Abstract
The function of microbes can be inferred from knowledge of genes specifically expressed in natural environments. Here, we report the in vivo transcriptome of the entomopathogenic bacterium Yersinia entomophaga MH96, captured during initial, septicemic, and pre-cadaveric stages of intrahemocoelic infection in Galleria mellonella. A total of 1285 genes were significantly upregulated by MH96 during infection; 829 genes responded to in vivo conditions during at least one stage of infection, 289 responded during two stages of infection, and 167 transcripts responded throughout all three stages of infection compared to in vitro conditions at equivalent cell densities. Genes upregulated during the earliest infection stage included components of the insecticidal toxin complex Yen-TC (chi1, chi2, and yenC1), genes for rearrangement hotspot element containing protein yenC3, cytolethal distending toxin cdtAB, and vegetative insecticidal toxin vip2. Genes more highly expressed throughout the infection cycle included the putative heat-stable enterotoxin yenT and three adhesins (usher-chaperone fimbria, filamentous hemagglutinin, and an AidA-like secreted adhesin). Clustering and functional enrichment of gene expression data also revealed expression of genes encoding type III and VI secretion system-associated effectors. Together these data provide insight into the pathobiology of MH96 and serve as an important resource supporting efforts to identify novel insecticidal agents.
Keywords: transcriptome, entomopathogenic bacteria, insecticidal toxins, Galleria mellonella, Yersinia virulence factors
Introduction
For successful invasion, colonization, and bioconversion of host tissues, bacterial pathogens must acclimate to the host environment via alterations in patterns of gene expression (Crawford et al. 2010; Tran et al. 2013; Heroven and Dersch 2014; Fang et al. 2016; Reniere 2018). Knowledge of those genes activated specifically within host tissues allows insight into the genetic determinants of in vivo colonization and pathogenicity. An often used strategy involves transcriptional analysis of pathogen gene expression (transcriptomics) based on patterns of mRNA transcript abundance during infection (Deng et al. 2018; Valenzuela-Miranda and Gallardo-Escárate 2018; Nobori et al. 2018; Arenas et al. 2019; Griesenauer et al. 2019; Haueisen et al. 2019; Ibberson and Whiteley 2019; Sun et al. 2019; Tang et al. 2019; Fan et al. 2020; Luo et al. 2020).
In vivo transcriptomic approaches have been widely applied to human pathogenic bacteria using cell lines and model hosts (Yan et al. 2013; Avican et al. 2015; Baddal et al. 2016; Damron et al. 2016; Westermann et al. 2016; Nuss et al. 2017). Additionally, time-resolved in vivo transcriptomics have increased understanding of the function of bacterial pathogens during natural infection processes (Aprianto et al. 2016; Westermann et al. 2019; Li et al. 2020; Luo et al. 2020; Tang et al. 2020). However, similar strategies are yet to be applied to pathogens infecting insects, despite the obvious potential to identify new insect-active agents.
Yersinia entomophaga MH96 is an entomopathogenic bacterium that was isolated from Costelytra giveni (Coleoptera: Scarabaeidae) (Hurst et al. 2011a), an endemic and economically significant pasture pest in New Zealand (Ferguson et al. 2019). Development of MH96 as a biopesticide has proven consistent pathogenesis by per os challenge against C. giveni, as well as a wide range of coleopteran, lepidopteran, and orthopteran species (Hurst et al. 2011a, 2014, 2015, 2019; Marshall et al. 2012). The key virulence determinant of MH96 is an insecticidal toxin complex (TC) called Yen-TC, which facilitates ingress of MH96 through the insect midgut (Hurst et al. 2011b, 2014; Marshall et al. 2012) and is constitutively produced and secreted by MH96 when grown in broth culture at temperatures of 25°C or lower (Hurst et al. 2011b). In addition to the orally active insecticidal activities of Yen-TC, structural characterization and understanding the molecular mechanisms of pore-formation and translocation of cytotoxin has been the focus of much research (Landsberg et al. 2011; Busby et al. 2012, 2013; Piper et al. 2019).
Despite the importance of Yen-TC in MH96’s ability to breach the insect midgut, additional investigations have suggested that as yet unknown virulence factors (VFs) are deployed during intrahemocoelic infection of Galleria mellonella. As a prime example, Hurst et al. (2015) showed that both wild-type MH96 and ΔTC (a Yen-TC deficient strain) have the same median lethal dose (LD50) of only ∼3 cells and were equally as effective at stopping the host’s phenoloxidase immune response when injected directly into the hemocoel of G. mellonella (Hurst et al. 2015). Genes for many other diverse putative VFs, including known insecticidal toxins and effectors, secretion and iron acquisition systems, proteolytic enzymes, and adhesins have also been identified in the MH96 draft genome (Hurst et al. 2016). The extensive repertoire of putative VF-encoding genes represents an opportunity to utilize in vivo transcriptomics to further characterize this entomopathogen’s mode of action against insects, especially during intrahemocoelic infection of G. mellonella.
Larvae of the greater wax moth G. mellonella (Lepidoptera) have emerged as a valuable model host for study of microbial infection and innate immunity (Desbois and McMillan 2015; Champion et al. 2016; Tsai et al. 2016). Yet, to date, pathogen in vivo transcriptome studies from G. mellonella have been confined to fungal pathogens only, including Candida albicans (Amorim-Vaz et al. 2015) and entomopathogenic Beauveria bassiana (Chen et al. 2018). Since the immune system of G. mellonella consists of both cellular (i.e., hemocytes) and humoral components (Wojda 2017; Singkum et al. 2019), the intrahemocoelic infection is useful to characterize host-dependent genome-wide transcriptional modulation by MH96 during three different stages of infection using a time-resolved approach.
Here, we focus on the interaction between MH96 and G. mellonella, using an in vivo transcriptomic strategy to identify pathogen genes activated under initial insect immunity directly following intrahemocoelic infection, as well as later septicemic and pre-cadaver stages of infection. Differential expression (DE) analysis comparing MH96 transcriptome under in vivo and in vitro conditions identified pathogen genes, including known and putative VFs, that significantly responded either at specific or multiple timepoints throughout the infection process. To further understand the pathobiology of MH96, time-resolved in vivo versus in vitro fold-changes were clustered and functionally annotated using a database of known bacterial VFs. Particularly notable was upregulation of multiple genes encoding predicted adhesins, toxins, and type III secretion system (T3SS) and T6SS translocated effectors.
Materials and methods
Intrahemocoelic infection of G. mellonella and pathogen RNA enrichment
Larval G. mellonella (Biosuppliers, Auckland, New Zealand) was provided with fresh diet of liquid honey, glycerol, Farex® brand baby rice cereal, and granular yeast for up to 1 week. Healthy final instar larvae of similar lengths (>20 mm, between 0.15 and 0.30 g) were selected and immobilized on ice. For intrahemocoelic infection, MH96 was grown overnight (18 h) in 3-ml Luria Broth Base (Invitrogen) with shaking (250 rpm) at 30°C, and then cultures were diluted in phosphate buffer solution (Sigma) and placed on ice prior to injection. Larvae were surface sterilized with 70% ethanol and then injected below the third right leg with 10 µl of inoculum using a 30 gauge 12-mm needle on a 1-ml tuberculin syringe (Terumo) with a microinjector.
Dilutions of MH96 ranging from ∼157 up to 2 × 107 colony forming units (CFUs) were injected into G. mellonella depending on the stage of infection for each in vivo treatment (i.e., early, middle, or late) (Supplementary Table S1). To ensure sufficient quantity of pathogen RNA for in vivo transcriptomics, early in vivo treatments (i.e., initial insect immunity) required that larvae were injected with a greater amount of pathogen cells than would be expected to naturally occur during early intrahemocoelic infection. Following injection, hemolymph from early in vivo treatment larvae was collected within 75- to 90-min post-intrahemocoelic infection, which was considered sufficient time to trigger a measurable host-dependent transcriptional response by MH96. To first define the respective 18–19 and 26–30 h incubation times for the middle (i.e., septicemic) and late infection treatments (i.e., pre-cadaver) a 48 h post-inoculation (hpi) growth curve was obtained following injection of ∼345 CFUs (Supplementary Figure S1). MH96 CFU/larva were then enumerated on Yersinia semi-selective medium from dilutions of homogenized whole infected larvae collected at specific timepoints post-inoculation. Consistent with the 48 hpi growth curve, larvae for middle, and late in vivo treatments were injected on separate days with either ∼157 or 693 CFUs and collected after 18–19 and 26–30 hpi, respectively.
Following injection, the larvae were placed in petri-dishes and incubated at 25°C within sealed plastic bags with moistened paper towel. After incubation, hemolymph was obtained by puncturing the dorsal cuticle near the second segment with a needle. Hemolymph was pooled from five to six individuals, yielding ∼150-µl total volume. Four biological replicate samples were collected from each of the three in vivo infection stages (12 in vivo samples total). Pooled hemolymph samples were collected directly into microcentrifuge tubes with 300 µl of RNAprotect Bacterial Reagent (Qiagen), vortexed for 5 s, incubated at room temperature for 5 min, and then centrifuged at 300 × g for 5 min at 4°C to pellet hemocytes and cell debris. The supernatant was carefully separated from the host cell pellet by pipette and bacterial cells were then pelleted by centrifugation (10 min at 5000 × g at ambient temperature of ∼22°C). Following supernatant removal, the pellets were stored at −80°C. To estimate the CFUs per larva, serial dilutions from G. mellonella homogenate were plated on a Yersinia semi-selective medium and all Yersinia-positive colonies were counted within 24 h (five separate larva per treatment; surface sterilized with 70% ethanol). The weight of each larva was measured enabling determination of CFU per gram larval homogenate.
Control in vitro RNA samples were collected separately from broth cultures and two independent biological replicate samples were collected for each of the in vitro treatments—lag, exponential, and stationary growth phases (6 in vitro samples total). For in vitro growth MH96 was grown overnight in 3-ml broth at 30°C with shaking (250 rpm). The next day, flasks containing 50-ml fresh broth were inoculated with 1% overnight culture and grown at 25°C until samples were collected after 4 h 45 min (2 ml), 6 h (1 ml), and 9 h (1 ml) corresponding to specific cell densities of 3.2 ± 0.1 × 107, 2.9 ± 0.9 × 108, and 4.4 ± 0.1 × 109 CFU ml−1 (comparable to CFU g−1 assessed from in vivo G. mellonella hemolymph RNA-seq samples), respectively. In vitro RNA samples were stabilized in RNAprotect Bacterial Reagent (Qiagen) according to the manufacturer’s protocol and also stored at −80°C.
RNA extraction, library preparation, and sequencing
Within 1 week of collection, RNA samples were re-suspended in 100-µl Tris-EDTA buffer (10-mM Tris-HCl, 1-mM EDTA; pH 8.0) (Ambion) containing 15-mg ml−1 lysozyme (Roche or Sigma-Aldrich) and 10-µl proteinase K (Roche). The samples were incubated for 10 min, interrupted by 10 s vortex every 2 min to lyse the cells. The RNeasy mini kit (Qiagen) was used for RNA extraction and RNA clean-up, which included two separate DNA digestions (on-column then off-column) with DNAse (Qiagen), all according to manufacturer’s guidelines. RNA samples were then precipitated at −20°C overnight in three volumes 100% cold isopropanol with 1/10 volumes 3 M sodium acetate (pH 5.2), followed by two 70% cold ethanol washes. Purified RNA was re-suspended in RNAse-free water and stored at −80°C.
RNA extract from early infection hemolymph contained approximately half the amount of total RNA required for sequencing, so early infection RNA samples were doubled such that each sample was comprised from a pool of 10–12 larvae, while middle and late infection RNA samples were from five or six pooled individuals. Sufficient removal of genomic DNA from the samples was determined by confirming lack of PCR amplification from in vivo RNA template using primers RecA_352F 5ʹ—TCTCAGCCAGATACCGGTGA and RecA_987R 5ʹ-CAGCAACATTTCACGCAGCT for the housekeeping gene recA. Middle and late infection in vivo RNA samples containing ∼7.2-µg total RNA and early infection samples containing <7.2 µg were stabilized on RNAstable 1.5 microcentrifuge tubes (Biomātrica) by SpeedVac (Savant) at ambient temperature (∼22°C) for 1.5 h.
Macrogen Korea carried out library preparation and sequencing. Following recovery from the RNAstable tubes, RNA quantity and Integrity Number were determined using the 2200 TapeStation (Agilent Technologies). Host and bacterial ribosomal RNA (rRNA) was depleted from the in vivo samples using the ScriptSeq Complete Gold Kit (Epidemiology) (Illumina) then strand-specific cDNA libraries were prepared using the ScriptSeq RNA-Seq Library Preparation Kit (Illumina) according to the manufacturer’s guidelines. The in vitro samples were prepared using the TruSeq stranded mRNA (Illumina) with the Ribo-Zero rRNA removal kit for Bacteria (Epicentre) according to standard protocol. To verify the size of the PCR enriched fragments, the template size distribution was assessed on the 2100 Bioanalyzer using a DNA 1000 chip (Agilent Technologies). The libraries were sequenced on the HiSeq2500 platform (Illumina) (Version HCS v2.2) to generate 101-bp paired-end reads.
Bioinformatic analysis
Adapter and barcode contamination were filtered using Flexbar (v2.4) against Illumina adapter and barcode reference sequences (Supplementary Tables S2 and S3). Reads were trimmed in “ANY” mode with minimum overlap of 5, maximum uncalled base of 1, and minimum quality threshold of 20 (Dodt et al. 2012). The bbsplit.sh function of the BBMap package was used to quantify and remove contaminating rRNA sequences with minimum ratio of 0.5, minimum hit number of one, and maximum insertion/deletion length of 500 bp (Bushnell 2015). All libraries were screened against full-length 5S, 16S, and 23S rRNA sequence from the MH96 genome (see accession information below). To remove host sequences, in vivo libraries were also mapped against the G. mellonella partial 18S and 28S rRNA sequence retrieved from GenBank (Accessions: U65198.1, U65138.1, AF286298.1, AF423921.1, and X89491.1) and the G. mellonella immune cDNA library (JG394435–JG406465) (Vogel et al. 2011).
Remaining paired-end reads were aligned to the MH96 genome (Accession: CP010029.1, GI: 1034308998) using Rockhopper (v2.03) in verbose mode. Alignment stringencies were increased from default values, such that allowable mismatch threshold was decreased from 15% to 10% of read length and the minimum seed was increased from 33% to 50% of read length. Any residual count data aligning to 5S, 16S, and 23S rRNA genes or any short (<50 bp) non-coding RNA or anti-sense RNA were not considered further in the analysis.
Due to low alignment rates to the reference genome, most of the early infection in vivo libraries contained less than the minimal threshold of between 2 and 3 million transcripts per library determined by Haas et al. (2012) as necessary for adequate detection of differentially expressed genes in bacteria. Accordingly, the early infection in vivo libraries were investigated using the iNEXT R package (nboot = 50) (v2.0.18) (Hsieh et al. 2016) whether the sequence depth adequately captured the majority of MH96 low-abundance transcripts. The RUVseq R package (v1.16.0) was used to generate relative log expression (RLE) plots and conduct principle component analysis (PCA) (Risso et al. 2014). Upper-quartile normalized (Bullard et al. 2010) libraries were filtered for transcripts with greater than five aligned reads in at least two libraries.
Normalized expression data were then converted to log2-counts-per-million (Log2CPM) using the R package limma (v3.38.3) (Smyth 2004; Ritchie et al. 2015) and the mean–variance relationship was modeled using the “voom” method (Law et al. 2014). Genome-wide host-dependent responses by MH96 to both in vitro and in vivo conditions were visualized with Circos (v0.69) (Krzywinski et al. 2009) and putative genomic islands (GIs) were assigned using the webserver IslandViewer 4 (Bertelli et al. 2017). To determine MH96 genes that significantly responded to in vivo conditions, a DE analysis was undertaken by comparing average transcript abundance between in vivo and in vitro libraries sharing equivalent cell densities (i.e., early infection vs lag growth in broth, middle infection vs exponential growth in broth, and late infection vs stationary growth in broth). Genes with significantly DE were defined by multiple testing with false discovery rate of 0.05 using the Benjamini and Yekutieli multiple test correction method (Benjamini and Yekutieli 2001) using limma.
Time-resolved average in vivo vs in vitro log2CPM fold-change values for all genes with significantly DE identified between the in vivo and in vitro treatments (DE analysis described above) were clustered using the fuzzy c-means (FCM) algorithm (Futschik and Carlisle 2005) with the MFuzz R package (v2.42.0) (Kumar and Futschik 2007). A heuristic qualitative assessment was used to identify both the optimal FCM parameter (i.e., “fuzzifier”) and number of clusters (Kumar and Futschik, 2007). The assessment considered both predicted operon structure (output from Rockhopper) and functional annotations assigned using homology-based search (BLASTx, e-value cutoff = 1 × 10−8) against the virulence factor database (VFDB) (accessed July 2018) (Chen et al. 2016).
MH96 host-specific factors were identified among clusters containing transcripts with comparably greater median log2CPM in vivo fold-change values (≥ ∼2) for at least one stage of growth during intrahemocoelic infection. Next, these host-specific factors were functionally characterized as putative VFs when significant sequence homology was identified against an already characterized VF found in the VFDB. Finally, functional categories of virulence were assigned to all of these putative VFs primarily based on the VFDB-derived annotation, in accordance to our definitions found in Supplementary Table S4. When VFDB-derived annotations were not informative, additional information, such as top-scoring BLASTx hits from the GenBank non-redundant protein database, was used to assign a possible function or otherwise these hypotheticals were left unclassified.
Data availability
Sequence data and both raw and normalized count tables were deposited in the GenBank Geo Submission Omnibus under accession: GSE142509. All genes with significantly DE detected between in vivo vs in vitro time-resolved contrasts (early infection vs lag growth in vitro; middle infection vs exponential growth in vitro; late infection vs stationary growth in vitro) are provided in Supplementary Table S5. All functionally annotated putative VFs identified from host-specific fuzzy clusters are presented in Supplementary Table S6. All Supplementary figures, tables, and datasets (i.e., Supplementary Tables S5 and S6) associated with this study have been deposited in figshare: https://doi.org/10.25387/g3.13206221. Open-source R code for statistical analysis is available on github.com/damselflywingz/In_vivo_infection_series_fuzzy_RNA-seq.
Supplementary material is available at figshare DOI: https://doi.org/10.25387/g3.13206221.
Results
Experimental infection and RNA sequencing
MH96 was enriched from hemolymph at cell densities of 4.4 × 107, 4.4 × 108, and 5.0 × 109 CFU g−1 (Table 1). In order to distinguish host-specific gene induction by MH96 from response to general growth conditions, in vitro samples were collected across three growth phases at approximately equivalent cell densities to the in vivo samples (Table 1).
Table 1.
MH96 cell density under in vivo and in vitro conditions used for DE analysis
| In vivo infection stage/in vitro growth | Mean cell density in vivo (i.e., G. mellonella homogenate) (CFU g−1 ± SEM) |
Mean cell density in vitro (i.e., Luria Broth Base broth) (CFU ml−1 ± SEM) |
|---|---|---|
|
Early infection/ lag-growth |
4.4 ± 0.5 × 107 | 3.2 ± 0.1 × 107 |
|
Middle infection/ exponential growth |
4.4 ± 1.7 × 108 | 2.9 ± 0.9 × 108 |
|
Late infection/ stationary growth |
5.0 ± 2.2 × 109 | 4.4 ± 0.1 × 109 |
CFU, colony forming units; SEM, standard error of the mean.
After quality trimming, adapter removal of host and rRNA sequences, the paired-end reads were aligned to the draft MH96 genome. Only an average of 9% of reads from early infection libraries could be aligned to the reference, which was lower by comparison to the average reads aligned from middle (58%) and late infection (59%), or in vitro libraries (84–93%) (Supplementary Figure S2 and Table S7). In order to determine if transcripts expressed in lower abundances were adequately captured within the early infection libraries, these libraries were analyzed by rarefaction, which predicted only a slight improvement in total transcript detection if sequencing depth were increased (Supplementary Figure S3). Based on this analysis, the in vivo early infection libraries were determined to have captured most MH96 transcripts produced across a dynamic range of conditions expected under these conditions and were considered saturated.
Preliminary data analysis explored the libraries for unwanted technical variation arising from batch effects and potential outliers using both RLE plots and PCA (Supplementary Figures S4 and S5), respectively. Variation in sequencing depth between libraries was found to have affected the distribution of RLE values (Supplementary Figure S4A), which was largely controlled for by upper-quartile normalization (Supplementary Figure S4B). Even after normalization, the variation in RLE values from the stationary growth in vitro libraries was greater when compared to all other libraries indicating residual unwanted technical variation arose from this treatment. A clear separation between in vitro and in vivo libraries was determined by PCA of normalized count data (Supplementary Figure S5). Libraries consistently clustered by treatment, except one in vivo late infection library, which clustered more closely to the middle infection in vivo libraries and was identified as a biological outlier and removed from subsequent analysis.
DE of MH96's transcriptome during early, middle, and late infection stages
Comparison of average transcript abundances across three stages of growth identified 2397 differentially expressed genes that were either upregulated or downregulated between in vivo and in vitro libraries (Supplementary Figure S6 and Table S5). MH96 upregulated 1285 genes during intrahemocoelic infection of G. mellonella; 829 genes responded to in vivo conditions during at least one stage of infection, 289 responded during two stages of infection and 167 transcripts responded throughout all three stages of infection. While the majority of MH96 transcripts significantly upregulated during in vivo conditions represented protein-coding genes, 72 non-coding and 19 anti-sense RNAs were also more highly expressed in vivo. MH96 also upregulated 1243 genes under in vitro conditions, of which 131 were also found to be upregulated under in vivo conditions at different cell densities.
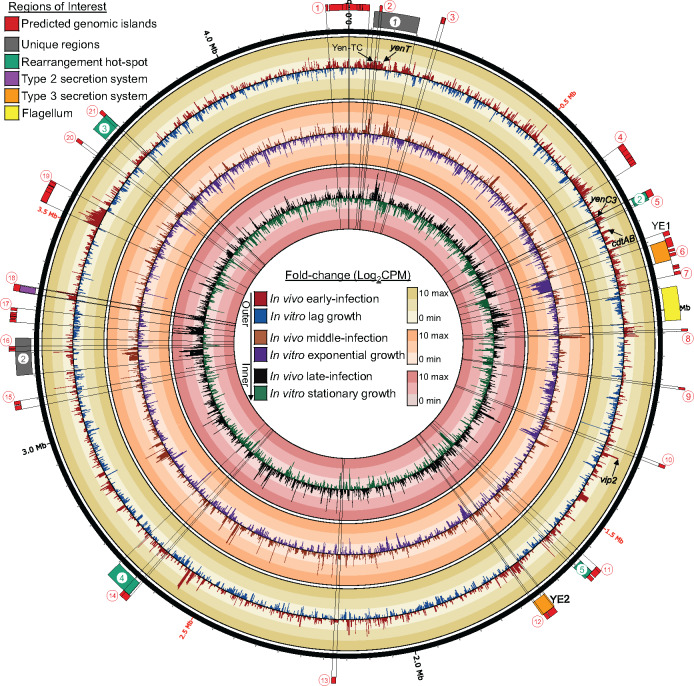
Regions of interest from the MH96 draft genome either known or suspected to encode VFs (Supplementary Table S8) were explored with respect to time-resolved genome-wide host-dependent transcriptional responses during intrahemocoelic infection (Figure 1). Several predicted GIs partially or fully overlapped with previously identified genomic regions of interest (i.e., unique region 1 overlaps Is.2, Rhs2 overlaps Is.5, T3SSYE1 overlaps Is.6, Rhs5 overlaps Is.11, T3SSYE2 overlaps Is.12, Rhs4 overlaps Is.14, unique region 2 overlaps Is.16, T2SS overlaps Is.18, and Rhs3/T6SS overlaps Is.21).
Figure 1.
Circos plot of Y. entomophaga MH96 genome-wide transcriptional response to in vivo and in vitro conditions during early, middle, and late intrahemocoelic infection and lag, exponential and stationary growth, respectively. CPM, counts-per-million. Mean fold-change in log counts-per-million (Log2CPM) (in vivo early and middle infection: n = 4, in vivo late infection: n = 3, and all in vitro: n = 2). Red circles numbered 1–21 represent predicted GIs identified using IslandViewer 4 webserver (refer to Supplementary Table S8). Regions of interest/loci previously identified from the draft genome are numbered or labeled as required (Hurst et al. 2016).
Wide-spread host-dependent upregulation of multigene clusters located within several regions of interest/predicted GIs, including unique regions 1 and 2, Rhs2/Is.5, T3SSYE2/Is.12, and T2SS/Is.18, was observed in MH96. Significantly higher expression for the entire T2SS island was measured during early and/or late infection stages compared to in vitro growth conditions. Significantly higher responses by genes for several putative T3SSYE2 structural components to in vivo growth conditions compared to in vitro during early and/or late infection were also identified. In contrast, most of the T3SSYE1 structural component genes were significantly upregulated by MH96 either in response to in vitro conditions or both in vitro and in vivo conditions depending on cell density, except putative T3SSYE1-associated chaperone (PL78_18170) that was more highly expressed in vivo during early intrahemocoelic infection only. Multiple genes located within a predicted phage-like islands Is.4, Is.8, and Is.19 were also significantly upregulated during early stage of intrahemocoelic infection, compared to growth in vitro.
Genes for other putative VFs from MH96 genomic regions of interest were more responsive during late stages of intrahemocoelic infection compared to other time points. Many of the iron acquisition gene clusters located within unique region 2 were found to have higher expression under in vivo conditions, especially during later stages of infection. Similarly, many flagellum genes were also more highly expressed mostly during the later infection stage. Furthermore, phage-related and hypothetical genes encoded within the predicted GI Is.17 were also found to have significant transcriptional response to in vivo conditions compared to growth in vitro during late infection stages as well.
An important region in the MH96 genome is Unique Region 1 (contains PAIYE96), where genes for hypothetical proteins yenU (PL78_03775) and yenW (PL78_03790), putative heat-stable enterotoxin yenT (PL78_03785), as well as several iron-siderophore biosynthesis/transport and serralysin genes were significantly upregulated in the host at various or throughout all stages of infection compared to in vitro treatments. Also located in Unique region 1, Yen-TC genes chi1 (PL78_03740), chi2 (PL78_03755), and yenC1 (PL78_03765) were found to have significantly higher expression during early intrahemocoelic infection only, while DE was not detected for other Yen-TC component genes.
Functional enrichment of in vivo host clusters
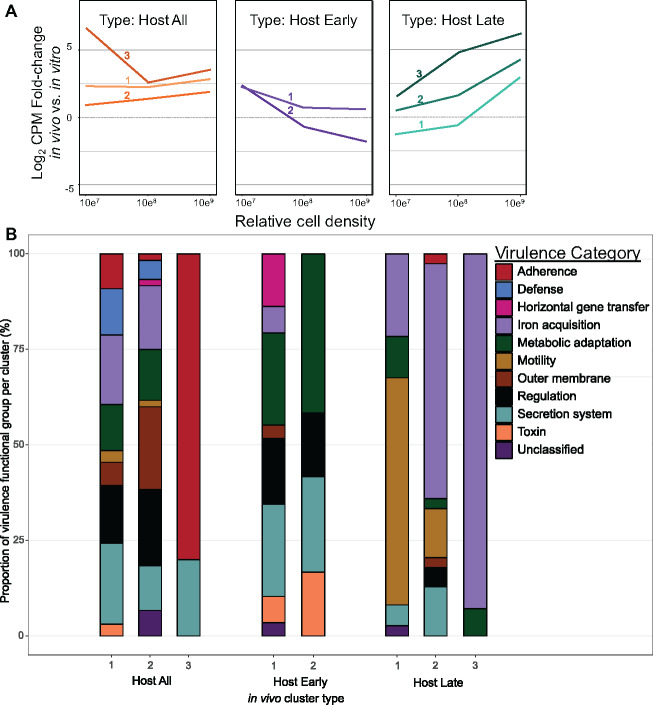
Qualitative assessment of fuzzy clustering results showed that 20 clusters with fuzzifier value of 1.5 were optimal for the analysis of fold-change log2CPM values comparing MH96 transcriptome across three different stages of intrahemocoelic infection to in vitro growth controls. Among these 20 clusters, 8 clusters (containing 798 transcripts) exhibited the most striking responses to in vivo conditions compared to in vitro growth (Supplementary Figure S7). These eight “in vivo clusters” were further organized into three main “in vivo cluster types” based on shared expression profiles across multiple in vivo clusters (Figure 2A). In vivo clusters containing transcripts with elevated abundances across all three stages of infection were identified as “Host All” type clusters, while “Host Early” and “Host Late” type clusters were characterized by containing mostly transcripts with elevated abundances during early or late stages of infection only, respectively.
Figure 2.
Yersinia entomophaga MH96 putative host-specific VFs sharing significant sequence similarity to known VFs from the VF database identified among host-specific in vivo clusters. (A) Median log2 counts-per-million-fold-change between in vivo and in vitro libraries organized by in vivo cluster type (each line represents a separate host-specific in vivo cluster, refer to Supplementary Figure S7). (B) The proportion of genes organized by virulence function classification among in vivo cluster types. Refer to Supplementary Table S6.
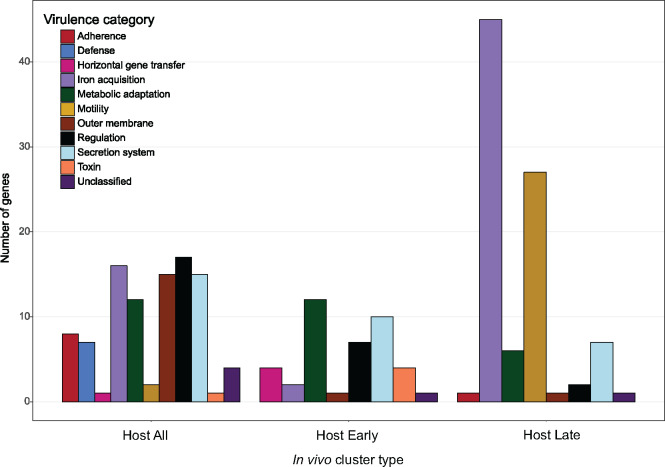
Figure 3.
Number of genes assigned to each virulence category for putative VFs identified from in vivo cluster types. In vivo clusters combined by type: Host All = constitutively higher in vivo expression during early, middle, and late infection; Host Early = higher in vivo during early infection only; Host Late = higher in vivo expression during late infection only. See Figure 2A for Host All, Host Early, and Host Late median expression profiles and Supplementary Table S6 for more details.
To further classify the role of putative MH96 VFs in pathogenesis, those transcripts from the eight in vivo clusters were functionally annotated using VFDB. Using this approach, 28.7% of the transcripts from in vivo clusters could be further categorized by potential role in pathogenicity (i.e., adherence, defense response, horizontal gene transfer, iron acquisition, metabolic adaptation, mobility, outer membrane, regulation, secretion system, and toxin/effector or unclassified) (Figures 2B and 3 and Supplementary Table S6). Transcripts related to iron acquisition, secretion systems, regulation, and metabolic adaptation were commonly identified among each of the three different in vivo cluster types but an obvious temporal shift in the expression of iron acquisition and flagellar motility vs toxins, effectors, and adhesions were identified when comparing early and late in vivo clusters.
Four genes encoding an usher-chaperone fimbrial cluster (PL78_12465, PL78_12470, PL78_12475, and PL78_12480) coexpressed with putative T3SS lipoprotein chaperone YscW-like (PL78_14485) were identified as some of the most host-responsive genes in the MH96 genome, especially at the early stage of intrahemocoelic infection (in vivo early infection vs in vitro growth fold-change values ranging from greater than 4.6 up to 7.1× Log2CPM). Genes for filamentous hemagglutinin (PL78_11060, previously annotated as a hemolysin) and its cognate two-partner secretion system (TPS) (PL78_11055) also had significantly higher expression during early and middle stages of intrahemocoelic infection. A predicted fimbrial gene (PL78_08275) and chitin-binding protein (PL78_08295) both associated with the Chi-Fim region (Hurst et al. 2016) were also captured from in vivo Host all type clusters but the relative expression levels of the Chi-Fim-associated fimbrial and chitin-binding protein genes were orders of magnitude lower than genes for other putative MH96 adhesions that were upregulated in vivo.
Transcripts for several putative toxins and effectors were identified among in vivo clusters (Table 2). The aforementioned heat-stable enterotoxin yenT was the only putative toxin gene highly expressed in vivo at two timepoints during infection and the transcript encoding putative AidA-like type V secretion system (T5SS) secreted adhesin clustered with other MH96 transcripts with higher in vivo gene expression throughout infection (Table 2).
Table 2.
Putative MH96 toxin and effector genes from host-specific gene expression clusters with significant sequence homology to other known toxins or effectors from VFDB
| Toxin/effector type | Locus tag | Designation | Host cluster type | Region of interest/GI/locusa |
|---|---|---|---|---|
| Toxin | PL78_03785 | yenT | Host all | Unique region 1-PAIYE96 |
| Toxin | PL78_18780 | yenC3 | Host early | Rhs-associated region 2/Is. 5 |
| Toxin | PL78_18440 | cdtA | Host early | cdtAB |
| Toxin | PL78_18445 | cdtB | Host early | cdtAB |
| Toxin | PL78_16145 | vip2 | Host early | vip2 |
| Effector—T3SS | PL78_14610 | sicC-like | Host all | T3SSYE2/Is.12 |
| Effector—T3SS | PL78_07345 | ipgD/sopB-like | Host all | — |
| Effector—T3SS | PL78_14615 | sicD-like | Host early | T3SSYE2/Is.12 |
| Effector—T3SS | PL78_18085 | ipaB/evcA-like | Host early | T3SSYE1/Is.6 |
| Effector—T3SS | PL78_14295 | sopD-like | Host early | — |
| Tube/Effector—T6SS | PL78_02751 | hpc | Host all | — |
| Tube/Effector—T6SS | PL78_00900 | hpc | Host early | Rhs-associated region 3 |
| Effector—T6SS | PL78_01040 | rhs-containing | Host early | Rhs-associated region 3/Is.21 |
| Tube/Effector—T6SS | PL78_03598 | hpc | Host late | Is.1 |
| Tube/Effector—T6SS | PL78_09211 | hpc | Host late | Is.17 |
| Effector—T5SS | PL78_10240 | aidA | Host all | — |
Regions of interest (see Figure 1): MH96 pathogenicity-associated island (PAIYE96), rearrangement-hotspot (Rhs), MH96 type 3 secretion systems (T3SSs) T3SSYE1 and T3SSYE2 (Hurst et al. 2016); predicted GIs see Figure 1 and Supplementary Table S8; loci: cyto-lethal distending toxin components (cdtA, cdtB), vegetative insecticidal protein 2 (vip2) (Hurst et al. 2016).
T3SS, type III secretion system; T5SS, type V secretion system; T6SS, type VI secretion system.
Several other transcripts for putative toxins were identified from Host Early in vivo clusters, including putative cytolethal distending toxin subcomponents cdtA and cdtB, yenC3 (a YenC-homolog also known as rhsA encoded within Rhs2/Is.5) and putative vegetative insecticidal protein component, vip2 (Table 2). Transcripts for known, and putative Yen-TC components, chi1 and yenC3, respectively, were captured within a Host Early in vivo cluster, but only yenC3, not chi1, shared any significant homology against known VFs from the VFDB using BLASTx homology search.
In addition to predicted toxins, transcripts for T2SS and T3SSYE2 structural components were also identified as putative VFs from in vivo clusters (Supplementary Table S6). Genes for T3SSYE2 co-located putative effectors were represented from in vivo clusters and found to have significantly greater expression under in vivo conditions compared to in vitro controls, including sicC-like and sicD-like, with the former being more highly expressed across all cell densities and the latter more highly expressed during early infection only. In addition to T3SSYE2-associated effector genes, two other genes for putative T3SS effectors were also found to be more highly expressed in vivo during early infection including T3SSYE1-associated ipaB/evcA-like and orphan sopD-like. Another gene for putative T3SS orphan effector IpgD/SopB-like represents a fifth possible T3SS effector gene, with significantly increased expression throughout all three stages of infection, compared to in vitro controls.
With respect to T6SS, genes for structural machinery components were not identified as having significantly greater expression in vivo compared to in vitro controls. MH96 genes for putative T6SS effectors were however identified from several of the in vivo cluster types, including genes for Rhs-containing protein and five different hcp-type structural tube/effector proteins. Also, the gene encoding the putative T6SS effector that shares sequence similarity to the eukaryotic transcription elongation factor domain Spt4 (PL78_00995) was found to have higher in vivo gene expression throughout infection but was not assigned a VFDB annotation based on amino acid sequence.
Not all MH96 predicted toxins and effectors were highly expressed under in vivo conditions though; genes for putative PirAB binary toxin (PL78_09590 and PL78_09595), RTX toxin (PL78_16910), and adenylate cyclase (PL78_08395) were found to have higher expression during exponential and/or stationary growth in vitro, with no significant response to in vivo conditions observed. Furthermore, despite having significantly greater abundance during early infection of G. mellonella compared to growth in vitro, transcripts for some other known or suspected MH96 VFs, including insecticidal TC components Chi2 and YenC1, and putative T3SS effector LopT (PL78_18760) were not found to cluster among the eight in vivo clusters, instead clustering with other transcripts associated with comparatively lower overall median in vivo vs in vitro fold-change values.
Discussion
MH96 in vivo transcriptome revealed wide-spread and coordinated pathogen response to different stages of intrahemocoelic infection
Our in vivo transcriptome analysis has shed light on MH96 genes activated during intrahemocoelic infection of G. mellonella. Major shifts in gene expression by MH96 were observed between in vivo and in vitro conditions across all three stages of growth. The mosaic nature of the MH96 genome was highlighted by widespread coexpression of genes located in horizontally acquired regions that positively responded to host, including genes for Yen-TC components, putative toxins (YenT, YenC3), translocated effectors, secretion system component (T2SS and T3SSYE2), iron acquisition systems, and phage-encoded hypotheticals. These findings provide evidence that complex genome-wide responses by MH96 occur throughout different stages of intrahemocoelic infection and reveal important insights into host-dependent transcriptional responses by entomopathogenic bacteria in nature.
Following disassociation of host midgut tissues (Marshall et al. 2012), MH96 gains access to the hemocoel where it encounters the cellular and humoral components of the insect host immune system (Nielsen-LeRoux et al. 2012). Within lepidopteran larvae, the hemolymph contains different hemocytes involved in phagocytosis, nodulation, and encapsulation as well as humoral factors like prophenoloxidase, antimicrobial peptides, reactive nitrogen, and oxidative species or lysozyme (Jiang et al. 2010; Casanova-Torres and Goodrich-Blair 2013; Wojda 2017). In order to occupy the insect hemocoel, MH96 produces VFs that combat the host immune defenses encountered during early intrahemocoelic infection, including anti-hemocytic factors such as toxins or translocated effectors. Upon incapacitation of the host immune system, MH96 produces additional VFs that function to mobilize and acquire essential nutrients, like iron, from insect cells to fuel pathogen metabolism and proliferation, ultimately leading to septicemia and host death (Nielsen-LeRoux et al. 2012).
MH96 upregulated toxins, secretion systems, effectors, and adhesions during intrahemocoelic infection of G. mellonella
During early stages of intrahemocoelic infection MH96 upregulates expression of Yen-TC components chi1, chi2, and yenC1 as well as putative Yen-TC component yenC3 and toxins cdtAB and vip2, which are known or suspected to be anti-hemocytic factors based on functional enrichment. CdtAB is a predicted genotoxin, which are known to interfere with normal hemocyte cell cycle (Lara-Tejero and Galán 2000; Heywood et al. 2005), while Vip2 and YenC3 may act on host cytoskeleton through (ADP)-ribosyltransferase activity targeting globular (G)-actin (Barth et al. 2004) or Rho GTPAses (Lang et al. 2010), respectively. While Yen-TC is established as the primary orally active virulence determinant of MH96 (Hurst et al. 2011b; Marshall et al. 2012), purified Yen-TC was also found to cause ruffling of G. mellonella hemocytes (Hurst et al. 2015) and nuclear and actin condensation and apoptosis of Spodoptera frugiperda insect cell line Sf9 (Marshall et al. 2012). Taken together, Yen-TC should be considered both an anti-hemocytic factor as well as an orally active insecticidal toxin.
Unlike putative toxin genes that were upregulated only during the initial intrahemocoelic infection, the MH96 heat-stable enterotoxin, yenT, was highly expressed during both the septicemic and pre-cadaveric stages of infection compared in vitro control treatments. Similar in size YenT shares N-terminal amino acid sequence homology to the heat-stable enterotoxins YstA and YstB (Hurst et al. 2011b), encoded by gastrointestinal-disease causing strains of Yersinia enterocolitica (Delor et al. 1990; Ramamurthy et al. 1997; Grant et al. 1998; Singh and Virdi 2004), Yersinia frederiksenii, Yersinia intermedia, and Yersinia kristensenii (Imori et al. 2017). Recently, the heat-stable enterotoxin, YacT, produced by Y. frederiksenii has been implicated in host immune modulation following intrahemocoelic injection of G. mellonella. The Escherichia coli recombinant YacT, altered the morphology and increased the aggregation of G. mellonella hemocytes (Springer et al. 2018). It is plausible that, like YacT of Y. frederiksenii, MH96 YenT also functions as an anti-hemocytic factor given the responsiveness of yenT to intrahemocoelic conditions at multiple stages of infection.
Among pathogenic Yersinia, T3SSs are known to target host cells through tight attachment and translocate effectors into the cytoplasm to disrupt cytoskeleton, trigger apoptosis, prevent phagocytosis, or interfere with immune pathways (Bliska et al. 2013; Plano and Schesser 2013; Pha and Navarro 2016). The Y. enterocolitica T3SS Ysa (encoded by ysa found in biotype 1B strains) (Foultier et al. 2002; Thomson et al. 2006) shares the same gene order with the MH96 T3SSYE2 locus (Hurst et al. 2016) of the predicted island Is.12, suggesting a common origin in both pathogens. In addition to interacting with mammalian cells (Matsumoto and Young 2006; Bent et al. 2013), Ysa is essential for replication within Drosophila melanogaster S2 cells (Walker et al. 2013) and secretes effectors at temperatures of <26°C in nutrient-rich broth with high salt concentrations (Venecia and Young 2005). Based on the similar gene order to ysa and the upregulation during intrahemocoelic infection, T3SSYE2 is likely another anti-hemocytic mechanism used by MH96 to overcome the insect immune system.
Also, similar to Ysa, which secretes effectors encoded by genes within distal regions of the Y. enterocolitca genome (Walker and Miller 2009), the MH96 in vivo transcriptome revealed evidence for coordinated host-dependent regulation of genes for T3SSYE2 structural components and several predicted effectors, some of which were not co-located within the T3SSYE2 region of the genome. For example, MH96 encodes a gene for a putative LopT-like effector, which is homologous to LopT, a T3SS effector (similar to YopT of Y. pestis) with known insecticidal activity in Photorhabdus luminescens (Brugirard-Ricaud et al. 2005). Expression of lopT was significantly upregulated by MH96 during initial intrahemocoelic infection. Notably, lopT and yenC3 are both co-located within an effector-island (i.e., Rhs-associated region 2/Is.5), and are possibly coopted by respective translocation machineries T3SSYE2 and Yen-TC and reflect the multiple diverse modes of action deployed by MH96 against the host during early stages of intrahemocoelic infection.
In contrast to T3SSYE2 and its predicted effectors that were upregulated under in vivo conditions, genes for T3SSYE1 of the predicted island Is.6 responded mainly to in vitro conditions. The differing transcriptional responses by the MH96 T3SSs depending on growth under in vivo and in vitro conditions allude to a specific role of each T3SS in MH96 pathogenicity. It was also noted that MH96 genes encoding the predicted PirAB binary toxin, RTX toxin, and adenylate cyclase had significantly higher expression in vitro compared to in vivo conditions. Clearly, some yet unknown environmental factor appears to control expression of T3SSYE1 and other insect-active genes. It is plausible that abiotic factors like pH and concentration of oxygen or nutrients occurring during in vitro growth could mimic signals that MH96 is likely to encounter in its natural environment, such as intracellular replication within G. mellonella hemocytes (Hurst et al. 2015; A. R. Paulson, personal communication), infection of insect midgut or different insect hosts (Hurst et al. 2011b, 2019) or even the occupation of the human urinary tract during asymptomatic catheter infection (Le Guern et al. 2018).
Along with T3SSs, another major class of well-studied VFs known from pathogenic Yersinia is adhesins, including T5SS-secreted adhesins and fimbriae (Chauhan et al. 2016). Putative adhesin genes upregulated by MH96 throughout or during specific stages of intrahemocoelic infection included T5SS-secreted AidA-like and filamentous hemagglutinin, and four-gene usher-chaperone fimbrial cluster, the latter representing some of the most responsive genes to in vivo conditions in the entire MH96 genome. In addition to host cell attachment or biofilm formation, in other pathogenic Yersinia species, adhesins have also been linked to effector translocation through enhanced attachment to host cells (Maldonado-arocho et al. 2013; Keller et al. 2015; Mühlenkamp et al. 2015; Deuschle et al. 2016). Perhaps adhesins also contribute to the effective anti-hemocytic activities of MH96 during intrahemocoelic infection of G. mellonella by enhancing effector translocation efficiency like other pathogenic Yersinia, yet such mechanisms remain unknown among entomopathogenic bacteria.
Transcriptional activation of numerous genes for known and putative VFs supports insecticidal activities of MH96 by “over-kill” strategy
The MH96 in vivo transcriptome revealed a high degree of functional redundancy, with multiple toxins, T3SS and T6SS effectors, and adhesins all likely to target hemocytes during early and/or throughout multiple stages of infection. Moreover, greater transcriptional activation of hypothetical genes encoded within several putative phage-related GIs suggests additional factors may further contribute to MH96 highly insecticidal activities. Such functional redundancy has also been observed in another highly entomopathogenic bacterium, P. luminescens (Wilkinson et al. 2009), that like MH96, produces many toxins, including insecticidal TC (ffrench-Constant et al. 2007), makes caterpillar floppy (Daborn et al. 2002), PirAB binary toxin (Waterfield et al. 2005; Wu and Yi 2016), and several RTX-like toxins (Tobias et al. 2017). Such abundance of toxins carried within the genomes of obligate entomopathogenic bacteria is thought to have arisen by adaptation for over-kill of a wide variety of hosts (ffrench-Constant et al. 2003). This is likely also true for MH96, which has broad insecticidal activities against diverse insect hosts (Hurst et al., 2011b, 2014, 2015, 2019). Also, MH96 is highly virulent against G. mellonella during intrahemocoelic infection but the LD50 of a Yen-TC deficient strain (ΔTC) was comparable to that of wild type (∼3 cells). It was also found that both wild type and ΔTC had the same ability to reduce the phenoloxidase activity of G. mellonella hemolymph 24 hpi, findings that suggest the presence of additional hemocytic VFs during intrahemocoelic infection by MH96 (Hurst et al. 2015). Transcriptional response by MH96 to in vivo conditions, including widespread and significant host-dependent upregulation of genes for Yen-TC components and numerous other predicted toxins, secretion systems, effectors, adhesins, and GI-encoded hypotheticals is another indicator that the highly insecticidal nature of MH96 stems from multiple diverse modes of action.
Beyond primary VFs that can act directly to modulate or destroy host cells, functional enrichment of the MH96 in vivo transcriptome also revealed changes in the temporal expression of genes for other putative in-host fitness factors (i.e., motility, metabolism, iron acquisition, response to host-induced stress, etc.) vs toxins, effectors, and adhesins from early to late infection. Notably, during the late stage of infection, MH96 significantly increased expression of genes related to flagellum and iron acquisition, compared to early and middle infection stages, which supports a role for iron acquisition and flagellum-related motility in MH96 in-host fitness during pre-cadaveric stages of insect infection.
Specific to entomopathogenic bacteria, both the flagellum and iron acquisition systems have already been implicated in intrahemocoelic infection of G. mellonella by triphasic nematode symbionts P. luminescens and closely related Xenorhabdus nematophila. Flagellum-driven motility provided a selective fitness advantage during colonization of G. mellonella by P. luminescens (Easom and Clarke 2008). Also, iron sensing by X. nematophila was shown to regulate the expression of flagellin gene fliC and hemolysin genes xaxAB, which significantly increased during late stage intrahemocoelic infection of G. mellonella and reached maximum levels following host death (Jubelin et al. 2011). Beyond entomopathogenic bacteria, iron homeostasis and hemoglobin utilization genes of C. albicans were found to be upregulated during later infection of G. mellonella and murine models compared to an earlier infection; however, in this study, the early to late increase in iron-related genes was much greater in G. mellonella compared to the mouse model (Amorim-Vaz et al. 2015). Taken together, pathogens infecting G. mellonella, whether bacterial or fungal, appear to shift into an iron scavenging mode during late stage (i.e., pre-cadaveric) infection, indicating that pathogen sensing and response to in vivo iron fluxes during insect infection are likely an important component of in-host fitness or represent an interspecific competitive strategy.
Acknowledgements
Research associate, Mitchell Weston, performed sample collection and RNA extraction for one of the in vitro treatments used in this experiment. We would like to acknowledge Trystan Whang and Elena Kim for coordinating library preparation, quality control, and sequencing at Macrogen Korea. Finally, AgResearch Ltd. bioinformaticians Dr. Aurelie Laugraud, Dr. Charles Hefer, and Paul Maclean helped with job execution on high-performance computing clusters.
Funding
This research aligned to and was supported by the Next Generation Biopesticides Programme, funded by the New Zealand Ministry for Business, Innovation and Employment (Contract C10X1310). Canadian National Science and Engineering Council for Postgraduate Scholarship-Doctoral Program and Universities New Zealand—Te Pōkai Tara via Massey University provided support in the form of student scholarships.
Conflicts of interest: None declared.
Literature cited
- Amorim-Vaz S, Tran VDT, Pradervand S, Pagni M, Coste AT, et al. 2015. RNA enrichment method for quantitative transcriptional analysis of pathogens in vivo applied to the fungus Candida albicans. MBio. 6:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprianto R, Slager J, Holsappel S, Veening JW.. 2016. Time-resolved dual RNA-seq reveals extensive rewiring of lung epithelial and pneumococcal transcriptomes during early infection. Genome Biol. 17:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas J, Bossers-de Vries R, Harders-Westerveen J, Buys H, Ruuls-van Stalle LMF, et al. 2019. In vivo transcriptomes of Streptococcus suis reveal genes required for niche-specific adaptation and pathogenesis. Virulence. 10:334–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avican K, Fahlgren A, Huss M, Heroven AK, Beckstette M, et al. 2015. Reprogramming of Yersinia from virulent to persistent mode revealed by complex in vivo RNA-seq analysis. PLoS Pathog. 11:e1004600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddal B, Muzzi A, Censini S, Calogero RA, Torricelli G, et al. 2016. Dual RNA-seq of nontypeable Haemophilus influenzae and host cell transcriptomes reveals novel insights into host-pathogen cross talk. MBio. 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H, Aktories K, Popoff MR, Stiles BG.. 2004. Binary bacterial toxins: biochemistry, biology and applications of common Clostridium and Bacillus Proteins. Microbiol Mol Biol Rev. 68:373–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D.. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 29:1165–1188. [Google Scholar]
- Bent ZW, Branda SS, Young GM.. 2013. The Yersinia enterocolitica Ysa type III secretion system is expressed during infections both in vitro and in vivo. Microbiologyopen. 2:962–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Simon Fraser University Research Computing Group, et al. 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45:W30–W35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska JB, Wang X, Viboud GI, Brodsky IE.. 2013. Modulation of innate immune responses by Yersinia type III secretion system translocators and effectors. Cell Microbiol. 15:1622–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugirard-Ricaud K, Duchaud E, Givaudan A, Girard PA, Kunst F, et al. 2005. Site-specific antiphagocytic function of the Photorhabdus luminescens type III secretion system during insect colonization. Cell Microbiol. 7:363–371. [DOI] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S.. 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby JN, Landsberg MJ, Simpson RM, Jones SA, Hankamer B, et al. 2012. Structural analysis of Chi1 chitinase from Yen-Tc: The multisubunit insecticidal ABC toxin complex of Yersinia entomophaga. J Mol Biol. 415:359–371. [DOI] [PubMed] [Google Scholar]
- Busby JN, Panjikar S, Landsberg MJ, Hurst MRH, Lott JS.. 2013. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature. 501:547–550. [DOI] [PubMed] [Google Scholar]
- Bushnell B. 2015. BBMap short-read aligner, and other bioinformatics tools. http://sourceforge.net/projects/bbmap/ (Accessed September 2018).
- Casanova-Torres ÁM, Goodrich-Blair H.. 2013. Immune signaling and antimicrobial peptide expression in Lepidoptera. Insects. 4:320–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion OL, Wagley S, Titball RW.. 2016. Galleria mellonella as a model host for microbiological and toxin research. Virulence. 7:840–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N, Wrobel A, Skurnik M, Leo JC.. 2016. Yersinia adhesins: an arsenal for infection. Prot Clin Appl. 10:949–963. [DOI] [PubMed] [Google Scholar]
- Chen A, Wang Y, Shao Y, Zhou Q, Chen S, et al. 2018. Genes involved in Beauveria bassiana infection to Galleria mellonella. Arch Microbiol. 200:541–552. [DOI] [PubMed] [Google Scholar]
- Chen L, Zheng D, Liu B, Yang J, Jin Q.. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 44:D694–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Kontnik R, Clardy J.. 2010. Regulating alternative lifestyles in entomopathogenic bacteria. Curr Biol. 20:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn PJ, Waterfield N, Silva CP, Au CPY, Sharma S, et al. 2002. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc Natl Acad Sci U S A. 99:10742–10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Oglesby-Sherrouse AG, Wilks A, Barbier M.. 2016. Dual-seq transcriptomics reveals the battle for iron during Pseudomonas aeruginosa acute murine pneumonia. Sci. Rep. 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delor I, Kaeckenbeeck A, Wauters G, Cornelis GR.. 1990. Nucleotide sequence of yst, the Yersinia enterocolitica gene encoding the heat-stable enterotoxin, and prevalence of the gene among pathogenic and nonpathogenic Yersiniae. Infect Immun. 58:2983–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng ZL, Sztajer H, Jarek M, Bhuju S, Wagner-Döbler I.. 2018. Worlds apart—transcriptome profiles of key oral microbes in the periodontal pocket compared to single laboratory culture reflect synergistic interactions. Front Microbiol. 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois AP, McMillan S.. 2015. Paving the way to acceptance of Galleria mellonella as a new model insect. Virulence. 6:410–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle E, Keller B, Siegfried A, Manncke B, Spaeth T, et al. 2016. Role of β1 integrins and bacterial adhesins for Yop injection into leukocytes in Yersinia enterocolitica systemic mouse infection. Int J Med Microbiol. 306:77–88. [DOI] [PubMed] [Google Scholar]
- Dodt M, Roehr J, Ahmed R, Dieterich C.. 2012. FLEXBAR—flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel). 1:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easom CA, Clarke DJ.. 2008. Motility is required for the competitive fitness of entomopathogenic Photorhabdus luminescens during insect infection. BMC Microbiol. 8:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ma L, Zhao C, Yan J, Che S, et al. 2020. Transcriptome of Pectobacterium carotovorum subsp. carotovorum PccS1 infected in calla plants in vivo highlights a spatiotemporal expression pattern of genes related to virulence, adaptation, and host response. Mol Plant Pathol. 21:871–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC, Frawley ER, Tapscott T, Vázquez-Torres A.. 2016. Bacterial stress responses during host infection. Cell Host Microbe. 20:133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson CM, Barratt BIP, Bell N, Goldson SL, Jackson M, et al. 2019. Quantifying the economic cost of invertebrate pests to New Zealand’s pastoral industry. New Zeal. J Agric Res. 62:255–315. [Google Scholar]
- ffrench-Constant RH, Dowling A, Waterfield NR.. 2007. Insecticidal toxins from Photorhabdus bacteria and their potential use in agriculture. Toxicon. 49:436–451. [DOI] [PubMed] [Google Scholar]
- ffrench-Constant R, Waterfield N, Daborn P, Susan J, Bennett H, et al. 2003. Photorhabdus: towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol Rev. 26:433–456. [DOI] [PubMed] [Google Scholar]
- Foultier B, Troisfontaines P, Müller S, Opperdoes FR, Cornelis GR.. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J Mol Evol. 55:37–51. [DOI] [PubMed] [Google Scholar]
- Futschik ME, Carlisle B.. 2005. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 3:965–988. [DOI] [PubMed] [Google Scholar]
- Grant T, Bennett-Wood V, Robins-Browne RM.. 1998. Identification of virulence-associated characteristics in clinical isolates of Yersinia enterocolitica lacking classical virulence markers. Infect Immun. 66:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesenauer B, Tran TM, Fortney KR, Janowicz DM, Johnson P, et al. 2019. Determination of an interaction network between an extracellular bacterial pathogen and the human host. MBio. 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Chin M, Nusbaum C, Birren BW, Livny J.. 2012. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics. 13:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haueisen J, Möller M, Eschenbrenner CJ, Grandaubert J, Seybold H, et al. 2019. Highly flexible infection programs in a specialized wheat pathogen. Ecol Evol. 9:275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroven AK, Dersch P.. 2014. Coregulation of host-adapted metabolism and virulence by pathogenic yersiniae. Front Cell Infect Microbiol. 4:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heywood W, Henderson B, Nair SP.. 2005. Cytolethal distending toxin: creating a gap in the cell cycle. J Med Microbiol. 54:207–216. [DOI] [PubMed] [Google Scholar]
- Hsieh TC, Ma KH, Chao A.. 2016. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol. 7:1451–1456. [Google Scholar]
- Hurst MRH, Beattie A, Altermann E, Moraga RM, Harper LA, et al. 2016. The draft genome sequence of the Yersinia entomophaga entomopathogenic type strain MH96T. Toxins (Basel). 8:143–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst MRH, Beattie AK, Jones SA, Hsu P-C, Calder J, et al. 2015. Temperature-dependent Galleria mellonella mortality as a result of Yersinia entomophaga infection. Appl Environ Microbiol. 81:6404–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst MRH, Becher SA, Young SD, Nelson TL, Glare TR.. 2011a. Yersinia entomophaga sp. nov., isolated from the New Zealand grass grub Costelytra zealandica. Int J Syst Evol Microbiol. 61:844–849. [DOI] [PubMed] [Google Scholar]
- Hurst MRH, Jones SA, Beattie A, van Koten C, Shelton AM, et al. 2019. Assessment of Yersinia entomophaga as a control agent of the diamondback moth Plutella xylostella. J Invertebr Pathol. 162:19–25. [DOI] [PubMed] [Google Scholar]
- Hurst MRH, Jones SA, Binglin T, Harper LA, Jackson TA, et al. 2011b. The main virulence determinant of Yersinia entomophaga MH96 is a broad-host-range toxin complex active against insects. J Bacteriol. 193:1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst MRH, van Koten C, Jackson TA.. 2014. Pathology of Yersinia entomophaga MH96 towards Costelytra zealandica (Coleoptera; Scarabaeidae) larvae. J Invertebr Pathol. 115:102–107. [DOI] [PubMed] [Google Scholar]
- Ibberson CB, Whiteley M.. 2019. The Staphylococcus aureus transcriptome during cystic fibrosis lung infection. MBio. 10:e02774–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imori PFM, Passaglia J, Souza RA, Rocha LB, Falcão JP.. 2017. Virulence-related genes, adhesion and invasion of some Yersinia enterocolitica-like strains suggests its pathogenic potential. Microb Pathog. 104:72–77. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR.. 2010. Immunity in lepidopteran insects. In: Söderhäll K, editor. Invertebrate Immunity. Advances in experimental medicine and biology. New york, NY: Landes Bioscience and Springer Science+Business Media. p. 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelin G, Pagès S, Lanois A, Boyer M, Gaudriault S, et al. 2011. Studies of the dynamic expression of the Xenorhabdus FliAZ regulon reveal atypical iron-dependent regulation of the flagellin and haemolysin genes during. Environ Microbiol. 13:1271–1284. [DOI] [PubMed] [Google Scholar]
- Keller B, Mühlenkamp M, Deuschle E, Siegfried A, Mössner S, et al. 2015. Yersinia enterocolitica exploits different pathways to accomplish adhesion and toxin injection into host cells. Cell Microbiol. 17:1179–1204. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Futschik M.. 2007. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2:5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg MJ, Jones SA, Rothnagel R, Busby JN, Marshall SDG, et al. 2011. 3D structure of the Yersinia entomophaga toxin complex and implications for insecticidal activity. Proc Natl Acad Sci U S A. 108:20544–20549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AE, Schmidt G, Schlosser A, Hey TD, Larrinua IM, et al. 2010. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science. 327:1139–1143. [DOI] [PubMed] [Google Scholar]
- Lara-Tejero M, Galán JE.. 2000. Cell cycle progression as a deoxyribonuclease I–like protein. Science. 290:354–357. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK.. 2014. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guern AS, Savin C, Brémont S, Payro G, Bon D, et al. 2018. First isolation of Yersinia entomophaga in human urinary tract. New Microbes New Infect. 26:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lin L, Chong L, Gu S, Wen S, et al. 2020. Time-resolved mRNA and miRNA expression profiling reveals crucial coregulation of molecular pathways involved in epithelial—pneumococcal interactions. Immunol Cell Biol. 98:726–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Sun Y, Huang L, Su Y, Zhao L, et al. 2020. Time-resolved dual RNA-seq of tissue uncovers Pseudomonas plecoglossicida key virulence genes in host-pathogen interaction with Epinephelus coioides. Environ Microbiol. 22:677–693. [DOI] [PubMed] [Google Scholar]
- Maldonado-Arocho FJ, Green C, Fisher ML, Paczosa MK, Mecsas J.. 2013. Adhesins and host serum factors drive yop translocation by Yersinia into professional phagocytes during animal infection. PLoS Pathog. 9:e1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall SDG, Hares MC, Jones SA, Harper LA, Vernon JR, et al. 2012. Histopathological effects of the Yen-Tc toxin complex from Yersinia entomophaga MH96 (Enterobacteriaceae) on the Costelytra zealandica (Coleoptera: Scarabaeidae) larval midgut. Appl Environ Microbiol. 78:4835–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Young GM.. 2006. Proteomic and functional analysis of the suite of Ysp proteins exported by the Ysa type III secretion system of Yersinia enterocolitica Biovar 1B. Mol Microbiol. 59:689–706. [DOI] [PubMed] [Google Scholar]
- Mühlenkamp M, Oberhettinger P, Leo JC, Linke D, Schütz MS.. 2015. Yersinia adhesin A (YadA)—beauty & beast. Int J Med Microbiol. 305:252–258. [DOI] [PubMed] [Google Scholar]
- Nielsen-LeRoux C, Gaudriault S, Ramarao N, Lereclus D, Givaudan A.. 2012. How the insect pathogen bacteria Bacillus thuringiensis and Xenorhabdus/Photorhabdus occupy their hosts. Curr Opin Microbiol. 15:220–231. [DOI] [PubMed] [Google Scholar]
- Nobori T, Velásquez AC, Wu J, Kvitko BH, Kremer JM, et al. 2018. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc Natl Acad Sci U S A. 115:E3055–E3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss AM, Beckstette M, Pimenova M, Schmühl C, Opitz W, et al. 2017. Tissue dual RNA-seq allows fast discovery of infection-specific functions and riboregulators shaping host–pathogen transcriptomes. Proc Natl Acad Sci U S A. 114:E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pha K, Navarro L.. 2016. Yersinia type III effectors perturb host innate immune responses. World J Biol Chem. 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper SJ, Brillault L, Rothnagel R, Croll TI, Box JK, et al. 2019. Cryo-EM structures of the pore-forming A subunit from the Yersinia entomophaga ABC toxin. Nat Commun. 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plano GV, Schesser K.. 2013. The Yersinia pestis type III secretion system: expression, assembly and role in the evasion of host defenses. Immunol Res. 57:237–245. [DOI] [PubMed] [Google Scholar]
- Ramamurthy T, Yoshino KI, Huang X, Balakrish Nair G, Carniel E, et al. 1997. The novel heat-stable enterotoxin subtype gene (ystB) of Yersinia enterocolitica: nucleotide sequence and distribution of the yst genes. Microb Pathog. 23:189–200. [DOI] [PubMed] [Google Scholar]
- Reniere ML. 2018. Reduce, induce, thrive: bacterial redox sensing during pathogenesis. J Bacteriol. 200:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, Dudoit S.. 2014. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 32:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, et al. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I, Virdi JS.. 2004. Production of Yersinia stable toxin (YST) and distribution of yst genes in biotype 1A strains of Yersinia enterocolitica. J Med Microbiol. 53:1065–1068. [DOI] [PubMed] [Google Scholar]
- Singkum P, Suwanmanee S, Pumeesat P, Luplertlop N.. 2019. A powerful in vivo alternative model in scientific research: Galleria mellonella. Acta Microbiol Immunol Hung. 66:31–55. [DOI] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 3:1–25. [DOI] [PubMed] [Google Scholar]
- Springer K, Sänger P-A, Moritz C, Felsl A, Rattei T, et al. 2018. Insecticidal toxicity of Yersinia frederiksenii involves the novel enterotoxin YacT. Front Cell Infect Microbiol. 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhuang Z, Wang X, Huang H, Fu Q, et al. 2019. Dual RNA-seq reveals the effect of the flgM gene of Pseudomonas plecoglossicida on the immune response of Epinephelus coioides. Fish Shellfish Immunol. 87:515–523. [DOI] [PubMed] [Google Scholar]
- Tang Y, Xin G, Zhao L, Huang L, Qin Y, et al. 2020. Novel insights into host-pathogen interactions of large yellow croakers (Larimichthys crocea) and pathogenic bacterium Pseudomonas plecoglossicida using time- resolved dual RNA-seq of infected spleens. Zool Res. 41:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Zhao L, Xu X, Huang L, Qin Y, et al. 2019. Dual RNA-Seq uncovers the function of an ABC transporter gene in the host-pathogen interaction between Epinephelus coioides and Pseudomonas plecoglossicida. Fish Shellfish Immunol. 92:45–53. [DOI] [PubMed] [Google Scholar]
- Thomson NR, Howard S, Wren BW, Holden MTG, Crossman L, et al. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias NJ, Heinrich AK, Eresmann H, Wright PR, Neubacher N, et al. 2017. Photorhabdus-nematode symbiosis is dependent on hfq-mediated regulation of secondary metabolites. Environ Microbiol. 19:119–129. [DOI] [PubMed] [Google Scholar]
- Tran SL, Guillemet E, Lereclus D, Ramarao N.. 2013. Iron regulates Bacillus thuringiensis haemolysin hlyII gene expression during insect infection. J Invertebr Pathol. 113:205–208. [DOI] [PubMed] [Google Scholar]
- Tsai CJY, Loh JMS, Proft T.. 2016. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 7:214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Miranda D, Gallardo-Escárate C.. 2018. Dual RNA-Seq uncovers metabolic amino acids dependency of the intracellular bacterium Piscirickettsia salmonis infecting Atlantic salmon. Front Microbiol. 9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venecia K, Young GM.. 2005. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect Immun. 73:5961–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H, Altincicek B, Glöckner G, Vilcinskas A.. 2011. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics. 12:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KA, Maltez VI, Hall JD, Vitko NP, Miller VL.. 2013. A phenotype at last: essential role for the Yersinia enterocolitica Ysa: type III secretion system in a Drosophila melanogaster S2 cell model. Infect Immun. 81:2478–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KA, Miller VL.. 2009. Synchronous gene expression of the Yersinia enterocolitica Ysa type III secretion system and its effectors. J Bacteriol. 191:1816–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield N, Kamita SG, Hammock BD, Ffrench-Constant R.. 2005. The Photorhabdus Pir toxins are similar to a developmentally regulated insect protein but show no juvenile hormone esterase activity. FEMS Microbiol Lett. 245:47–52. [DOI] [PubMed] [Google Scholar]
- Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, et al. 2016. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature. 529:496–501. [DOI] [PubMed] [Google Scholar]
- Westermann AJ, Venturini E, Sellin ME, Förstner KU, Hardt WD, et al. 2019. The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica serovar Typhimurium. MBio. 10:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson P, Waterfield NR, Crossman L, Corton C, Sanchez-Contreras M, et al. 2009. Comparative genomics of the emerging human pathogen Photorhabdus asymbiotica with the insect pathogen Photorhabdus luminescens. BMC Genomics. 10:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojda I. 2017. Immunity of the greater wax moth Galleria mellonella. Insect Sci. 24:342–357. [DOI] [PubMed] [Google Scholar]
- Wu G, Yi Y.. 2016. Haemocoel injection of PirA1B1 to Galleria mellonella larvae leads to disruption of the haemocyte immune functions. Nat Publ Gr. 6:34996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Su S, Meng X, Ji X, Qu Y, et al. 2013. Determination of sRNA expressions by RNA-seq in Yersinia pestis grown in vitro and during infection. PLoS One. 8:e74495–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data and both raw and normalized count tables were deposited in the GenBank Geo Submission Omnibus under accession: GSE142509. All genes with significantly DE detected between in vivo vs in vitro time-resolved contrasts (early infection vs lag growth in vitro; middle infection vs exponential growth in vitro; late infection vs stationary growth in vitro) are provided in Supplementary Table S5. All functionally annotated putative VFs identified from host-specific fuzzy clusters are presented in Supplementary Table S6. All Supplementary figures, tables, and datasets (i.e., Supplementary Tables S5 and S6) associated with this study have been deposited in figshare: https://doi.org/10.25387/g3.13206221. Open-source R code for statistical analysis is available on github.com/damselflywingz/In_vivo_infection_series_fuzzy_RNA-seq.
Supplementary material is available at figshare DOI: https://doi.org/10.25387/g3.13206221.