Abstract
Background
Identification of HER2-positive breast cancers with high anti-HER2 sensitivity could help de-escalate chemotherapy. Here, we tested a clinically applicable RNA-based assay that combines ERBB2 and the HER2-enriched (HER2-E) intrinsic subtype in HER2-positive disease treated with dual HER2-blockade without chemotherapy.
Methods
A research-based PAM50 assay was applied in 422 HER2-positive tumors from five II–III clinical trials (SOLTI-PAMELA, TBCRC023, TBCRC006, PER-ELISA, EGF104090). In SOLTI-PAMELA, TBCRC023, TBCRC006, and PER-ELISA, all patients had early disease and were treated with neoadjuvant lapatinib or pertuzumab plus trastuzumab for 12–24 weeks. Primary outcome was pathological complete response (pCR). In EGF104900, 296 women with advanced disease were randomized to receive either lapatinib alone or lapatinib plus trastuzumab. Progression-free survival (PFS), overall response rate (ORR), and overall survival (OS) were evaluated.
Results
A total of 305 patients with early and 117 patients with advanced HER2-positive disease were analyzed. In early disease, HER2-E represented 83.8% and 44.7% of ERBB2-high and ERBB2-low tumors, respectively. Following lapatinib and trastuzumab, the HER2-E and ERBB2 (HER2-E/ERBB2)-high group showed a higher pCR rate compared to the rest (44.5%, 95% confidence interval [CI] = 35.4% to 53.9% vs 11.6%, 95% CI = 6.9% to 18.0%; adjusted odds ratio [OR] = 6.05, 95% CI = 3.10 to 11.80, P < .001). Similar findings were observed with neoadjuvant trastuzumab and pertuzumab (pCR rate of 66.7% in HER2-E/ERBB2-high, 95% CI = 22.3% to 95.7% vs 14.7% in others, 95% CI = 4.9% to 31.1%; adjusted OR = 11.60, 95% CI = 1.66 to 81.10, P = .01). In the advanced setting, the HER2-E/ERBB2-high group was independently associated with longer PFS (hazard ratio [HR] = 0.52, 95% CI = 0.35 to 0.79, P < .001); higher ORR (16.3%, 95% CI = 8.9% to 26.2% vs 3.7%, 95% CI = 0.8% to 10.3%, P = .02); and longer OS (HR = 0.66, 95% CI = 0.44 to 0.97, P = .01).
Conclusions
Combining HER2-E subtype and ERBB2 mRNA into a single assay identifies tumors with high responsiveness to HER2-targeted therapy. This biomarker could help de-escalate chemotherapy in approximately 40% of patients with HER2-positive breast cancer.
In patients with primary or metastatic HER2-positive breast cancer, dual HER2-blockade without chemotherapy has shown high activity in a subgroup of patients (1–9). In HER2-positive disease, three neoadjuvant studies [TBCRC006 (3), TBCRC023 (4), and SOLTI-PAMELA (5)] with a total of 314 patients with early disease reported pathological complete response (pCR) rates in the breast of approximately 30% following neoadjuvant trastuzumab and lapatinib. These results suggest that a subgroup of patients with early-stage HER2-positive tumors might achieve similar outcomes if treated with less or no chemotherapy. However, beyond hormone receptor (HR) status, no predictive biomarker of response to dual HER2-blockade has shown clinical utility, to date.
HER2-positive disease is biologically heterogeneous when assessed by gene expression profiling and can be divided into four main intrinsic molecular subtypes (Luminal A, Luminal B, HER2-enriched [HER2-E], and basal-like) (10–14). Among them, the HER2-E subtype is characterized by higher expression of ERBB2, ERBB2-amplicon genes (eg, GRB7), and receptor tyrosine kinases including FGFR4 and EGFR, and lower expression of luminal-related genes compared to the luminal subtypes. Thus, HER2-E tumors are likely to have the highest activation of the EGFR and/or HER2 pathway and the ones to benefit the most from dual HER2 blockade.
The primary results from SOLTI-PAMELA revealed that the HER2-E subtype within HER2-positive disease was associated with a higher likelihood of achieving a pCR following neoadjuvant trastuzumab and lapatinib (5). Interestingly, HR status was not found to be statistically significantly associated with pCR once intrinsic subtype and other clinical-pathological variables were considered in multivariable analysis. Based on SOLTI-PAMELA trial primary results, four questions needed to be addressed: Can this finding be recapitulated in an independent cohort? Can HER2-E subtype predict anti-HER2 sensitivity beyond single ERBB2 mRNA expression? Can these findings be applied to treatment with trastuzumab and pertuzumab without chemotherapy? Can identification of anti-HER2 sensitivity predict survival outcome following dual HER2-blockade without chemotherapy?
Methods
Study Designs and Participants
Baseline tumor samples from four HER2-positive neoadjuvant studies [TBCRC006 (3), TBCRC023 (4), SOLTI-PAMELA (5), and PER-ELISA (6)] were evaluated. The main inclusion criteria of the four studies have been previously reported (3–6).
Briefly, in TBCRC006 and TBCRC023 (TBCRC006/023) and SOLTI-PAMELA, all patients received lapatinib and trastuzumab. Patients with HR-positive tumors received letrozole with or without a luteinizing hormone-releasing hormone agonist according to menopausal status. In SOLTI-PAMELA, premenopausal patients received tamoxifen.
TBCRC006 was a single-arm phase II study of 66 patients with stage II–III HER2-positive disease. Patients received 12 weeks of neoadjuvant treatment. TBCRC023 was a randomized, open-label phase II study of 128 patients with stage II–III HER2-positive disease. Patients were randomized to receive 12 vs 24 weeks of neoadjuvant treatment. Finally, the SOLTI-PAMELA was a single-arm phase II neoadjuvant study of 151 patients with stage I–III HER2-positive disease (5) treated for 18 weeks.
PER-ELISA was a single-arm phase II study of 64 patients with stage I–III HER2-positive and HR-positive disease (6). After diagnostic core-biopsy including baseline Ki67 evaluation, the patients started letrozole for 2 weeks followed by a core-biopsy for Ki67 central evaluation. Patients defined as molecular responders (Ki67 relative reduction >20% from baseline) started therapy with the combination of letrozole, trastuzumab, and pertuzumab. Trastuzumab and pertuzumab were administered for five courses, and letrozole was continued until surgery. Patients defined as molecular nonresponders discontinued letrozole and received weekly paclitaxel combined with pertuzumab and trastuzumab. In this study, we only selected molecular responders.
EGF104900 was a randomized phase III clinical trial (2, 9) of 296 women with HER2-positive advanced disease, who experienced progression on prior trastuzumab-containing regimens, to receive either lapatinib alone or lapatinib and trastuzumab. The primary endpoint was progression-free survival (PFS). Secondary endpoints included overall response rate (ORR) and overall survival (OS).
The studies were undertaken in accordance with Good Clinical Practice guidelines and the World Medical Association Declaration of Helsinki. All patients provided written informed consent. Approvals for the studies were obtained from independent ethics committees. This study is reported according to REMARK recommendations (15).
Tumor Samples and Gene Expression
Methods for RNA extraction, quantification, quality assessment, and gene expression analysis can be found in Supplementary Methods (available online).
Outcomes
For all neoadjuvant studies, except PER-ELISA, the primary endpoint was pCR in the breast defined as the absence of invasive neoplastic cells at microscopic examination of the primary tumor at surgery (ypT0/Tis). The primary endpoint in PER-ELISA was pCR in the breast and axilla. In EGF104900, the primary endpoint was PFS defined as the time from randomization until objective tumor progression or death. The secondary endpoints included ORR (confirmed complete response plus partial response) and OS (time from randomization until death because of any cause).
Statistical Analysis
To compare distribution of variables between two groups, we used the Fisher exact test. Proportions and 95% confidence interval (CI) were also provided. Univariate and multivariable logistic regression analyses were performed to investigate the association of each variable with pCR. Odds ratios (ORs) and 95% CIs were calculated for each variable. Monte Carlo cross-validation (16) was conducted (see Supplementary Methods, available online). Univariate and multivariable Cox proportional hazard regression analyses were performed to investigate the association of each variable with PFS or OS. The proportionality assumption was tested through evaluation of Schoenfeld residuals, which indicated a modest association with time (P= .046). The PFS model was fit with an additional time-dependent stratification variable to account for change in proportional hazards that occurs after 3 months of follow-up. Hazard ratio estimates from the early stratum of the model were in close agreement with both the unstratified model and a model where outcomes were censored at 3 months. The statistical significance level was set to a two-sided α of 0.05. All statistical tests were two-sided.
Results
Clinical-Pathological Characteristics of the Lapatinib and Trastuzumab Neoadjuvant Studies
Baseline tumor samples from a total of 265 of 314 patients (84.4%) were gene expression profiled (Supplementary Figure 1, available online). No major statistically significant clinical-pathological differences were observed between the TBCRC006/023 and SOLTI-PAMELA cohorts (Table 1). Overall, most patients had tumor stage II (138 [52.1%]), clinically node-negative (160 [60.4%]), and HR-positive (147 [55.5%]) disease and were postmenopausal (155 [58.5%]). A pCR in the breast was noted in 70 of 265 women (26.4%, 95% CI = 21.2 to 32.2).
Table 1.
Patient demographics of the neoadjuvant cohorts at baseline*
| Variable | TBCRC 006, | TBCRC 023, | SOLTI-PAMELA, | All, |
|---|---|---|---|---|
| No. (%) | No. (%) | No. (%) | No. (%) | |
| No. | 29 | 85 | 151 | 265 |
| Age, y, mean (SD) | 52.4 (10.0) | 52.5 (11.2) | 54.8 (12.9) | 53.8 (12.2) |
| Menopausal status | ||||
| Premenopausal | 12 (41.4) | 37 (43.5) | 61 (40.4) | 110 (43.0) |
| Postmenopausal | 17 (58.6) | 48 (56.5) | 90 (59.6) | 155 (57.0) |
| Tumor stage | ||||
| T1 | 1 (3.4) | 2 (2.4) | 60 (39.7) | 63 (23.8) |
| T2 | 10 (34.5) | 49 (57.6) | 79 (52.3) | 138 (52.1) |
| T3–T4 | 18 (62.1) | 34 (40.0) | 12 (7.9) | 64 (24.2) |
| Nodal status | ||||
| Negative | 10 (34.5) | 52 (61.2) | 98 (64.9) | 160 (60.4) |
| Positive | 17 (58.6) | 31 (36.5) | 53 (35.1) | 101 (38.1) |
| Missing | 2 (6.9) | 2 (2.4) | NA | 4 (1.5) |
| Treatment duration | ||||
| 12 weeks | 29 (100.0) | 31 (36.5) | NA | 60 (22.6) |
| 18 weeks | NA | NA | 151 (100.0) | 151 (57.0) |
| 24 weeks | NA | 54 (63.5) | NA | 54 (20.4) |
| Hormone receptor | ||||
| Positive | 19 (65.5) | 51 (60.0) | 77 (51.01) | 147 (55.5) |
| Negative | 10 (34.5) | 34 (40.0) | 74 (49.0) | 118 (44.5) |
| PAM50 | ||||
| Luminal A | NA | 4 (4.7) | 22 (14.6) | 26 (9.8) |
| Luminal B | 4 (13.8) | 7 (8.2) | 16 (10.6) | 27 (10.2) |
| HER2-E | 22 (75.8) | 51 (60.0) | 101 (66.9) | 174 (65.6) |
| Basal-like | 1 (3.4) | 11 (12.9) | 9 (5.9) | 21 (7.9) |
| Normal-like | 2 (6.9) | 12 (14.1) | 3 (2.0) | 17 (6.4) |
| pCR rate in the breast | 7 (24.1) | 17 (20.0) | 46 (30.5) | 70 (26.4) |
pCR = pathological complete response; NA = not applicable.
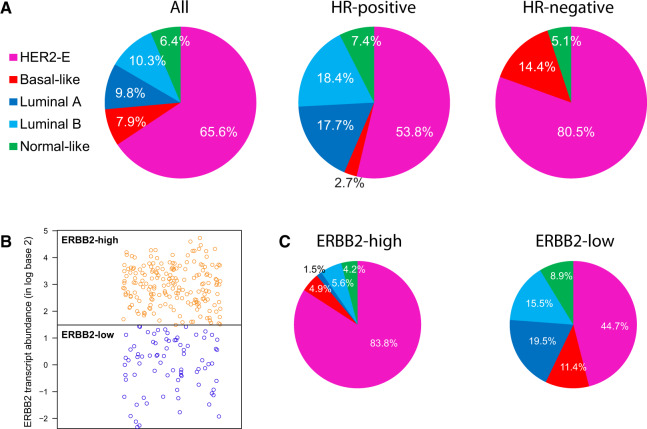
According to intrinsic molecular subtyping, most tumors were identified as HER2-E (174 [65.7%]), followed by Luminal B (27 [10.2%]), Luminal A (26 [9.8%]), basal-like (21 [7.9%]), and normal-like (17 [6.4%]) (Figure 1A). No statistically significant difference in the percentage of HER2-E tumors was observed between the TBCRC006/023 and SOLTI-PAMELA cohorts (P = .695). As expected, Luminal A and B subtypes were only identified in HR-positive disease, and most basal-like tumors were identified in HR-negative disease (P < .001). However, HER2-E was identified in both groups in a substantial proportion of patients (54.0% and 81.0% in HR-positive and HR-negative disease, respectively) (Figure 1A).
Figure 1.
ERBB2 vs intrinsic subtypes in the combined neoadjuvant HER2-positive dataset (n = 265). A) Distribution of the intrinsic subtypes in all patients and based on hormone receptor status. B) Distribution of tumor samples based on ERBB2 mRNA levels. C) Distribution of HER2-E subtype within ERBB2-high and ERBB2-low groups. HER2-E= HER2-enriched; HR + hormone receptor.
HER2-Enriched Association with Response in TBCRC006/023 Cohort
Twenty of 73 patients with the HER2-E subtype (27.4%, 95% CI = 17.6% to 39.1%) and 4 of 41 patients with non-HER2-E subtypes (9.8%, 95% CI = 2.7% to 23.1%) achieved a pCR at the time of surgery (OR = 3.49, 95% CI = 1.10 to 11.05, P = .03) (data not shown). No other clinical-pathological variable was statistically significantly associated with pCR in the univariate or multivariable model (data not shown), although the study was not powered for this purpose.
ERBB2 mRNA Association with Response in the SOLTI-PAMELA Cohort
A large range of ERBB2 mRNA expression was observed (ie, 14-fold change between the lowest and highest expressing tumor). The pCR rates of ERBB2 expression across tertiles (T) in SOLTI-PAMELA (ie, T1–T3) were 8.2% (T1), 23.5% (T2), and 58.8% (T3) (P < .001). The OR of the T2–T3 group for achieving a pCR was 7.88 (95% CI = 2.63–23.56, P < .001) compared to the T1 group. The OR of the T3 group for achieving a pCR was 4.72 (95% CI = 2.69 to 8.42, P < .001) compared to the T1–T2 group.
The ERBB2 expression cutoff to define T1 (ERBB2-low) vs T2-T3 (ERBB2-high) in SOLTI-PAMELA was chosen based on its higher OR compared to the T3 vs T1–T2 comparison (ie, OR = 7.88, 95% CI = 2.63 to 23.56 vs OR = 4.72, 95% CI = 2.69 to 8.42). To characterize the stability and expected performance of our threshold-defining rule, we performed a Monte Carlo cross-validation (16) (Supplementary Methods, available online). Results of cross-validation demonstrated the stability of the cutoff (interquartile range [IQR] = 0.18) relative to the dynamic range of ERBB2 expression (IQR = 2.22) (Supplementary Figure 2A, available online). The final cutoff determined from the complete cohort and used throughout the article was very close to the mean of all the selected cutoffs during cross-validation (difference = 0.02). Performance estimates of the chosen ERBB2 cutoff resulted in 92% sensitivity, 45% specificity, 42% positive predictive value, and 93% negative predictive value (Supplementary Figure 2B, available online).
ERBB2 mRNA Association with Response in the TBCRC006/023 Cohort
The derived cutoff value was applied in 114 tumor samples from the TBCRC006/023 cohort blinded from clinical data. The pCR rates in T1 and T2-T3 were 8.3% (95% CI = 2.3% to 20.0%) and 30.3% (95% CI = 19.6% to 42.9%, P < .001). The OR of the T2–T3 group for achieving a pCR was 4.78 (95% CI = 1.51 to 15.11, P = .008) compared to the T1 group (data not shown).
Analyses in the Lapatinib and Trastuzumab Combined Neoadjuvant Cohort
HER2-E subtype was found statistically significantly associated with pCR compared to non-HER2-E (35.1% vs 9.9%; OR = 4.92, 95% CI = 2.31 to 10.50, P < .001) (data not shown). At the same time, the ERBB2-high group (defined as ERBB2 expression above T1 [percentile 33rd] in SOLTI-PAMELA) was also found to be statistically significantly associated with pCR compared to the ERBB2-low group (36.1% vs 8.2%; OR = 6.51, 95% CI = 2.96 to 14.31, P < .001).
Like previous studies, a statistically significant higher ERBB2 mRNA expression was found in HER2-E disease compared to non-HER2-E disease. However, a large range of ERBB2 expression was observed within HER2-E and non-HER2-E subtypes, and overlap between the two groups was also evident (Figure 1B). Indeed, the proportion of HER2-E disease within ERBB2-high and ERBB2-low groups was 83.8% and 44.7%, respectively (P < .001) (Figure 1C). Thus, although there was a clear relationship between HER2-E and ERBB2 levels, the discordance rate at the individual level was 21.1% (56 of 265).
Previous data have suggested that HER2-E subtype and ERBB2 levels should not be considered the same and that a combination of both allows better identification of anti-HER2 sensitivity than either one alone. To test it, we evaluated the pCR rates when samples are classified by subtype (ie, HER2-E vs non-HER2-E) and ERBB2 levels (ie, ERBB2-low and ERBB2-high). The HER2-E and ERBB2 (HER2-E/ERBB2)-high group represented 44.9% of all samples. According to this classification, HER2-E/ERBB2-high tumors showed the highest pCR rate (44.5%, 95% CI = 35.4% to 53.9%), followed by non-HER2-E/ERBB2-high (16.1%, 95% CI = 5.3% to 34.1%), HER2-E/ERBB2-low (10.8%, 95% CI = 3.2% to 24.9%), and non-HER2-E/ERBB2-low (6.7%, 95% CI = 2.3% to 14.8%) groups (Supplementary Figure 3, available online). The HER2-E/ERBB2-high group showed a higher pCR rate compared to the rest (44.5%, 95% CI = 35.4% to 53.9% vs 11.6%, 95% CI = 6.9% to 18.0%). The OR of the HER2-E/ERBB2-high group for achieving a pCR was 6.09 (95% CI = 3.27 to 11.35, P < .001) compared to the rest (Table 2). Finally, a multivariable analysis, including type of study, confirmed the independent association of the HER2-E/ERBB2-high group with pCR (adjusted OR = 6.05, 95% CI = 3.10 to 11.80, P < .001) (Supplementary Figure 4, available online).
Table 2.
Logistic regression model analyses of treatment pathological response in the combined neoadjuvant cohort
| Variable | No. | pCR, % | Univariate |
Multivariable |
||
|---|---|---|---|---|---|---|
| OR (95% CI) | P * | OR (95% CI) | P * | |||
| Trial | ||||||
| TBCRC 006 | 29 | 24.1 | 1.00 (Reference) | — | 1.00 (Reference) | — |
| TBCRC 023 | 85 | 20.0 | 0.78 (0.29 to 2.14) | .263 | 0.61 (0.19 to 2.03) | .385 |
| SOLTI-PAMELA | 151 | 30.5 | 1.37 (0.55 to 3.45) | .157 | 0.78 (0.24 to 2.51) | .973 |
| Tumor stage | ||||||
| T1–T2 | 201 | 30.3 | 1.00 (Reference) | — | 1.00 (Reference) | — |
| T3–T4 | 64 | 14.1 | 0.38 (0.17 to 0.81) | .012 | 0.32 (0.12 to 0.83) | .019 |
| Menopausal status | ||||||
| Premenopausal | 110 | 29.1 | 1.00 (Reference) | — | 1.00 (Reference) | — |
| Postmenopausal | 155 | 24.5 | 0.79 (0.46 to 1.37) | .406 | 0.73 (0.39 to 1.36) | .32 |
| Nodal status | ||||||
| Negative | 160 | 30.6 | 1.00 (Reference) | — | 1.00 (Reference) | — |
| Positive | 101 | 19.8 | 0.56 (0.31 to 1.01) | .055 | 0.55 (0.28 to 1.08) | .081 |
| Treatment duration | ||||||
| 12 weeks | 60 | 18.3 | 1.00 (Reference) | — | — | |
| 18 weeks | 151 | 30.5 | 1.95 (0.93 to 4.09) | .088 | — | — |
| 24 weeks | 54 | 24.1 | 1.41 (0.57 to 3.49) | .976 | — | — |
| Hormone receptor | — | |||||
| Positive | 147 | 19.0 | 1.00 (Reference) | — | 1.00 (Reference) | — |
| Negative | 118 | 35.6 | 2.35 (1.34 to 4.10) | .003 | 1.58 (0.83 to 2.99) | .1649 |
| PAM50+ERBB2 | ||||||
| Others | 146 | 11.6 | 1.00 (Reference) | — | 1.00 (Reference) | — |
| HER2-E/ERBB2- high | 119 | 44.5 | 6.09 (3.27 to 11.35) | <.001 | 6.05 (3.10 to 11.80) | <.001 |
Univariate and multivariable logistic regression analyses were done to investigate the association of each variable with pathological complete response (pCR). Odds ratios (ORs) and 95% confidence interval (CIs) were calculated for each variable. The statistical significance level was set to a two-sided α of 0.05. HER2-E = HER2-enriched.
Independent Validation of the HER2-E/ERBB2-High Biomarker in per-ELISA
To further test the ability of the combined biomarker to predict pCR, 40 of 44 (91.0%) HER2-positive and HR-positive tumor samples treated with pertuzumab, trastuzumab, and letrozole in the PER-ELISA trial (17) were profiled blinded from clinical data using the same exact assay and ERBB2 cut point (Supplementary Table 1, available online). The proportion of HER2-E disease within the ERBB2-high and ERBB2-low groups was 46.2% (6 of 13) and 18.5% (5 of 27), respectively. The discordance rate at the individual level was 30.0% (12 of 40). A total of 6 (15.0%) and 34 (85.0%) samples were HER2-E/ERBB2-high and others, respectively. The pCR rate of HER2-E/ERBB2-high was 66.7% (95% CI = 22.3% to 95.7%, 4 of 6), and the pCR rate of the others group was 14.7% (95% CI = 4.9% to 31.1%, 5 of 34). The OR of the HER2-E/ERBB2-high group for achieving a pCR was 11.60 (95% CI = 1.66 to 81.10, P = .01) compared to the other groups. No other clinical-pathological variable was statistically significantly associated with pCR, although the study was not powered for this purpose.
Independent Validation of the HER2-E/ERBB2-High Biomarker in EGF104900
To test the ability of the combined biomarker to predict survival outcome, 117 (39.5%) of tumor samples from the EGF104900 trial were profiled blinded from clinical data using the same exact assay and ERBB2 cut point as in the previous studies (Supplementary Figure 5, available online). Subtype distribution was 58.2% HER2-E, 15.3% basal-like, 10.2% Luminal B, 2.3% Luminal A, and 14.1% normal-like. Within ERBB2-high and ERBB2-low, 70.8% and 31.0% of tumors were HER2-E, respectively. The PAM50 population retained similar clinical-pathological features and survival outcomes as the original trial population (Supplementary Table 2, available online).
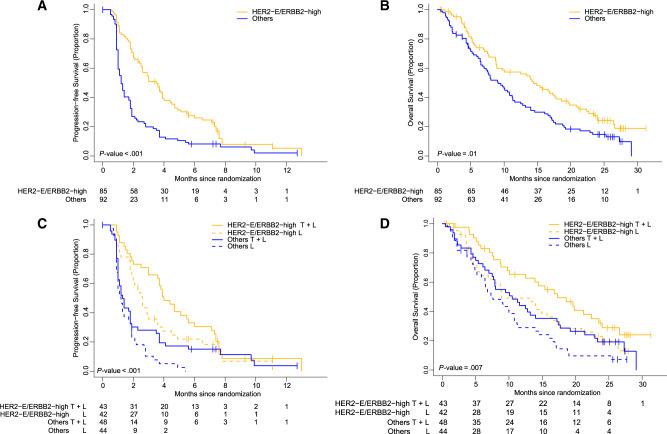
The HER2-E/ERBB2-high group represented 48.0% of all samples. The adjusted PFS hazard ratio of the HER2-E/ERBB2-high group vs others was 0.52 (95% CI = 0.35 to 0.79, P < .001). Median PFS of the HER2-E/ERBB2-high group was 3.5 months (95% CI = 2.6 to 5.4 months) compared to 1.2 months (95% CI = 1.0 to 1.7 months) in others (Figure 2, A and C). The ORR of the HER2-E/ERBB2-high group was statistically significantly higher compared to others (ORR = 16.3%, 95% CI = 8.9% to 26.2% vs 3.7%, 95% CI = 0.8% to 10.3%, P = .02). For OS, the adjusted hazard ratio for HER2-E/ERBB2-high vs others was 0.66 (95% CI = 0.44 to 0.97, P = .01). Median OS was higher in the HER2-E/ERBB2-high group (14.4 months; 95% CI = 8.9 to 17.8 months) compared to others (9.1 months; 95% CI = 7 to 11.3 months) (Figure 2, B and D). Finally, the study was not powered to detect an interaction between HER2-E/ERBB2-high and the treatment arm in relation to PFS or OS, and a statistically significant interaction effect was not observed.
Figure 2.
HER2-E/ERBB2-high biomarker in the EGF104900 trial of advanced HER2-positive breast cancer. A) Progression-free survival (PFS). B) Overall survival (OS). C) PFS according to treatment arm. D) OS according to treatment arm. Estimates of PFS and OS were from Kaplan–Meier curves and tests of differences by two-sided log-rank test. HER2-E = HER2-enriched; HER2-E/ERBB2-high = HER2-enriched and ERBB2-high group; T = trastuzumab; L = lapatinib.
Distribution of the HER2-E/ERBB2-High Biomarker in HER2-Positive Disease
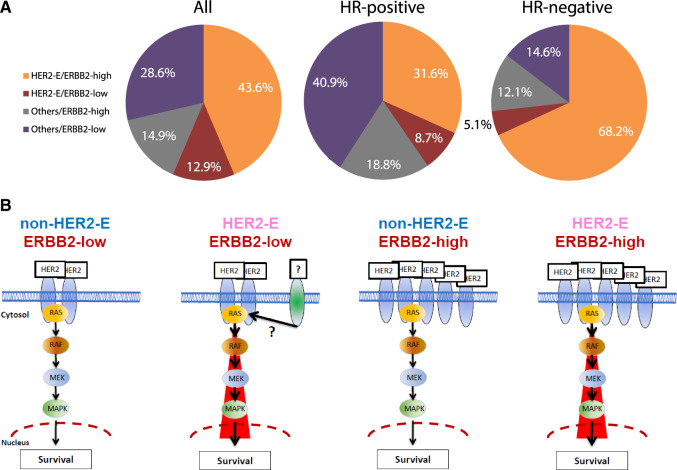
To evaluate the proportion of patients with HER2-E/ERBB2-high disease, we combined all the available data from the five trials, 305 patients with early and 117 patients with advanced HER2-positive disease (Figure 3A, Supplementary Figure 6, available online). Overall, HER2-E/ERBB2-high disease represented 43.6% of the patients with HER2-positive disease, being 68.1% in HR-negative disease and 31.7% in HR-positive disease.
Figure 3.
HER2-E/ERBB2-high biomarker in patients with HER2-positive breast cancer. A) Proportion of patients according to intrinsic subtype and ERBB2 levels. B) Schematic view of the four potential cell signaling scenarios (HER2 and its downstream signaling activation, such as the RAS/MAPK pathway) identified within HER2-positive breast cancer based on ERBB2 mRNA levels and intrinsic subtype (HER2-E vs non-HER2-E). In HER2-E/ERBB2-low disease, it is plausible that another transmembrane growth factor receptor with kinase activity such as FGFR4 is driving the HER2-E phenotype either alone or in combination with HER2. HER2-E = HER2-enriched; HER2-E/ERBB2 = HER2-E and ERBB2 group; HR + hormone receptor; MAPK = mitogen-activated protein kinase.
Discussion
In the SOLTI-PAMELA trial (5), the HER2-E subtype was associated with a higher probability of achieving a pCR than non-HER2-E disease following dual HER2-blockade without chemotherapy. Here, we validated this same observation in an independent dataset composed of 114 baseline tumor samples from two TBCRC trials (ie, TBCRC006 and 023) (3, 4). In addition, the combined analysis of the SOLTI-PAMELA and TBCRC studies allowed us to evaluate the predictive value of HER2-E subtype beyond ERBB2 mRNA levels. Furthermore, we tested the ability of the biomarker (HER2-E/ERBB2-high) to predict response and survival in the metastatic setting following anti-HER2 therapy without chemotherapy in EGF104900. Overall, HER2-E is a consistent biomarker of pathological response following lapatinib and trastuzumab, and both HER2-E and ERBB2 mRNA levels provide additional information from each other, and their combination into a single variable is better. To our knowledge, this is the first study to report that a combined score based on two different mRNA-based variables tracking the HER2 signaling pathway can identify high anti-HER2 sensitivity.
At first glance, the lack of high concordance between HER2-E subtype and ERBB2 levels might seem counterintuitive. However, approximately 35% of HER2-E tumors are HER2-negative by standard American Society of Clinical Oncology (ASCO) and College of American Pathologists (CAP) criteria (14). Thus, both variables provide different and complementary information. Although a HER2-E profile might indicate the level of activation of the HER2 and/or EGFR downstream signaling pathway, ERBB2 mRNA levels ultimately determine the amount of target present in the tumor cells. Thus, tumors that have high expression of the main drug target and high activation of the pathway (ie, ERBB2-high and HER2-E) are potentially the tumors most “HER2-addicted” and most sensitive to anti-HER2-targeted therapies (Figure 3B). Interestingly, tumors that meet only one criterion (ie, the drug target is highly expressed but the downstream signaling pathway is lowly activated, or vice versa) have an anti-HER2 sensitivity that is very similar to non-HER2-E/ERBB2-low disease. This could help explain the variable response to HER2-targeted therapy in HER2-positive tumors defined by the ASCO/CAP criteria (14). Hence, further studies to elucidate the driver(s) of the HER2-E subtype within HER2-negative disease (18–20) and the non-HER2-E subtype within HER2-positive disease could, in the future, better refine therapeutic options in breast cancer beyond current methods.
The ultimate goal of TBCRC006/023 (3, 4), PER-ELISA (6), and SOLTI-PAMELA (5) studies is the promised land of nonchemotherapy in early HER2-positive disease. The combination of anti-HER2 doublets with optimal chemotherapy regimens is providing pCR rates in the range of 60%, increasing up to 75% in HER2-positive and HR-negative tumors (21–25). The approximate 45% pCR rate in our study for the HER2-E/ERBB2-high group is still far from this result. However, weekly paclitaxel plus trastuzumab, a well-accepted regimen for patients with stage I HER2-positive disease (26), has provided pCR rates ranging from 29% to 46% (7, 10, 27). Our result suggests that a statistically significant fraction of patients with HER2-positive tumors may not need chemotherapy at all. If these patients could be identified before the start of treatment, optimal response could potentially be achieved with HER2-targeted therapy alone. Although dual anti-HER2 blockage without chemotherapy is still an exploratory approach, the molecular HER2-E profile, together with ERBB2 mRNA levels, may be a helpful tool to guide patients with sensitive tumors to nonchemotherapy or less chemotherapy. Future, well-designed prospective clinical trials should help establish the clinical utility of the combined biomarker in specific scenarios and with particular cut points in both the early and the advanced settings.
Our study has several limitations. First, the clinical cohorts in this study were powered for heterogeneous primary endpoints, which have been evaluated in primary publications. The analysis presented here used all available subjects from these studies, but the lack of formal design through pre-planned analysis prohibits inference of negative results. Instead, we focus our tests on variables with high precedent in these cohorts, namely, ERBB2 expression and HER2-E subtype. Additionally, we restrict our inference to statistically significant results, which are repeatably and independently assessed across all cohorts in the study. Second, the HER2-E subtype can be identified to date using the PAM50 standardized nCounter-based assay from tumor samples. Regarding the ERBB2 gene, which is included in the PAM50 assay, we have proposed a particular cut point to define high vs low based on tertile expression. Although the nCounter platform allows the clinical implementation of highly reproducible assays (28, 29), further analytical validation is needed. Third, we assessed clinical efficacy in the early-stage setting based on pCR rates instead of survival. In addition, our data are mostly based on the combination of trastuzumab plus lapatinib, and our findings will require confirmation in additional studies that test the combination of trastuzumab plus pertuzumab. However, a retrospective ERBB2 qRT-PCR mRNA-based analysis of 102 baseline tumor samples from the chemotherapy-free arm of the NeoSphere neoadjuvant HER2-positive trial with trastuzumab and pertuzumab showed pCR rates in ERBB2-high and ERBB2-low groups (using median expression as the cutoff) of 23.4% and 10.9%, respectively (30). Fourth, other promising molecular biomarkers have not been researched in this study. Finally, several studies have suggested that constitutive activation of the PI3K and AKT pathway is associated with anti-HER2 resistance (31–33).
Our study establishes the potential clinical validity of the HER2-E subtype and ERBB2 mRNA levels to predict anti-HER2 sensitivity. Our findings should now be externally validated, and research groups from the National Clinical Trial Network in the United States are now actively discussing the adoption of standardized and analytically validated assays to allow the prospective testing of this approach as an enrichment strategy for the primary endpoint in prospective trials for early-stage HER2-positive disease.
Funding
This work was supported by GlaxoSmithKline (to SOLTI), Instituto de Salud Carlos III - PI16/00904 (to AP), Banco Bilbao Vizcaya Argentaria Foundation (to AP), Pas a Pas (to AP), Save the Mama (to AP), Breast Cancer Research Foundation (to AP), Career Catalyst Grant CCR13261208 from the Susan Komen Foundation (to AP), Fundación Científica Asociación Española Contra el Cáncer (Ayuda Postdoctoral AECC 2017; to FB-M), NIH: SPORE Grants P50 CA058183 and CA186784 (to RS, CKO, and MFR), Cancer Center Grant P30 CA125123 (to CKO and BCM), and the Department of Defense grants W81XWH-17–1-0579 (to MFR) and W81XWH-17–1-0580 (to RS).
Notes
Participated in the design and/or interpretation of the reported experiments or results: AP, TP, CdA, CG, AL-C, TW, JC, LP, PN, FC, RS, VG, CKO, MFR. Participated in the acquisition and/or analysis of data: all authors. Participated in drafting and/or revising the manuscript: all authors. Provided administrative, technical, or supervisory support: AP, TP, JC, LP, PG, JV, RS, CHO, MFR.
AP reports consulting fees from Nanostring Technologies, outside the submitted work. TP reports consulting fees from Roche/Genentech. JC reports personal fees from Roche, Novartis, and Eisai and consulting fees from Roche/Genentech, Celgene, Astra Zeneca, Biothera Pharmaceutical, Merus, and Seattle Genetics, outside the submitted work. ALC reports personal fees from Novartis and Roche/Genentech, outside the submitted work. MFR reports personal fees from GlaxoSmithKline and consulting fees from Roche/Genentech, Macrogenics, Novartis, and Daiichi Sankyo, outside the submitted work. MO reports consulting fees from Roche/Genentech and GlaxoSmithKline, outside the submitted work. VG reports research funding from Roche/Genentech. PC reports consulting fees from Roche/Genentech. JP is a co-inventor on patents related to the PAM50, licensed to Nanostring Technologies, Inc. RS reports personal fees from Macrogenics, consulting to Eli Lily, and research funding (to institute) form GlaxoSmithKline, Eli Lily, Gilead, and AstraZeneca, outside the submitted work.
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication. We thank all the patients and their family members for participating in the three studies. We would also like to show our gratitude to Guillem Clot for sharing his knowledge in predictive models.
Supplementary Material
References
- 1. Swain S, Baselga J, Kim S-B, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372(8):724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blackwell KL, Burstein HJ, Storniolo AM, et al. Overall survival benefit with lapatinib in combination with trastuzumab for patients with human epidermal growth factor receptor 2–positive metastatic breast cancer: final results from the EGF104900 study. J Clinc Oncol. 2012;30(21):2585–2592. [DOI] [PubMed] [Google Scholar]
- 3. Rimawi MF, Mayer IA, Forero A, et al. Multicenter phase II study of neoadjuvant lapatinib and trastuzumab with hormonal therapy and without chemotherapy in patients with human epidermal growth factor receptor 2–overexpressing breast cancer: TBCRC 006. J Clinc Oncol. 2013;31(14):1726–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rimawi MF, Niravath PA, Wang T, et al. Abstract S6-02: TBCRC023: a randomized multicenter phase II neoadjuvant trial of lapatinib plus trastuzumab, with endocrine therapy and without chemotherapy, for 12 vs. 24 weeks in patients with HER2 overexpressing breast cancer. Cancer Res. 2015;75(suppl 9):S6-02-S6-02. [DOI] [PubMed] [Google Scholar]
- 5. Llombart-Cussac A, Cortés J, Paré L, et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017;18(4):545–554. [DOI] [PubMed] [Google Scholar]
- 6. Guarneri V, Dieci M, Bisagni G, et al. De-escalated therapy for HR+/HER2+ breast cancer patients with Ki67 response after 2 weeks letrozole: results of the PerELISA neoadjuvant study. Annals of Oncology 2019. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baselga J, Bradbury I, Eidtmann H, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379(9816):633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. von Minckwitz G, Procter M, de Azambuja E, et al. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer. N Engl J Med. 2017;377(2):122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blackwell KL, Burstein HJ, Storniolo AM, et al. Randomized study of lapatinib alone or in combination with trastuzumab in women with ErbB2-positive, trastuzumab-refractory metastatic breast cancer. J Clin Oncol. 2010;28(7):1124–1130. [DOI] [PubMed] [Google Scholar]
- 10. Carey LA, Berry DA, Cirrincione CT, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol. 2016;34(6):542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prat A, Bianchini G, Thomas M, et al. Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2-positive breast cancer in the NOAH study. Am Assoc Cancer Res. 2014;20(2):511–521. [DOI] [PubMed] [Google Scholar]
- 12. Prat A, Perou CM.. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prat A, Pineda E, Adamo B, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(1):S26–S35. [DOI] [PubMed] [Google Scholar]
- 14. Prat A, Carey LA, Adamo B, et al. Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast cancer. J Natl Cancer Inst. 2014;106(8):dju152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97(16):1180–1184. [DOI] [PubMed] [Google Scholar]
- 16. Xu Q-S, Liang Y-Z.. Monte Carlo cross validation. Chemometr Intell Lab Syst. 2001;56(1):1–11. [Google Scholar]
- 17. Guarneri V, Dieci MV, Bisagni G, et al. De-escalated treatment with trastuzumab-pertuzumab-letrozole in patients with HR+/HER2+ operable breast cancer with Ki67 response after 2 weeks letrozole: final results of the PerELISA neoadjuvant study. J Clinc Oncol. 2018;36(suppl 15):507–507. [Google Scholar]
- 18. Prat A, Fan C, Fernández A, et al. Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy. BMC Med. 2015;13(1):303.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cejalvo JM, Pascual T, Fernandez MA, et al. Clinical implications of the non-luminal intrinsic subtypes in hormone receptor-positive breast cancer. Cancer Treat Rev. 2018;67(1):63–70. [DOI] [PubMed] [Google Scholar]
- 20. Prat A, Cheang MC, Galván P, et al. Prognostic value of intrinsic subtypes in hormone receptor–positive metastatic breast cancer treated with letrozole with or without lapatinib. JAMA Oncol. 2016;2(10):1287–1294. [DOI] [PubMed] [Google Scholar]
- 21. Schneeweiss A, Chia S, Hickish T, et al. Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: a randomized phase II cardiac safety study (TRYPHAENA). Ann Oncol. 2013;24(9):2278–2284. [DOI] [PubMed] [Google Scholar]
- 22. Robidoux A, Tang G, Rastogi P, et al. Lapatinib as a component of neoadjuvant therapy for HER2-positive operable breast cancer (NSABP protocol B-41): an open-label, randomised phase 3 trial. Lancet Oncol. 2013;14(12):1183–1192. [DOI] [PubMed] [Google Scholar]
- 23. Guarneri V, Frassoldati A, Bottini A, et al. Preoperative chemotherapy plus trastuzumab, lapatinib, or both in human epidermal growth factor receptor 2–positive operable breast cancer: Results of the Randomized Phase II CHER-LOB Study. J Clin Oncol 2012;30(16):1989-1995. [DOI] [PubMed] [Google Scholar]
- 24. Gavilá J, Oliveira M, Pascual T, et al. Safety, activity, and molecular heterogeneity following neoadjuvant non-pegylated liposomal doxorubicin, paclitaxel, trastuzumab, and pertuzumab in HER2-positive breast cancer (Opti-HER HEART): an open-label, single-group, multicenter, phase 2 trial. BMC Med. 2019;17(1):8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Swain S, Ewer M, Viale G, et al. Pertuzumab, trastuzumab, and standard anthracycline-and taxane-based chemotherapy for the neoadjuvant treatment of patients with HER2-positive localized breast cancer (BERENICE): a phase II, open-label, multicenter, multinational cardiac safety study. Ann Oncol. 2018;29(3):646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tolaney SM, Barry WT, Dang CT, et al. Adjuvant paclitaxel and trastuzumab for node-negative, HER2-positive breast cancer. N Engl J Med. 2015;372(2):134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gianni L, Pienkowski T, Im Y-H, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25–32. [DOI] [PubMed] [Google Scholar]
- 28. Nielsen T, Wallden B, Schaper C, et al. Analytical validation of the PAM50-based Prosigna breast cancer prognostic gene signature assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer. 2014;14(1):177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prat A, Galván P, Jimenez B, et al. Prediction of response to neoadjuvant chemotherapy using core needle biopsy samples with the prosigna assay. Clin Cancer Res. 2016;22(3):560–566. [DOI] [PubMed] [Google Scholar]
- 30. Bianchini G, Kiermaier A, Bianchi GV, et al. Biomarker analysis of the NeoSphere study: pertuzumab, trastuzumab, and docetaxel versus trastuzumab plus docetaxel, pertuzumab plus trastuzumab, or pertuzumab plus docetaxel for the neoadjuvant treatment of HER2-positive breast cancer. Breast Cancer Res. 2017;19(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Veeraraghavan J, De Angelis C, Reis-Filho JS, et al. De-escalation of treatment in HER2-positive breast cancer: determinants of response and mechanisms of resistance. Breast. 2017;34(suppl 1):S19–S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chandarlapaty S, Sakr RA, Giri D, et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res. 2012;18(24):6784–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.