ABSTRACT
Bispecific antibodies are an important and growing segment in antibody therapeutics, particularly in the immuno-oncology space. Manufacturing of a bispecific antibody with two different heavy chains is greatly simplified if the light chains can be the same for both arms of the antibody. Here, we introduce a strain of common light chain chickens, called OmniClic®, that produces antibody repertoires largely devoid of light chain diversity. The antibody repertoire in these chickens is composed of diverse human heavy chain variable regions capable of high-affinity antigen-specific binding and broad epitope diversity when paired with the germline human kappa light chain. OmniClic birds can be used in immunization campaigns for discovery of human heavy chains to different targets. Subsequent pairing of the heavy chain with a germline human kappa light chain serves to facilitate bispecific antibody production by increasing the efficiency of correct pairing.
Abbreviations: AID: activation-induced cytidine deaminase; bsAb: bispecific antibody; CDR: complementarity-determining region; CL: light chain constant region; CmLC: common light chain; D: diversity region; ELISA: enzyme-linked immunosorbent assay; FACS: fluorescence-activated cell sorting; Fc: fragment crystallizable; FcRn: neonatal Fc receptor; FR: framework region; GEM: gel-encapsulated microenvironment; Ig: immunoglobulin; IMGT: the international ImMunoGeneTics information system®; J: joining region; KO: knockout; mAb: monoclonal antibody; NGS: next-generation sequencing; PBS: phosphate-buffered saline; PCR: polymerase chain reaction; PGC: primordial germ cell; PGRN: progranulin; TCR: T cell receptor; V: variable region; VK: kappa light chain variable region; VL: light chain variable region; VH: heavy chain variable region
KEYWORDS: Bispecific, common light chain, transgenic chicken, human antibodies, antibody engineering
Introduction
Bispecific antibodies (bsAbs) comprise a diverse collection of novel molecules in various formats and configurations designed to bind two distinct epitopes or targets at the same time. The pipeline of bsAbs in pharmaceutical development is quite large and diverse, with goals such as retargeting T effector cells to tumor cells, chaperoning proteins across the blood-brain barrier, anti-inflammatory effects, and increasing avidity by binding multiple epitopes on the same cell.1 In the case of retargeting of T cells to tumor cells, one arm of the bsAb is often specific for the CD3 delta or epsilon chains of the T cell receptor complex, and the other arm specific for a receptor on the tumor cells. To date, three bsAbs have been granted marketing approvals,2 although currently only two are on the market (blinatumomab (Blincyto®), for acute B-cell lymphoblastic leukemia, and emicizumab (Hemlibra®), for treating hemophilia A). With over 85 potential bsAbs in clinical development, this number is likely to rise.1
Using the modular domain structure of immunoglobulins, bispecific molecules have been formatted in myriad creative ways.3,4 These formats can be roughly divided into those that include the Fc domain and those that do not. Of those that include the Fc, some maintain an IgG-like structure that is similar to native IgG, whereas others take more creative structures not seen in nature such as multiple binding domains in tandem. Conforming to a more native IgG-like format offers advantages in patients, such as binding to FcRn and longer serum half-life, and potentially desired effector functions mediated by the intact Fc. More unusual formats may suffer from aggregation, longer development lead times, and require further engineering,5–7 whereas IgG-like formats are well-studied and have a long history in patients. However, bsAbs with an IgG-like format pose their own engineering challenges for manufacturing.2,3 Normal antibodies consist of a homodimer of a heavy-light chain pair, forming the two arms that both have the same antigen-binding site. In contrast, a bispecific antibody in IgG-like format must be a heterodimer with two distinct arms targeting the two distinct epitopes. If two different heavy chains and two different light chains were to be expressed in the same cell, then the various chains could assemble in 10 different combinations, only one of which would be the desired bispecific configuration.8 This challenging expression problem can be separated into the heavy chain pairing problem, in which the goal is an obligate heavy chain heterodimer with no homodimerization, and the light chain pairing problem, in which the correct light chain must pair with the correct heavy chain. By using two heavy chains that can pair with a common light chain, one of these two problems is solved, leaving only the heavy chain pairing problem. Various strategies at promoting heterodimerization or purifying heterodimers of the Fc domains of heavy chains have been developed.9–11 The common light chain strategy is most useful in IgG and IgG-like formats and eliminates a substantial hurdle in bispecific antibody production.
Discovery of heavy chains that can pair with a common light chain has been pursued either in display systems12–15 or in transgenic rodents.16–18 Conserved human targets may not be amenable to antibody discovery in rodent platforms due to the high degree of sequence homology, which can result in low immunogenicity. For targeting such antigens, chickens represent an evolutionarily divergent host in which most human proteins will induce a robust immune response, enabling antibody discovery when campaigns in rodents have failed.19–23 The chicken also readily produces antibodies that are cross-reactive to the mouse ortholog of the human target, obviating the need to identify surrogate antibodies for pre-clinical studies. As with campaigns for monospecific antibodies, bispecific antibody campaigns can benefit from the ability to identify mouse cross-reactive antibodies.24 In addition, as described below, the mechanisms of antibody generation in the chicken are uniquely suited to the engineering of a common light chain transgenic animal.
In chickens, development of the antibody repertoire is initiated when V(D)J recombination leads to expression of the functional B cell receptor on the surface of early B cells.25 However, unlike in mammals, very little diversity is produced at the recombination step. Chickens contain only a single functional V gene and a single J gene at the heavy and light chain loci,26,27 and the heavy chain contains a cluster of D segments that are highly related to each other.28 No terminal deoxynucleotidyl transferase activity is present in chicken B cells, so no N addition can occur.29 Recombination thus produces essentially the same clonotype in every B cell, with the same V framework and nearly the same complementarity-determining regions (CDRs). As soon as recombination has produced a functional receptor, variable region sequences begin to mutate by the process of gene conversion, which leads to a highly diverse repertoire. Gene conversion incorporates sequences from an array of upstream pseudogenes in a homologous recombination-based process, leading to changes in the functional V. Multiple rounds of overlapping gene conversion occur from different pseudogenes, and indels can also be incorporated. In wild-type chickens, the light chain contains 25 pseudogenes and the heavy chain about 80 pseudogenes (the international ImMunoGeneTics information system, IMGT30). Pseudogene sequences contain diversity mainly in the CDRs, while the sequence of the VL/VH frameworks is largely maintained. However, in addition to gene conversion, random somatic hypermutation can also occur and adds to the sequence diversity in the repertoire.31
We have engineered chickens to express human V region antibodies (OmniChickens®).20,32 The constructs in these birds are designed with human pseudogenes containing diverse CDRs, to produce large repertoires of human sequences in the B cell population of these birds.20,33,34 Here, we pursued the opposite goal, at least for the light chain: a chicken with a non-mutating, common light chain that can pair with any heavy chain in the bird, for discovery of antibodies that can be developed into bispecifics. To design a light chain transgene in the chicken that does not produce a diverse sequence repertoire, we took advantage of gene conversion, but instead of inserting diverse pseudogenes, we inserted an array of identical pseudogenes to the functional human VK3-15/JK1 gene. The process of gene conversion and single framework architecture in the chicken is perfectly suited to this type of active suppression of antibody diversity, as compared to the typical mammalian locus with its multiple V genes that undergo recombination. In this common light chain chicken (called OmniClic®), not only will gene conversion not provide sequence diversity, it is designed to actively “cleanse” any mutations that may arise by random somatic hypermutation by reverting them to the wild-type sequence. Since we are taking advantage of the normal gene conversion process used by the chicken B cell, it is not necessary to make any other genetic or physiological changes to developing B cells, and diversity generation in the heavy chains should be unimpeded or possibly even encouraged given the lack of diversity in the light chain.
Here we show that the OmniClic chicken produces minimally mutated light chains. Despite the lack of light chain diversity, OmniClic raises robust immune responses upon immunization and produces high-affinity antibodies with diverse human heavy chains. Comparison to normal OmniChickens expressing diverse human heavy/light pairs shows that epitope binning and affinity are essentially normal when diversity is restricted to the heavy chain. The OmniClic platform can be used for antibody discovery of heavy chain sequences to be used with the germline VK3-15 light chain for bispecific applications.
Results
Generation of OmniClic birds
We set out to design a transgene construct that would express a human variable region light chain in chicken B cells that would remain unmutated, yet pair with diversified heavy chains to produce antibodies with high affinity and broad epitope coverage. This common light chain bird we call OmniClic, to distinguish it from our “standard” OmniChicken birds, which produce diverse repertoires in both light and heavy chains through gene conversion by upstream human-based pseudogenes. In OmniChickens, gene conversion events in the mature repertoire can be attributed to specific pseudogenes, showing that the pseudogenes are participating in gene conversion, even though most CDR sequences in mature antibodies are highly mutated and cannot be ascribed to identifiable gene conversion.20 To design OmniClic, we reasoned that an array of upstream pseudogenes identical to the germline functional V region could use the mechanisms of gene conversion to the opposite effect, i.e., to revert any mutations that might occur randomly by non-templated somatic hypermutation back to the germline sequence. This strategy effectively uses gene conversion to maintain the germline sequence, rather than producing a diverse repertoire of mutations. By using a pre-rearranged light chain and full-length reverting pseudogenes, we include all three CDRs in the reversion strategy. The enzymatic machinery for generating sequence diversity in immunoglobulins is conserved in chickens35,36 and depends on an initiating lesion by the activation-induced cytidine deaminase (AID) enzyme, leading to somatic hypermutation, gene conversion, or class switch recombination.37–39 These AID-based mechanisms remain intact in OmniClic, which is crucial because they are necessary to produce a diverse heavy chain repertoire and are also necessary for the reverting gene conversion strategy in the light chain.
The common light chain construct (CmLC1, Figure 1) in OmniClic is inserted in the chicken light chain locus, as are all the human light chain transgenes in OmniChicken, thereby utilizing the endogenous transcriptional regulatory elements and non-translated regions for optimal lineage-specific expression. This location also ensures that gene conversion in the B cell lineage will be focused on the light chain transgene.40,41 The variable region is a pre-rearranged VK3-15/JK1 region made by joining the germline gene segments together, with no chewing back of the ends. This rearranged sequence is expressed in humans, as it is found in the National Center for Biotechnology Information’s Expressed Sequence Tags database. The V region splices to the chicken CL constant region, producing a chimeric light chain. In the OmniClic locus, the chicken VL pseudogenes remain upstream of the human pseudogenes (Figure 1).
Figure 1.
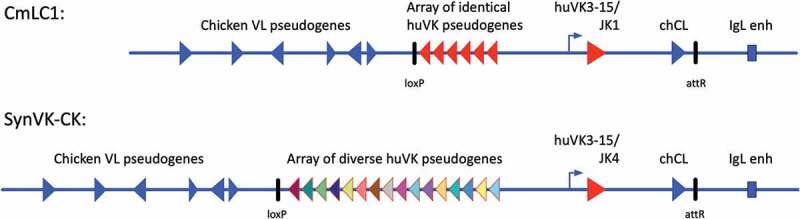
CmLC1 transgene structure
Top line, the CmLC1 transgene was inserted into the chicken light chain locus using phiC31 integrase, as previously described for the V-kappa and V-lambda OmnChicken light chains.20,32 The transgene contains a human pre-rearranged variable region (VK3-15/JK1) transcriptionally driven by the chicken IgL promoter and enhancer elements (IgL enh). Upstream of the promoter lie six pseudogenes with the same sequence as the functional V region, in opposite orientation as shown by the direction of the arrowhead shapes. The human V region splices to the chicken CL to produce a chimeric light chain. Upstream of the human pseudogenes lies the chicken light chain pseudogenes. For comparison, the V-kappa transgene in normal OmniChickens (SynK-CK) is shown, with its array of diverse human pseudogenes. The SynVK-CK transgene also contains a JK4 segment, rather than JK1 in CmLC1, and the first amino acid of the constant region (which is partly encoded by the V region exon) is R in SynVK-CK and G in CmLC1. The rest of the constant region is the chicken CL in both. Human sequences are shown in red, chicken sequences in blue. After plasmid insertion into the light chain locus, selectable markers are removed with Cre recombination, leaving behind a single loxP site and an attR site.
In OmniClic chickens, the CmLC1 light chain can be paired with either of the two human heavy chain transgenes that we use in OmniChicken lines, SynVH-C (pre-rearranged VH3-23/JH4) and SynVH-SD (rearranging VH3-23/Ds/JH6). In OmniChicken lines, these heavy chains are paired with the human V-kappa transgene (SynVK-CK; pre-rearranged VK3-15/JK4),20 or a human V-lambda transgene (SynVL-E-CL; VL1-44/JL3).32 The heavy chains are designed in a similar fashion to the light chains, each with a single functional human variable region, an array of upstream human pseudogenes for gene conversion, and splicing to the chicken constant regions for efficient engagement of Fc receptors and B cell receptor complexes in the bird. Chickens contain one light chain and one heavy chain. The human heavy and light transgenes are inserted at the endogenous heavy or light chain loci, and the other allele at each locus in the birds is a knockout (KO), so the only functional allele is the human transgene.26,27,42,43
B cell development in OmniClic birds
OmniClic birds were evaluated for B cell development (Fig. S1). Peripheral blood lymphocytes were stained with antibodies to the chicken B cell marker Bu-1, surface IgM, IgL, T cell markers, and human VK3-15/VH3-23. Normal proportions of B cells were observed in OmniClic, and expression of human VK3-15/VH3-23 was detected. Since the constant regions of both the light and heavy chains are chicken, we can detect surface or plasma Ig using anti-chicken reagents and directly compare our transgenic birds to wild-type birds. Titers of total IgY levels in plasma were assessed and compared to wild-type chickens and the kappa and lambda OmniChicken strains (Fig. S2). The OmniClic chickens contained plasma IgY levels comparable to those in the V-lambda OmniChickens, which is about twofold below the wild-type level (Fig. S2). In contrast, V-kappa OmniChickens with the SynVK-CK construct express levels that are at least 10-fold lower than wild-type (Fig. S2).20 The fact that the CmLC1 construct expresses a higher level of serum Ig than the SynVK-CK construct, even though both contain human V-kappa, may stem from sequence differences in the contact residues of the V and C domains in these chimeric proteins. Based on structural modeling, the V and C domains make contacts at their interface, and the residue in the framework region (FR)4-CL junction is predicted to have contacts with the CL domain (Fig. S3). This residue is in a connecting loop between the domains, and its codon is split between the V exon and the C exon. This loop residue is Gly in CmLC1 and wild-type chicken light chain, but in SynVK-CK and wild-type human kappa it is Arg. The large, charged side chain of Arg in this position could hamper the ability of the human V region to fold efficiently with the chicken C region in SynVK-CK. There are a few other differences in FR1 and in the germline CDRs between CmLC1 and SynVK-CK, but they are not predicted to have an effect on the V-C interaction. The OmniChicken V-lambda construct is fully human (not chimeric), and therefore should have no inter-domain folding issues. Thus, the junction region between FR4 and the CL appears to be important for the efficient expression of a chimeric light chain with human VK and chicken CL.
Immunization of OmniClic birds and evaluation of heavy and light chain repertoires produced
Birds were immunized with human progranulin protein (PGRN), a modular protein that we have used for comparison of immune responses across various genotypes of OmniChickens and wild-type chickens.19,20,32 PGRN is composed of 7.5 domains that can be used for identifying epitope binning patterns in a cohort of antibodies. Six OmniClic birds were immunized with human PGRN, three with the SynVH-C heavy chain and three with the SynVH-SD heavy chain, on a two-week interval of intramuscular boosts, with serum titers determined on the in-between weeks.20 High titers were obtained after the first boost (Fig. S4) and were comparable to those observed in OmniChickens.20 After reaching a sufficient titer, spleen lymphocytes were harvested and cryopreserved for either bulk sequence analysis or monoclonal antibody (mAb) recovery.
To assess overall levels of diversity in the antibody repertoire, next-generation sequencing (NGS) of CmLC and VH amplicons was performed on the six spleen samples from PGRN-immunized OmniClic birds and compared to similar data from normal OmniChickens also immunized with PGRN (heavy chains reported in Leighton et al.34). The regular OmniChickens served as a control for the level of diversity to be expected in light chains, as they were immunized with the same immunogen and the sequencing was performed the same way. In addition, the heavy chain transgenes are the same in all of the birds, which allowed us to compare the heavy chain repertoires produced when paired with either the common light chain or a mutating light chain. Primers for amplification of the V regions were chosen in the 5ʹ UTR and either the JK (for light chain) or IgY constant region (for heavy chain), to specifically analyze class-switched heavy chain sequences. Because there is only one functional framework (germline gene) for light chain and one for heavy chain, only a single primer pair is necessary to capture all of the expressed light (or heavy) variable region sequences, which obviates the risk of amplification bias from mixtures of V-family primers that are required to amplify the different V family members in mammalian species. In addition, the diversity analysis is simplified because we are comparing mutation frequencies in a single FR across various genotypes and individual animals. Paired-end reads were assembled and preprocessed for length and V region open reading frame. Singleton sequences were eliminated, and sequences were translated into the predicted protein sequence. As shown in Figure 2a, 95% of the total sequence reads (n = 4.8x106 reads) of CmLC from the immunized birds contained 0–2 changes at the amino acid level, including 87% with no changes at all. In contrast, the normal OmniChicken light chain bulk repertoire contained only 0.8% sequences with 0–2 amino acid changes and 0.3% with no changes (n = 7.2 x106 reads from 6 birds). The mean number of light chain changes in OmniClic was 0.4 compared to 7.4 for OmniChickens. The frequencies of changes at the DNA level were similar: 94% vs. 0.6% for 0–2 changes in OmniChicken versus OmniClic. In contrast to the reduced levels of mutation in the light chains, heavy chains showed increased mutation levels in OmniClic compared to OmniChicken (Figure 2a). The mean number of changes in heavy chain was 13.3 in OmniClic, as compared to 11.5 in OmniChicken. The graphs in Figure 2a were calculated based on the number of changes in FR1 through FR3 and did not include CDR3, since there is no germline CDR3 sequence in the case of the rearranging heavy chain SynVH-SD that could be used to calculate the changes.
Figure 2.

Analysis of bulk NGS sequencing data from OmniClic compared to OmniChicken
(a) Frequency of mutations. Bulk repertoire sequences were obtained by amplicon sequencing of RT-PCR products obtained from splenocytes of immunized birds. The number of amino acid changes compared to the germline sequence present in the birds in the light chain (in blue) and heavy chain (in red) of OmniClic and normal V-kappa OmniChickens is shown. Data from birds carrying both heavy chain transgenes are combined (SynVH-C and SynVH-SD). For each bin on the histogram (0 changes, 1 change, etc.), the number of sequence reads (duplicates) with the indicated number of changes were added up and divided by the total number of reads (total number N is shown for each plot). This gives a normalized frequency of the abundance of each bin. The mean and SD for each V region is indicated in the figure. The V region from FR1-FR3 was used. All sequences were filtered for human sequence (some heavy chain repertoires, but not light chain, included a small amount of chicken sequence34). All genotype combinations had three birds each except for SynVK-CK/SynVH-C which had six birds. (b). Frequency of unique CDR-H3 (left, in red) and unique FR1-FR3 sequences (right, in blue) are shown for the light (CmLC1 or VK) and heavy (VH; combined SynVH-C and SynVH-SD) chains of OmniClic and OmniChicken. The number of unique CDR-H3s found in each bird (determined at the amino acid level) was divided by the number of sequence reads, to give the ratios shown. The number of unique FR1-FR3 sequences (determined at the DNA sequence level) was divided by the total number of sequence reads to give the ratios shown.
To assess clonotype diversity based on CDR3 sequences, the numbers of unique CDR3s per genotype were counted and normalized by dividing by the total number of sequence reads (Figure 2b). The frequency of unique CDR-H3s was strikingly higher in OmniClic heavy chains, indicating that more clonotypes were generated when the heavy chains were paired with CmLC1. This observation is consistent with the idea that the VH underwent enhanced selection for diverse sequences, not only in CDRs 1–2 as indicated in Figure 2a, but also in CDR3. To compare the results in Figure 2a using this type of analysis, the frequency of unique sequences was also calculated for the non-CDR3 portions of the V region (FR1 through FR3; Figure 2b right panel). OmniClic light chain had a much lower ratio of unique sequences than any of the other chains (Figure 2b), likely reflecting the fact that so many of the sequence reads were germline and would thus be counted as a single unique sequence. This analysis also confirmed that the VH sequences from the OmiClic birds had the highest ratio of unique sequences overall. These results showed that there was no restriction in the B cell repertoire caused by the limitation of light chain diversity, but rather suggest that the lack of diversity in the light chains, and CDR3 in particular, leads to a compensatory increase in frequency of mutations in the heavy chain CDR3.
Even though the overall frequency of light chain mutations was much reduced in OmniClic, amino acid changes seemed to be clustered in the CDRs, suggesting that they were undergoing positive selection in the immune response (Fig. S5 shows plots of mutation by position in light chains). Interestingly, the amino acid diversity observed in the CDR-L3 NGS data appeared higher in OmniClic than in OmniChicken, as shown by the additional colors seen in the bars at CDR3 positions, even though the absolute frequency of these mutations was much lower in OmniClic. This result would indicate that some rare, mutated sequences were produced in OmniClic, and they were rarer than in OmniChicken.
Identification of antigen-specific antibodies from OmniClic birds
To further characterize the antibodies that can be obtained from immunized OmniClic animals, mAbs were identified by single B cell screening, using gel-encapsulated microenvironment (GEM) technology.44 Antibodies were cloned in a single-chain variable fragment (scFv)-human Fc format. Two types of cloning strategies were pursued for antibodies from the OmniClic splenocytes. In one set of experiments, the native light chain sequence from the B cell was amplified and cloned with its paired VH, to capture and observe the frequency of mutations occurring in a cohort of antigen-specific antibodies produced in vivo. These experiments were done with one CmLC1/SynVH-C bird and one CmLC1/SynVH-SD bird. In other experiments, we amplified only the heavy chains from individual antigen-specific B cells and directly cloned them with the fully germline light chain. This direct cloning strategy with germline light chain would be the preferred route during an OmniClic antibody discovery campaign, as it would by definition produce only candidates with the desired attribute of working with the germline light chain. Cloning VH with the germline light chain is a more streamlined process requiring fewer amplification steps and is more efficient in the long run if one is only interested in heavy chains that can pair with the germline light chain. Any clones that require the mutations that might be present in their native light chains would drop out during the screening process, but our results show that we expect the number of such clones to be low.
In the first experiments, native light chains with their paired heavy chains were cloned. A cohort of 41 unique antibodies was obtained from 2 birds, and these were confirmed to bind PGRN by enzyme-linked immunosorbent assay (ELISA). Sequence analysis of the light chains showed that nearly one-fifth (4 of 16 for bird 45211 and 3 of 25 for bird 45331) of the light chains in these clones were fully germline in sequence (Figure 3a), and about 37% of the total had only 1 mutation. In the whole cohort, the median number of amino acid changes per V region was 1, and the average number of amino acid changes per clone was 2.4 (Figure 3a). These numbers are higher than in the bulk non-selected NGS data, indicating that selection for antigen binding enriched to a certain degree for mutated sequences, although much less than in regular OmniChickens (Figure 3a). The changes appear more likely to occur in the CDRs (Figure 3b), further indicating that they are functionally selected. The heavy chains in these antibodies showed levels of mutation similar to normal OmniChicken-derived antibodies to the PGRN target, unlike the increased mutation levels we observed in the NGS data.
Figure 3.

Frequency of mutations in anti-PGRN mAbs cloned with native light chains
(a). Frequencies of amino acid changes in mAbs cloned with native light chain from OmniClic were calculated and compared to mAbs from V-kappa OmniChickens.20 Clones from SynVH-C and SynVH-SD birds were combined. The mean, SD and N for VK and VH regions are indicated in the figure. (b). The levels of amino acid diversity at each position across the V regions are shown. Changes from the germline-encoded residue at each position are indicated by the colored bars, with the frequency shown by the height of the bar and different colors indicated for the various amino acids. The CDRs are indicated by the boxes. Although levels of mutation are low, the CDRs tend to be the focus of most of the sequence variation in OmniClic light chains. Only the SynVH-SD-derived mAbs are shown here, for simplicity (the results with SynVH-C were similar). OmniClic, N = 25 antibodies; OmniChicken, N = 57 antibodies.
In the second set of experiments, heavy chains were amplified from single cells in GEMs and cloned directly with the germline light chain. A total of 51 antibodies with unique VHs were obtained from 3 birds (the same 2 birds from above plus one additional bird), cloned into a vector containing the germline light chain and confirmed to bind PGRN by ELISA. In the two birds that were used for both native and germline light chain cloning, 14 antibodies were found with the same VH in both groups. When these VHs were paired with their native light chain, most of them (12, or 86%) contained a least one mutation in the light chain, usually in one of the CDRs. However, these mutations were not necessary for antigen binding since the same VH with germline light chain was still able to bind the same PGRN domain, and there was no significant change in affinity. The other two VHs in this group were paired with germline light chain in the native cloning, and were never found paired with mutant light chains.
Binding affinities and epitope binning of OmniClic-derived antibodies
All the antibodies were analyzed on the Carterra LSA instrument for kinetics and epitope binning, as done previously on cohorts of OmniChicken-derived mAbs.20,32 Figure 4a shows a sequence dendrogram for the clones lined up with a column indicating the PGRN subdomain targeted by each clone, and a column indicating whether the clone binds to the mouse ortholog of PGRN. Clone names highlighted in green contain germline light chain. For the seven full-length PGRN domains, multiple clones targeting each domain were identified. Bins are populated by germline light chain-containing clones in each case, showing that the lack of light chain diversity has not affected antigen recognition, and suggesting that paratope determinants provided by the VH alone are sufficient. Mouse cross-reactivity is also found in the group of germline light chain clones. Closely related clones are always found to target the same domain, but a given domain may be targeted by groups of unrelated clones across the dendrogram, indicating that diverse VH clonotypes have arisen that target the same domain. The nodal plot of the binning results in Figure 4b shows that at least one PGRN domain (CD in this example) can be targeted at more than one distinct epitope in OmniClic birds.
Figure 4.

Sequence dendrogram, epitope binning and mouse cross-reactivity of all anti-PGRN clones (native and germline light chain)
(a). The sequence dendrogram is based on the combined VK-VH sequence for each clone and is lined up next to columns indicating the progranulin domain that is targeted by the clone and whether the mAb binds to the mouse ortholog of progranulin (labeled ‘mouse cross’). The clones highlighted in green have the germline light chain sequence. All of the PGRN domains were targeted by both germline and native light chain-containing clones except for P, which is a half-size domain that typically elicits fewer antibodies than the other domains.20 (b). Nodal plot of epitope binning relationships. PGRN domains targeted by known standards from previous campaigns in wild-type19 and OmniChickens20 were binned on the Carterra LSA with all of the OmniClic antibodies. Three different epitopes on the CD-domain were identified as indicated by the three nodes. OmniClic antibodies are colored blue and standards are red.
Binding kinetics were determined for the cohorts and plotted in Figure 5. The isoaffinity plot (Figure 5a) shows binding affinities colored by PGRN domain. For most of the domains (A, B, CD and E), clones showed a wide range of affinities, with some high-affinity clones in each group. Within the cohort, a significant amount of kinetic diversity was observed (Figure S6). While the majority of antibodies bound rapidly, off-rates varied, with some antibodies even remaining stable throughout the 20-minute dissociation period. In Figure 5b, clones were separated into those with a mutated light chain from the native cloning (labeled “native” in the figure) and those with germline light chain. Although the germline light chain-containing clones showed a slightly lower median affinity (10.3 nM) compared to the native light chain clones (7.8 nM), the difference of the means was not significant (t-test, P = .32) and the range of affinities was quite similar with numerous clones in the sub-nanomolar category in both groups. The fact that the native, mutated clones had such a similar median affinity suggests that the mutations in the native light chains are playing only a minor role in determining binding affinity.
Figure 5.

Binding kinetics
(a). Isoaffinity plot of all the anti-PGRN mAbs, with native and germline light chain, colored by epitope bin. Antibodies to most domains displayed a wide range of affinities. Antibodies to domains F and G did not attain as high affinities as the others, although, the sample size was low. (b). Dot plot of affinity of the cohort of mAbs to human progranulin. Most clones were run in duplicate and both values were included. mAbs were grouped based on whether the light chain (OmniClic VK) was native (excluding those clones that happened to have germline sequence), or had the germline light chain sequence. Clones with native light chain had a median KD of 7.8 nM, whereas the clones with germline light chain had a median KD of 10.3 nM, indicating slightly less tight binding. Native light chain, N = 27 clones, germline light chain, N = 47 clones.
In order to directly compare the binding kinetics of the same VH with either its native light chain or the germline light chain, some clones, with a range of kinetics and unique heavy chains that were initially identified with their native light chain, were chosen for germlining of the light chain. Heavy chains were re-amplified from the original plasmid and cloned into a vector containing the CmLC1 germline light chain sequence in order to express in the same scFv-Fc format as before. The native and germlined antibodies were analyzed on the Carterra LSA side-by-side. The binding kinetics were plotted (Figure 6) with the native KD on the x-axis versus the germlined KD on the y-axis. Most of the clones had very similar binding kinetics after germlining, as they fall close to the line drawn at a slope of 1. One VH was found twice with two different native light chains, and had affinities of 2.8 and 102 nM with those two light chains. When cloned with the germline light chain, this VH clone dropped to a KD of 187 nM. All of the clones retained the same binning and mouse cross-reactivity characteristics.
Figure 6.

Pairing the same VH with its native VK or germline VK
Seven mAbs with native OmniClic VK that contained somatic mutations were reformatted with the germline VK. The affinities of both versions to PGRN were determined, and each dot on the plot is the germline VK affinity vs. the native VK affinity for a given VH clone. The line represents a slope of 1. Five of the seven antibodies fall close to the line, indicating the affinity did not change for those VH when paired with germline VK. One clone increased its affinity and one clone reduced its affinity when paired with germline VK.
Discussion
When considering an antibody discovery platform for a bispecific application, in vitro systems might have some appeal because there is no need to pair native heavy and light chain V regions, since the light chain can be a fixed sequence. However, one loses the advantages of the powerful in vivo affinity maturation and selection processes provided by the intact immune system, which remains fully in effect on the heavy chain in OmniClic. We have exploited the unique characteristics of the chicken immune system to express a common light chain paired with a diverse repertoire of human heavy chains. The human V regions derived from these birds can be used in a traditional IgG-like format, or they could be re-formatted into other types of structures using the V region-building blocks, if desired. The OmniClic birds retain the antigen-recognition properties of the chicken host, namely that conserved mammalian targets will be more immunogenic than in mammalian hosts, and species cross-reactivity will be expanded to the murine (and other mammalian) orthologs of human proteins.
OmniClic birds were designed to minimize sequence diversity in the light chain, and analysis of NGS data sets and panels of antigen-specific clones demonstrated that they in fact do show a high proportion of germline and near-germline sequences in the bulk antibody repertoires and in the antigen-specific mAbs. OmniClic birds are thus well-equipped for discovery of human heavy chains that can be formatted into bispecifics in an IgG-like format with germline kappa light chain. These data were in contrast to “regular” OmniChickens in which a high degree of light chain diversity is produced and germline light chains are very rare in the antibody repertoire. In terms of the light chain construct design, the main feature that distinguishes the two genotypes is the pseudogene array in each: one array contains no diversity (OmniClic) and the other contains diversity (OmniChickens). Although we speculate that the identical pseudogenes actively revert any randomly arising somatic mutations back to germline, it is also possible that the reduced mutational frequency observed in OmniClic could simply be a result of the lack of contribution by diverse pseudogene sequences, and not to any active cleansing of mutations. The low levels of diversity in OmniClic would in that case be caused by infrequent non-templated somatic mutation. To prove that the identical pseudogenes in OmniClic have a cleansing effect on the antibody repertoire by erasing diversity through gene conversion, we would need to generate a bird that contains no pseudogenes at all and compare the sequences that are produced. However, there is evidence that non-templated somatic hypermutation would actually produce much higher levels of diversity in the chicken if there were no reverting pseudogenes. We have made a heavy chain transgene, SynVH-A7, which contained only Y, W and S residues in the CDRs of the pseudogene array upstream of a pre-rearranged VH3-23 gene.20 When we immunized these birds with PGRN and analyzed either bulk NGS data or sequences of antigen-specific antibodies, the CDRs were found to contain diverse sequences incorporating many different amino acids, not just YWS, suggesting that somatic hypermutation alone can produce a diverse repertoire in the chicken. The pseudogenes in SynVH-A7 were found to donate YWS sequences only rarely to sequences in the repertoire. In the chicken B cell line DT40, a similar experiment showed that diversity accumulates in the VH when the pseudogene array contains only limited amino acid content, indicating efficient non-templated mutation.33 Data from several other studies in DT40 corroborate this conclusion: deletion of the pseudogene array still allowed non-templated mutations to accumulate.39,45–47 Based on these data, we believe that gene conversion with the identical pseudogenes is playing an active role to minimize sequence diversity of the antibody repertoire in OmniClic birds.
Despite the lack of light chain diversity, antibodies of high affinity could be obtained from OmniClic birds. These results show that the human heavy chain transgenes SynVH-C and SynVH-SD (used for both OmniClic and OmniChickens) are capable of producing high-affinity binding paratopes when paired with germline light chains. Both SynVH-C and SynVH-SD heavy chains produced antibodies with a range of affinities and diverse repertoires, and there is no evidence of a difference in performance between the two in the context of OmniClic, at least for this target. Although the OmniClic light chains showed little mutation, their germline or near-germline CDRs may still make contact with antigen. It would be interesting to analyze structural data from OmniClic and OmniChicken mAbs to see if any specific epitopes are bound in new ways in OmniClic-derived antibodies as a result of the germline light chain CDRs. When the same VH was paired with either its native light chain or the germline light chain, most antibodies retained their affinities and binding characteristics, probably because the native light chains that came from OmniClic had so few changes to begin with and those mutations did not significantly contribute to affinity. In addition, the heavy chain repertoires produced by these transgenes are sufficiently diverse for robust antibody responses and affinity maturation in these birds. In the bulk sequencing data, the OmniClic heavy chain diversity appeared higher than in OmniChicken, although this effect was not apparent in the PGRN mAbs. In an OmniClic antibody discovery campaign, cloning would be done directly with the germline light chain with the expectation of little or no loss in terms of number of binders and their affinities. Cloning VH with the germline light chain is a more streamlined process requiring fewer amplification steps and is more efficient in the long run if one is only interested in heavy chains that can pair with the germline light chain. Any clones that require the mutations that might be present in their native light chains would drop out during the screening process, but our results show that we can expect the number of such clones to be low. More broadly, OmniFlic rats with a fixed light chain16 use essentially the same human VK3-15 and JK1 genes as OmniClic (they differ by one amino acid change in CDR-L3), so heavy chains discovered in OmniFlic and OmniClic could be used in combination to develop a bispecific antibody.
In summary, OmniClic chickens offer the advantages of an in vivo host system to deliver high-quality human heavy chains paired with a common light chain, thereby enabling the development of therapeutic bispecific antibodies.
Materials and methods
Constructs: The common light chain construct CmLC1 was built with a human germline VK3-15/JK1 gene in pre-rearranged configuration. Six pseudogenes with the same sequence, in reverse orientation to the functional V, were inserted upstream, with ~100 base pair spacers from the chicken pseudogene locus between each human pseudogene pair. A unique primer binding site for sequencing of the final construct was included in each spacer region. The chicken light chain promoter (the 2.4 kilobase pair region between the chicken VL and the first upstream pseudogene), J-C intron, and chicken CL were included, so that all of the regulatory elements and untranslated transcript sequences were chicken. An attB site was included for insertion into an attP site previously targeted to the IgL locus in chicken primordial germ cells.42 PGC line 229–92 was transfected with CmLC1, producing ~20 single clones per transfection of 5 × 106 cells.
Breeding: All animal experiments were done in accordance with protocols approved by Ligand Pharmaceuticals’ Institutional Animal Care and Use Committee. The animal facility is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. CmLC1 chimeras were produced by injecting embryos with either single clones, or pooled cells from several clones.48 Germline transmission from chimeras made with pooled clones (average rate, 30% from 13 chimeras) was very similar to that from chimeras made with single clones (average rate, 31% in 24 chimeras made with 4 clones). Transgenic chicks were confirmed for the correct insertion of the CmLC1 construct by PCR upon hatch. Founder CmLC1 transgenics were obtained by breeding to Cre/IgL KO/IgH KO birds49 to remove selectable markers. To obtain OmniClic birds with the genotype CmLC1/IgL KO; SynVH/IgH KO, crosses between CmLC1/+; IgH KO/+ and IgL KO/+; SynVH/+ birds were performed. Progeny was genotyped by PCR using DNA obtained from comb biopsy. Males and females were kept for analysis.
Flow cytometry: Blood samples were collected from 5-week-old birds into ethylenediaminetetraacetic acid-containing collection tubes. For single-cell analysis, peripheral blood mononuclear cells were isolated using histopaque-1077 (Sigma, 10771), labeled on ice for 1 hour with the following antibodies from Southern Biotech: ms anti-Bu1 (8395), ms anti-ch IgM (1020), ms anti-ch IgL(8340), ms anti-ch ΤCR-γδ (8230), ms anti-TCRαβ/Vβ1 (8240), ms anti-TCRαβ/Vβ2 (8250) or a custom polyclonal antibody raised against VH3-23/VK3-15. All antibodies were diluted in 1% bovine serum albumin-phosphate-buffered saline (PBS). Following primary antibody incubation, the cells were washed and then incubated with either AF-647 anti-ms IgG (Jackson ImmunoResearch, 715–605-150) or AF-647 anti-rb IgG (Thermo, A-21244). Data were collected using an Attune Acoustic Focusing Cytometer (Thermo Fisher) and single lymphocytes (gated on FSH v. FSA) were analyzed using FloJo. The plots shown are representative of several independent experiments with several different cohorts of animals. A two-tailed Student’s T-test was used for statistical analysis using GraphPad Prism.
Immunization: Animals were initially immunized intramuscularly with 200 μg recombinant human PGRN (Sino Biological, 10826-H08H) emulsified with 250 μL Freund’s Complete Adjuvant (Thermo Fisher, 77140). Two weeks later, animals began a bi-weekly intramuscular boosting schedule of 100 μg protein with 250 μL Freund’s Incomplete Adjuvant (Thermo Fisher, 77145). Antigen-specific titer was assessed by ELISA (see below) on the off week. Four days before euthanization, animals received a final boost of 200 μg of protein intravenously with no adjuvant.
ELISA: Blood samples were taken from birds and plasma isolated by centrifugation. For antigen-specific ELISAs, plates were coated with 2 μg/mL human PGRN (Sino Biologicals, 10826-H08H) in PBS at 4°C overnight. For assessment of total IgY titer, plates were coated with rabbit anti-chicken IgY (H + L) at 2.0 μg/mL (Sigma, C2288) in PBS at 4°C overnight. After coating, plates were blocked with 3% nonfat dry milk-PBS-T for 1 hour, then incubated with plasma at the indicated dilutions. After one hour, plates were washed in PBS-T and incubated with rabbit anti-chicken IgY-horseradish peroxidase (Sigma, A9046) for an additional hour. Plates were washed and developed with 3,3ʹ,5,5ʹ-tetramethylbenzidine (Sigma, 002023) and absorbance at 450 nm read on a BioTek plate reader.
Single-cell screening: A single-cell screening method, the GEM assay44 (US Patents 8030095 and 84151738) was employed to identify antigen-specific B cells. 5 μm aldehyde-latex beads (Thermo Fisher, A37306) were coated with streptavidin followed by biotinylated antigen, overnight, blocked with 3% milk-PBS, and tested by labeling with plasma from immunized animals. GEMs were prepared containing a single secreting B cell and antigen-coated beads and incubated for 3 hours at 37°C in RPMI/10% fetal calf serum containing 2 μg/mL AF-594 anti-ch IgY (Thermo Fisher, A11042) as previously described.44
scFvFc cloning: Single antigen-specific B cells were lysed and mRNA isolated. VH and VK regions were amplified in a two-step semi-nested strategy previously described.44 For native VH/VK pairing, the variable regions were assembled into an scFv by overlap extension PCR and inserted into a mammalian expression vector containing human Fc (IgG1 CH2-CH3) using InFusion (Takeda Bio, 638911). For expression with the germline CmLC, amplified VH were inserted into a vector containing both the germline CmLC and human Fc (IgG1 CH2-CH3).
Expression of scFvFc antibodies: Recombinant antibodies were expressed in the Expi293 expression system (Thermo Fisher, A14525) as previously described.44
Biosensor analysis: Binding kinetics and interaction analysis studies were performed by high-throughput surface plasmon resonance (SPR) on Carterra’s LSA platform equipped with HC-30 M or HCX-30 M (pre-activated) sensor chips. The method has been previously described in detail elsewhere.32 As before, subdomain specificity was determined by merging the antibody cohort with a panel of previously characterized standards.19 To calculate binding affinity, a chip coupled with goat anti-human IgG Fc-specific polyclonal (Southern Biotech 2047–01) was used as the capture reagent.
Supplementary Material
Disclosure of Potential Conflicts of Interest
Kathryn H. Ching, Kimberley Berg, Kevin Reynolds, Darlene Pedersen, Alba Macias, William D. Harriman, and Philip A. Leighton were employed by Ligand Pharmaceuticals or its subsidiary Vernalis while performing the work described in the manuscript. Funding was provided by Ligand.
Yasmina N. Abdiche reports no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Labrijn AF, Janmaat ML, Reichert JM, Parren PWHI.. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov. 2019;18:585–11. [DOI] [PubMed] [Google Scholar]
- 2.Suurs FV, Lub-de Hooge MN, EGE DV, de Groot DJA. A review of bispecific antibodies and antibody constructs in oncology and clinical challenges. Pharmacol Ther. 2019;201:103–19. doi: 10.1016/j.pharmthera.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Spiess C, Zhai Q, Carter PJ. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol. 2015;67:95–106. doi: 10.1016/j.molimm.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs. 2017;9:182–212. doi: 10.1080/19420862.2016.1268307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manikwar P, Mulagapati SHR, Kasturirangan S, Moez K, Rainey GJ, Lobo B. Characterization of a novel bispecific antibody with improved conformational and chemical stability. J Pharm Sci. 2020;109:220–32. doi: 10.1016/j.xphs.2019.06.025. [DOI] [PubMed] [Google Scholar]
- 6.Miller BR, Demarest SJ, Lugovskoy A, Huang F, Wu X, Snyder WB, Croner LJ, Wang N, Amatucci A, Michaelson JS, et al. Stability engineering of scFvs for the development of bispecific and multivalent antibodies. Protein Eng Des Sel. 2010;23:549–57. doi: 10.1093/protein/gzq028. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Demarest SJ. Building blocks for bispecific and trispecific antibodies. Methods. 2019;154:3–9. doi: 10.1016/j.ymeth.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Klein C, Sustmann C, Thomas M, Stubenrauch K, Croasdale R, Schanzer J, Brinkmann U, Kettenberger H, Regula JT, Schaefer W. Progress in overcoming the chain association issue in bispecific heterodimeric IgG antibodies. MAbs. 2012;4:653–63. doi: 10.4161/mabs.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridgway JB, Presta LG, Carter P. “Knobs-into-holes” engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617–21. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 10.Ha J-H, Kim J-E, Kim Y-S. Immunoglobulin Fc heterodimer platform technology: from design to applications in therapeutic antibodies and proteins. Front Immun. 2016;7:394. doi: 10.3389/fimmu.2016.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Nardis C, Hendriks LJA, Poirier E, Arvinte T, Gros P, Bakker ABH, de Kruif J. A new approach for generating bispecific antibodies based on a common light chain format and the stable architecture of human immunoglobulin G1. Journal of Biological Chemistry. 2017;292:14706–17. doi: 10.1074/jbc.M117.793497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merchant AM, Zhu Z, Yuan JQ, Goddard A, Adams CW, Presta LG, Carter P. An efficient route to human bispecific IgG. Nat Biotechnol. 1998;16:677–81. doi: 10.1038/nbt0798-677. [DOI] [PubMed] [Google Scholar]
- 13.Jackman J, Chen Y, Huang A, Moffat B, Scheer JM, Leong SR, Lee WP, Zhang J, Sharma N, Lu Y, et al. Development of a two-part strategy to identify a therapeutic human bispecific antibody that inhibits IgE receptor signaling. J Biol Chem. 2010;285:20850–59. doi: 10.1074/jbc.M110.113910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krah S, Schröter C, Eller C, Rhiel L, Rasche N, Beck J, Sellmann C, Günther R, Toleikis L, Hock B, et al. Generation of human bispecific common light chain antibodies by combining animal immunization and yeast display. Protein Eng Des Sel. 2017;30:291–301. [DOI] [PubMed] [Google Scholar]
- 15.Van Blarcom T, Lindquist K, Melton Z, Cheung WL, Wagstrom C, McDonough D, Valle Oseguera C, Ding S, Rossi A, Potluri S, et al. Productive common light chain libraries yield diverse panels of high affinity bispecific antibodies. MAbs. 2018;10:256–68. doi: 10.1080/19420862.2017.1406570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris KE, Aldred SF, Davison LM, Ogana HAN, Boudreau A, Bruggemann M, Osborn M, Ma B, Buelow B, Clarke SC, et al. Sequence-based discovery demonstrates that fixed light chain human transgenic rats produce a diverse repertoire of antigen-specific antibodies. Front Immun. 2018;9:889. doi: 10.3389/fimmu.2018.00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith EJ, Olson K, Haber LJ, Varghese B, Duramad P, Tustian AD, Oyejide A, Kirshner JR, Canova L, Menon J, et al. A novel, native-format bispecific antibody triggering T-cell killing of B-cells is robustly active in mouse tumor models and cynomolgus monkeys. Sci Rep. 2015;5:17943–12. doi: 10.1038/srep17943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Logtenberg T, Throsby M, Kramer RA, Pinto RD, de Kruif CA, Houtzager E. Antibody producing non-human mammals. 2017; US Patent:US9765133.
- 19.Abdiche YN, Harriman R, Deng X, Yeung YA, Miles A, Morishige W, Boustany L, Zhu L, Izquierdo SM, Harriman W. Assessing kinetic and epitopic diversity across orthogonal monoclonal antibody generation platforms. MAbs. 2016;8:264–77. doi: 10.1080/19420862.2015.1118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ching KH, Collarini EJ, Abdiche YN, Bedinger D, Pedersen D, Izquierdo S, Harriman R, Zhu L, Etches RJ, M-C VDL, et al. Chickens with humanized immunoglobulin genes generate antibodies with high affinity and broad epitope coverage to conserved targets. MAbs. 2018;10:71–80. doi: 10.1080/19420862.2017.1386825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Könitzer JD, Pramanick S, Pan Q, Augustin R, Bandholtz S, Harriman W, Izquierdo S. Generation of a highly diverse panel of antagonistic chicken monoclonal antibodies against the GIP receptor. MAbs. 2017;9:536–49. doi: 10.1080/19420862.2016.1276683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bednenko J, Harriman R, Mariën L, Nguyen HM, Agrawal A, Papoyan A, Bisharyan Y, Cardarelli J, Cassidy-Hanley D, Clark T, et al. A multiplatform strategy for the discovery of conventional monoclonal antibodies that inhibit the voltage-gated potassium channel Kv1.3. MAbs. 2018;10:636–50. doi: 10.1080/19420862.2018.1445451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sim J, Sockolosky JT, Sangalang E, Izquierdo S, Pedersen D, Harriman W, Wibowo AS, Carter J, Madan A, Doyle L, et al. Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRPα. MAbs. 2019;11:1036–52. doi: 10.1080/19420862.2019.1624123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Tsai JC, Davis JH, Chau B, Dong J, West SM, Hogan JM, Wheeler ML, Bee C, Morishige W, et al. Design and characterization of mouse IgG1 and IgG2a bispecific antibodies for use in syngeneic models. MAbs. 2020;12:1685350. doi: 10.1080/19420862.2019.1685350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratcliffe MJH. Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken B cell development. Dev Comp Immunol. 2006;30:101–18. doi: 10.1016/j.dci.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 26.Reynaud C, Anquez V, Dahan A, Weill J. A single rearrangement event generates most of the chicken immunoglobulin light chain diversity. Cell. 1985;40:283. doi: 10.1016/0092-8674(85)90142-4. [DOI] [PubMed] [Google Scholar]
- 27.Reynaud C, Dahan A, Anquez V, Weill J. Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell. 1989;59:171–83. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- 28.Reynaud C, Anquez V, Weill J. The chicken D locus and its contribution to the immunoglobulin heavy chain repertoire. Eur J Immunol. 1991;21:2661–70. doi: 10.1002/eji.1830211104. [DOI] [PubMed] [Google Scholar]
- 29.Yang B, Gathy KN, Coleman MS. T-cell specific avian TdT: characterization of the cDNA and recombinant enzyme. Nucleic Acids Res. 1995;23:2041–48. doi: 10.1093/nar/23.11.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefranc M-P. IMGT, the International ImMunoGeneTics Information System. Cold Spring Harb Protoc. 2011;2011:595–603. doi: 10.1101/pdb.top115. [DOI] [PubMed] [Google Scholar]
- 31.Parvari R, Ziv E, Lantner F, Heller D, Schechter I. Somatic diversification of chicken immunoglobulin light chains by point mutations. Proc Natl Acad Sci USA. 1990;87:3072–76. doi: 10.1073/pnas.87.8.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ching KH, Berg K, Morales J, Pedersen D, Harriman WD, Abdiche YN, Leighton PA. Expression of human lambda expands the repertoire of OmniChickens. PLoS ONE. 2020;15:e0228164. doi: 10.1371/journal.pone.0228164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leighton PA, Schusser B, Yi H, Glanville J, Harriman W. A diverse repertoire of human immunoglobulin variable genes in a chicken B cell line is generated by both gene conversion and somatic hypermutation. Front Immun. 2015;6:126. doi: 10.3389/fimmu.2015.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leighton PA, Morales J, Harriman WD, Ching KH. V(D)J rearrangement is dispensable for producing CDR-H3 sequence diversity in a gene converting species. Front Immun. 2018;9:1317. doi: 10.3389/fimmu.2018.01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arakawa H, Hauschild J, Buerstedde J. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–06. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 36.Harris RS, Sale JE, Petersen-Mahrt SK, Neuberger MS. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Current Biology. 2002;12:435–38. doi: 10.1016/S0960-9822(02)00717-0. [DOI] [PubMed] [Google Scholar]
- 37.Fugmann S, Schatz D. Immunology. One AID to unite them all. Science. 2002;295:1244–45. doi: 10.1126/science.1070023. [DOI] [PubMed] [Google Scholar]
- 38.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/S0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 39.Sale J, Calandrini D, Takata M, Takeda S, Neuberger M. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–26. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 40.Buerstedde J-M, Alinikula J, Arakawa H, McDonald JJ, Schatz DG. Targeting of somatic hypermutation by immunoglobulin enhancer and enhancer-like sequences. PLoS Biol. 2014;12:e1001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinesh RK, Barnhill B, Ilanges A, Wu L, Michelson DA, Senigl F, Alinikula J, Shabanowitz J, Hunt DF, Schatz DG. Transcription factor binding at Ig enhancers is linked to somatic hypermutation targeting. Eur J Immunol. 2020;50:380–95. doi: 10.1002/eji.201948357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schusser B, Collarini EJ, Pedersen D, Yi H, Ching K, Izquierdo S, Thoma T, Lettmann S, Kaspers B, Etches RJ, et al. Expression of heavy chain-only antibodies can support B-cell development in light chain knockout chickens. Eur J Immunol. 2016;46:2137–48. doi: 10.1002/eji.201546171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schusser B, Collarini EJ, Yi H, Izquierdo SM, Fesler J, Pedersen D, Klasing KC, Kaspers B, Harriman WD, van de Lavoir MC, et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc Natl Acad Sci USA. 2013;110:20170–75. doi: 10.1073/pnas.1317106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mettler-Izquierdo S, Varela S, Park M, Collarini EJ, Lu D, Pramanick S, Rucker J, Lopalco L, Etches R, Harriman W. High-efficiency antibody discovery achieved with multiplexed microscopy. Microscopy (Oxf). 2016;65:341–52. doi: 10.1093/jmicro/dfw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arakawa H, Saribasak H, Buerstedde J. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2004;2:E179. doi: 10.1371/journal.pbio.0020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arakawa H, Buerstedde J-M. Activation-induced cytidine deaminase-mediated hypermutation in the DT40 cell line. Philos Trans R Soc Lond B Biol Sci. 2009;364:639–44. doi: 10.1098/rstb.2008.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longerich S, Orelli BJ, Martin RW, Bishop DK. Brca1 in immunoglobulin gene conversion and somatic hypermutation. DNA Repair (Amst). 2008;7:253–66. doi: 10.1016/j.dnarep.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collarini EJ, Leighton PA, van de Lavoir M-C. Production of transgenic chickens using cultured primordial germ cells and gonocytes. Methods Mol Biol. 2019;1874:403–30. [DOI] [PubMed] [Google Scholar]
- 49.Leighton PA, Pedersen D, Ching K, Collarini EJ, Izquierdo S, Jacob R, van de Lavoir M-C. Generation of chickens expressing Cre recombinase. Transgenic Res. 2016;25:609–16. doi: 10.1007/s11248-016-9952-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


