Abstract
The factors that drive the rapid changes in abundance of tandem arrays of highly repetitive sequences, known as satellite DNA, are not well understood. Drosophila virilis has one of the highest relative amounts of simple satellites of any organism that has been studied, with an estimated >40% of its genome composed of a few related 7-bp satellites. Here, we use D. virilis as a model to understand technical biases affecting satellite sequencing and the evolutionary processes that drive satellite composition. By analyzing sequencing data from Illumina, PacBio, and Nanopore platforms, we identify platform-specific biases and suggest best practices for accurate characterization of satellites by sequencing. We use comparative genomics and cytogenetics to demonstrate that the highly abundant AAACTAC satellite family arose from a related satellite in the branch leading to the virilis phylad 4.5–11 Ma before exploding in abundance in some species of the clade. The most abundant satellite is conserved in sequence and location in the pericentromeric region but has diverged widely in abundance among species, whereas the satellites nearest the centromere are rapidly turning over in sequence composition. By analyzing multiple strains of D. virilis, we saw that the abundances of two centromere-proximal satellites are anticorrelated along a geographical gradient, which we suggest could be caused by ongoing conflicts at the centromere. In conclusion, we illuminate several key attributes of satellite evolutionary dynamics that we hypothesize to be driven by processes including selection, meiotic drive, and constraints on satellite sequence and abundance.
Keywords: centromeres, comparative genomics, repetitive DNA, long-read sequencing
Introduction
Repetitive DNA is abundant in most eukaryotic genomes and is now understood to be correlated with the manifold variation in genome size across the tree of life (Elliott and Gregory 2015). For most species, transposable elements (TEs) dominate the repeat landscape, including in humans, plants, and Drosophila melanogaster. Satellite DNA, which is characterized by tandem repeats spanning long arrays, very rarely has dominated a genome to a similar extent as TEs. An unprecedented case is that of Drosophila virilis, the Drosophila species with the largest estimated genome size (up to 389 Mb) (Bosco et al. 2007), where some 40% of the genome (estimated in the pericentromeric region) comprised just three simple 7-mer satellites: AAACTAC, AAACTAT, and AAATTAC (Gall et al. 1971; Gall and Atherton 1974). Since the 1970s, there has been no follow-up to validate the amount of 7-mers with modern techniques, or evolutionary studies to understand how and why these satellite repeats expanded so explosively. The genomic composition of simple satellites in D. virilis provides an excellent model for an investigation of the evolutionary dynamics involved in their expansion in the genome as well as the technical challenges facing simple satellite analysis.
Satellites are rapidly evolving in sequence and copy number, and there is a high level of variation in satellite content among and within species (Wei et al. 2014, 2018). The reasons for such dramatic variation are not well understood, and cannot be fully explained by current models. The consequences of having varying satellite composition near the centromere are also unclear. To date, we have very little knowledge about how pericentromeric satellites evolve on a population-wide and species-wide scale.
Satellites have been long hypothesized to be slightly deleterious and therefore governed primarily by the strength of negative selection (Ohno 1972). However, the abundance of satellite repeats in the genome that would incur a fitness cost depends on many factors and cannot be easily predicted (Charlesworth et al. 1994; Gregory 2001). The fact that most organisms have satellite repeats in or near centromeres suggests that they are important for centromere function. Satellite repeats can also be important for maintenance of the chromocenter and packaging of chromosomes in the nucleus (Jagannathan et al. 2018, 2019), and the transcripts of some satellites may be essential for fertility (Mills et al. 2019). In heterozygotes with alleles that differ in pericentromeric satellite sequence or abundance, one allele may assemble a stronger kinetochore during female meiosis I, increasing its probability of transmission into the egg (rather than polar bodies). This transmission advantage, known as centromere drive, allows satellites to rapidly change in composition in the population, regardless of their whole-organism fitness effects (Henikoff et al. 2001). If satellite DNA is an essential component of the genome or is only selfish, it is still not clear why some species have almost no pericentromeric satellite DNA while others, like D. virilis, possess pericentromeric satellites that make up almost half of the genome.
Comparing the satellites of D. virilis to those of its sister species can elucidate when the abundant satellites arose, and how rapidly their copy numbers and sequences evolved. Drosophila virilis is 4.5 My diverged from its sister species D. novamexicana and D. americana, which are both restricted to North America, unlike globally distributed D. virilis (Caletka and McAllister 2004). Drosophila novamexicana and D. americana have a smaller estimated genome size than D. virilis (∼250 vs. 389 Mb), suggesting these species may have less satellite content (Bosco et al. 2007). Additionally, using intraspecies comparisons across global populations can give indications about factors that may be influencing satellite dynamics. For example, in D. melanogaster, patterns of abundance of the Prodsat satellite closely mirror the migration patterns of species, suggesting an ongoing expansion of this satellite (Wei et al. 2014). Genetic drift or meiotic drive may contribute to patterns of geographical gradients of satellite abundance. We can also use intraspecies data to pose hypotheses about nonneutral processes that may be driving satellite content. Previous work has shown evidence for conflicts or tradeoffs between satellites within the genome, and these constraints can be illuminated by analyzing satellites in several strains (Flynn et al. 2017, 2018).
Genome-wide characterization of satellites has taken off since high-throughput sequencing has become widely available. We have learned from several informative studies about the sequences and relative abundances of satellites in various species (Pavlek et al. 2015; de Lima et al. 2017; Flynn et al. 2017; Wei et al. 2018), but technical challenges may prevent accurate quantitative estimates. Satellites may be more prone to errors or biases in the sequencing process that do not affect the better studied regions of the genome. Satellites are difficult to assemble even with long-read sequencing (Chang and Larracuente 2019). The genome assembly of D. virilis is approximately half its estimated genome size by flow cytometry (∼200 vs. 389 Mb; Bosco et al. 2007), and it is likely that much of what is missing is simple satellite DNA. However, even assembly free raw read quantification methods have not produced satellite DNA estimates that approach the amount that was estimated from early work (Gall et al. 1971; Gall and Atherton 1974; Wei et al. 2018). Now, as long-read sequencing is also being exploited to study satellites, we must evaluate satellite DNA abundance estimates to assess if there are platform-specific biases that may affect evolutionary analysis of satellite DNA.
The purpose of this article is 2-fold; first to explore the technical biases that pose a challenge to accurate characterization and quantification of simple satellites, and second to use a comparative approach to understand the evolutionary dynamics of the extremely abundant 7-mers in the D. virilis group. First, we characterize satellites in D. virilis sequencing data from different platforms and assess biases that affect accurate satellite characterization. We then use comparative genomics and cytogenetics in D. virilis and its sister species to understand the composition and changes in the highly abundant simple satellites. Finally, we sequence multiple strains of D. virilis and sister species to estimate polymorphism in satellite abundance and infer processes that may be influencing their evolution. From this, we infer that there are likely multiple processes affecting satellite DNA in this organism, including positive selection, meiotic drive, and constraints and tradeoffs between satellites.
Results
Technical Biases in Characterizing Simple Satellites from Sequencing
Long-Read Genome Assemblies Have an Underrepresentation of Simple Satellites
Long-read sequencing technologies have high error rates prompting a need for extensive alignments for error-correction and assembly, which may result in collapsing satellite arrays. First we asked whether assemblies from long-read technologies can better assemble simple satellite reads than the previous Sanger assembly. We compared the amount of simple 7-mer satellites (AAACTAC, AAACTAT, AAATTAC, and AAACAAC) in three D. virilis genome assemblies: the CAF1 assembly produced from Sanger sequencing (Drosophila 12 Genomes Consortium et al. 2007), a PacBio assembly produced by our group by ∼100× coverage (available at https://www.ncbi.nlm.nih.gov/bioproject/? term=txid7214), and a Nanopore assembly produced from ∼20× sequencing coverage (Miller et al. 2018). All assemblies were approximately the same size at ∼200 Mb and were produced from the same strain (supplementary table S1, Supplementary Material online). The PacBio and Nanopore assemblies contained a similarly low amount of simple 7-mer satellites, 29 and 28 kb, respectively. The CAF1 assembly however contained 7.36 Mb of these satellites. This discrepancy is likely largely due to the difference in assembly algorithms used for short- and long-read data. Long reads must be thoroughly clustered and aligned to be incorporated into the assembly, whereas Sanger satellite reads are included as unplaced scaffolds. Use of modified methods can improve assemblies of repetitive regions (Chang and Larracuente 2019), but for highly homogeneous simple satellites, whose arrays span 10–100× longer than the current maximum read length, it is practically impossible to produce a continuous assembly. Additionally, part of the discrepancy between the Sanger CAF1 assembly may be due to the tissue used. The CAF1 assembly was produced from DNA extracted from homogeneous diploid embryos, whereas the long-read assemblies were produced from fly carcasses, which contain polytene cells (supplementary table S1, Supplementary Material online). We discuss issues with polyteny further below.
Simulations to Assess Simple Repeat Quantification from Long-Read Sequencing Data
Due to assembly issues of simple satellites, they must be quantified from raw unassembled reads. Long-read sequencing data poses a significant challenge because of the high error rate including a high indel rate in the raw reads. We therefore used two different approaches along with simulations to assess their accuracy. The first approach used k-Seek (Wei et al. 2014) to select repeat-rich reads and then Phobos (https://www.ruhr-uni-bochum.de/ecoevo/cm/cm_phobos.htm, last accessed February 5, 2019) to quantify satellites. This approach discovers and quantifies satellites de novo (details in Materials and Methods section). We used Noise-Cancelling Repeat Finder (NCRF, Harris et al. 2019) for our second approach, which can quantify previously defined satellites. Phobos was designed to have high sensitivity in detecting tandem repeats with sequence errors in them, but was not specifically designed for long-read data as NCRF was.
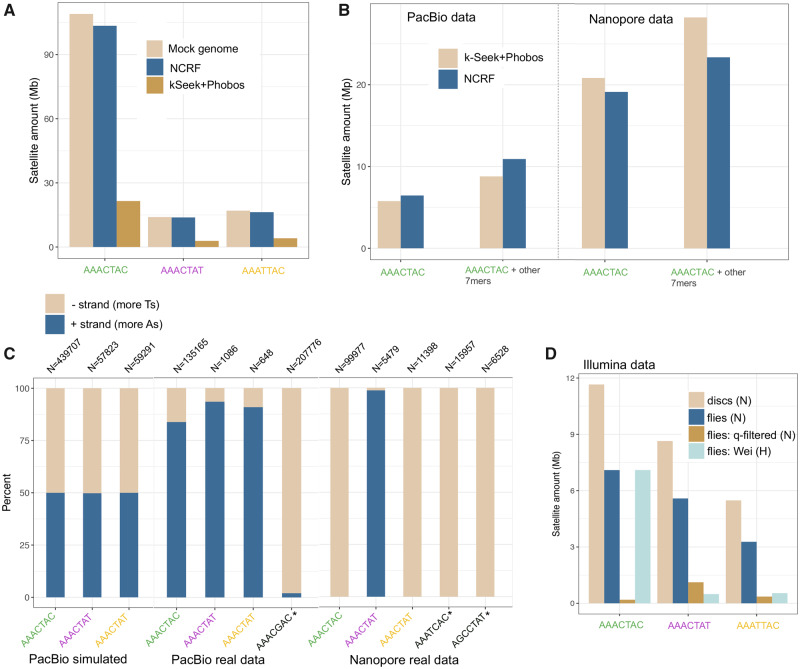
To evaluate our approaches, we created a mock D. virilis-like genome containing the estimated amounts of pericentromeric and centromeric satellites on each of five chromosomes. Satellite DNA composed 40% of this 351 Mb mock genome, and we used our cytogenetic data (discussed below) to guide the relative organization of the satellites (see Materials and Methods section). We then simulated 10× coverage PacBio reads from the mock genome, incorporating errors with PBSIM (Ono et al. 2013). We quantified satellites from the simulated long reads using both approaches. NCRF found almost the same amount of satellites that truly existed in the mock genome whereas the k-Seek + Phobos method only found about 20% (fig. 1A).
Fig. 1.
Issues in quantifying simple satellites in sequencing data (all data shown are Drosophila virilis). (A) Cumulative stacked barplot comparing the performance of the two tested approaches on PacBio data simulated with PBSim from a mock genome. (B) Comparing the results of the two approaches on the PacBio and Nanopore data; “other” refers to additional satellites in the family, including suspected artifactual ones (AAAGCAC for PacBio and AAATCAC + AGCCTAT for Nanopore). (C) Strand biases in the sequenced satellites in long-read sequencing data. Satellites with asterisks are suspected artifactual ones. N refers to the number of read fragments used for the calculation. (D) Amount of satellites quantified in Illumina data sets: imaginal discs (pure diploid), compared with flies (some polyteny), fly data that has been quality filtered (this study), and fly data from a previous study by Wei et al. (2018). N indicates NextSeq platform and H indicates HiSeq platform.
The Amounts and Biases in Simple 7-mer Repeats Differ between Nanopore and PacBio Sequencing Reads
Next, we quantified simple satellites in the long-read data generated from our PacBio sequencing and the publicly available Nanopore sequencing using the two approaches mentioned above. Both data sets were produced from fly carcasses (male in the case of the PacBio data and female in the case of the Nanopore data). Unlike in the simulations, both approaches produced very similar (but lower than expected) estimates at 8.8–10.9 Mb for the PacBio data (fig. 1B). This observation suggests an interaction between algorithmic efficiency and satellite sequencing efficiency such that the algorithm is the limiting factor for the simulated reads, but not so for the real reads where the satellites are much less abundant. The Nanopore data contained almost three times as much 7-mer satellites compared with PacBio, with 23.4–28.2 Mb (fig. 1C). This cannot be accounted for by male–female differences, which differ in satellite content only by 0.7% (Wei et al. 2018), and so it likely indicates a platform-specific difference in the ability to sequence long arrays of simple tandem repeats. PacBio reads contained a similar normalized abundance of AAACTAC as Illumina reads both from this study and a previous study (Wei et al. 2018; fig 1B and D). However, both PacBio and Nanopore reads were greatly depleted of AAACTAT and AAATTAC compared with Illumina NextSeq (but not compared with Illumina HiSeq). No technology contained satellite normalized abundance estimates approaching the estimated >100 Mb in the genome (fig 1B and D).
Both the PacBio and Nanopore reads contained large amounts of what we expect to be artifactual repeats, which were found with the k-Seek + Phobos approach, and validated with NCRF. NCRF found 4.4 Mb (normalized to 1× genome coverage) of AAACGAC in the PacBio reads. This satellite was not found in the Nanopore or Illumina data (this and previous studies) or in previous studies that characterized the most abundant satellites in D. virilis. Manual inspection indicated that the AAACGAC satellite was the true consensus found in long arrays in the reads and did not represent an error in our approaches’ characterization of satellites. Similarly, AAATCAC, AGCCTAT, ACAGGCT, and AATGG were found in megabase quantities (after normalization) in the Nanopore data—whereas these satellites were not found in Illumina or PacBio data. We suggest these satellites are also technical artifacts introduced at the base-calling level.
In the PacBio data, the relative amounts of 7-mer satellites (AAACTAC, AAACTAT, and AAATTAC) were lower than expected. This additional evidence led us to hypothesize that there were context-specific errors in our PacBio data affecting our particular satellites. If the sequencing were unbiased, we would expect to have an equal amount of satellites being detected on reads coming from both DNA strands. We evaluated the strand bias in the simulated and real long-read data for the three most abundant true satellites, as well as some artifactual satellites. We arbitrarily label the positive strand as AAACTAC and the negative strand as GTAGTTT, etc. In the simulated data, the positive and negative strands of satellites were detected in equal amounts (fig. 1C). However, there was a strong strand bias for all satellites in both the PacBio and Nanopore data (fig. 1C). For PacBio, the real satellites AAACTAC, AAACTAT, and AAATTAC had a positive strand bias, whereas the artifactual satellite had a negative strand bias: 98% of the reads with this satellite were from the negative strand. Based on communication with PacBio representatives, this issue seemed to be caused by context-specific issues with base-calling algorithms used for this sequencing run. As base-calling algorithms improve, these issues will likely begin to be remedied. In fact, we performed PacBio circular consensus sequencing (CCS) or “HiFi” sequencing for a closely related species, D. americana, and the base-calling issue was remedied. In the Nanopore data, strand biases were even more extreme: the negative strand was sequenced almost exclusively for real satellites AAACTAC and AAATTAC and suspect satellite AAATCAC, whereas the positive strand was sequenced for real satellite AAACTAT. For Nanopore, strand biases may be caused by unsequenceable secondary structures developing more frequently on one strand of the satellite DNA than the other. We analyzed Illumina NextSeq reads for D. virilis (data described in the following two sections), and no such strand bias was found.
Drosophila virilis Whole-Flies Have 40% Less Pericentromeric Satellites than Nonpolytene Tissue
Polyteny occurs in all differentiated tissues of Dipterans, and is characterized by multiple rounds of local DNA replication within the same nucleus and without cell division, a process known as endoreduplication (Smith and Orr-Weaver 1991; Kim et al. 2011; Yarosh and Spradling 2014). However, the pericentromeric heterochromatin, where most satellite DNA is located, is underreplicated (Belyaeva et al. 1998). It has never been tested if the level of polyteny in an adult fly makes a difference in the estimate of satellites per genome. Thus, we sequenced adult male flies (which have multiple polytene tissues) and imaginal discs (which are diploid) from male larvae and compared the amount of simple satellites in these data sets. We used Illumina sequencing and PCR-free library preparations to reduce known PCR bias (Wei et al. 2018). We found that for each of the four most abundant 7-mer satellites in the D. virilis genome, there was ∼40% less in the flies compared with the imaginal discs (fig. 1D). This pattern is not observed for microsatellites which are known to localize outside of pericentromeric heterochromatin (supplementary fig. S2A, Supplementary Material online). We also analyzed publicly available D. melanogaster data, including flies, imaginal discs, and salivary glands (which are the most extreme in polyteny), and observed this same pattern of underreplication of pericentromeric heterochromatin satellite repeats in polytene tissues (supplementary fig. S2B and C Supplementary Material online).
Reads with Satellites Had Lower Quality Scores in Illumina Data
Because extensive quality filtering is often performed on Illumina sequences before analysis, we sought to investigate the distribution of quality scores on satellite-containing reads produced from Illumina platforms. Upon inspection of our data with FastQC from the polyteny analysis, we found a bimodal distribution of quality scores, with one peak at 22 and another at 37 (supplementary fig. S3A, Supplementary Material online). After filtering low-quality reads, the majority of reads with simple satellites were removed (supplementary fig. S3, Supplementary Material online). The quantity of satellites was reduced by ∼15-fold after quality filtering (fig. 1D). It is apparent that in our data set, simple satellite-containing reads were highly enriched for low-quality scores. We examined other published D. virilis Illumina data sets to evaluate if this issue existed in other sequencing runs. Two other data sets were available and the one that was produced on the Illumina NextSeq platform like our data (Miller et al. 2018) showed the same pattern of biased quality scores in repetitive reads (supplementary fig. S4, Supplementary Material online). The data set produced on the HiSeq platform (Wei et al. 2018) did not show this pattern. It should be noted however that the amounts of 7-mer satellites sequenced in the NextSeq data sets were higher than the HiSeq data set (fig 1D). Our libraries were multiplexed with other non-D. virilis group samples from unrelated projects and only represented ∼20% of the total sequenced lane so that we would not have issues related to low complexity. We also noticed this pattern (but less dramatically) in our Illumina sequencing of multiple strains.
AAACTAC-Related Satellite Abundances in Related Species
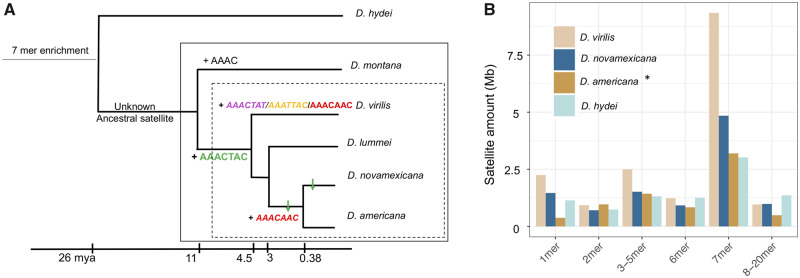
Drosophila novamexicana and D. americana which are 0.38 My diverged from each other, are sister species of D. virilis, which is ∼4.5 My diverged (Caletka and McAllister 2004; fig. 2A). We sequenced these species with high coverage PacBio runs and characterized and quantified satellites with k-Seek + Phobos and NCRF (see Materials and Methods section). We emphasize the comparison of relative satellite amounts since all are likely underrepresented. Drosophila americana was sequenced with PacBio HiFi reads, which eliminated artifactual satellites, but make quantitative comparisons difficult since different chemistries have different efficiencies of sequencing satellites. Nevertheless, we also found a high enrichment of 7-bp satellites in D. novamexicana and D. americana (fig. 2B). Interestingly, we found the most abundant satellite in D. virilis, AAACTAC, is also the most abundant in D. novamexicana and D. americana, albeit with about half the total amount. The second and third most abundant repeats, AAACTAT and AAATTAC, however were not present in long tandem arrays in D. novamexicana. The second most abundant satellite in D. novamexicana and americana was AAACAAC, whereas in D. virilis there is only a few kilobases.
Fig. 2.
Comparative analysis of simple satellites in the Drosophila virilis group. (A) Phylogeny demonstrating when satellites arose (+). AAACAAC may have emerged once and was lost once, or emerged twice as we illustrate here. ↓ indicate decreases in abundance of respective satellites. Centromere-proximal satellites are italicized. Dashed box: virilis phylad; solid box: virilis group. (B) Total amount of satellites of different unit lengths (k-mers) across four related species. The asterisk for D. americana indicates that it was sequenced with a different sequencing chemistry version (3.0 vs. 2.0 chemistry).
By analyzing sequencing data in more diverged species, we can infer when the AAACTAC satellite family arose. Drosophila hydei is ∼26 My diverged from D. virilis (Izumitani et al. 2016), and we had PacBio long-read data for this species. Here 7-bp satellites are again the most enriched (fig. 2B), but the sequences are unrelated to those in D. virilis (ACCCATG, AAAGGTC from PacBio data). We analyzed Illumina data for D. montana, another member of the virilis group that is 7–11 My diverged from D. virilis (Ostrega and Thompson 1986; Spicer and Bell 2002; fig. 2A). This species does not have any AAACTAC family satellites, and in fact no enrichment of 7-bp satellites. The most abundant satellite in D. montana is AAAC. From these data, we infer that the AAACTAC family of satellites arose in the clade leading to the D. virilis phylad 4.5–11 Ma. We also analyzed Illumina sequencing data for Drosophila lummei, which is 3 My diverged from D. novamexicana/D. americana (fig. 2A). AAACTAC is conserved in D. lummei, but it is the only enriched 7-bp satellite in this species (table 1).
Table 1.
Summary of Comparative Analysis of Simple Satellites in the Drosophila virilis group.
| Species | Technologies Used | Main Findings |
|---|---|---|
| Drosophila virilis | Illumina, PacBio, Nanopore, FISH | AAACTAC pericentromeric and extremely abundant, AAACTAT and AAATTAC are centromere proximal and highly abundant |
| Drosophila novamexicana | Illumina, PacBio, FISH | AAACTAC pericentromeric and much lower abundance than D. virilis, except on putative Chr 5. AAACAAC is centromere proximal |
| Drosophila americana | Illumina, PacBio CCS, FISH | AAACTAC pericentromeric and intermediate abundance between D. virilis and novamexicana. AAACAAC is centromere proximal |
| Drosophila lummei | Illumina, FISH | AAACTAC highly abundant and pericentromeric |
| Drosophila montana | Illumina | No abundant 7-mers present; most abundant satellite is AAAC |
Note.—CCS, circular consensus sequencing; FISH, fluorescence in situ hybridization. Centromere-proximal satellites are italicized.
Fluorescence In Situ Hybridization Reveals Evolutionary Dynamics of 7-bp Repeats
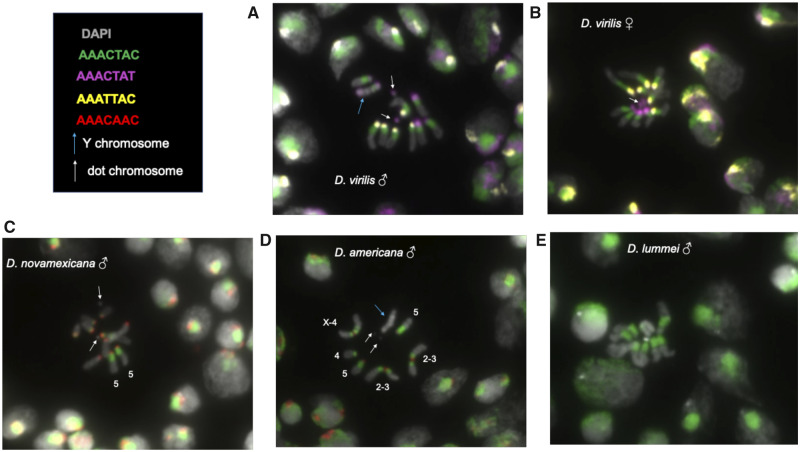
We present the first visualization to our knowledge of the 7-bp satellites on metaphase chromosomes in the D. virilis group. From our sequencing data, we know that the AAACTAC satellite is conserved between D. virilis, D. novamexicana, D. americana, and D. lummei, but the abundance varies by ∼2-fold. The second most abundant satellites have turned over between D. virilis and novamexicana/D. americana (table 1). We used fluorescence in situ hybridization (FISH) of the most abundant 7-mers (AAACTAC, AAACTAT, AAATTAC, and AAACAAC) in these four sister species. We also constructed a probe for the putative artifactual satellite AAAGCAC, which appeared to be the second most abundant satellite in D. virilis according to the PacBio data. However, it did not produce a hybridization signal with the conditions we used for the other satellites. Drosophila virilis and D. novamexicana have the same karyotype with five acrocentric chromosomes plus the very small F element or “dot chromosome.” The strain of D. americana we used has centromere–centromere fusions between the X and fourth chromosomes and the second and third chromosomes.
The FISH results in D. virilis show that the most abundant satellite determined by sequencing, AAACTAC, is clearly the most abundant and occurs in approximately equal amounts in the pericentromeric region on the five pairs of large chromosomes. The Y chromosome appears to have slightly less AAACTAC satellite. The second and third most abundant satellites, AAATTAC and AAACTAT, are localized more proximally, near or at the centromere. There are five single chromosomes having each of these satellites, indicating that one chromosome pair has different satellite content—which we hypothesized to be the X and Y. Based on differences between male and female FISH results (fig. 3A and B), we suggest the Y chromosome has AAACTAT at both distal ends of the chromosome and AAACTAC only flanking one end, whereas the X chromosome has the other centromeric repeat, AAATTAC. We were also able to visualize the dot chromosomes in D. virilis, which we find is mostly composed of AAACTAT. The AAACAAC satellite is present in small amounts in D. virilis, very likely on a single chromosome (supplementary fig. S5, Supplementary Material online).
Fig. 3.
DNA FISH of larval neuroblast nuclei. Up to three fluorescent probes were hybridized per experiment. Arrows indicate the common features of the Y chromosome and the dot chromosome, where they are clearly distinguishable. (A) Drosophila virilis male; (B) Drosophila virilis female; (C) Drosophila novamexicana male; (D) Drosophila americana male, chromosomes are distinguishable and thus are labeled; (E) Drosophila lummei male.
We estimated from sequencing that D. novamexicana has approximately half the AAACTAC as D. virilis, and visualizing it with FISH reveals a pattern that suggests aspects of its evolution. Its pericentromeric localization is conserved. One chromosome pair has the same amount of AAACTAC as D. virilis, whereas all other chromosomes have a very small amount (fig. 3C). Based on the FISH images, it appears that it is the fifth chromosome in D. novamexicana that has the greatest amount of pericentromeric AAACTAC conserved. The centromeric repeat on all major chromosomes is AAACAAC in D. novamexicana and D. americana. Our images illustrate clearly the centromere-centromere fusion between chromosome X-4 and 2-3 in D. americana with the satellites being maintained on both sides of the fusion (fig. 3D). None of the four simple satellite probes bound to the Y chromosome of D. novamexicana or D. americana. Based on the images, we suggest that D. americana has an intermediate amount of pericentromeric AAACTAC satellite compared with D. virilis and novamexicana. Drosophila lummei only contains AAACTAC, but at very high amounts, similar to D. virilis, in the pericentromeric region (fig. 3E).
Complex Satellites Are Also Abundant in D. virilis Group Genomes
We searched the high-quality genome assemblies for complex satellites (defined here as unit lengths greater than 20 bp). In D. virilis, we found a 36-bp satellite AAAACGACATAACTCCGCGCGGAGATATGACGTTCC making up ∼800 kb of the assembly. This satellite was found in previous studies and is thought to be associated with the possibly mobile element pDv (Zelentsova et al. 1986; Heikkinen et al. 1995). In D. novamexicana, we found a 32-bp satellite AAAAGCTGATTGCTATATGTGCAATAGCTGAC along with a related 29-bp satellite. The 32-bp satellite spanned over 1.1 Mb on a single 3-Mb contig in the D. novamexicana assembly. The nonsatellite portion of the contig had similarity to chromosome 6 (dot chromosome/Muller element F; supplementary fig. S6, Supplementary Material online). In D. americana, we found this identical 32-bp satellite, but in total its span was only ∼150 kb. In all D. virilis group species, we also found a series of similar satellites varying in size (150–500 bp) related to the previously described helitron central repeat DINE-1 (Drosophila INterspersed Elements) that has expanded to tandem repeats in the virilis group (Dias et al. 2015).
Variation in D. virilis Group Global Strains
Drosophil a virilis is globally distributed while its sister species are localized to North America, with D. novamexicana more restricted than D. americana. Patterns of variation in satellites may reveal potential mechanisms that can be hypothesized to be driving satellite evolution. Additionally, D. americana has a polymorphic fusion between the X and fourth chromosomes, so we may be able to identify differences in satellite composition associated with the fusion. This fusion has been shown to be currently undergoing meiotic drive, potentially mediated by a larger total centromere or pericentromere size in the fused strains compared with the nonfused strains (Stewart et al. 2019). On the other hand, chromosome fusions are often caused by Robertsonian translocations with loss of some nonessential DNA, which might include pericentromeric satellites (Schubert and Lysak 2011).
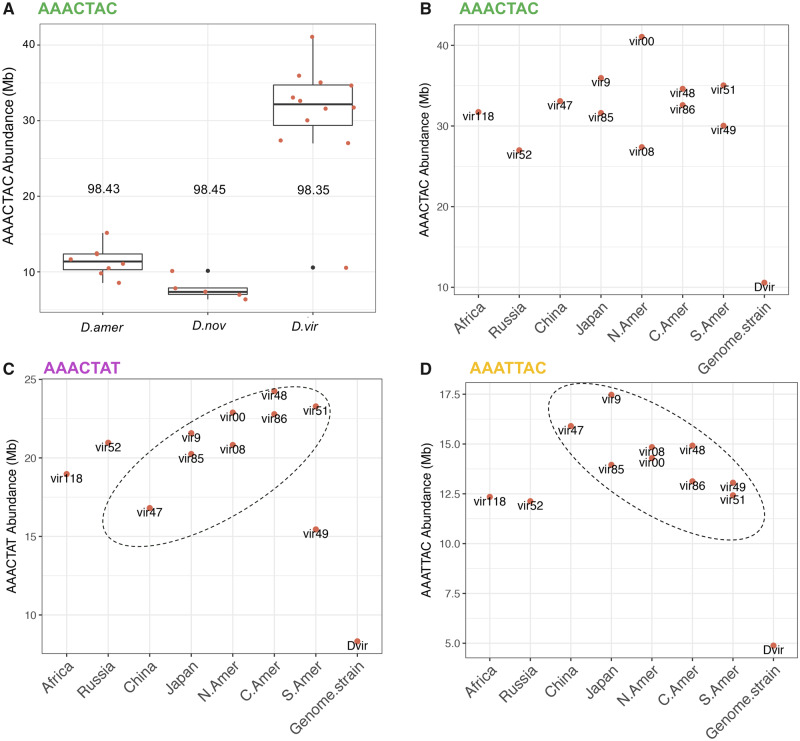
We used Illumina sequencing with PCR-free library preparation and k-Seek to estimate the abundance of 7-mer satellites across 12 worldwide strains of D. virilis, 8 strains of D. americana (including 4 strains that have the X-4 fusion and 4 that do not), and five strains of D. novamexicana (supplementary table S2, Supplementary Material online). All sequenced strains were male except a female of the D. virilis inbred genome strain 87 as a comparison. A PCA (Principal Components Analysis) using only the four most abundant 7-mers shows clustering of the three species, but the separation is much more dramatic in the PCA using the 20 most abundant simple satellites (supplementary fig. S7, Supplementary Material online). Overall, D. virilis had the highest AAACTAC satellite content as well as the highest variation, with D. americana intermediate between D. virilis and D. novamexicana (fig. 4A). Using different normalization procedures including mapping and GC correction (see Materials and Methods section), produced the same relative ranking of satellite abundances between species. In all cases, the inbred strain from which the genome sequence was produced had the lowest abundance of AAACTAC. In the case of D. virilis, this difference was very high. This was not due to a normalization bias as we did mapping-free normalization.
Fig. 4.
Variation in satellites across species and strains. (A) AAACTAC total abundance across the three species. The number above the box plots is the average sequence identity for arrays of AAACTAC. (B) AAACTAC, (C) AAACTAC, and (D) AAATTAC, abundance across Drosophila virilis strains originating from different localities (x axis). The strain Dvir is strain 87, the inbred strain used for genome assemblies. All data were generated with NextSeq.
Satellite abundances in D. virilis displayed a pattern that appeared to be correlated to the geographic location from which strains were collected. For the centromeric satellite AAATTAC, there was a linear decrease in abundance from West to East then South following probable migration from Beringia (Throckmorton 1982) beginning in China (fig. 4C). For the centromeric satellite AAACTAT, the pattern was the opposite; a linear increase in abundance from West to East then South (fig. 4D).
Satellite arrays are expected to randomly accumulate sequence mutations, which could indicate their relative age. Low sequence variation could indicate the satellites were recently formed, or that concerted evolution maintains high sequence identity. On average, the centromeric satellite arrays were very homogeneous in sequence in D. virilis (average above 99% sequence identity in Illumina reads). This means in a 100-bp section of an array, there is only one SNP on average altering one unit of (AAACTAT)n. The pericentromeric satellite AAACTAC has almost identical average sequence divergence across the three species (∼98.5%; fig. 4A). There was a greater magnitude of variation in average sequence identity between males and females of D. virilis strain 87 then there was across all global strains, with 98.6% sequence identity in the female and 97.8% identity in the male.
There was no detectable difference in centromeric or pericentromeric satellite abundance in D. americana strains with versus without the polymorphic centromere-centromere fusion. We conclude that molecular events surrounding the fusion did not produce any changes in satellite abundance (supplementary fig. S8, Supplementary Material online).
Discussion
Here, we used the satellite DNA-rich genome of D. virilis to highlight three previously uncharacterized mechanisms for biases that occur in sequencing and analyzing satellite DNA. We find that there is currently no sequencing method that can accurately measure simple satellite abundance. We emphasize that comparing satellite DNA amounts between different platforms (e.g., Illumina, PacBio, Nanopore, and even different versions of each) should be done with caution as each technology has its own biases. We have found that issues arise when long arrays of simple satellite DNA are attempted to be sequenced by long-read platforms. In the case of PacBio, systematic errors in base calling may be introduced when sequencing through long arrays of satellites. This issue is not specific to our satellites, as a recent study has also found systematic errors and strand biases in shorter arrays of human satellites in both PacBio and Nanopore reads (Mitsuhashi et al. 2019). CCS or “HiFi,” a type of sequencing offered by PacBio which allows an accurate consensus to be produced after multiple rounds of sequencing the same molecule, may be more appropriate for sequencing analysis of satellite DNA. No systematic errors in satellite sequences resulted with the new CCS platform after collaboration with PacBio representatives. In the case of Nanopore, it is possible that similar satellite-specific base-calling errors exist, or that there is a strand-specific difference in secondary or tertiary structures that occur in long strands of simple satellite DNA. We caution readers in interpreting composition and abundance of simple satellite DNA from long-read sequencing data and suggest validation with satellites of known sequence and abundance, if available, or by use of Illumina sequencing (without quality filtering) as we have demonstrated here. Long-read platforms are already improving their chemistry and software for better satellite characterization. Because long reads are likely to cross boundaries of different repetitive regions, long-read sequencing proved useful in understanding the length of the satellite arrays and TE insertions into them. Moreover, we demonstrate that the abundance of satellites in pericentromeric heterochromatin are underestimated when sequencing whole adult Drosophila compared with pure diploid tissue because of polyteny. We caution readers in performing quality filtering of Illumina reads before simple satellite analysis, as satellite-containing reads may be enriched for lower quality scores.
Because we used validation from multiple types of data, we are confident in our comparative analysis of satellite sequences and relative abundances. We found the abundant AAACTAC family of satellites arose in the branch leading to the virilis phylad 4.5–11 Ma (fig. 2A). Interestingly, the most abundant satellite in D. montana, 7–11 My diverged, is AAAC. The AAACTAC and AAAC satellites were likely derived from a common ancestor satellite (fig. 2A). From both FISH and sequencing analysis, we found that D. virilis has the highest total amount of AAACTAC family satellites, D. novamexicana has about half of D. virilis, and D. americana intermediate between the two species. Drosophila lummei has the lowest relative satellite content, and its only high-abundance simple satellite is AAACTAC. Unlike the pericentromeric satellite, the centromere-proximal satellite sequence has turned over between D. virilis and D. americana/D. novamexicana. The AAACAAC satellite could have evolved once in the branch leading to the virilis group and was lost in D. lummei, or it could have evolved twice—once as a low abundance satellite in D. virilis and again as the centromere-proximal satellite in D. americana and D. novamexicana. The latter possibility is shown in figure 2A. The AAACTAT and AAATTAC satellites are unique to D. virilis and occupy the centromeric region. The emerging pattern is that the centromere-proximal satellites have turned over more rapidly than the pericentromeric satellite. This is likely due to satellites participating in conflicts at centromeres (Malik and Bayes 2006, and discussed below). Although sequencing quantified only up to 30 Mb of the AAACTAC family of satellites, FISH confirmed that these satellites are extremely abundant in D. virilis and the 40% of the genome estimate seems realistic. By exploiting interspecific crosses of virilis group species with different satellite compositions we could begin to elucidate the roles of individual satellites.
We can make hypotheses about how and why the satellites expanded in D. virilis. We know that mutation rates for changes in copy number of satellite DNA are high, and potentially have a tendency to expand rather than contract in the absence of selection (Flynn et al. 2017, 2018). High rates of mutation must be accompanied by a regime that would allow a satellite copy number increase to sweep the population—which could be mediated by positive selection if there is a benefit of the satellite increase, or centromere drive if the phenomenon is at play. Alternatively, in a situation where satellites are slightly deleterious, small effective population sizes in isolated populations or continued bottlenecks could allow satellites to expand in the genome without being removed by selection. However, D. novamexicana has the lowest effective population size of the virilis phylad, and yet it has the lowest amount of satellite DNA. We already know that the centromere-to-centromere fusions in D. americana have undergone meiotic drive hypothesized to be mediated by the increase in centromere total size with the fusion (Stewart et al. 2019). The mechanism allowing drive in D. americana may have been at play in the branch leading to D. virilis or may be currently occurring. Why have satellites not expanded to this extent in the other species? Drosophila virilis might have some attributes about its biology that made the satellite expansion favorable or allowable. For example, genome size is positively correlated with development time in Drosophilidae (Gregory and Johnston 2008). Drosophila virilis has a slow development time, and this may have evolved in concert with the expansion in satellite abundance in its genome.
We can use data from multiple strains to make hypotheses about factors driving satellite DNA evolution in D. virilis. Ancestrally, D. virilis had a relatively small effective population size in an isolated range in Asia, and has undergone a recent population and range expansion (Mirol et al. 2008). The amount of the most abundant pericentromeric satellite AAACTAC does not show a geographical pattern across the global strains. Assuming we sequenced a strain from the ancestral range, this suggests that population bottlenecks were not what allowed AAACTAC to expand, and the satellite expansion likely occurred before the population expansion.
Our observation of rapid evolution and enrichment of AAACTAC in D. virilis in a short evolutionary time period (a few million years) is consistent with the centromere-drive model to account for the evolution of centromere complexity in genetic conflict (Malik and Bayes, 2006). In this model, the asymmetric female meiosis can cause competition between the centromeres with or without newly formed satellites or with more or less satellites, to be included into the oocyte to pass to next generation. A consequence of the competition would be runway expansions of centromeric satellites, and rapid replacements by novel satellites. We hypothesize that the pattern of the centromere-proximal satellite AAACTAT increasing on a geographical gradient while AAATTAC decreases along the same gradient is driven by centromeric conflicts. AAACTAT may be starting to occupy centromeres that AAATTAC occupied, benefitting from a transmission advantage (centromere drive), while the AAATTAC satellite may be decreasing in parallel because of selection “pushing back,” for example, because of a maximum limit on satellite amount in the centromeric region. Another line of evidence that centromere-related conflicts are playing a role is the rapid rate of turnover of the centromere-proximal satellites compared with the pericentromeric satellite.
Interestingly, in D. novamexicana, AAACTAC was greatly reduced in the pericentromeric regions on all chromosome pairs except one. Based on the FISH images in D. novamexicana and D. americana, we hypothesize that it is the 5th chromosome that has the high amount of AAACTAC satellite. This is interesting because previous work has shown that the fifth chromosome contains a high amount of DINE-1 helitron satellite in D. virilis but not in D. americana (Dias et al. 2015). This may be evidence of past and ongoing competition and tradeoffs between the DINE-1 satellite and AAACTAC. We have seen evidence for this tradeoff, or appearance of competitive exclusion, being invoked under selection in our previous studies in Daphnia (Flynn et al. 2017, 2018). There may have been a similar conflict on Chr5 of D. novamexicana, where AAACTAC retained a high copy number to prevent DINE-1 from expanding. Interestingly, the opposite has occurred on the D. novamexicana and D. americana Y chromosome, where AAACTAC family satellites are absent but DINE repeats are abundant. A potential mechanism mediating apparent stabilizing selection on total satellite abundance is that satellites can act as a sink for heterochromatin factors, with their abundance affecting chromatin state (Lemos et al. 2010).
The AAACTAC satellite has remained conserved in sequence and location in the virilis phylad. It has also maintained high levels of sequence identity that is equal in the three species we sequenced (98.5% based on Illumina reads). The conservation may reflect a constraint due to selection or a pervasive mechanism of concerted evolution. The periodicity of the sequence may stabilize the DNA helix wrapping around nucleosomes, or it may be constrained by coevolution of an important satellite DNA binding protein (Maio et al. 1977; Jagannathan et al. 2018). Additionally, within the AAACTAC family, the position and identity of the four A-nucleotides are conserved in all four satellites (AAACTAC, AAATTAC, AAACTAT, and AAACAAC)—which may indicate constraint based on the above mechanisms. Conservation of particular satellite unit lengths and “AA” periodicities has been found in other divergent species (Lowman and Bina 1990). Concerted evolution of satellites could be achieved by repeated recycling of units by copy number changes associated with replication slippage or unequal recombination or gene conversion (Walsh 1987; Elder and Turner 1995). However, recombination in the pericentromeric heterochromatin has never been detected in wild-type flies (Mehrotra and McKim 2006; Hughes et al. 2018). On the other hand, if recombination were occurring, satellite arrays will eventually be lost unless they are conserved by selection (Charlesworth et al. 1986). Clearly, we are still lacking in understanding how and why long simple satellite arrays maintain their homogeneity, and whether recombination plays a role in their dynamics. Concordant with the hypothesis that recombination is playing a role, males have lower average sequence identity in the 7-bp satellites than females, which could indicate increased decay on the Y chromosome where there no homologous recombination (fig. 4A).
An interesting observation from the sequencing of multiple strains of the three species was that in all cases, the inbred strain that the reference genome was made from had the lowest amount of AAACTAC. For D. virilis, this difference was extreme. It is tempting to speculate that the process of inbreeding and/or long periods in the lab may have driven the reduction in pericentromeric satellite abundance.
In conclusion, our results show very rapid dynamics in the abundant satellites of the D. virilis group that are likely explained by various cellular and population-level forces that are not yet understood. Further studies can test if there is a species-specific upper limit to satellite amount per genome or per chromosome upon which negative fitness effects occur, which may result in tradeoffs or competition between satellites. Centromere drive may be an important process affecting satellite evolution in this species group, and might partially explain why the satellites expanded 4.5–11 Ma, why satellite sequences at the centromere turned over more rapidly, and why there is a gradient of increasing satellite content related to geographical distribution of strains. A more extensive study to determine if inbreeding or extended periods in the lab drives a reduction in satellite abundance will help illuminate the processes that are important for maintaining satellite content. Determining the frequency of recombination in the large pericentromeric heterochromatin blocks in species like D. virilis will be challenging but important for understanding how the satellites maintain homogeneity in their sequence. To understand the role of satellites and the importance of their sequence, unit length, and abundance, researchers can strive to develop methods to engineer satellites by modifying specific bases and their abundances.
Materials and Methods
All scripts for analyzing the data and to produce the results we show are here: https://github.com/jmf422/D_virilis_satellites. Illumina sequencing reads for all species and PacBio CCS data of D. americana generated for this study are deposited in NCBI SRA under accession PRJNA548201. Raw PacBio reads for D. virilis, D. hydei, and D. novamexicana are deposited under accession PRJNA475270.
PacBio Sequencing
High-molecular weight DNA was gently extracted from 50 inbred male flies in a single 1.6 ml microcentrifuge tube using a modified CTAB procedure. Strains used were as follows: D. virilis: 15010-1051.87, D. novamexicana: 15010-1031.14, D. americana: G96, D. hydei: 15805-1641.58. Sequencing libraries were then constructed using manufacturer’s recommended protocols. Libraries were sequenced to ∼100× genome coverage on a PacBio Sequel instrument with chemistry version 2.0. Drosophila americana CCS libraries were prepared at PacBio headquarters using chemistry version 3.0 and sequenced at ∼10× coverage on a Sequel instrument.
Characterizing Satellite DNA from Genome Assemblies
All scripts and R markdown files used for this analysis are provided in https://github.com/jmf422/D_virilis_satellites/tree/master/Genome_assembly_analysis.
We used the D. virilis genome assembly produced by the PacBio sequencing project (https://www.ncbi.nlm.nih.gov/bioproject/? term=txid7214). We also downloaded the D. virilis genome produced by Nanopore sequencing from (Miller et al. 2018), and the CAF1 assembly from (Drosophila 12 Genomes Consortium et al. 2007). We used Phobos (https://www.ruhr-uni-bochum.de/spezzoo/cm/cm_phobos.htm) and Tandem repeats finder (Benson 1999) to characterize simple and complex satellites in these genome assemblies. To identify the chromosomal linkage of complex satellites in the genome assembly, we produced a dotplot with D-GENIES (Cabanettes and Klopp 2018).
Characterizing Satellite DNA from Raw Long Reads
Characterizing and quantifying satellites from long reads is a challenge because of the sequencing high error rate. We used two approaches to characterize satellites from raw long reads. The first approach, we call k-Seek + Phobos, in which we first broke the reads into 100-bp subreads and ran k-Seek on them. k-Seek is very efficient for analyzing many reads, however is not very sensitive for reads with a high error rate since it was designed for Illumina reads (Wei et al. 2014). If k-Seek found satellites on at least one subread, we would run the complete parent read through Phobos. Phobos is more sensitive to imperfect repeats and error rates, but cannot handle huge quantities of data; thus why we only ran the portion of reads identified by k-Seek to have tandem repeats. This approach allowed us to characterize satellites de novo and quantify them. All scripts for the analysis of long reads with the k-Seek + Phobos approaches are located here: https://github.com/jmf422/D_virilis_satellites/tree/master/LongRead_kseek_Phobos. The second approach we used is NCRF (Harris et al. 2019). This program was designed to quantify satellites from long reads with high error rates by aligning target satellites to the reads. However, it cannot identify satellites de novo and requires specific satellite sequences to search for. NCRF also requires a “max divergence allowed” parameter, which we tuned with simulations (see below). Scripts used for the NCRF approach are located here: https://github.com/jmf422/D_virilis_satellites/tree/master/LongRead_NCRF.
We performed simulations to assess both approaches: https://github.com/jmf422/D_virilis_satellites/tree/master/Simulations. First, we generated a simplified mock D. virilis genome with a satellite DNA composition based on our FISH results. The current genome assembly would not have been suitable because it contains a very low abundance of satellite DNA. We generated each mock chromosome with a centromeric satellite of either AAATTAC or AAACTAC, flanked by the pericentromeric AAACTAC satellite sequence. The total satellite abundance was 40% of the 351-Mb mock genome sequence (109-Mb AAACTAC, 14-Mb AAACTAT, and 17-Mb AAATTAC). The nonsatellite DNA portion of the genome was generated with random sequence making up a 40% GC content. We then used PBSIM (Ono et al. 2013) to simulate PacBio reads with error (parameters: –data-type CLR –depth 10 –model_qc model_qc_clr). We used these simulated reads for several analyses. First, we used them to determine the most appropriate maximum divergence parameter value for NCRF by evaluating a range of values for this parameter (18–30%). We found that the amount of satellites found, particularly the most abundant one, leveled off at 25% max divergence (supplementary fig. S1, Supplementary Material online). This is the parameter value we used moving forward. We also used these simulated reads to quantify satellites with both approaches and compare them. Finally, we used these simulated reads to assess strand biases in long-read sequencing data (see below).
Identification of Biases in Simple Satellites in Long-Read Data
We suspected that there were biases in the satellite DNA found in the D. virilis group PacBio (and Nanopore) data because we found high-abundance satellites that had never been found before with other types of data, and so we suspected they were artifactual. These artifactual satellites were found with both kSeek + Phobos and NCRF approaches, but were not found in the simulated data. We tried testing for a strand bias in reads that contained satellite DNA. Using both the summarized output from NCRF and validated with a custom script (LongRead_NCRF/which_strand_pacbio_script.sh), we counted the satellite DNA stretches that originated from each the positive and negative strand. The positive strand is defined as the one that contains the satellite AAACTAC and derivatives (more As than Ts), and the negative strand is the one that contains the reverse complement (e.g. GTAGTTT, more Ts than As). We did this for the three satellites used in the simulated data and real and artifactual satellites found in the PacBio and Nanopore data. Detailed analysis and visualization of the biases is shown here: LongRead_kseek_Phobos/longread_analysis.html
Sequencing of Polytene and Nonpolytene Tissue
To acquire D. virilis pure diploid tissue, we dissected male third instar larvae and collected imaginal discs including the eye-antennal disc and wing discs. Approximately 100 larvae were required to get enough DNA (>1 μg). We also collected ∼5 adult flies for fly libraries. We used the inbred genome assembly strain 87 for these libraries. DNA was extracted with Qiagen DNeasy blood and tissue kit and PCR-free libraries were prepared. Libraries were run on an Illumina NextSeq with 1 × 150 bp reads, and each sample took up ∼7% of the flowcell. The other libraries run on this flowcell were from an unrelated project including RNAseq from other species. Reads were analyzed with k-Seek both before and after filtering with Trimmomatic (Bolger et al. 2014). FastQC was run to evaluate the quality of the reads. Scripts are here: https://github.com/jmf422/D_virilis_satellites/tree/master/Polyteny. We also analyzed publicly available D. melanogaster data from the same strain and same sequencing platform of embryos (nonpolytene), salivary glands (extreme polyteny) from (Yarosh and Spradling 2014), and adult flies (varied levels of polyteny) from (Gutzwiller et al. 2015).
FISH of Satellite DNAs
We followed the protocol of (Larracuente and Ferree 2015) for satellite DNA FISH. We ordered the following probes from IDT with 5’ modifications: (AAACTAC)6 with alexa-488 fluorophore, (AAACTAT)6 with Cyanine5 fluorophore, (AAATTAC)6 with Cyanine3 fluorophore, (AAACAAC)6 with Cyanine3 fluorophore, and (AAACGAC)6 with Cyanine5 fluorophore. We hybridized three probes at a time, to allow for similar probes to compete to result in specific hybridization with the rationale shown in (Beliveau et al. 2015). Hybridization temperature was 32°C. We imaged on an Olympus fluorescent microscope and Metamorph capture system at the Cornell Imaging Facility. Composite images were produced with ImageJ.
Sequencing of Multiple D. virilis Group Strains
We obtained as many strains of D. virilis that have information about where they were collected as possible. This included 12 strains as live stocks we obtained either from stocks in our lab or from the Drosophila species stock center (supplementary table S2, Supplementary Material online). We also prepared a female library for the genome strain 87 to identify male and female differences in satellite composition. We also obtained five strains of D. novamexicana and eight strains of D. americana. All were obtained from live stocks and the inbred genome strains were included for both species as well (strain 14 and G96, respectfully). For D. americana, we included four strains that have the chromosome X-4 fusion and four strains that do not have it, based on communication with the Bryant McAllister lab. DNA was extracted as above from five flies each and samples were prepared identically as above and sequenced on 50% of three flowcells of Illumina NextSeq 1 × 150 bp reads. We dispersed the samples from each species between multiple flowcells. Our samples took up only half the flowcell with the other half being occupied by RNAseq libraries from an unrelated project.
All scripts used to analyze these data are located here: https://github.com/jmf422/D_virilis_satellites/tree/master/Intra_inter_species_sequencing.
Reads were evaluated with FastQC and not filtered for quality based on the potential bias of Illumina quality scores on satellites. We used k-Seek to quantify satellites. We tried several normalization strategies but decided the most appropriate was a mapping-free normalization. We estimated average depth by dividing the total number of bases sequenced by the estimated genome size by flow cytometry (Bosco et al. 2007). We believe this was the best option in this case because: 1) we were concerned about a mapping bias because for each species the strain that the genome assembly was made from may have more reads map to it; and 2) after masking the genome from the 7-mer satellites and also excluding the X and Y contigs (because we had male and female strains, and the Y chromosome contained more low GC regions)—there was little difference in coverage based on GC content. We include results when we used a mapping based GC normalization in the subdirectory “AlternativeNormalization.”
We used NCRF with modified parameters (minlength = 100, maxdiv = 10) to characterize the average sequence identity of satellite arrays from the Illumina data. We also analyzed Illumina DNA sequencing reads of D. montana (Parker et al. 2018) and D. lummei (Ahmed-Braimah et al. 2017) with k-Seek to identify the most abundant satellites and whether or not the AAACTAC satellite family was present.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Yasir Ahmed-Braimah for helpful discussions and advice for some analyses. We also thank Bryant McAllister for providing Drosophila americana strains along with their fusion status. We are grateful for Elissa Cosgrove’s help with some computational trouble-shooting and Asha Jain’s help in preparing DNA sequencing libraries. We also thank Danny Miller for useful discussions and for providing the raw Nanopore reads from his study. Jianwei Zhang helped with PacBio raw data transfers. Sarah Kingan, Jane Landolin, and Greg Young from Pacific Biosciences were very helpful in exploring the causes for the artifactual repeats and in producing the HiFi data. We thank Amanda Larracuente for advice on FISH protocols and the Cornell Imaging Facility for use of their microscope. This project was funded by National Institutes of Health Grant Number GM116113 to R.A.W., M.L., and A.G.C. and Grant Number GM119125 to A.G.C. and Daniel Barbash. J.M.F. was supported by an Natural Sciences and Engineering Research Council of Canada Doctoral Scholarship.
References
- Ahmed-Braimah YH, Unckless RL, Clark AG.. 2017. Evolutionary dynamics of male reproductive genes in the Drosophila virilis subgroup. G3 (Bethesda) 7:3145–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Boettiger AN, Avendaño MS, Jungmann R, McCole RB, Joyce EF, Kim-Kiselak C, Bantignies F, Fonseka CY, Erceg J, et al. 2015. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun. 6:7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva ES, Zhimulev IF, Volkova EI, Alekseyenko AA, Moshkin YM, Koryakov DE.. 1998. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc Natl Acad Sci U S A. 95(13):7532–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Campbell P, Leiva-Neto JT, Markow TA.. 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177(3):1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanettes F, Klopp C.. 2018. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ. 6:e4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caletka BC, McAllister BF.. 2004. A genealogical view of chromosomal evolution and species delimitation in the Drosophila virilis species subgroup. Mol Phylogenet Evol. 33(3):664–670. [DOI] [PubMed] [Google Scholar]
- Chang C-H, Larracuente AM.. 2019. Heterochromatin-enriched assemblies reveal the sequence and organization of the Drosophila melanogaster Y chromosome. Genetics 211(1):333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH, Stephan W.. 1986. The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112(4):947–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W.. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371(6494):215–220. [DOI] [PubMed] [Google Scholar]
- de Lima LG, Svartman M, Kuhn GCS.. 2017. Dissecting the satellite DNA landscape in three cactophilic sequenced genomes. G3 (Bethesda) 7:2831–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias GB, Heringer P, Svartman M, Kuhn GCS.. 2015. Helitrons shaping the genomic architecture of Drosophila: enrichment of DINE-TR1 in α- and β-heterochromatin, satellite DNA emergence, and piRNA expression. Chromosome Res. 23(3):597–613. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes ConsortiumClark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 450:203–218. [DOI] [PubMed] [Google Scholar]
- Elder JF Jr, Turner BJ.. 1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol. 70:297–320. [DOI] [PubMed] [Google Scholar]
- Elliott TA, Gregory TR.. 2015. What’s in a genome? The C-value enigma and the evolution of eukaryotic genome content. Philos Trans R Soc B. 370(1678):20140331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Caldas I, Cristescu ME, Clark AG.. 2017. Selection constrains high rates of tandem repetitive DNA mutation in Daphnia pulex. Genetics 207:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Lower SE, Barbash DA, Clark AG.. 2018. Rates and patterns of mutation in tandem repetitive DNA in six independent lineages of Chlamydomonas reinhardtii. Genome Biol Evol. 10(7):1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J, Cohen E, Polan M.. 1971. Repetitive DNA sequences in Drosophila. Chromosoma 33(3):319. [DOI] [PubMed] [Google Scholar]
- Gall JG, Atherton DD.. 1974. Satellite DNA sequences in Drosophila virilis. J Mol Biol. 85(4):633–664. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev. 76(1):65–101. [DOI] [PubMed] [Google Scholar]
- Gregory TR, Johnston JS.. 2008. Genome size diversity in the family Drosophilidae. Heredity 101(3):228–238. [DOI] [PubMed] [Google Scholar]
- Gutzwiller F, Carmo CR, Miller DE, Rice DW, Newton ILG, et al. 2015. Dynamics of Wolbachia pipientis gene expression across the Drosophila melanogaster life cycle. G3 (Bethesda) 5:2843–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Cechova M, Makova KD.. 2019. Noise-cancelling repeat finder: uncovering tandem repeats in error-prone long-read sequencing data. Bioinformatics 35:4809–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen E, Launonen V, Müller E, Bachmann L.. 1995. The pvB370 BamHI satellite DNA family of the Drosophila virilis group and its evolutionary relation to mobile dispersed genetic pDv elements. J Mol Evol. 41(5):604–614. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS.. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1102. [DOI] [PubMed] [Google Scholar]
- Hughes SE, Miller DE, Miller AL, Hawley RS.. 2018. Female meiosis: synapsis, recombination, and segregation in Drosophila melanogaster. Genetics 208(3):875–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumitani HF, Kusaka Y, Koshikawa S, Toda MJ, Katoh T.. 2016. Phylogeography of the subgenus Drosophila (Diptera: Drosophilidae): evolutionary history of faunal divergence between the old and the new worlds. PLoS One 11(7):e0160051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Cummings R, Yamashita YM.. 2018. A conserved function for pericentromeric satellite DNA. Elife 7:e34122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Cummings R, Yamashita YM.. 2019. The modular mechanism of chromocenter formation in Drosophila. Elife 8:e43938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Nordman J, Xie F, Kashevsky H, Eng T, Li S, MacAlpine DM, Orr-Weaver TL.. 2011. Integrative analysis of gene amplification in Drosophila follicle cells: parameters of origin activation and repression. Genes Dev. 25(13):1384–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, Ferree PM.. 2015. Simple method for fluorescence DNA in situ hybridization to squashed chromosomes. J Vis Exp. 95:e52288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Branco AT, Hartl DL.. 2010. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci U S A. 107(36):15826–15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman H, Bina M.. 1990. Correlation between dinucleotide periodicities and nucleosome positioning on mouse satellite DNA. Biopolymers 30(9–10):861–876. [DOI] [PubMed] [Google Scholar]
- Malik HS, Bayes JJ.. 2006. Genetic conflicts during meiosis and the evolutionary origins of centromere complexity. Biochem Soc Trans. 34(4):569–573. [DOI] [PubMed] [Google Scholar]
- Maio JJ, Brown FL, Musich PR.. 1977. Subunit structure of chromatin and the organization of eukaryotic highly repetitive DNA: recurrent periodicities and models for the evolutionary origins of repetitive DNA. J Mol Biol. 117(3):637–655. [DOI] [PubMed] [Google Scholar]
- Mehrotra S, McKim KS.. 2006. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2(11):e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DE, Staber C, Zeitlinger J, Hawley RS.. 2018. Highly contiguous genome assemblies of 15 Drosophila species generated using nanopore sequencing. G3 (Bethesda) 8:3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills WK, Lee YCG, Kochendoerfer AM, Dunleavy EM, Karpen GH.. 2019. RNA transcribed from a simple-tandem repeat is required for sperm maturation and male fertility in D. melanogaster. Elife 8:e48940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirol PM, Routtu J, Hoikkala A, Butlin RK.. 2008. Signals of demographic expansion in Drosophila virilis. BMC Evol Biol. 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S, Frith MC, Mizuguchi T, Miyatake S, Toyota T, Adachi H, Oma Y, Kino Y, Mitsuhashi H, Matsumoto N.. 2019. Tandem-genotypes: robust detection of tandem repeat expansions from long DNA reads. Genome Biol. 20(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1972. So much “junk” DNA in our genome. Brookhaven Symp Biol. 23:366–370. [PubMed] [Google Scholar]
- Ono Y, Asai K, Hamada M.. 2013. PBSIM: pacBio reads simulator—toward accurate genome assembly. Bioinformatics 29(1):119–121. [DOI] [PubMed] [Google Scholar]
- Ostrega MS, Thompson V.. 1986. Mitochondrial DNA Restriction site polymorphism in Drosophila montana and Drosophila virilis. Biochem Syst Ecol. 14(5):515–519. [Google Scholar]
- Parker DJ, Wiberg RAW, Trivedi U, Tyukmaeva VI, Gharbi K, Butlin RK, Hoikkala A, Kankare M, Ritchie MG.. 2018. Inter and intraspecific genomic divergence in Drosophila montana shows evidence for cold adaptation. Genome Biol Evol. 10(8):2086–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlek M, Gelfand Y, Plohl M, Meštrović N.. 2015. Genome-wide analysis of tandem repeats in Tribolium castaneum genome reveals abundant and highly dynamic tandem repeat families with satellite DNA features in euchromatic chromosomal arms. DNA Res. 22(6):387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I, Lysak MA.. 2011. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 27(6):207–216. [DOI] [PubMed] [Google Scholar]
- Smith AV, Orr-Weaver TL.. 1991. The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development 112(4):997–1008. [DOI] [PubMed] [Google Scholar]
- Spicer GS, Bell CD.. 2002. Molecular phylogeny of the Drosophila virilis species group (Diptera: Drosophilidae) inferred from mitochondrial 12S and 16S ribosomal RNA genes. Ann Entomol Soc Am. 95(2):156–161. [Google Scholar]
- Stewart BS, Ahmed-Braimah YH, Cerne DG, McAllister BF.. 2019. Female meiotic drive preferentially segregates derived metacentric chromosomes in Drosophila Unpublished data, Biorxiv. doi: 10.1101/638684. Accessed August 12, 2019. [DOI]
- Throckmorton LH. 1982. The Genetics and Biology of Drosophila. Vol. 3b New York: Academic Press. [Google Scholar]
- Walsh JB. 1987. Persistence of tandem arrays: implications for satellite and simple-sequence DNAs. Genetics 115(3):553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KH-C, Grenier JK, Barbash DA, Clark AG.. 2014. Correlated variation and population differentiation in satellite DNA abundance among lines of Drosophila melanogaster. Proc Natl Acad Sci U S A. 111(52):18793–18798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KH-C, Lower SE, Caldas IV, Sless TJS, Barbash DA, Clark AG.. 2018. Variable rates of simple satellite gains across the Drosophila phylogeny. Mol Biol Evol. 35(4):925–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarosh W, Spradling AC.. 2014. Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev. 28:1840–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelentsova ES, Vashakidze RP, Krayev AS, Evgen'ev MB.. 1986. Dispersed repeats in Drosophila virilis: elements mobilized by interspecific hybridization. Chromosoma 93(6):469–476. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.