cell cycle progression was analyzed by a flow cytometer (FACScan, BD Biosciences).
Cell apoptosis was detected using Annexin V-FITC/PI apoptosis detection kit (BD Biosciences). Briefly, transfected cells (1 × 106) were double-stained with 10 µl of Annexin V-FITC (0.5 µg/mL) and 5 µl of PI (5 µg/mL) for 15 min in the dark, followed by the measurement of cell apoptotic rate by the FACScan flow cytometer within 60 min.
2.5. Cell proliferation assay
The Cell Counting Kit-8 (CCK-8) assay was used to evaluate cell proliferation as per the manufacturing protocols (Dojindo, Kumamoto, Japan). Briefly, transfected cells (2 × 103 cells/well) were plated in 96-well plates and incubated for the indicated time points before adding 10 µl of CCK-8 solution into each well for 2 h at 4°C. The number of viable cells was proportional to the absorbance at 450 nm by a microplate reader (Bio-Tek instruments, Bad Friedrichshall, Germany).
2.6. Transwell assay of cell migration and invasion
For migration assay, transfected cells (2 × 104 cells/well) in serum-free DMEM were seeded into the upper chamber of 24-well transwell plates with the non-coated membrane (8 µm pore size, BD Biosciences). For invasion assay, transfected cells (1 × 105 cells/well) were added into the upper chamber of the transwell chamber with the Matrigel-coated membrane (BD Biosciences). In both assays, 700 µl of growth medium containing 10% FBS was added into the lower chamber. After 48 h incubation, the cells that had migrated or invaded through the membrane to the lower surface were fixed with paraformaldehyde and stained with crystal violet. The images were captured with a microscope (Nikon, Shinagawa, Tokyo, Japan) and the number of migrated or invaded cells were counted in five random fields.
2.7. Bioinformatic analysis
The potential interacted miRNAs of XIST were predicted by starBase v.2 (http://starbase.sysu.edu.cn/starbase2/index.php), DIANA tool-lncBase v.2 database (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index) and miRcode (http://www.mircode.org/?gene=XIST&mirfam=&class=&cons=&trregion=). The molecular targets of miR-129-5p were predicted by starBase v.2 software.
2.8. Dual-luciferase reporter assay
XIST luciferase reporter plasmid (XIST-WT) harboring the putative complementary site for miR-129-5p, CCND1 3ʹ-UTR wild-type reporter plasmid (CCND1-WT) containing the putative binding site for miR-129-5p, and empty luciferase plasmid were obtained from Applied Biosystems. The mutations (XIST-MUT and CCND1-MUT) in putative miR-129-5p-binding sequences were generated using the QuickChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the instructions of manufacturers. Cells (1 × 105) were cotransfected with 20 ng of XIST-WT, XIST-MUT, CCND1-WT or CCND1-MUT, together with 20 nM of miR-129-5p mimics or miR-NC mimics. After 48 h post-transfection, luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Relative luciferase activity was determined based on the ratio of Renilla to firefly luciferase.
2.9. RNA immunoprecipitation (RIP) and RNA pull-down assays
RIP assay was performed using the Magna RIP Immunoprecipitation Kit (Millipore, Bedford, MA, USA) according to the protocols of manufacturers. To be brief, cells were transfected with miR-129-5p mimics or miR-NC mimics and then lysed with RIPA lysis buffer. Then, the lysates were incubated with RIP buffer containing A/G magnetic beads conjugated with anti-Argonaute2 (anti-Ago2, Cell Signaling Technology, Danvers, MA, USA) or anti-IgG (Cell Signaling Technology) overnight at 4°C. Following the incubation with Proteinase K, the enrichment levels of XIST and CCND1 were detected by qRT-PCR in total RNA from the beads.
For RNA pull-down analysis, cell lysates were incubated with chemically modified miR-129-5p probe (Applied Biosystems) and control probe for 4 h at 4°C before adding the streptavidin beads (Sigma-Aldrich, Toyko, Japan) for 2 h. Beads were harvested and total RNA was extracted for qRT-PCR analysis.
2.10. Subcellular fractionation
A Cytoplasmic & Nuclear RNA Purification Kit was used to extract cytoplasmic and nuclear RNA as per the manufacturing protocols (Norgen Biotek, Thorold, ON, Canada). β-actin and U6 were used as the cytoplasmic and nuclear controls, respectively.
2.11. Western blot analysis of CCND1
Total cell extracts (100 µg) were loaded and separated on 10% SDS polyacrylamide gel and then transferred onto a PVDF membrane (Millipore). The PVDF membranes were incubated with anti-CCND1 (Cell Signaling Technology; dilution 1:1000) and anti-β-actin (Abcam, Cambridge, UK; dilution 1:2000), followed by the incubation with HRP-conjugated secondary antibodies (Abcam; dilution 1:5000). The protein bands were analyzed using the enhanced chemiluminescence (ECL, GE Healthcare, Madison, WI, USA) and quantified by LAS 4000 Image Reader (Fujifilm, Tokyo, Japan) with MultiGauge V3.2 software.
2.12. Lentivirus vector transduction
Lentiviruses containing XIST-shRNA or nontarget-shRNA (sh-XIST or sh-NC) were designed and synthesized by Applied Biosystems, and empty lentivirus particles were used as the blank control. Eca-109 cells were transduced with blank control, sh-NC, or sh-XIST. After 48 h transfection, cells with positive transduction were screened with puromycin at a final concentration of 10 µg/ml.
2.13. In vivo assay
Female BALB/c nude mice (6–8 weeks, 18–22 g) were purchased from Henan Research Center of Laboratory Animal (Zhengzhou, Henan, China). Approximately 4.0 × 106 Eca-109 cells stably transfected with sh-NC, sh-XIST, or blank control were subcutaneously inoculated into the nude mice to generate ESCC xenograft in vivo (n = 8 each group). After 7 days, tumor volume was measured with a digital caliper every 7 days and calculated by the formula: (length × width2) × 0.5. 35 days later, all mice were euthanized and tumors were excised and weighted. The levels of XIST, miR-129-5p and CCND1 were determined by qRT-PCR. Besides, CCND1 level in the xenograft tissues was detected by immunohistochemistry using anti-CCND1 antibody as previously described [13]. Our study was approved by the Animal Care and Use Committee of the Affiliated Cancer Hospital of Zhengzhou University.
2.14. Statistical analysis
All experiments were repeated twice and performed in triplicates each time. All data were presented as mean ± standard deviation (SD) using Prism 7.0 software (GraphPad Software Inc., La Jolla, CA, USA). Statistical significance for experiments was detected by a Student’s t-test, Mann-Whitney U test or one-way ANOVA with Tukey’s test or Dunnett-t test. The overall survival of ESCC patients was analyzed using Kaplan-Meier method and log-rank tests. The correlation between XIST and miR-129-5p expression levels in tumor tissues was determined by Spearman test. The associations between the levels of XIST and miR-129-5p and clinicopathologic features of ESCC patients were analyzed by χ2 test. P values at 0.05 or smaller were considered significant.
3. Results
3.1. XIST was upregulated in ESCC tissues and cells and associated with poor prognosis
Firstly, we examined the expression of XIST in ESCC tissues and cells by qRT-PCR assay. In contrast to the adjacent noncancerous tissues, XIST expression was significantly elevated in ESCC tissues (Figure 1a). Moreover, the expression of XIST was higher in TNM stage III–IV group than that in stage I–II group (Figure 1b). Additionally, qRT-PCR data revealed a significant upregulation of XIST level in EC9706, KYSE150, KYSE450 and Eca-109 ESCC cells compared with normal human esophageal epithelial cell line Het-1A (Figure 1c).
Figure 1.
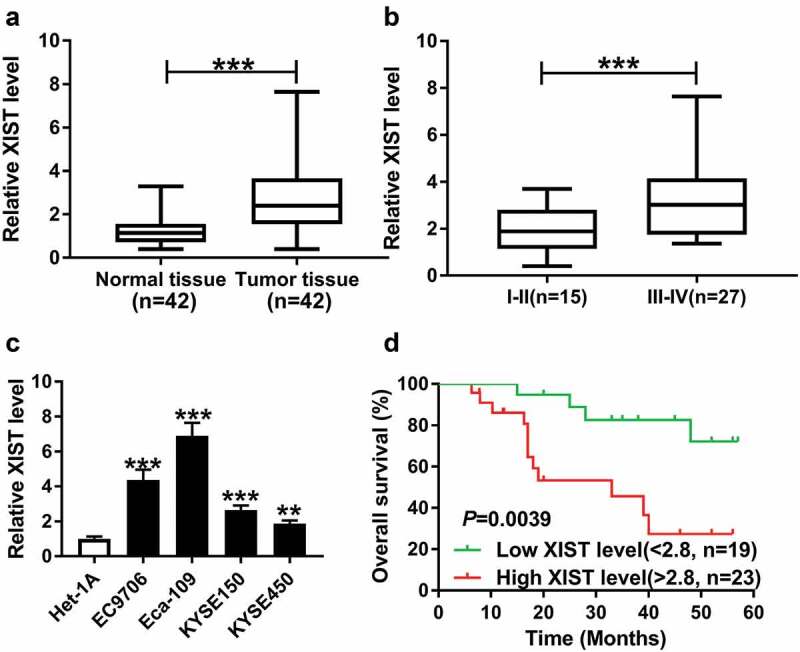
XIST was upregulated in ESCC tissues and cells and associated with poor prognosis. (a) XIST expression was detected in 42 pairs of ESCC tissues and adjacent noncancerous tissues by qrt-PCR assay. data were expressed as mean ± SD of three independent replicates. ***P < 0.001 by Mann-Whitney U test. (b) XIST expression was determined in ESCC patients grouped according to TNM pathological grades: stage I–II group (n = 15) and stage III–IV group (n = 27). Data were expressed as mean ± SD of three independent replicates. ***P < 0.001 by student’s t-test. (c) The expression of XIST was examined in human esophageal epithelial cell line (Het-1A) and ESCC cell lines (EC9706, KYSE150, KYSE450 and Eca-109). Data were expressed as mean ± SD of three independent replicates. **P < 0.01 or ***P < 0.001 by ANOVA with dunnett-t test. (d) Kaplan-Meier survival curves and log-rank tests were used to evaluate the survival rate of ESCC patients with high or low XIST level. **P < 0.01 by log-rank test
To observe the correlation between XIST and prognosis of ESCC patients, the 42 ESCC patients were divided into two groups: high XIST level group (>2.8, n = 23) and low XIST level group (<2.8, n = 19) according to the mean value (relative XIST expression level: 2.8) of XIST expression. Kaplan-Meier survival curves showed that the patients in high XIST level group (median survival: 39 months) had a shorter survival time than those in low XIST level group (median survival: >39 months) (Figure 1d). Additionally, we determined the relationship between XIST expression and the clinicopathologic features of ESCC patients. As demonstrated in Table 1. XIST expression was prominently correlated with TMN stage (P = 0.006).
Table 1.
The association between the clinicopathologic features of ESCC patients and XIST expression
| Clinicopathologic features | N(%) | XIST level |
P value | |
|---|---|---|---|---|
| High(%) | Low(%) | |||
| Age(years) | P = 0.8792 | |||
| ≥55 | 26 (61.9) | 14 (53.8) | 12 (46.2) | |
| <55 | 16 (38.1) | 9 (56.3) | 7 (43.7) | |
| Gender | P = 0.9364 | |||
| Male | 29 (69.0) | 16 (55.2) | 13 (44.8) | |
| Female | 13 (31.0) | 7 (53.8) | 6 (46.2) | |
| TNM stage | P = 0.0064 | |||
| Ⅰ-Ⅱ | 15 (35.7) | 4 (26.7) | 11 (73.3) | |
| III-Ⅳ | 27 (64.3) | 19 (70.4) | 8 (29.6) | |
| Tumor location | P = 0.9287 | |||
| Middle | 18 (42.9) | 10 (55.6) | 8 (44.4) | |
| Lower | 24 (57.1) | 13 (54.2) | 11 (45.8) | |
3.2. XIST silencing repressed cell cycle progression, proliferation, migration, invasion, and promoted the apoptosis in ESCC cells
To investigate the effect of XIST on ESCC progression, loss-of-function experiments were performed by si-XIST transfection. Transfection of si-XIST (si-XIST-1/-2/-3), but not the si-NC control, significantly reduced the level of XIST in both EC9706 and Eca-109 cells (Figure 2a). Flow cytometric analysis revealed that XIST silencing led to a decrease in the population of cells that were in S phase in the two cells (Figure 2b). The results of CCK-8 assay showed that XIST silencing remarkably inhibited cell proliferation (Figure 2c). Transwell assay showed XIST silencing markedly suppressed the migration and invasion of EC9706 and Eca-109 cells (Figure 2(d,e)). Conversely, the apoptosis ability was strikingly promoted by XIST silencing in both EC9706 and Eca-109 cells (figure 2f).
Figure 2.
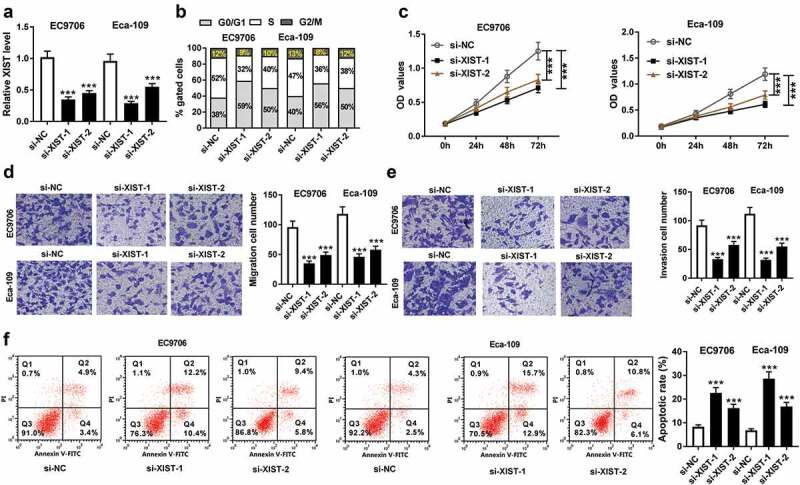
XIST silencing repressed cell cycle progression, proliferation, migration, invasion, and promoted the apoptosis in ESCC cells. (a) relative XIST level by qRT-PCR assay in EC9706 and Eca-109 cells transfected with si-NC, si-XIST-1 (also called si-XIST) or si-XIST-2. Data were expressed as mean ± SD of three independent replicates. ***P < 0.001 by ANOVA with Tukey’s test. EC9706 and Eca-109 cells were transfected with si-NC, si-XIST-1 or si-XIST-2, followed by the measurement of cell cycle progression by flow cytometric analysis (b), cell proliferation by CCK-8 assay (c), cell migration (d) and invasion (e) by transwell assay, cell apoptosis by flow cytometric analysis (f). Data were expressed as mean ± SD of three independent replicates. ***P < 0.001 by ANOVA with Tukey’s test
3.3. XIST directly interacted with miR-129-5p
To further explore the underlying mechanism by which XIST affected ESCC cell progression, we used three computational databases to predict the interacted miRNAs of XIST. The predicted data revealed that XIST contained a putative target sequence for miR-129-5p (Figure 3a). To confirm whether XIST directly interacted with miR-129-5p, XIST luciferase reporter plasmid (XIST-WT) harboring the putative complementary site for miR-129-5p was transfected into EC9706 and Eca-109 cells, together with miR-129-5p mimics. These data showed that the transfection of miR-129-5p mimics, but not miR-NC mimics, significantly reduced the luciferase activity of XIST-WT (Figure 3(b,c)). However, the site-directed mutant of the seed region prominently abrogated the repressive effect of miR-129-5p (Figure 3(b,c)). Ago2, a component of RNA-induced silencing complex (RISC), plays a vital role in the mature processes of miRNAs. Hence, to validate the endogenous interaction between XIST and miR-129-5p, RIP assay was performed using anti-Ago2 antibody. In contrast to the negative control, XIST was substantially enriched by miR-129-5p overexpression in EC9706 and Eca-109 cells (Figure 3(d,e)). Additionally, subcellular localization assays revealed that XIST and miR-129-5p in the two ESCC cells were mainly localized in the cytoplasm (figure 3f and 3g).
Figure 3.
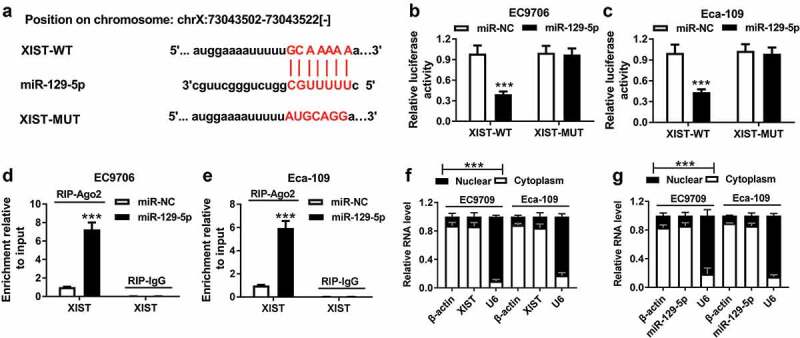
XIST directly interacted with miR-129-5p. (a) Nucleotide resolution of the predicted miR-129-5p binding site in XIST: seed sequence and mutated miR-129-5p binding sequence. (b and c) The luciferase activity was detected in EC9706 and Eca-109 cells transfected with XIST-WT or XIST-MUT and miR-NC mimics or miR-129-5p mimics. Data were expressed as mean ± SD of three independent replicates. ***P < 0.001 by student’s t-test. (d and e) RIP assay was performed using anti-Ago2 or anti-IgG in EC9706 and Eca-109 cells transfected with miR-NC mimics or miR-129-5p mimics. Data were expressed as mean ± SD of three independent replicates. ***P < 0.001 by student’s t-test. (f and g) subcellular localization assays in EC9706 and Eca-109 cells. Data were expressed as mean ± SD of three independent replicates
3.4. MiR-129-5p was downregulated in ESCC and its expression was repressed by XIST
Then, we determined the expression of miR-129-5p in ESCC tissues and cells. By contrast, qRT-PCR results revealed a significant downregulation of miR-129-5p level in ESCC tissues and cells (Figure 4(a,b)). More interestingly, our data showed that miR-129-5p level was inversely correlated with XIST expression in ESCC tissues (Figure 4c). Additionally, miR-129-5p expression was significantly associated with TMN stage of these ESCC patients (Table 2).
Figure 4.
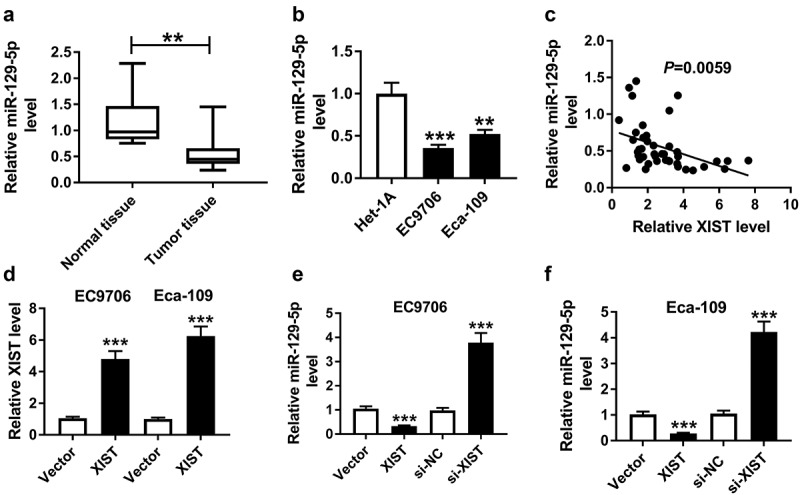
MiR-129-5p was downregulated in ESCC and its expression was repressed by XIST. The expression of miR-129-5p was detected by qRT-PCR assay in ESCC tissues and adjacent noncancerous tissues (a), human esophageal epithelial cell line (Het-1A) and ESCC cell lines (EC9706 and Eca-109) (b). Data were expressed as mean ± SD of three independent replicates. **P < 0.01 or ***P < 0.001 by Mann-Whitney U test or ANOVA with Dunnett-t test. (c) The correlation between miR-129-5p and XIST was determined in ESCC tissues. (d) Relative XIST level by qRT-PCR in EC9706 and Eca-109 cells transfected with vector or vector-XIST. Data were expressed as mean ± SD of three independent replicates. ***P < 0.001 by student’s t-test. MiR-129-5p expression was assessed in EC9706 (e) and Eca-109 (f) cells transfected with vector, vector-XIST, si-NC or si-XIST. Data were expressed as mean ± SD of three independent replicates. ***P < 0.001 by ANOVA with Tukey’s test
Table 2.
The association between the clinicopathologic features of ESCC patients and miR-129-5p expression
| Clinicopathologic features | N(%) | miR-129-5p level |
P value | |
|---|---|---|---|---|
| Low(%) | High(%) | |||
| Age(years) | P = 0.4736 | |||
| ≥55 | 26 (61.9) | 15 (57.7) | 11 (42.3) | |
| <55 | 16 (38.1) | 11 (68.8) | 5 (31.2) | |
| Gender | P = 0.4715 | |||
| Male | 29 (69.0) | 19 (65.5) | 10 (34.5) | |
| Female | 13 (31.0) | 7 (53.8) | 6 (46.2) | |
| TNM stage | P = 0.0293 | |||
| I-II | 15 (35.7) | 6 (40.0) | 9 (60.0) | |
| III-IV | 27 (64.3) | 20 (74.1) | 7 (25.9) | |
| Tumor location | P = 0.5821 | |||
| Middle | 18 (42.9) | 12 (66.7) | 6 (33.3) | |
| Lower | 24 (57.1) | 14 (58.3) | 10 (41.7) | |
Given the data that XIST directly interacted with miR-129-5p, we further observed whether XIST modulated miR-129-5p expression. The transfection efficiency of XIST overexpression plasmid (Vector-XIST) was gauged by qRT-PCR (Figure 4d). As expected, miR-129-5p expression was markedly repressed when introduction of Vector-XIST, while it was dramatically elevated by si-XIST silencing in EC9706 and Eca-109 cells (Figure 4(e,f)).
3.5. MiR-129-5p overexpression repressed cell cycle progression, proliferation, migration, invasion, and enhanced the apoptosis in ESCC cells
Further, to investigate the role of miR-129-5p in ESCC cell progression, we manipulated miR-129-5p expression by miR-129-5p mimics. Transfection of miR-129-5p mimics, but not miR-NC mimics, resulted in a 5.2-fold increase of miR-129-5p expression in EC9706 cells and a 4.1-fold increase in Eca-109 cells (Figure 5a). Subsequently, functional experiments revealed that in comparison to their counterparts, the enforced miR-129-5p expression led to a significant suppression in cell cycle progression, proliferation, migration and invasion, as well as a striking promotion in cell apoptosis (Figure 5(b,c,d,e,f)).
Figure 5.
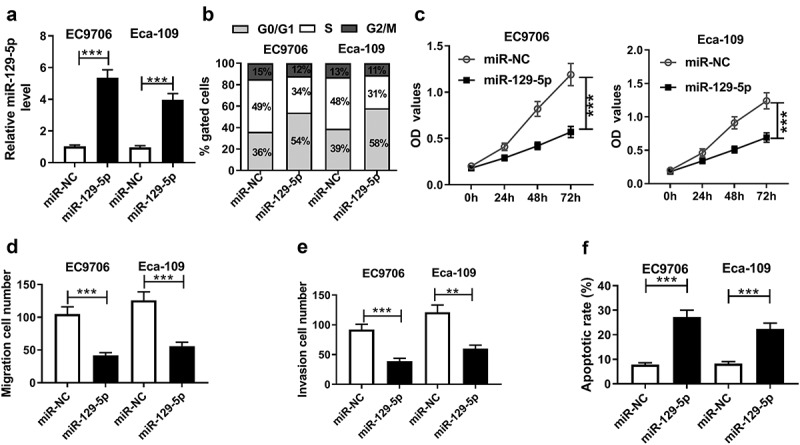
MiR-129-5p overexpression repressed cell cycle progression, proliferation, migration, invasion, and enhanced the apoptosis in ESCC cells. EC9706 and Eca-109 cells were transfected with miR-NC mimics or miR-129-5p mimics. (a) qRT-PCR assay for miR-129-5p expression in transfected cells. (b) Flow cytometric analysis for cell cycle progression. (c) CCK-8 assay for cell proliferation. Transwell assay for cell migration (d) and invasion (e) in transfected cells. (f) Flow cytometric analysis for cell apoptosis in treated cells. Data were expressed as mean ± SD of three independent replicates. **P < 0.01 or ***P < 0.001 by Student’s t-test
3.6. MiR-129-5p mediated the regulatory effect of XIST on ESCC cell cycle progression, proliferation, migration, invasion and apoptosis
To provide further mechanistic insight into the link between XIST and miR-129-5p on ESCC cell progression, EC9706 and Eca-109 cells were cotransfected with si-XIST and anti-miR-129-5p. qRT-PCR assay showed that XIST silencing-mediated miR-129-5p upregulation was sharply abated by the cotransfection with anti-miR-129-5p (Figure 6(a,b)). Subsequent experiments demonstrated that si-XIST-mediated inhibitory effects on cell cycle progression, proliferation, migration and invasion were drastically abolished by miR-129-5p down-regulation compared with the negative control (Figure 6(c,d,e,f,g,h,i,j)). Moreover, si-XIST-mediated pro-apoptosis effect was significantly reversed by the reduced expression of miR-129-5p in EC9706 and Eca-109 cells (Figure 6(k,l)).
Figure 6.
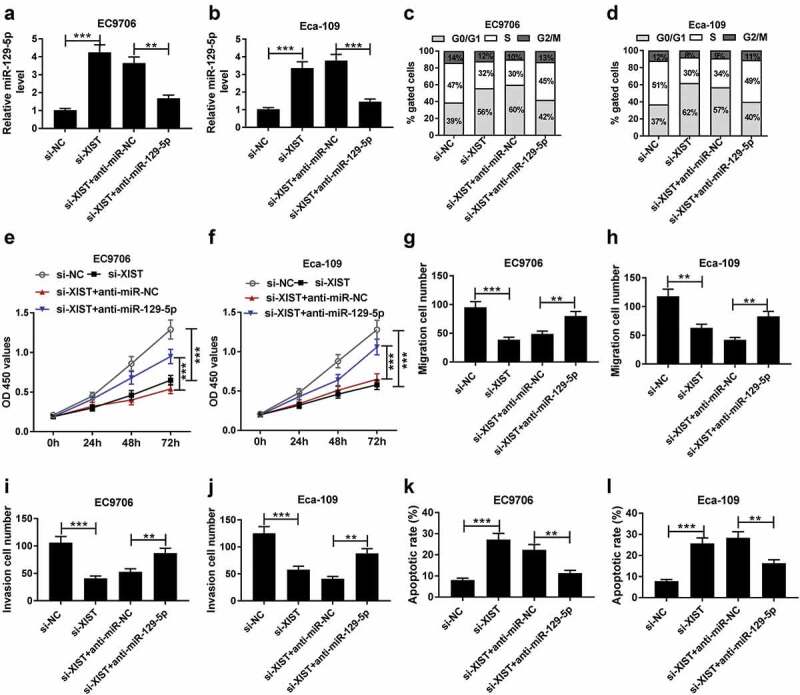
XIST exerted its regulatory effect on ESCC cell progression by miR-129-5p. EC9706 and Eca-109 cells were transfected with si-NC, si-XIST, si-XIST+anti-miR-NC or si-XIST+anti-miR-129-5p, followed by the determination of miR-129-5p expression by qRT-PCR assay (a and b), cell cycle progression by flow cytometric analysis (c and d), cell proliferation by CCK-8 assay (e and f), cell migration (g and h) and invasion (i and j) by transwell assay, cell apoptosis by flow cytometric analysis (k and l). Data were expressed as mean ± SD of three independent replicates. **P < 0.01 or ***P < 0.001 by ANOVA with Tukey’s test
3.7. XIST promoted CCND1 expression by sponging miR-129-5p in ESCC cells
Then, we carried out a detailed analysis of the molecular targets of miR-129-5p using starBase v2.0 software. The predicted data revealed that three cell cycle-related genes (CCND1, CCND2 and CDK6) harbored a putative target sequence for miR-129-5p in the 3ʹ-UTR of CCND1 mRNA, and we found that the incubation with miR-129-5p probe led to the most significant increase in the enrichment level of CCND1 in both EC9706 and Eca-109 cells (Supplement Figure 1(a-c)). We thus selected CCND1 for further analysis. To verify whether CCND1 was a direct target of miR-129-5p, we cloned the segment of CCND1 3’-UTR encompassing the target region into a luciferase reporter (CCND1-WT) and mutated the target sequence (CCND1-MUT, Figure 7a). The transfection of CCND1-WT in the presence of miR-129-5p mimics induced about a 70% downregulation of relative luciferase activity, and the effect was significantly abolished by CCND1-MUT (Figure 7(b,c)). Subsequent RIP experiment revealed that miR-129-5p overexpression led to a remarkable increase in the enrichment level of CCND1 in the two cells (Figure 7(d,e)).
Figure 7.
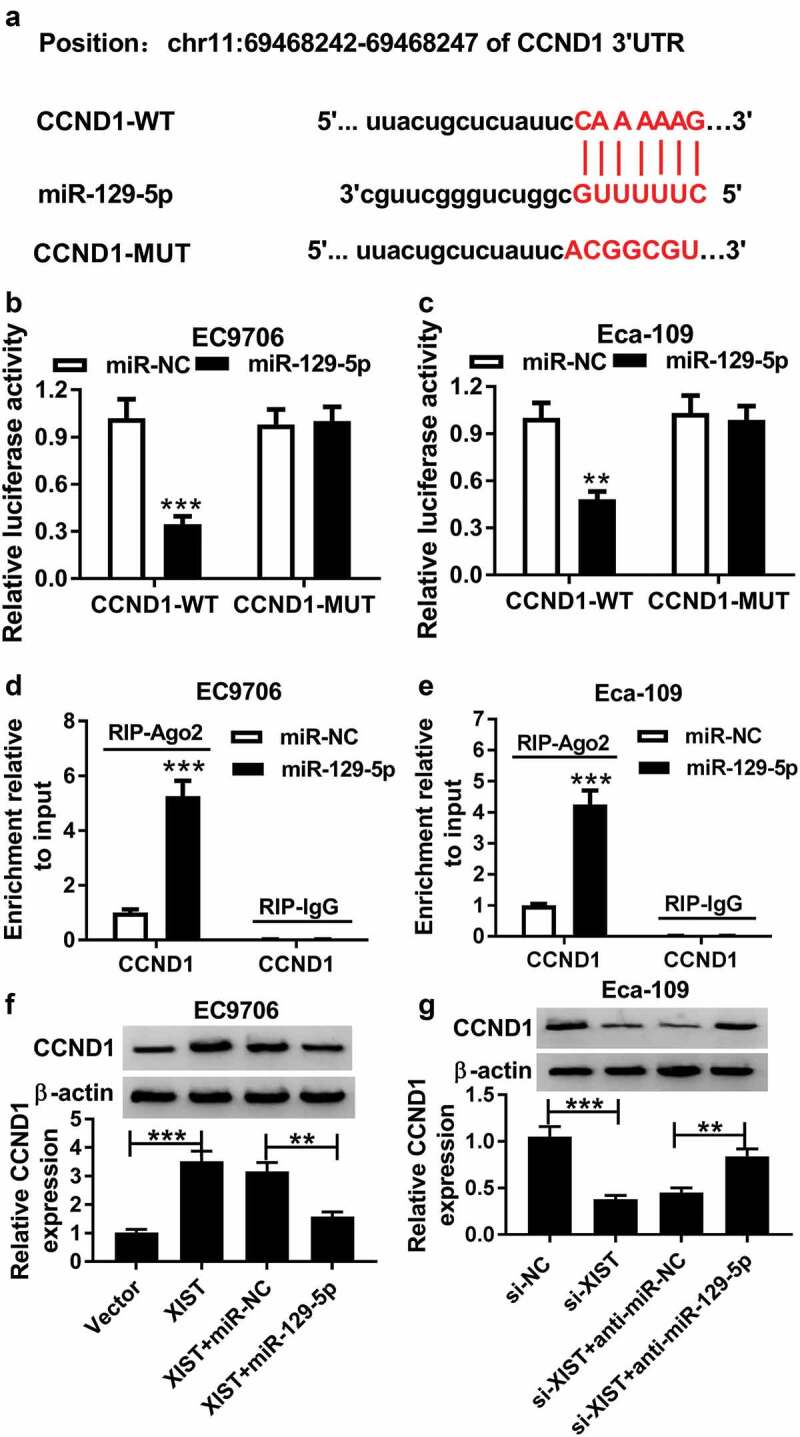
XIST promoted CCND1 expression by sponging miR-129-5p in ESCC cells. (a) Nucleotide resolution of the predicted miR-129-5p binding site in the 3ʹ-UTR of CCND1 mRNA: the seed sequence and mutated miR-129-5p-binding sequence. (b and c) Dual-luciferase reporter assay was performed to verify whether CCND1 was a direct target of miR-129-5p by transfecting with CCND1-WT or CCND1-MUT into EC9706 and Eca-109 cells together with miR-NC mimics or miR-129-5p mimics. Data were expressed as mean ± SD of three independent replicates. **P < 0.01 or ***P < 0.001 by Student’s t-test. (d and e) RIP assay was employed to confirm the interaction between XIST and miR-129-5p using anti-Ago2 in EC9706 and Eca-109 cells transfected with miR-NC mimics or miR-129-5p mimics. Data were expressed as mean ± SD of three independent replicates. ***P < 0.001 by student’s t-test. (f) EC9706 cells were transfected with vector, vector-XIST, vector-XIST+miR-NC mimics or vector-XIST+miR-129-5p mimics, and then CCND1 expression was detected by western blot analysis. Data were expressed as mean ± SD of three independent replicates. **P < 0.01 or ***P < 0.001 by ANOVA with Tukey’s test. (g) The expression of CCND1 was assessed by western blot analysis in Eca-109 cells transfected with si-NC, si-XIST, si-XIST+anti-miR-NC or si-XIST+anti-miR-129-5p. Data were expressed as mean ± SD of three independent replicates. **P < 0.01 or ***P < 0.001 by ANOVA with Tukey’s test
Then, we asked whether, if so, how XIST regulated CCND1 expression in ESCC cells. As expected, in comparison to their counterparts, CCND1 expression was significantly enhanced by XIST upregulation, and it was markedly inhibited when XIST silencing (figure 7(f,g)). Nevertheless, these effects were remarkably abolished by the alteration of miR-129-5p expression (figure 7(f,g)).
3.8. XIST silencing inhibited tumor growth in vivo
Given our data that XIST depletion repressed ESCC cell progression in vitro, we further explored the effect of XIST on tumor growth in vivo. These results revealed that transduction of sh-XIST significantly inhibited tumor growth, as presented by the decrease of tumor volume and weight (Figure 8(a,b)). Additionally, qRT-PCR assay presented that sh-XIST transduction resulted in a significant decrease of XIST expression and an increase of miR-129-5p expression in the xenograft tissues (Figure 8(c,d)). Moreover, XIST silencing led to a distinct inhibition of CCND1 expression in tumor tissues (Figure 8(e,f)).
Figure 8.
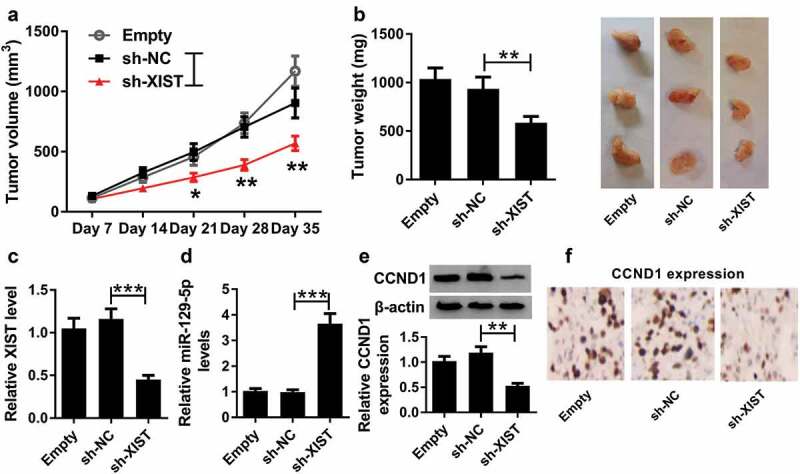
XIST silencing inhibited tumor growth in vivo. Eca-109 cells stably transfected with sh-NC or sh-XIST were subcutaneously implanted into nude mice (n = 6 each group). After 35 days implantation, all mice were sacrificed and the xenograft tissues were excised. (a) After 7 days, tumor volume was measured with a digital caliper every 7 days. (b) Representative images were photographed and tumor average weight was calculated. (c) qRT-PCR assay for XIST expression in xenograft tissues. (d) qRT-PCR assay for miR-129-5p level in excised tissues. (e) Western blot analysis for CCND1 expression in xenograft tissues. (f) Immunohistochemistry assay for CCND1 expression in excised tissues. Data were expressed as mean ± SD of six independent replicates. *P < 0.05 or **P < 0.01 or ***P < 0.001 by ANOVA with Dunnett-t test
4. Discussion
Growing amount of evidence has suggested that the dysregulation of lncRNAs is involved in the carcinogenesis of human cancers, including ESCC [14,15]. Previous documents identified that XIST functioned as a tumor driver in many human cancers, such as pancreatic cancer and gastric cancer [16,17]. In the present study, we pointed to the role of XIST as an oncogenic lncRNA in ESCC, consistent with previous work [12]. The findings by Chen et al. demonstrated the precise parts of XIST in ESCC cell line TE-1 and esophageal adenocarcinoma cell line SKGT-4 [18], which was different from the present study in two ESCC cells. Additionally, compared with the research by Wu et al. [12], we were first to uncover that XIST knockdown hampered ESCC cell cycle progression and enhanced cell apoptosis. Similar to our findings, several other lncRNAs, such as FOXD2-AS1 and BANCR, had been reported to be upregulated in ESCC tissues and predicted poor prognosis of ESCC patients [19,20]. Previous studies had uncovered that XIST played a key role in human carcinogenesis by functioning as a miRNA sponge [12,16,18]. For example, Chen et al. reported the oncogenic role of XIST in esophageal cancer through regulating miR-494 [18]. Wu et al. identified XIST as a tumor driver in ESCC by sponging miR-101 [12]. These reports described in this paper demonstrated XIST as an oncogenic lncRNA in cancer biology by targeting many different miRNAs, and these miRNAs might be parallel in terms of XIST functions.
Here, we confirmed that XIST functioned as a molecular sponge of miR-129-5p in ESCC cells. MiR-129-5p was reported to inhibit tumor growth and metastasis of many human cancers, including gastric cancer [21], laryngeal squamous cell carcinoma [22] and non-small cell lung cancer [23], highlighting its role as a tumor suppressor in cancer progression. In addition, a previous document reported that miR-129-5p was involved in the antitumor activity of histone deacetylase inhibitors and provided a miRNA-driven cell death mechanism in papillary thyroid cancer cells [24]. Moreover, miR-129 was showed to participate in the regulatory network of lnc-ROR-miRNA-SOX9 and NEAT1-miRNA-CTBP2 in ESCC [25,26]. In this study, our data indicated that miR-129-5p was downregulated in ESCC tissues and cells, and the enforced level of miR-129-5p repressed ESCC progression in vitro. More importantly, we firstly elucidated that miR-129-5p was a functional mediator in XIST in regulating on ESCC cell progression in vitro. Combining with the findings by Wu et al. [12], XIST accelerated ESCC progression through sponging miR-129-5p and miR-101, which might be two parallel pathways or interacted results.
It is widely acknowledged that lncRNAs regulate gene expression by functioning as sponges of miRNAs [8]. Among these miR-129-5p targets predicted by starBase v.2 software, CCND1 was of interest in this study, considering its protooncogene role in multiple cancers, such as bladder cancer [27], prostate cancer [28], and ovarian cancer [29]. CCND1 was reported to act as a key regulator of the G1/S phase of the cell cycle in cancer cells [30]. Moreover, CCND1 was involved in the progression of ESCC by mediating the regulation of miR-1 [31]. Additionally, CCND1 expression was highly elevated in ESCC and CCND1 silencing repressed ESCC progression in vitro [32,33]. In the present study, we first highlighted that XIST promoted CCND1 expression by sponging miR-129-5p in ESCC cells. Similar to our findings, several other miRNAs, such as miR-503 and miR-99, were demonstrated to mitigate ESCC cell progression in vitro through inhibiting CCND1 expression [32,34]. Ren et al. reported that lncRNA HOTAIR contributed to ESCC progression through sponging miR-1 and regulating CCND1 expression [33]. Lastly, in vivo assays further revealed that XIST silencing repressed tumor grown possibly through regulating the miR-129-5p/CCND1 axis in vivo. The regulatory networks of the ceRNAs were complicated, and there might be other pro-progression mechanism of XIST on ESCC waiting to be uncovered. In conclusion, our study indicated that XIST silencing repressed the progression of ESCC at least partly through regulating the miR-129-5p/CCND1 axis. Targeting XIST might be a potential therapeutic strategy for ESCC treatment.
Supplementary Material
Acknowledgments
Not applicable
Funding Statement
The authors have no funding to report.
Authors’ Contributions
Conception and design: Haoran Wang and Yin Li; Development of methodology: Haomiao Li, Yongkui Yu; Acquisition of data: Qingfeng Jiang; Analysis and interpretation of data: Ruixiang Zhang, Haibo Sun; Writing, review, and revision of article: Haoran Wang and Wenqun Xing; Study supervision: Yin Li. All coauthors have reviewed and approved of the article before submission.
Availability of data and materials
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable
Disclosure statement
No financial and/or non-financial conflicts should be declared by all named co-authors.
Ethics approval and consent to participate
Written informed consent was obtained from all patients and the study was approved by the Ethics Committee of the Affiliated Cancer Hospital of Zhengzhou University.
Our study was approved by the Animal Care and Use Committee of the Affiliated Cancer Hospital of Zhengzhou University.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Abnet CC, Arnold M, Wei WQ.. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154(2):360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rice TW, Apperson-Hansen C, DiPaola LM, et al. Worldwide esophageal cancer collaboration: clinical staging data. Dis Esophagus. 2016;29(7):707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–638. [DOI] [PubMed] [Google Scholar]
- [7].Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cheng Y, Chang Q, Zheng B, et al. LncRNA XIST promotes the epithelial to mesenchymal transition of retinoblastoma via sponging miR-101. Eur J Pharmacol. 2019;843:210–216. [DOI] [PubMed] [Google Scholar]
- [10].Zhu H, Zheng T, Yu J, et al. LncRNA XIST accelerates cervical cancer progression via upregulating fus through competitively binding with miR-200a. Biomed Pharmacother. 2018a;105:789–797. [DOI] [PubMed] [Google Scholar]
- [11].Zhu J, Zhang R, Yang D, et al. Knockdown of long non-coding RNA xist inhibited doxorubicin resistance in colorectal cancer by upregulation of miR-124 and downregulation of SGK1. Cell Physiol Biochem. 2018b;51(1):113–128. [DOI] [PubMed] [Google Scholar]
- [12].Wu X, Dinglin X, Wang X, et al. Long noncoding RNA XIST promotes malignancies of esophageal squamous cell carcinoma via regulation of miR-101/EZH2. Oncotarget. 2017;8(44):76015–76028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu T, Liu Y, Wei C, et al. LncRNA HULC promotes the progression of gastric cancer by regulating miR-9-5p/MYH9 axis. Biomed Pharmacother. 2019;121:109607. [DOI] [PubMed] [Google Scholar]
- [14].Lin C, Wang Y, Wang Y, et al. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene. 2017;36(38):5392–5406. [DOI] [PubMed] [Google Scholar]
- [15].Song W, Zou SB, Prognostic role of lncRNA HOTAIR in esophageal squamous cell carcinoma. Clin Chim Acta. 2016;463: 169–173. [DOI] [PubMed] [Google Scholar]
- [16].Chen DL, Ju HQ, Lu YX, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ma L, Zhou Y, Luo X, et al. Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancer. Oncotarget. 2017a;8(3):4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen Z, Hu X, Wu Y, et al. Long non-coding RNA XIST promotes the development of esophageal cancer by sponging miR-494 to regulate CDK6 expression. Biomed Pharmacother. 2019b;109:2228–2236. [DOI] [PubMed] [Google Scholar]
- [19].Bao J, Zhou C, Zhang J, et al. Upregulation of the long noncoding RNA FOXD2-AS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Biomark. 2018;21(3):527–533. [DOI] [PubMed] [Google Scholar]
- [20].Liu Z, Yang T, Xu Z, et al. Upregulation of the long non-coding RNA BANCR correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Biomed Pharmacother. 2016;82:406–412. [DOI] [PubMed] [Google Scholar]
- [21].Yu X, Song H, Xia T, et al. Growth inhibitory effects of three miR-129 family members on gastric cancer. Gene. 2013;532(1):87–93. [DOI] [PubMed] [Google Scholar]
- [22].Li M, Tian L, Wang L, et al. Down-regulation of miR-129-5p inhibits growth and induces apoptosis in laryngeal squamous cell carcinoma by targeting APC. PLoS One. 2013;8(10):e77829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma Z, Cai H, Zhang Y, et al. MiR-129-5p inhibits non-small cell lung cancer cell stemness and chemoresistance through targeting DLK1. Biochem Biophys Res Commun. 2017b;490(2):309–316. [DOI] [PubMed] [Google Scholar]
- [24].Brest P, Lassalle S, Hofman V, et al. MiR-129-5p is required for histone deacetylase inhibitor-induced cell death in thyroid cancer cells. Endocr Relat Cancer. 2011;18(6):711–719. [DOI] [PubMed] [Google Scholar]
- [25].Li Y, Chen D, Gao X, et al. LncRNA NEAT1 regulates cell viability and invasion in esophageal squamous cell carcinoma through the miR-129/CTBP2 axis. Dis Markers. 2017;(2017:5314649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang L, Yu X, Zhang Z, et al. Linc-ROR promotes esophageal squamous cell carcinoma progression through the derepression of SOX9. J Exp Clin Cancer Res. 2017;36(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen Z, Chen X, Xie R, et al. DANCR promotes metastasis and proliferation in bladder cancer cells by enhancing il-11-stat3 signaling and CCND1 expression. Mol Ther. 2019a;27(2):326–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Long B, Li N, Xu XX, et al. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem Biophys Res Commun. 2018;505(2):561–568. [DOI] [PubMed] [Google Scholar]
- [29].Dai J, Wei RJ, Li R, et al. A study of CCND1 with epithelial ovarian cancer cell proliferation and apoptosis. Eur Rev Med Pharmacol Sci. 2016;20:4230–4235. [PubMed] [Google Scholar]
- [30].Gennaro VJ, Stanek TJ, Peck AR, et al. Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc Natl Acad Sci U S A. 2018;115(40):E9298–e9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jiang S, Zhao C, Yang X, et al. miR-1 suppresses the growth of esophageal squamous cell carcinoma in vivo and in vitro through the downregulation of MET, cyclin D1 and CDK4 expression. Int J Mol Med. 2016;38(1):113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jiang L, Zhao Z, Zheng L, et al. Downregulation of miR-503 promotes escc cell proliferation, migration, and invasion by targeting cyclin D1. Genomics Proteomics Bioinformatics. 2017;15(3):208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ren K, Li Y, Lu H, et al. Long noncoding RNA hotair controls cell cycle by functioning as a competing endogenous RNA in esophageal squamous cell carcinoma. In: Transl Oncol 2016;9:489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mei LL, Qiu YT, Huang MB, et al. mir-99a suppresses proliferation, migration and invasion of esophageal squamous cell carcinoma cells through inhibiting the IGF1R signaling pathway. Cancer Biomark. 2017;20(4):527–537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.


