ABSTRACT
Cryptochrome (CRY) is a blue light receptor that is widely distributed in animals, plants, and microorganisms. CRY as a coding gene of cryptochrome that regulates the organism gene expression and plays an important role in organism growth and development. In this study, we identified four photolyase/cryptochrome (PHR/CRY) members from the genome of Ginkgo biloba. Phylogenetic tree analysis showed that the Ginkgo PHR/CRY family members were closely related to Arabidopsis thaliana and Solanum lycopersicum. We isolated a cryptochrome gene, GbCRY1, from G. biloba and analyzed its structure and function. GbCRY1 shared high similarity with AtCRY1 from A. thaliana. GbCRY1 expression level was higher in stems and leaves and lower in roots, male strobili, female strobili. GbCRY1 expression level fluctuated periodically within 24 h, gradually increased in the dark, and decreased under blue light. The newly germinated ginkgo seedlings were cultured under dark, white light, and blue light conditions. The blue light normally induced photomorphogenesis of ginkgo seedlings, which included hypocotyl elongation inhibition, leaf expansion inhibition, and chlorophyll formation. Treating dark-adapted ginkgo leaves with blue light could induce stomatal opening. At the same time, blue light reduced the expression level of GbCRY1 in the process of inducing photomorphogenesis and stoma opening. Our results provide evidence that GbCRY1 expression is affected by space, circadian cycle and light, and also proves that GbCRY1 is related to ginkgo circadian clock, photomorphogenesis and stoma opening process.
KEYWORDS: Ginkgo biloba, CRY1, blue light, expression
Introduction
Light is an important factor for organisms that adjust their own functions in response to changes in the environment. Some organisms can adjust their functions by sensing changes in light through photoreceptors. Three kinds of photoreceptors exist in organisms, namely, phytochrome (PHY),1 phototropin (PHOT),2–4 and cryptochrome (CRY).5,6 Cryptochromes are members of the photolyase/cryptochrome (PHR/CRY) family, which have various functions that existed widely in organisms ranging from bacteria to humans. The cryptochromes share structural homology with DNA photolyases but are distinguished by the presence of a C-terminal extension.7 Functionally, DNA photolyases repair DNA damage caused by UV-B radiation by exposure to UV-A/blue light simultaneously or subsequently.
Cryptochromes are photoreceptor proteins that regulate circadian clock, morphogenesis, phototaxis, and other responses to UV and blue light in various organisms,8 but they have no photolyase activity.5,9–11 The cryptochromes have a molecular weight of 70–80 kDa and have two identifiable regions. One is the photolyase-related N-terminal PHR that has a homologous sequence with the photolyase but does not possess the DNA repair activity of the photolyase. The other is the C-terminal extension region that the photolyase does not have.12 Cryptochrome does not have the function of binding DNA and repairing pyrimidine dimer, but the FAD binding region of cryptochrome can bind ATP, while photolyase cannot.13 In addition to capturing light signals, the PHR region can also form homodimers, which are essential for the normal function of cryptochromes.14 Cryptochromes mediate the various blue light responses of plants,10,11 including altering transcription process,15 inhibition of hypocotyl elongation,16 stimulation of cotyledon expansion,17 promotion of floral initiation,18 entrainment of the circadian clock,19 stimulation of stomata opening,20 fostering pathogen resistance,21 suppression of leaf senescence,22 inhibition of dormant grain germination,23 stomatal development regulation,24 shade avoidance,25 and light-dependent stress responses.20,26–28
Sancar et al.29,30 cloned the PHR gene from E. coli in 1978 and determined that PHR enzymes repair DNA dimers formed through the action of ultraviolet (UV) light from sunlight for the first time. Ahmad et al.16 discovered the cryptochrome gene in Arabidopsis thaliana for the first time and named it CRY1. Then, two cryptochrome homologous genes were found in A. thaliana.31 Lin et al.32 and Lin et al.17 successively isolated CRY2 from A. thaliana. CRY3 without photomorphogenesis function was found in A. thaliana.31,33,34 Further research led to the successive isolation of CRY1 from dicotyledonous plants such as Solanum lycopersicum,35 Pisum sativum L.,36 and Brassica napus37 and monocotyledonous plants such as Oryza sativa L.39 and Triticum aestivum L.33 At the same time, the CRY1 gene was also isolated from lower plants, such as Chlamydomonas,38 Adiantum capillus-veneris L.,39 and Physcomitrella patens.40 Cryptochromes are common in insect,41 mammals,42 birds,43 and other organisms. Cryptochromes are widely distributed in organisms and gymnosperms are a larger group in the botanical classification, but the functions of cryptochrome in gymnosperms are less widely characterized.
Ginkgo biloba is a gymnosperm known as a living fossil, as it has a history of more than 300 million years. Its leaf and fruit extracts contain many different medicinally active ingredients. It is an economic tree species that is both medicinal and edible. Cryptochromes have multiple functions in plant growth and development. However, few reports exist on the function of cryptochrome in G. biloba growth and development. Isolation, identification, and analysis of the potential function of G. biloba cryptochrome genes will help understand the expression pattern of these genes and improve cryptochrome gene function in ginkgo. In this paper, to study the potential role of cryptochrome in G. biloba growth and development, we isolated and identified members of the PHR/CRY family from the ginkgo genome, cloned the full length of the GbCRY1, and analyzed the temporal and spatial expression of the GbCRY1. We studied and analyzed the potential functions of blue light-mediated GbCRY1 in the ginkgo circadian clock, photomorphogenesis, and stimulation of stomata opening. The research on the function of ginkgo cryptochrome has reference value for regulating ginkgo growth and development.
Materials and methods
Plant material
Ginkgo “Jiafoshou” 31-year-old trees were used in this study. They were selected from the ginkgo Germplasm Repository of Yangtze University, China (N30.35, E112.14). Ginkgo seeds were harvested from the 31-year-old trees of ginkgo ‘Jiafoshou’ and stored in a refrigerator at 4°C for 2–3 months. After the seed embryo matures, washed it with clean water, disinfected with 0.1% potassium permanganate for 5 minutes, and then put it in a porcelain dish, cover it with gauze and keep it at 24°C. After the seed shell ruptured, the embryos were transplanted in plastic flowerpots (12 × 10 cm), and the cultivation was continued in the dark.
Phylogenetic, classification and identification of GbCRY1 in Ginkgo PHR/CRY family
Ginkgo genome, nucleic acid, and protein sequences were downloaded from the GIGADB database. The available hidden Markov model (HMM) model (Pfam03441) of the PHR/CRY family was retrieved from the Pfam database44 and was used as a query to scan the proteome file via HMMER software with a default E-value. Candidate proteins were submitted to the Pfam, SMART, and NCBI Conserved Domains45 to verify the existence of the conserved domain. The physical and chemical properties of ginkgo PHR/CRY family are predicted by ProtParam.46 The PHR/CRY gene family members in other organisms were downloaded from NCBI database by referring to the Nuri Ozturk research results.47 Then, the protein sequences of putative PHR/CRY genes in ginkgo and other organisms were aligned with ClustalX software48 with default parameters. Phylogenetic trees were constructed using the aligned result with MEGA-X software49 via the Neighbor-Joining method (Parameter setting: Bootstrap method-1,000 replicates).
CRY1 protein-protein interaction network prediction
CRY1 is ubiquitous in animals, plants, and microorganisms, and the protein interaction network and functions are very complex. We searched the protein-protein interaction network in the STRING database with CRY1 as the key word and with plants and mammals as the search group. CRY1 protein-protein interaction network results were imported into Cytoscape_3.7.2 for visual analysis. By analyzing the protein interaction network of CRY1 in plants and animals and by comparing the similarities and differences of plant and animal protein interaction networks, we predicted the possible functions of GbCRY1.
Cloning of GbCRY1
Total RNA was isolated from frozen tissues of leaves from the 31-year-old ginkgo trees using the TaKaRa MiniBEST Plant RNA Extraction Kit. The first-strand cDNA was synthesized using PrimeScript™ 1st Strand cDNA Synthesis Kit. The specific primers, namely, G-F and G-R, as shown Table S1, were designed based on the GbCRY1 cDNA sequence from the ginkgo genome. The reverse transcribed cDNA was used as a template for RT-PCR with a reaction system of 25 µL. The reaction procedure was as follows: 94°C for 3 min; followed by 32 cycles of amplification at 94°C for 30 s, 55.4°C for 30 s, and 72°C for 90 s; and extension at 72°C for 10 min. The PCR amplification product was recovered and purified and then cloned into the pMD18-T vector before being transformed into Escherichia coli DH5α. The positive clones were verified by bacterial liquid PCR. The positive clones were sent to Sangon Biotechnology (Shanghai) Company for sequencing.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted from roots, stems, leaves, male strobili, female strobili, and fruits of the 31-year-old trees of ginkgo and reverse transcribed into cDNA. According to the cDNA sequence of GbCRY1 gene, the qRT-PCR primers (Table S1) of GbCRY1 was designed. The GAPDH was used as the reference gene for the qRT-PCR. The upstream and downstream primers were H-F and H-R, respectively (Table S1). The qRT-PCR was performed on a Bio-Rad CFX according to the BioEasy Master Mix (SYBR Green) kit instructions. Each sample was set with three biological replicates and three technical replicates. The relative expression fold of each sample was calculated through its Ct value, which was normalized to the Ct-value of reference gene using the 2−ΔΔCt method. Finally, the expression levels of GbCRY1 in different tissues of ginkgo were analyzed by combining RNA-seq and qRT-PCR.
Observation on the stomata opening of Ginkgo leaves
After the ginkgo seedlings germinated, they were cultured for 21 days under normal light conditions (12 h dark/12 h white light). Complete leaves (3–5) were selected from seedlings that were sturdy, showed consistent growth, had no pests and diseases, and were adapted for 2 days in the dark. Then, the ginkgo seedlings were transferred to blue light (2000 lx) for 12 h. A total of 45 ginkgo seedlings with consistent growth were selected and randomly divided into 15 groups, with three ginkgo seedlings in each group. Each of the three groups was used as a replicate. The first complete leaf of the seedling from top to bottom was selected to observe the stomata shape. The sampling interval was 3 h, and samples were taken five times in total. The leaves of the seedlings were stored in liquid nitrogen for the qRT-PCR analysis of GbCRY1.
Circadian rhythm analysis of Ginkgo
Germinated seedlings that were robust, grew uniformly, and had no pests and diseases were selected and cultured for 21 days under 12 h dark/12 h blue light (2000 lx) conditions. A total of 153 ginkgo seedlings with consistent growth were selected and randomly divided into 51 groups, with three ginkgo seedlings in each group. Each of the 3 groups was used as a replicate. The seedlings were sampled at 6 o’clock on the 22nd day, and the seedling leaves were sampled every 3 h for 48 h for a total of 17 times. The samples were quick-frozen in liquid nitrogen then were stored in a refrigerator at −80 °C until use. The changes of the expression of GbCRY1 were detected by qRT-PCR.
Ginkgo photomorphogenesis
After the germination of G. biloba, seedlings that were robust, grew uniformly, and had no pests and diseases were selected and transferred under the following light conditions: (1) dark conditions; (2) 12 h white light (2000 lx)/12 h dark; and (3) 12 h blue light (2000 lx)/12 h dark. A total of 27 ginkgo seedlings with consistent growth were selected and randomly divided into 9 groups, with three ginkgo seedlings in each group. Each seedling in the three groups was used as a repetition. For each treatment, seedlings were cultured for 7 days, and samples were taken for observation after 7 days. Vernier calipers were used to detect shoot length, internode length, and hypocotyl length. Paraffin section method was used to observe changes in hypocotyl cell size and chloroplast formation. Ginkgo seeding shoots were sampled and stored in liquid nitrogen for the qRT-PCR of GbCRY1.
Results
Phylogenetic, classification and identification of GbCRY1
Nuri Ozturk47 systematically analyzed the PHR/CRY family members in animals, plants, and microorganisms and divided the PHR/CRY family into 10 subgroups. This study referred to the research results of Nuri Ozturk47 to perform a cluster analysis on the ginkgo PHR/CRY family. The four PHR/CRY genes were identified from the ginkgo genome. We constructed a phylogenetic tree with the four PHR/CRY proteins and 50 PHR/CRY proteins from other organisms (Table S2). Four ginkgo PHR/CRY proteins were divided into three subgroups. Gb_13122 and Gb_39851 belonged to Plant CRY subgroup. Gb_25932 belonged to Class 0 PHR subgroup. Gb_03016 belonged to (6–4) PHR subgroup. According to the clustering relationship of ginkgo PHR/CRY family members in the phylogenetic tree, the four PHR/CRY protein were named as GbCRY1 (GenBank accession number MG25139), GbCRY2, Gb(6–4)PHR, and GbDASH (Figure 1(a)). The proteins of four PHR/CRY family members of ginkgo encoded 715, 434, 540 and 172 amino acids, respectively. The theoretical molecular weight of the deduced GbCRY1, GbCRY2, Gb(6–4)PHR, and GbDASH protein were 39.38 KDa, 81.06 KDa, 49.75 KDa, 62.37 KDa, 20.09 KDa, and the theoretical pI were 5.31, 6.66, 9.10, 8.73, respectively. Their total number of atoms were 11,256, 6,912, 8,730, 2,793, the aliphatic index of them were 80.21, 72.07, 76.19, 66.80, and the grand average of hydropathicity were −0.504, −0.644, −0.457, −0.396, respectively (Table 1).
Figure 1.
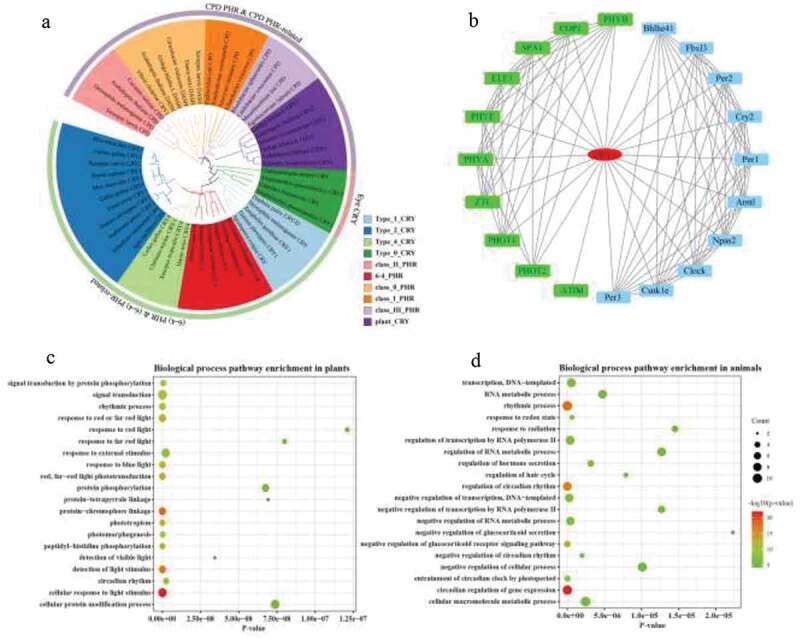
Phylogenetic tree analysis and function prediction of CRY1. (a) Phylogenetic analysis of ginkgo PHR/CRY family members. The color indicates the different subgroups. (b) Prediction of CRY1 protein-protein interaction network. Green represents the predicted results of protein-protein interaction in plants, and blue represents the predicted results of protein-protein interaction in animals. (c) GO enrichment analysis of CRY1 interacting protein in plants and (d) animals. The color indicates the q-value, with a lower q-value indicating more significant enrichment and the point size indicating the DEG number
Table 1.
Analysis of physicochemical properties of Ginkgo PHR/CRY family members
| Classification | Gene ID | Gene name | Number of amino acids | Molecular weight | Theoretical pI | Aliphatic index | Grand average of hydropathicity | Total number of atoms |
|---|---|---|---|---|---|---|---|---|
| Plant CRY | Gb_13122 | GbCRY1 | 715 | 81055.91 | 5.31 | 80.21 | −0.504 | 11256 |
| Plant CRY | Gb_39851 | GbCRY2 | 434 | 49754.12 | 6.66 | 72.07 | −0.644 | 6912 |
| Class 0 PHR | Gb_25932 | GbCRY_DASH | 540 | 62368.49 | 9.10 | 76.19 | −0.457 | 8730 |
| (6–4) PHR | Gb_03016 | Gb(6–4)PHR | 172 | 20085.95 | 8.73 | 66.80 | −0.396 | 2793 |
Prediction results of CRY1 protein-protein interaction network
Interaction between CRY1 and other proteins were predicted by the STRING database through co-expression, experimentally determined interaction, database annotated, automated text mining, and other information.50 Twenty interacting proteins were predicted in plant and animal populations (Figure 1(b)). The interacting proteins predicted by CRY1 included ATIM, COP1, PHYA, PHYB, PHYC, PHOT1, PHOT2, ELF3, SPA1, and ZIL in plant populations and Arntl, Bhlhe41, Fbxl3, Per2, Cry2, Per1, Npas2, Clock, Csnk1e, and Per3 in animal populations. Functional enrichment analysis of gene ontology biological process revealed that the interacting proteins were significantly enriched in the process of cellular response to light stimulus, protein-chromophore linkage, detection of light stimulus, phototropism, red/far-red light phototransduction, and peptidyl-histidine phosphorylation in plant populations (Figure 1(c)) and were significantly enriched in the process of circadian regulation of gene expression, rhythmic process, regulation of circadian rhythm, negative regulation of glucocorticoid receptor signaling pathway, photoperiodism, and entrainment of circadian clock by photoperiod in animal populations (Figure 1(d)). Circadian rhythm, Herpes simplex infection, and Circadian entrainment in animal populations and Circadian rhythm in plant populations were significantly enriched in the KEGG pathway.
Tissue expression analysis of GbCRY1
The qRT-PCR results showed that GbCRY1 expression level was higher in leaves and stems, and the difference was significant. The expression level in roots, male strobili, female strobili, and fruit was lower, and the difference was not significant (Figure 2). The RNA-seq results showed that the abundance level of GbCRY1 was higher in leaves, stems, and fruits, and significant differences were observed. The abundance level of GbCRY1 was lower in roots, male strobili, and female strobili, and no significant differences were found. The qRT-PCR results were significantly similar to the RNA-seq results (R2 = 0.80, p < .01; Figure 2), which indicated that the expression results of different tissues were reliable and accurate.
Figure 2.
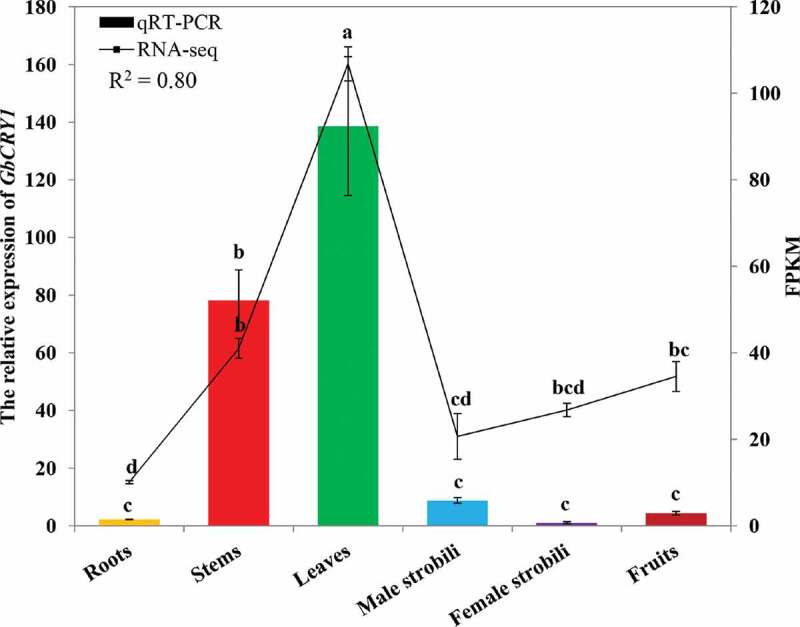
Abundance of CRY1 expression in different tissues of G. biloba. The error bars represent the standard error of three biological replicates. Bar and line charts represent the qRT-PCR and FPKM values of the genes, respectively. The expression in female strobili of G. biloba was set as 1 and the different letters indicates values are significantly different at P < .05. The R2 value represents the correlation between the qRT-PCR and FPKM values
Stomatal opening of ginkgo leaves and GbCRY1 expression under blue light
The dark-adapted G. biloba seedlings were treated with blue light for 12 h, and the stomatal conductance and GbCRY1 expression level were affected by blue light induction (Figure 3). The stomatal conductance of ginkgo seedling leaves reached the maximum after 3 h of blue light induction. With the extension of blue light treatment, the stomatal conductance remained open, but the stomatal conductance decreased slightly (Figure 3(a)). GbCRY1 expression level was highest in the dark, and it gradually decreased when induced by blue light. When ginkgo seedlings were treated with blue light for 3 h, GbCRY1 expression level decreased by 8.08%, but no significant difference was found with the expression under dark conditions. When ginkgo seedlings were induced by blue light for 6 h, the abundance level of GbCRY1 was significantly reduced and was 90.97% lower than that in the dark. After 6 h of blue light treatment, the abundance levels of GbCRY1 did not change significantly, but it was significantly lower in the 6 h blue light treatment than that in the dark, with a decrease of 93.84% and 97.87%, respectively (Figure 3(b)).
Figure 3.
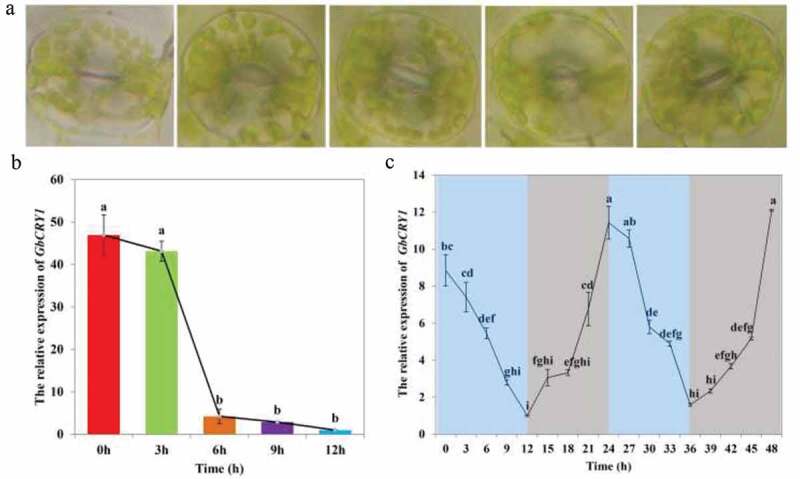
The effect of blue light mediated GbCRY1 on the stoma opening and Circadian clock of ginkgo seedlings. (a) Stomatal conductance changes. (b) The relative expression of GbCRY1 during blue light durations. (c) The relative expression of GbCRY1 during the periodic changes of blue light (12h blue light/12 dark). The different letters indicates values are significantly different at P < .05
Circadian clock and GbCRY1 expression
The circadian clock regulates many physiological and biochemical processes in plants that exhibit 24 h periodic rhythm changes. In this process, the circadian cycle is an important influencing factor.51 Plants adjust their physiological and biochemical functions by sensing changes in light and temperature during the day and night cycle. Photoreceptors, phytochromes, and cryptochromes are involved in light signal sensing and transmission.19,52 In this study, the newly germinated ginkgo seedlings were cultured under 12 h blue light/12 h dark cycles for 1 week. Total RNA was isolated from leaf samples harvested in 3 h intervals. The GbCRY1 expression level displayed clear circadian oscillations (Figure 3(c)). The qRT-PCR results showed that GbCRY1 expression level gradually decreased under blue light and gradually increased in the dark. The GbCRY1 expression level was lowest when seedlings were treated with blue light for 12 h, but reached its peak when seedlings were in the dark for 12 h. The above results indicated that GbCRY1 gene is involved in circadian clock regulation.
Photomorphogenesis of ginkgo seedlings and expression of GbCRY1 under blue light
The newly germinated ginkgo seedlings were cultured for 7 days in the dark and under blue and white lights. The photomorphological traits of seedlings showed great differences in leaf differentiation and expansion, de-etiolation, shoot length, internode length, hypocotyl length, stoma formation, and cell anatomy. Ginkgo seedlings differentiated to form leaf primordia under dark conditions, but the leaves did not expand and develop. By contrast, the leaves of the seedlings developed completely and gradually expanded under white and blue light conditions. The seedlings grown in the dark showed severe etiolation, whereas those grown under blue and white lights showed completely de-etiolation (Figure 4(a)). Observation of anatomical structure (Figure 4(g)) found that a large number of proplastids were formed in the seedling cells in the dark, but they failed to further differentiate to form organelles, such as chloroplasts. Under blue and white lights, the proplastids showed further differentiation, resulting in the de-etiolation of the seedlings. After the newly germinated seeds were treated with different light conditions, the plant height changed significantly. Seeding height was highest under dark conditions and was lowest under white light. The results of quantitative analysis (Figure 4(b)) showed that shoot length under dark conditions was significantly higher than that under blue light, and shoot length under blue light was significantly higher than that under white light. The measurement result of internode length is consistent with the change trend in shoot length. Hypocotyl length under dark conditions was significantly higher than that under white and blue lights, and no significant difference in hypocotyl length was observed under white and blue lights. The analysis of cell anatomy (Figure 4(g)) showed that the seedling cells were narrow and long under dark conditions, while they were thick and short under white and blue light conditions. Comparing the relative expression levels of GbCRY1 in seedlings under different culture conditions (Figure 4(c)), the results showed that expression under blue light was significantly lower than that in the dark but significantly higher than under white light conditions. The shoot (Figure 4(d)), internode (Figure 4(e)), hypocotyl (figure 4(f)) length, and the expression of GbCRY1 correlation analysis results showed that the hypocotyl length had the highest correlation with GbCRY1 expression, whereas the internode length had a lower correlation with GbCRY1 expression. In summary, GbCRY1 may be involved in the photomorphogenesis of ginkgo seedlings under blue light.
Figure 4.
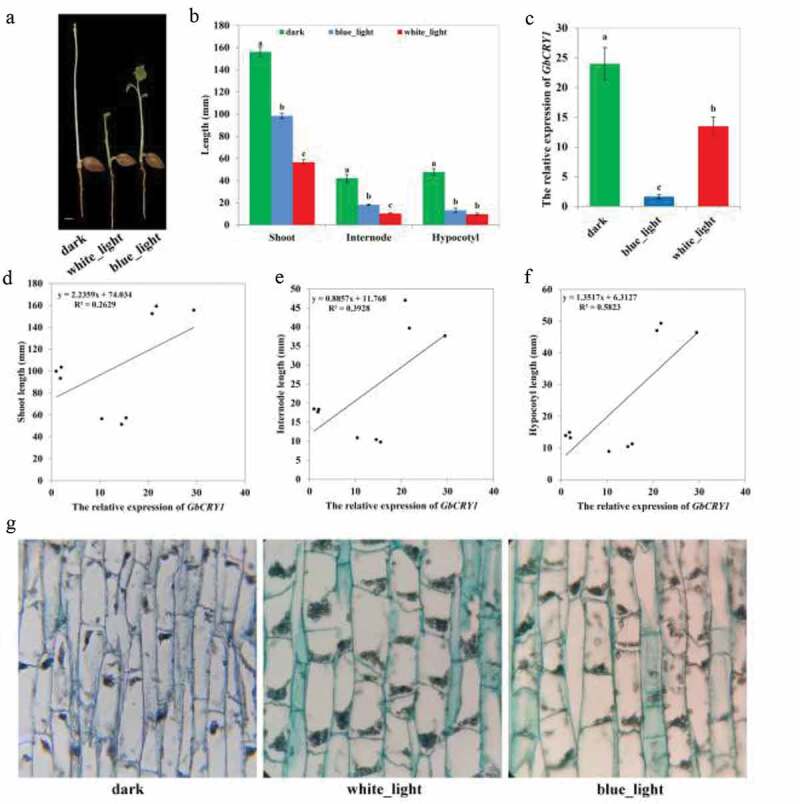
The effect of blue light mediated GbCRY1 on the photomorphogenesis of ginkgo seedlings. (a) Growth of ginkgo seedlings under different light conditions. (b) The length of shoot, internodes and hypocotyls of ginkgo seedlings under different light conditions. (c) Abundance of CRY1 expression under different light conditions. (d) Correlation plot of the shoot length and the relative expression of GbCRY1. (e) Correlation plot of the internode length and the relative expression of GbCRY1. (f) Correlation plot of the hypocotyl length and the relative expression of GbCRY1. (g) Observation on the anatomical structure of ginkgo seedling cells under different light conditions. The different letters indicates values are significantly different at P < .05
Discussion
Cryptochromes are present in all organisms from bacteria to humans and have a variety of functions. Cryptochromes are the core components of the circadian clock in humans, mammals, and insects.53–55 The cryptochrome of birds is a putative magnetoreceptor that can sense changes in the magnetic field,43 and studies on cryptochromes in plants are mostly focused on the model plant A. thaliana, in which the main function of cryptochromes was to mediate the suppression of seedling stem growth,16 the promotion of leaf and cotyledon expansion,56,57 the flowering time,18,58 the resetting of the circadian oscillator,19 the chlorophyll and anthocyanin synthesis,16 programmed cell death,59 and other processes. Whether the function of gymnosperm cryptochrome is similar to that of the model plant A. thaliana is still unknown. In this study, we identified four PHR/CRY family members from G. biloba and determined that ginkgo PHR/CRY family members are close to A. thaliana in the phylogenetic tree. We cloned a cryptochrome gene GbCRY1 and proved that GbCRY1 is related to ginkgo circadian clock, photomorphogenesis and stoma opening process.
Cryptochromes belong to the PHR/CRY family, which is classified based on the sequence similarity of the N-terminal photolyase/photolyase-relative region and the C-terminal extension region. Two types of structurally related DNA PHRs exist, as follows: cyclobutane pyrimidine dimer (CPD) PHRs that repair CPDs; and (6–4) PHRs that repair pyrimidine-pyrimidine (6–4) photoproducts.5 Yang et al.60 divided the PHR/CRY family into five major subgroups, namely, cyclobutane pyrimidine (CPD) photolyase, 6–4 photolyase, CRY-DASH, plant CRY, and animal CRY. Nuri Ozturk47 separated the PHR/CRY family into 10 major groups: Classes I, II, and III CPD PHRs; (6–4) PHRs; ssDNA PHRs (previously called DASH CRYs); plant CRYs; Types 1, 2, and 4 CRYs; and sponge CRYs (representing animal CRYs). Class I was found mostly in unicellular organisms; Class II was found in both unicellular and multicellular organisms; and Class III was found only in plants.61,62 CRY-DASH was found in bacteria and had no repair activities. This group have sequence homologs in Drosophila, Arabidopsis, Synechocystis, and human; thus, this group is named after their initials.63 Sponge CRYs were found in sponge.64 Plant CRYs are phylogenetically similar to CPD PHRs, whereas animal CRYs are similar to (6–4) PHRs.65 Animal CRYs were classified as Types 1 and 2. Type 4 CRYs exist in frogs, fishes, and birds.66 The phylogenetic tree classification results of the ginkgo PHR/CRY family were consistent with that of Nuri Ozturk.47 The four ginkgo PHR/CRY family members were divided into Plant CRY, Class 0 PHR, and (6–4) PHR (Figure 1(a)). GbCRY1 belongs to the Plant CRY subgroup, which was clustered in the same branch as A. thaliana CRY1 and S. lycopersicum CRY1. Researchers speculated that GbCRY1 and AtCRY1 have similar functions.
Cryptochromes are involved in a variety of biological processes; thus, interactions exist between cryptochromes and many other proteins. Yang et al.,67 Wang et al.,68 Yang et al.,69 and Yu et al.70 found that the CCE domains of CRY1 and CRY2 can bind to COP1. Lian et al.71 and Liu et al.72 found that CRY1 can bind to SPA1 and other SPA proteins in a blue light-dependent manner. Blue light-dependent CRY1 binding to SPA1 reduces COP1-SPA1 binding, which inactivates COP1, resulting in HY5 accumulation and the consequent transcriptional regulation of a number of genes.73,74 Ma et al.25 and Pedmale et al.28 demonstrated that CRY1 binds to PIF3, PIF4, and PIF5 in the presence of light. CRY1 physically interacts with phyA and phyB, which are major molecular species of phytochrome.64,75 CRY1 also physically binds with ZTL in yeast two-hybrid and in vitro pull-down assays.76 CRY1 and CRY2 likely regulate the entrainment of the circadian clock by regulating COP1-ELF3-mediated GI protein degradation.77 We predicted the protein-protein interaction relationship of CRY1 in animal and plant populations through the STRING database (Figure 1(b)). The protein interactions of CRY1 were independent of each other in plant and animal groups. The interacting proteins in plant populations include ATIM, COP1, PHYA, PHYB, PHYC, PHOT1, PHOT2, ELF3, SPA1, and ZIL, which play a role in the regulation of circadian rhythm and light response. The prediction results of these interacting proteins were consistent with those obtained in previous studies.67–77
Many organism activities from those of cyanobacteria to those of mammals are regulated through the circadian clock under the natural 24 h light/dark cycle. These organisms regulate the periodic changes in gene levels during transcription through negative feedback over a 24 h period. The clock proteins play an important role in the regulation of the circadian clock in Drosophila and mammals,78 but no homologous proteins have been found in plants. Many proteins found in plants exhibit circadian rhythm changes. Somers et al.19 showed that cryptochromes are involved in circadian clock regulation and are affected by blue light signals. Jarillo et al.76 isolated a circadian clock-related gene, ADAGIO1 (ADO1), which has a function similar to that of the animal clock gene from A. thaliana and can interact with cryptochrome. Genes that exhibit circadian rhythm expression are arrhythmic in the ado1 mutant.76 Toth et al.51 found that the expression levels of cryptochrome showed periodic fluctuations. CRY gene expression is induced by blue light and exhibits an oscillation period of almost 24 h in P. sativum.79 GbCRY1 expression level in ginkgo also showed an oscillation period under 12 h blue light/12 h dark. This finding showed that GbCRY1 was regulated by the ginkgo circadian clock, which was consistent with the results in A. thaliana and P. sativum. The relative expression of CRY1 in A. thaliana reached its peak after 6 h of light treatment. The peak and down-regulation time points of ginkgo GbCRY1 expression level under blue light were earlier than those of A. thaliana. The difference may be caused by species and light intensity, or blue light might be involved in the regulation of the circadian clock and thus affects GbCRY1 expression level.
Plant photomorphogenesis is regulated by CRYs, COP1, SPA, HY5, PIFs, AUX/IAA, and other proteins under blue light. The COP1/SPA complex recognizes and ubiquitinates HY5 protein in the dark, thereby degrading it via the 26S proteasome. Upon light exposure, CRYs and other photoreceptors inactivate the COP1/SPA complex, and HY5 protein consequently accumulated in the nucleus to induce photomorphogenesis.80 CRY1 more significantly contributes to de-etiolation phenotypes, such as cotyledon separation, cotyledon expansion, and chlorophyll accumulation.17,81 CRY1 regulates HY5 abundance by inactivating the COP1/SPA complex during CRY-mediated de-etiolation.71,72,82,83 Wang et al.68 and Yang et al.69 revealed that CRY1 and CRY2 interact with COP1 through their C terminus to inhibit COP1 activity and enhance the accumulation of HY5 to promote photomorphogenesis. Lian et al.71 and Liu et al.72 found that CRY1 and SPA also interact to inhibit the complex binding of COP1/SPA. Moreover, the endogenous auxin promotes hypocotyl elongation by promoting cell elongation.84 CRY1 can interact with AUX/IAA to inhibit A. thaliana auxin signal transmission, thereby inhibiting hypocotyl elongation.85,86 PIFs play an important role in the regulation of the phototropism of plants. Under suitable light conditions, CRY1 restricts PIF4 expression. Under low blue light conditions, CRY1 activity was inhibited, and the increase of PIF4 expression resulted in the increase of the phototropism of plants.87 In this study, under the white and blue lights, the processes of hypocotyl length inhibition, leaf expansion, and de-etiolation of G. biloba were completed normally, and GbCRY1 expression level in seedlings was significantly different under the two different lights. This finding indicated that GbCRY1 participated in the photomorphogenesis of G. biloba seedlings under blue light. The photomorphogenesis process of ginkgo seedlings under white light was more obvious than that under blue light, indicating that other photoreceptors are involved in the photomorphogenesis of ginkgo seedlings.
The stomata are the channels of gas exchange. Stomata are surrounded by a pair of guard cells, which control the stoma switch in response to environmental and internal signals, including light,88 humidity, drought stress, CO2, phytohormones, calcium, and reactive oxygen species.89 COP1 is a key protein regulating stoma opening and development.20,24 The stomata of WT A. thaliana were closed under dark conditions, and the stomata of cop1 mutant remained open under dark and light conditions, which proved that COP1 inhibits stomata opening. Further studies have found that cry1 mutant is less sensitive to light-induced stomatal regulation, and overexpression of CRY1 can promote A. thaliana stomatal opening, thereby indicating that CRY1 is involved in regulating blue light-induced stomatal opening in the upstream of COP1.20 In this study, the dark-adapted ginkgo stomata opened when blue light was induced for 3 h, and GbCRY1 expression level in leaves under blue light changed significantly compared those in the dark, indicating that GbCRY1 is involved in the blue light-induced stomatal opening of ginkgo leaves.
From the spatial point of view, CRY1 is expressed in the shoot of plants, and the expression is higher in the tender stems and leaves than in the other parts. A. thaliana CRY1 is expressed in shoot (stems, leaves, flowers, and fruit pods), especially in stems and leaves, but not in roots.51 The OsCRY1a and OsCRY1b of rice are expressed in the green tissues.90 GbCRY1 was expressed in roots, tender stems, leaves, male strobili, female strobili, and fruits, and the highest expression was observed in stems and leaves, which was consistent with the results in A. thaliana. However, CRY1 was expressed in ginkgo roots but not in A. thaliana roots. In addition, a high expression of GbCRY1 was detected in ginkgo yellow seedlings, which differed from the expression of CRY1 in rice (only in green tissues).90 These results showed that the expression of CRY gene appeared similar and different in gymnosperms, dicotyledons, and monocots. From the time point of view, CRY1 expression level was regulated by many factors, such as circadian rhythm, light, photoperiod, and the expression pattern was complicated. CRY1 and CRY2 in A. thaliana were both expressed in the circadian rhythm, and the expression level peaked at noon. Moreover, this circadian rhythm was affected by light, and the amplitude of the circadian rhythm decreased under continuous darkness. The expression of CRY was regulated by the circadian clock and photoperiod.51,79 The expressions of CRY1 in tomato,91 pea,79 Brassica,37 and apple92 were regulated by light. In addition, the 3′ UTR is widely known to function as a cis-acting element that regulated the mRNA half-life,93–95 leading to greater or lesser protein levels. The findings that the 3′ UTR of CRY1 gene in mice can increase the activity of CRY1 protein and reduce the expression level of CRY1 concluded that the rhythmical RNA-binding protein AUF1 on the 3′UTR recruits the 40S ribosomal subunit to the 5′ end of mRNA by associating with EIF3B, leading to time-dependent expression of CRY1.96 In this study, the peak and valley time of GbCRY1 expression levels in ginkgo and A. thaliana were different within 24 hours. In general, the results that the expression level of CRY1 was consistent with A. thaliana that showed a 24-hour cyclic fluctuation, and accumulated under dark, and decreased under light. Previous studies mostly focused on blue light activating CRY1 protein activity to regulate downstream genes. However, the molecular mechanism of blue light on cryptochrome transcription regulation is still unclear, and further research is needed.
Conclusion
In this study, four PHR/CRY family members were identified and classified. Among them, GbCRY1 was successfully isolated and cloned, and its response to blue light treatment was studied. The expression pattern of GbCRY1 and the phenotypic changes of ginkgo were analyzed under blue light. GbCRY1 shared a high similarity with the AtCRY1 of A. thaliana. The expression levels of GbCRY1 were higher in stems and leaves than in roots, male strobili, and female strobili of G. biloba. GbCRY1 expression level fluctuated periodically within 24 h, gradually increased in the dark, and decreased under blue light. The photomorphogenesis of ginkgo seedlings was induced by blue light. In addition, blue light treatment of dark-adapted ginkgo leaves could induce stomatal opening. Our results provided evidence that GbCRY1 is related to the circadian clock, photomorphogenesis, and stoma opening in ginkgo.
Supplementary Material
Funding Statement
This work was supported by the National Natural Science Foundation of China [No. 31670608] .
Author’s contribution
Gongping Nie and Feng Xu conceived and designed the experiments and drafted the manuscript. Gongping Nie, Xian Zhou, Qiling Song, Xiaomeng Liu performed the experiments. Gongping Nie, Xian Zhou, Qiling Song, Xiaomeng Liu, Xuefeng Wang and Mingyue Fu analyzed the data. All authors read an approved the manuscript.
Declaration of interest statement
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Rockwell NC, Su YS, Lagarias JC.. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2005;57(1):1–11. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs WR, Olney MA. Photoreceptors in plant photomorphogenesis to date. Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol. 2001;125(1):85–88. doi: 10.1104/pp.125.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liscum E, Hodgson DW, Campbell TJ. Blue light signaling through the cryptochromes and phototropins. So that’s what the blues is all about. Plant Physiol. 2003;133(4):1429–1436. doi: 10.1104/pp.103.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C. Plant blue-light receptors. Trends Plant Sci. 2000;5(8):337–342. doi: 10.1016/s1360-1385(00)01687-3. [DOI] [PubMed] [Google Scholar]
- 5.Lin C, Shalitin D. Cryptochrome structure and signal transduction. Annu Rev Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 6.Sancar A. Photolyase and cryptochrome blue-light photoreceptors. Adv Protein Chem. 2004;69:73–100. doi: 10.1016/S0065-3233(04)69003-6. [DOI] [PubMed] [Google Scholar]
- 7.Lin C, Todo T. The cryptochromes. Genome Biol. 2005;6(5):220. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vechtomova YL, Telegina TA, Kritsky MS. Evolution of Proteins of the DNA Photolyase/Cryptochrome Family. Biochemistry (Mosc). 2020;85:S131–S153. doi: 10.1134/S0006297920140072. [DOI] [PubMed] [Google Scholar]
- 9.Todo T. Functional diversity of the DNA photolyase/blue light receptor family. Mutat Res. 1999;434(2):89–97. doi: 10.1016/s0921-8777(99)00013-0. [DOI] [PubMed] [Google Scholar]
- 10.Cashmore AR. Cryptochromes: enabling plants and animals to determine circadian time. Cell. 2003;114(5):537–543. doi: 10.1016/j.cell.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103(6):2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 12.Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284(5415):760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 13.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101(33):12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sang Y, Li QH, Rubio V, Zhang Y-C, Mao J, Deng X-W, Yang H-Q. N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell. 2005;17(5):1569–1584. doi: 10.1105/tpc.104.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13(12):2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366(6451):162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 17.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci U S A. 1998;95(5):2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279(5355):1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 19.Somers DE, Devlin PF. Kay SA: phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. [DOI] [PubMed] [Google Scholar]
- 20.Mao J, Zhang YC, Sang Y, Li QH, Yang HQ. From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci U S A. 2005;102(34):12270–12275. doi: 10.1073/pnas.0501011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu L, Yang HQ. CRYPTOCHROME 1 is implicated in promoting R protein-mediated plant resistance to Pseudomonas syringae in Arabidopsis. Mol Plant. 2010;3(3):539–548. doi: 10.1093/mp/ssp107. [DOI] [PubMed] [Google Scholar]
- 22.Meng Y, Li H, Wang Q, Liu B, Lin C. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell. 2013;25(11):4405–4420. doi: 10.1105/tpc.113.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrero JM, Downie AB, Xu Q, Gubler F. A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell. 2014;26(3):1094–1104. doi: 10.1105/tpc.113.121830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ. Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell. 2009;21(9):2624–2641. doi: 10.1105/tpc.109.069765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedmale UV, Huang SC, Zander M, Cole B, Hetzel J, Ljung K, Reis PB, Sridevi P, Nito K, Nery J, et al. Cryptochromes Interact Directly with PIFs to Control Plant Growth in Limiting Blue Light. Cell. 2016;164(1–2):233–245. doi: 10.1016/j.cell.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu P, Xiang Y, Zhu H, Xu HB, Zhang ZZ, Zhang CQ, Zhang LX, Ma ZQ. Wheat cryptochromes: subcellular localization and involvement in photomorphogenesis and osmotic stress responses. Plant Physiol. 2009;149(2):760–774. doi: 10.1104/pp.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan S, Zhang ZW, Zheng C, Zhao Z-Y, Wang Y, Feng L-Y, Niu G, Wang C-Q, Wang J-H, Feng H, et al. Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc Natl Acad Sci U S A. 2016;113(27):7661–7666. doi: 10.1073/pnas.1602004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci U S A. 2016;113(1):224–229. doi: 10.1073/pnas.1511437113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sancar A. Regulation of the mammalian circadian clock by cryptochrome. J Biol Chem. 2004;279(33):34079–34082. doi: 10.1074/jbc.R400016200. [DOI] [PubMed] [Google Scholar]
- 30.Sancar A, Rupert CS. Cloning of the PHR gene and amplification of photolyase in Escherichia coli. Gene. 1978;4(4):295–308. doi: 10.1016/0378-1119(78)90047-1. [DOI] [PubMed] [Google Scholar]
- 31.Kleine T, Lockhart P, Batschauer A. An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J. 2003;35(1):93–103. doi: 10.1046/j.1365-313x.2003.01787.x. [DOI] [PubMed] [Google Scholar]
- 32.Lin C, Ahmad M, Chan J. Cashmore A.CRY2: aSecond member of the arabidopsis cryptochrome gene family. Plant Physiol. 1996;110:1047. [Google Scholar]
- 33.Huang Y, Baxter R, Smith BS, Partch CL, Colbert CL, Deisenhofer J. Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proc Natl Acad Sci U S A. 2006;103(47):17701–17706. doi: 10.1073/pnas.0608554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci U S A. 2006;103(47):17696–17700. doi: 10.1073/pnas.0607993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giliberto L, Perrotta G, Pallara P, Weller JL, Fraser PD, Bramley PM, Fiore A, Tavazza M, Giuliano G. Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 2005;137(1):199–208. doi: 10.1104/pp.104.051987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platten JD, Foo E, Foucher F, Hecht V, Reid JB, Weller JL. The cryptochrome gene family in pea includes two differentially expressed CRY2 genes. Plant Mol Biol. 2005;59(4):683–696. doi: 10.1007/s11103-005-0828-z. [DOI] [PubMed] [Google Scholar]
- 37.Chatterjee M, Sharma P, Khurana JP. Cryptochrome 1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol. 2006;141(1):61–74. doi: 10.1104/pp.105.076323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirose F, Shinomura T, Tanabata T, Shimada H, Takano M. Involvement of rice cryptochromes in de-etiolation responses and flowering. Plant Cell Physiol. 2006;47(7):915–925. doi: 10.1093/pcp/pcj064. [DOI] [PubMed] [Google Scholar]
- 39.Immeln D, Schlesinger R, Heberle J, Kottke T. Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome. J Biol Chem. 2007;282(30):21720–21728. doi: 10.1074/jbc.M700849200. [DOI] [PubMed] [Google Scholar]
- 40.Imaizumi T, Kanegae T, Wada M. Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capillus-veneris. Plant Cell. 2000;12(1):81–96. doi: 10.1105/tpc.12.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imaizumi T, Kadota A, Hasebe M, Wada M. Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell. 2002;14(2):373–386. doi: 10.1105/tpc.010388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshii T, Hermann-Luibl C, Kistenpfennig C, Schmid B, Tomioka K, Helfrich-Förster C. Cryptochrome-dependent and -independent circadian entrainment circuits in Drosophila. J Neurosci. 2015;35(15):6131–6141. doi: 10.1523/JNEUROSCI.0070-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gustafson CL, Partch CL. Emerging models for the molecular basis of mammalian circadian timing. Biochemistry. 2015;54(2):134–149. doi: 10.1021/bi500731f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H, Scholten A, Einwich A, Mouritsen H, Koch KW. Protein-protein interaction of the putative magnetoreceptor cryptochrome 4 expressed in the avian retina. Sci Rep. 2020;10(1):7364. doi: 10.1038/s41598-020-64429-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(1):427–432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(1):265–268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 48.Ozturk N. Phylogenetic and Functional Classification of the Photolyase/Cryptochrome Family. Photochem Photobiol. 2017;93(1):104–111. doi: 10.1111/php.12676. [DOI] [PubMed] [Google Scholar]
- 49.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tóth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognár L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127(4):1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12(12):2499–2510. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barinaga M. Clock photoreceptor shared by plants and animals. Science. 1998;282(5394):1628–1630. doi: 10.1126/science.282.5394.1628. [DOI] [PubMed] [Google Scholar]
- 55.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95(5):669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 56.Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282(5393):1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 57.Jackson JA, Jenkins GI. Extension-growth responses and expression of flavonoid biosynthesis genes in the Arabidopsis hy4 mutant. Planta. 1995;197(2):233–239. doi: 10.1007/BF00202642. [DOI] [PubMed] [Google Scholar]
- 58.Neff MM, Chory J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998;118(1):27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bagnall DJ, King RW, Hangarter RP. Blue-light promotion of flowering is absent in hy4 mutants of Arabidopsis. Planta. 1996;200(2):278–280. doi: 10.1007/BF00208319. [DOI] [PubMed] [Google Scholar]
- 60.Danon A, Coll NS, Apel K. Cryptochrome-1-dependent execution of programmed cell death induced by singlet oxygen in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2006;103(45):17036–17041. doi: 10.1073/pnas.0608139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Z, Liu B, Su J, Liao J, Lin C, Oka Y. Cryptochromes orchestrate transcription regulation of diverse blue light responses in plants. Photochem Photobiol. 2017;93(1):112–127. doi: 10.1111/php.12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oztürk N, Kao YT, Selby CP, Kavakli IH, Partch CL, Zhong D, Sancar A. Purification and characterization of a type III photolyase from Caulobacter crescentus. Biochemistry. 2008;47(39):10255–10261. doi: 10.1021/bi801085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oztürk N, Song SH, Ozgür S, Selby CP, Morrison L, Partch C, Zhong D, Sancar A. Structure and function of animal cryptochromes. Cold Spring Harb Symp Quant Biol. 2007;72(1):119–131. doi: 10.1101/sqb.2007.72.015. [DOI] [PubMed] [Google Scholar]
- 64.Rivera AS, Ozturk N, Fahey B, Plachetzki DC, Degnan BM, Sancar A, Oakley TH. Blue-light-receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin. J Exp Biol. 2012;215(Pt 8):1278–1286. doi: 10.1242/jeb.067140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1(7):939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- 66.Sancar A. Structure and function of photolyase and in vivo enzymology: 50th anniversary. J Biol Chem. 2008;283(47):32153–32157. doi: 10.1074/jbc.R800052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ozturk N, Selby CP, Song SH, Ye R, Tan C, Kao YT, Zhong D, Sancar A. Comparative photochemistry of animal type 1 and type 4 cryptochromes. Biochemistry. 2009. September 15;48(36):8585–8593. doi: 10.1021/bi901043s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103(5):815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 69.Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294(5540):154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- 70.Yang HQ, Tang RH, Cashmore AR. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell. 2001;13(12):2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C. Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci U S A. 2007;104(17):7289–7294. doi: 10.1073/pnas.0701912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011;25(10):1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011;25(10):1029–1034. doi: 10.1101/gad.2025011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003. November 1;17(21):2642–2647. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003;423(6943):995–999. doi: 10.1038/nature01696. [DOI] [PubMed] [Google Scholar]
- 76.Más P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408(6809):207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- 77.Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR. An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature. 2001. March 22;410(6827):487–490. doi: 10.1038/35068589. [DOI] [PubMed] [Google Scholar]
- 78.Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell. 2008;32(5):617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lakin-Thomas PL. Circadian rhythms: new functions for old clock genes. Trends Genet. 2000;16(3):135–142. doi: 10.1016/s0168-9525(99)01945-9. [DOI] [PubMed] [Google Scholar]
- 80.Platten JD, Foo E, Elliott RC, Hecht V, Reid JB, Weller JL. Cryptochrome 1 contributes to blue-light sensing in pea. Plant Physiol. 2005;139(3):1472–1482. doi: 10.1104/pp.105.067462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405(6785):462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 82.Lin C, Ahmad M, Cashmore AR. Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 1996;10(5):893–902. doi: 10.1046/j.1365-313x.1996.10050893.x. [DOI] [PubMed] [Google Scholar]
- 83.Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends in Plant Ence. 2011;16(12):684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu B, Yang Z, Gomez A, Liu B, Lin C, Oka Y. Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J Plant Res. 2016;129(2):137–148. doi: 10.1007/s10265-015-0782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2011. November 8;108(45):18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu F, He S, Zhang J, Mao Z, Wang W, Li T, Hua J, Du S, Xu P, Li L, et al. Photoactivated CRY1 and phyB Interact Directly with AUX/IAA Proteins to Inhibit Auxin Signaling in Arabidopsis. Mol Plant. 2018;11(4):523–541. doi: 10.1016/j.molp.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 87.Wang WX, Lian HL, Zhang LD, Mao ZL, Li XM, Xu F, Li L, Yang HQ. Transcriptome Analyses Reveal the Involvement of Both C and N Termini of Cryptochrome 1 in Its Regulation of Phytohormone-Responsive Gene Expression in Arabidopsis. Front Plant Sci. 2016. March 14;7:294. doi: 10.3389/fpls.2016.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boccaccini A, Legris M, Krahmer J, Allenbach-Petrolati L, Goyal A, Galvan-Ampudia C, Vernoux T, Karayekov E, Casal JJ, Fankhauser C. Low Blue Light Enhances Phototropism by Releasing Cryptochrome1-Mediated Inhibition of PIF4 Expression. Plant Physiol. 2020. August;183(4):1780–1793. doi: 10.1104/pp.20.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marten H, Hedrich R, Roelfsema MR. Blue light inhibits guard cell plasma membrane anion channels in a phototropin-dependent manner. Plant J. 2007;50(1):29–39. doi: 10.1111/j.1365-313X.2006.03026.x. [DOI] [PubMed] [Google Scholar]
- 90.Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. GUARD CELL SIGNAL TRANSDUCTION. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- 91.Facella P, Lopez L, Carbone F, Galbraith DW, Giuliano G, Perrotta G. Diurnal and circadian rhythms in the tomato transcriptome and their modulation by cryptochrome photoreceptors. PLoS One. 2008;3(7):e2798. doi: 10.1371/journal.pone.0002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li YY, Mao K, Zhao C, Zhang RF, Zhao XY, Zhang HL, Shu HR, Zhao YJ. Molecular cloning of cryptochrome 1 from apple and its functional characterization in Arabidopsis. Plant Physiol Biochem. 2013;67:169–177. doi: 10.1016/j.plaphy.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 93.Misquitta CM, Iyer VR, Werstiuk ES, Grover AK. The role of 3ʹ-untranslated region (3ʹ-UTR) mediated mRNA stability in cardiovascular pathophysiology. Mol Cell Biochem. 2001;224(1–2):53–67. doi: 10.1023/a:1011982932645. [DOI] [PubMed] [Google Scholar]
- 94.Von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181(2):189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13(4):246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee KH, Kim SH, Kim HJ, Kim W, Lee HR, Jung Y, Choi JH, Hong KY, Jang SK, Kim KT. AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res. 2014;42(6):3590–3606. doi: 10.1093/nar/gkt1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


