Abstract
Objectives
To develop classification algorithms that accurately identify axial SpA (axSpA) patients in electronic health records, and compare the performance of algorithms incorporating free-text data against approaches using only International Classification of Diseases (ICD) codes.
Methods
An enriched cohort of 7853 eligible patients was created from electronic health records of two large hospitals using automated searches (⩾1 ICD codes combined with simple text searches). Key disease concepts from free-text data were extracted using NLP and combined with ICD codes to develop algorithms. We created both supervised regression-based algorithms—on a training set of 127 axSpA cases and 423 non-cases—and unsupervised algorithms to identify patients with high probability of having axSpA from the enriched cohort. Their performance was compared against classifications using ICD codes only.
Results
NLP extracted four disease concepts of high predictive value: ankylosing spondylitis, sacroiliitis, HLA-B27 and spondylitis. The unsupervised algorithm, incorporating both the NLP concept and ICD code for AS, identified the greatest number of patients. By setting the probability threshold to attain 80% positive predictive value, it identified 1509 axSpA patients (mean age 53 years, 71% male). Sensitivity was 0.78, specificity 0.94 and area under the curve 0.93. The two supervised algorithms performed similarly but identified fewer patients. All three outperformed traditional approaches using ICD codes alone (area under the curve 0.80–0.87).
Conclusion
Algorithms incorporating free-text data can accurately identify axSpA patients in electronic health records. Large cohorts identified using these novel methods offer exciting opportunities for future clinical research.
Keywords: classification, phenotyping, electronic health records, free-text, natural language processing, machine learning, axial spondyloarthritis, ankylosing spondylitis, ICD code
Rheumatology key messages
Algorithms incorporating free-text data improve the accuracy for classifying axial SpA patients in electronic health records.
A robust axial SpA algorithm could be developed without training on chart-reviewed cases.
Large cohorts can be efficiently identified for clinical research even with evolving disease definitions.
Introduction
A key step in any clinical research is identifying a group of people with a disease of interest. This can be labour-intensive, particularly for relatively uncommon conditions such as axial SpA (axSpA). Due to its low prevalence (0.7% in the USA [1]), the majority of axSpA studies use specialized registries or small prospective cohorts. Rare outcomes, or those not recorded by registries, often cannot be studied in sufficient detail. Electronic health records (EHR) are increasingly used worldwide and contain vast amounts of real-world data on millions of patients. They provide opportunities to create relatively large cohorts that can meet these research needs.
Accurately identifying, or classifying, axSpA patients from EHR can be challenging. A common approach is rule-based, for example requiring a certain number of International Classification of Disease (ICD) or other diagnostic codes [2, 3]. Performance of these methods depends on the codes’ accuracy, which can vary substantially across healthcare systems [4]. Applying more strict rules may improve accuracy among those classified as cases, but will reduce the number and representativeness of patients identified. Moreover, ICD codes may not be well defined for evolving disease concepts such as axSpA where there are no specific codes up to the 10th version.
For several chronic diseases including RA, classification algorithms have been improved by supplementing codified data with information from free-text, or ‘narrative’, EHR data (e.g. healthcare provider notes) using natural language processing (NLP) [5–9]. NLP tools have been developed and successfully applied to identify clinical concepts [10]. In contrast to a search for keywords, NLP can distinguish positive/negative mentions of concepts (e.g. between ‘sacroiliitis’ and ‘no sacroiliitis’) and each concept can include several ways of expressing meaning. NLP-assisted algorithms were more accurate, and identified more patients, than those using codified data alone [11]. They also permit phenotyping of diseases without ICD codes.
We aimed to develop novel algorithms incorporating both codified and narrative data to identify axSpA patients in EHR, and to compare them against traditional approaches using only ICD codes.
Methods
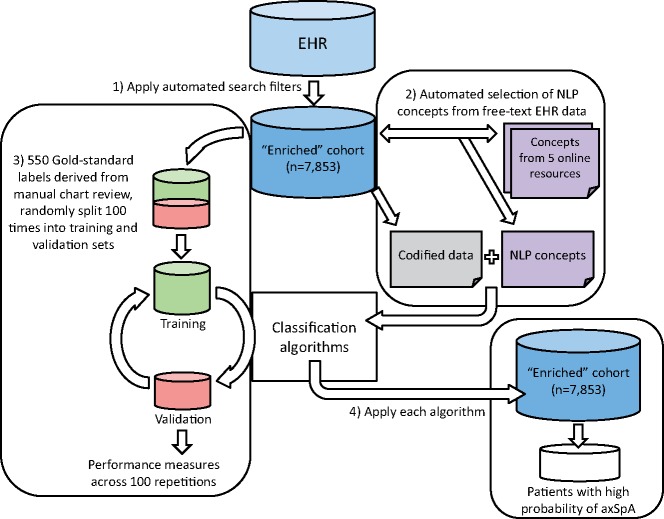
A flow-chart summary of the algorithm development process is shown in Fig. 1. Hereinafter, an algorithm is described as ‘supervised’ if it is trained using labelled data derived from manual chart review, and ‘unsupervised’ if it leverages inherent structures in the data without requiring labels.
Fig. 1.
Process of algorithm development for classifying axSpA patients in EHR
First, search filters were applied to create an enriched cohort of patients who may potentially have axSpA. Second, an automated process identified informative NLP disease concepts that, together with codified data, formed variables in the predictive models. Only the two supervised algorithms involved the third step: manual chart review of a random sample of patients to create gold-standard training labels. The unsupervised algorithm did not require labels for training. Finally, each of the three algorithms was applied to the enriched cohort to identify patients with high probability of having axSpA. axSpA: axial SpA; EHR: electronic health record; NLP, natural language processing.
Data source and enriched cohort
We used data from the Partners HealthCare EHR from two large tertiary care hospitals in Boston, USA. Both have been using EHR for approximately two decades, and through that time provided care for ∼7.4 million patients.
To develop the algorithms, we adapted a standardized phenotyping process using NLP and machine learning [12]. Including all patients in the EHR would substantially limit the accuracy of a classification algorithm, since positive predictive value (PPV) is dependent on prevalence. The process therefore starts by applying screening criteria to create an enriched cohort of patients who may potentially have axSpA, while excluding those with very low probability of the condition. We applied the following screening criteria: ⩾1 ICD-9 or -10 codes for ankylosing spondylitis (720.x or M45.x; collectively referred to as ‘ICD codes’ henceforth) or any mention of AS in the discharge or ambulatory notes. There are no ICD codes specifically for axSpA. Prior studies showed that ‘AS’ was used synonymously with axSpA in Partners’ EHR [13]. We also required patients to have ‘SI joint’, ‘syndesmophyte’ or words beginning with ‘sacroil-’ in clinical notes or radiology reports. Subjects under 18 years of age were excluded. Remaining individuals formed the enriched cohort, among whom algorithm development and evaluation were performed. This study was approved by the Partners Institute Review Board.
Codified data
The total number of ICD codes for AS ⩾7 days apart were counted for each patient. We also measured healthcare utilization as the number of medical encounters in each patient’s EHR, such as a physician visit or visit for an outpatient investigation.
Selecting informative disease concepts from narrative data
Healthcare professional use various terminologies and phrasing to express the same clinical meaning. For instance, ‘inflammation of the right sacroiliac joint’, ‘left sacroiliac joint arthritis’ and ‘bilateral sacroiliitis’ all refer to sacroiliitis. NLP can map these linguistic variations to the specific concept ‘sacroiliitis’ by linking to the Unified Medical Language System (UMLS) [14], while disregarding negative mentions such as ‘no evidence of sacroiliitis’ [15].
We followed the previously published Surrogate-Assisted Feature Extraction (SAFE) method to generate a list of candidate axSpA concepts to extract from narrative EHR notes [11]. The SAFE approach begins by identifying a list of potential concepts from online resources (e.g. MEDLINE and Medscape) that is equivalent to or better than lists curated by clinical domain experts for algorithm development [11]. We then processed free-text clinical notes using NLP to count the number of positive mentions of each axSpA concept for each patient. Healthcare provider notes, discharge summaries and radiology reports in typed format were used; scanned notes were not.
To select the most informative concepts from this list, least absolute shrinkage and selection operator (LASSO) penalized logistic regression was fitted to each of three surrogate labels against all candidate concepts repeatedly [11]. LASSO penalized regression performs selection by assigning the coefficients of non-informative concepts to zero. We constructed three surrogate labels using AS ICD code, AS NLP concept or a rule-based criterion with known high PPV [13] as proxies for the true axSpA label [11]. Concepts that were selected >50% of the time were retained and combined with codified data for subsequent supervised algorithm training. The SAFE method has been shown to reduce overfitting and improve model performance [11].
Gold-standard training labels
From the enriched cohort, we randomly selected 550 individuals whose records were reviewed by a rheumatologist. They were categorized into either (i) definite axSpA: meeting full or pragmatic versions [13] of the modified New York [16] or Assessment of Spondyloarthritis International Society (ASAS) classification criteria [17], or (ii) insufficient or no evidence from radiology reports or medical notes for axSpA classification criteria. This process has been previously described [13] and is shown in Supplementary Fig. 1, available at Rheumatology online. Algorithms were trained to identify cases of axSpA meeting research classification criteria.
Algorithm training and evaluation
We developed three algorithms using: (i) logistic regression including all variables selected by SAFE, which optimizes model fit at potential cost to overfitting, (ii) LASSO penalized logistic regression, which further reduces the number of variables in the model and optimizes external validity, and (iii) a multimodal automated phenotyping (MAP) approach [18]. The first two supervised algorithms were developed using the gold-standard labels. MAP is an unsupervised approach that classifies phenotypes in EHR data without requiring labels from manual chart review [18]. Instead, it is trained by combining information from three key variables (AS ICD code, AS NLP concept and healthcare utilization) using the entire enriched cohort.
Comparing to the chart-reviewed gold-standard, performance characteristics of each algorithm was reported using the area under the receiver operating characteristic curve, PPV, sensitivity, specificity and the F-score (harmonic mean of PPV and sensitivity). We chose 80% PPV as the threshold to allow consistent comparison across the algorithms, based on the maximum PPV in the majority of similar axSpA studies [2, 3, 19]. Cross-validation with 70: 30 splits averaged over 100 random partitions was used to correct for over-fitting bias in estimating accuracy measures.
The algorithms assigned each patient their probability of having axSpA; those with probabilities above a threshold that achieves 80% PPV were classified as having axSpA. Performance of the NLP-assisted algorithms was compared against classification using only ICD codes. A logistic regression model was created with the number of AS ICD codes as the only predictor. We also classified patients as having axSpA if they had ⩾2 or ⩾3 AS ICD codes. We used established phenotyping packages [12, 18] in R version 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria) for algorithm development, and Stata v14 (StataCorp, College Station, TX, USA) for all other analyses.
Results
A total of 7853 patients passed the initial screening criteria to form the enriched axSpA cohort. Of the 550 patients randomly sampled from this cohort, 127 (23%) were determined to have axSpA meeting classification criteria after manual chart review and 423 did not. Clinical characteristics and distributions of codified and narrative data are shown in Table 1. axSpA cases had higher counts of AS ICD codes and AS NLP concepts (P < 0.001). They were also more frequently male (83 vs 46%, P < 0.001); 263 patients had ⩾1 AS ICD codes, among whom 113 (43%) met classification criteria for axSpA.
Table 1.
Patient characteristics of the training set and distributions of their codified and narrative data
| axSpA cases | Controls | P-value | ||
|---|---|---|---|---|
| N | 127 | 423 | ||
| Age, years | 51.7 (18.4) | 55.4 (19.0) | 0.055 | |
| Male | 106 (83) | 196 (46) | <0.001 | |
| Race | White | 110 (87) | 356 (84) | 0.800 |
| African American | 12 (3) | 3 (2) | ||
| Other | 55 (13) | 14 (11) | ||
| HLA-B27 tested | 50 (39) | 89 (21) | <0.001 | |
| HLA-B27 positive | 40 (80) | 47 (53) | <0.001 | |
| Uveitis | 30 (24) | 23 (5) | <0.001 | |
| Psoriasis | 5 (4) | 38 (9) | 0.063 | |
| IBD | 10 (8) | 16 (4) | 0.057 | |
| Number of ICD codes for AS | 7 (2–13) | 0 (0–1) | <0.001 | |
| Number of AS concepts | 21 (8–59) | 1 (0–4) | <0.001 | |
| Number of sacroiliitis concepts | 1 (0–7) | 0 (0–1) | <0.001 | |
| Number of HLA-B27 concepts | 1 (0–6) | 0 (0–1) | <0.001 | |
| Number of spondylitis concepts | 2 (0–6) | 0 (0–1) | <0.001 | |
| Rheumatologist diagnosis of AS | 127 (100) | 62 (15) | — | |
Data shown as mean (s.d.), n (%) or median (interquartile range). axSpA: axial SpA; IBD: inflammatory bowel disease; ICD: International Classification of Diseases – versions 9 and 10.
Six disease concepts were selected by the SAFE procedure. The logistic regression model used all six concepts; the most informative was the NLP concept for AS, followed by the AS ICD code, and NLP concepts for sacroiliitis, HLA-B27 and spondylitis, in order of predictive value (Table 2). In the LASSO model, all variables except the NLP concept for spondylitis were retained. The MAP model by design requires the AS ICD code and NLP concept and does not provide coefficients.
Table 2.
Standardized model coefficients indicating each variable’s predictive value
| Logistic regression | LASSO | |
|---|---|---|
| ICD codes for AS | 0.99 | 0.99 |
| NLP for AS | 1.24 | 1.18 |
| NLP for sacroiliitis | 0.48 | 0.35 |
| NLP for HLA-B27 | 0.24 | 0.01 |
| NLP for spondylitis | 0.21 | |
| Healthcare utilization | −0.45 | −0.43 |
ICD: International Classification of Diseases – versions 9 and 10; LASSO: least absolute shrinkage and selection operator; NLP: concept derived from narrative data using natural language processing.
When all three algorithms were applied to the enriched cohort, MAP had the highest sensitivity and identified the greatest number of patients (Table 3). With probability threshold set to provide 80% PPV, MAP identified 1509 patients as having high probability of axSpA with 78% sensitivity and 94% specificity. Performances of the three NLP-assisted algorithms were otherwise similar and all out-performed classification using ICD codes (Table 3).
Table 3.
Performance characteristics of NLP-assisted and ICD-code-based classification methods
| Number of axSpA cases identified | AUC | PPV, % | Sensitivity, % | Specificity, % | F-score, % | |
|---|---|---|---|---|---|---|
| MAP | 1509 | 0.927 | 80a | 78 | 94 | 79 |
| LASSO | 1272 | 0.929 | 80a | 71 | 95 | 75 |
| Logistic regression | 1281 | 0.930 | 80a | 70 | 95 | 75 |
| ICD code count | 981 | 0.870 | 78a | 63 | 95 | 70 |
| ≥2 ICD codes | 1881 | 0.804 | 60 | 76 | 84 | 67 |
| ≥3 ICD codes | 1381 | 0.802 | 69 | 70 | 90 | 69 |
Performance characteristics were derived using cases fulfilling classification criteria as the gold-standard. aPPV was selected to be ∼80%; probability threshold can be adapted to provide PPVs according to study requirements. AUC: area under the receiver operating characteristic curve; axSpA: axial SpA; F-score: harmonic mean of PPV and sensitivity; ICD: International Classification of Diseases – versions 9 and 10; LASSO: least absolute shrinkage and selection operator; MAP: multimodal automated phenotyping; NLP: concept derived from narrative data using natural language processing; PPV: positive predictive value.
Characteristics of 1509 patients identified by the MAP algorithm are shown in Supplementary Table 1, available at Rheumatology online. These patients were younger (53 vs 57 years), more frequently male (71 vs 48%), and had higher counts of AS ICD codes (median 6 vs 0) and AS NLP concepts (median 25 vs 1), compared with those who were not classified as cases (all P < 0.001).
Discussion
The ability to accurately and efficiently classify diseases in EHR has significant implications for clinical research. We showed that evolving disease concepts such as axSpA, where no specific ICD codes are available, can be accurately identified in EHR by incorporating narrative data using NLP. The MAP algorithm demonstrated high sensitivity (78%) and specificity (94%) and, notably, was developed without the need for manually derived training labels or domain experts to identify predictive disease concepts. These algorithms allow large cohorts of even uncommon diseases to be generated from EHR to facilitate clinical research.
Comparing our algorithms against those from prior studies is challenging, since PPV (the main performance measure of interest) depends on prevalence of axSpA in each study and the disease definition used. The accuracy of AS ICD codes was assessed in 184 patients with ⩾2 rheumatology visits from the Veterans Affairs EHR. The prevalence of AS was 7%. Among 11 patients with ⩾1 AS ICD codes, 10 had rheumatologist-diagnosed AS; accordingly the PPV was 91% and sensitivity 83% [20]. As the authors noted, this was unexpectedly high compared with parallel studies in RA (PPV 66%); the small sample size may be one explanation. Two European studies reported performance characteristics only among classification-positive patients; those not classified as cases (and therefore prevalence/sensitivity) were not reported. Among 85 UK primary care patients who had ⩾1 Read codes for AS, 58 (PPV 68%) had AS according to rheumatologist diagnosis; PPV for classification criteria axSpA was 72% [3]. A Swedish study of 250 patients from rheumatology departments found that PPV for classification criteria axSpA was 79% among those with ⩾1 AS ICD codes [2].
While our cohort and case definitions were not directly comparable to the above studies, the accuracy of AS ICD codes in our EHR was generally lower. In the USA, 26–53% of axSpA diagnoses are made by primary care physicians, and up to 63% are diagnosed outside of rheumatology practices [21, 22]. Since the accuracy of these codes can be very low [21], the proportion of ICD codes assigned by non-rheumatologists will determine overall accuracy.
The need for improved methods of identifying axSpA patients in EHR was highlighted by the low estimated prevalence (23%) of axSpA despite the initial enrichment process, which used ICD codes and ‘keyword searches’ in clinical notes and radiology reports. Unlike NLP, the results of a keyword search cannot distinguish negative mentions, which may be more common; for example, absence of sacroiliitis is commonly reported during investigations for back pain. AS may also be included in lists of differential diagnoses and check lists (e.g. pre-operative anaesthetic assessment).
Compared with the above rule-based methods, an advantage of our algorithm is that the probability threshold to classify a case can be tailored according to the needs of the research question. Higher PPVs can be selected at the expense of sensitivity. Our classification process has several additional strengths through each step of its development. From the outset, records from all patient were used, not just those under rheumatologists. This is important since a significant proportion of axSpA patients are diagnosed and managed in primary care only [22, 23] but may attend hospital for other reasons. In forming the enriched cohort, we included not only individuals with AS ICD codes but also those with mention of the disease in notes, which is important since it was previously shown that 43% of axSpA patients did not have an AS ICD code [19]. In our chart-reviewed sample, 11% of those with axSpA did not have AS ICD codes. These features allow many more potential cases to be found.
One rate-limiting step in the development of any classification algorithm is identifying informative disease concepts. In a prior study involving complex and labour-intensive steps, the same axSpA concepts were found as in our study: HLA-B27, sacroiliitis and terms with the prefix ‘spond-’ [24]. We selected axSpA concepts using an automated process without dependence on expert input, with the final list reviewed by an expert for face validity. Furthermore, the MAP algorithm had superior performance without using these additional concepts at all. The ‘spondylitis’ concept may be of greater predictive value in healthcare systems where ‘axSpA’ is more widely used. Another major bottleneck in algorithm development is the time-consuming chart review required to create the gold-standard training labels. The MAP algorithm achieved similar predictive performance without the need for these labels.
Accurately and efficiently identifying diseases in EHR offers exciting opportunities for clinical research. Linking EHR and biobanks has allowed large-scale studies of biomarkers [5, 25] or genetic variants [26, 27] against a broad range of phenotypes. NLP can also improve identification of disease outcomes [7]. Large, real-world cohorts can be used to study healthcare utilization [13] or rarer outcomes overlooked by prospective registries.
Developing a classification algorithm in the context of evolving terminology is challenging. The concept of AS has expanded over the past decade to include non-radiographic, potentially early, forms of the disease [17]. The terms ‘axial spondyloarthritis’ and ‘non-radiographic axial spondyloarthritis’ were uncommon in our EHR because (i) EHR started before the ASAS criteria was introduced, and (ii) the 9th and 10th versions of ICD codes do not include axSpA. One limitation is that we used the NLP concept and ICD code for AS to identify axSpA patients, although our prior work showed that ‘AS’ was used synonymously with axSpA [13]. Equally, our approach has the advantage of allowing researchers to identify diseases that cannot be captured using ICD codes alone. Linguistic variations can be captured using concepts in the UMLS, which may have limited vocabulary coverage in other languages. Another limitation was that our algorithm was developed using EHR from a single healthcare system. Not all EHR systems may be able to provide data used in our development process, such as word searches in the initial enrichment process. The prevalence of axSpA may also be lower in smaller centres or higher in those providing specialist axSpA services. Although algorithms for RA developed using the same approach were shown to be portable [28], further studies to externally validate our axSpA algorithm in different EHR systems and other countries are needed. Chart review for the training set was performed by one rheumatologist. The purpose was to extract documented diagnoses and criteria components, rather than making subjective decisions on whether a case was axSpA that would necessitate a second reviewer.
In conclusion, we demonstrated that classification algorithms incorporating narrative EHR data and machine learning can accurately identify evolving disease concepts such as axSpA. The automated MAP algorithm allows large cohorts of even uncommon diseases to be efficiently created, offering exciting opportunities for future clinical research.
Supplementary Material
Acknowledgements
S.S.Z. was supported by awards from the Royal College of Physicians (John Glyn bursary) and Royal Society of Medicine (Kovacs fellowship). K.P.L. was supported by the NIH-P30-AR072577 (VERITY) grant. D.H.S. was supported by grants from the National Institutes of Health (NIH-P30-AR072577 and NIH-K24AR055989).
Funding: No funding was specifically obtained for this study.
Disclosure statement: The authors declare no conflicts of interest.
References
- 1. Strand V, Rao SA, Shillington AC. et al. Prevalence of axial spondyloarthritis in United States rheumatology practices: assessment of SpondyloArthritis International Society criteria versus rheumatology expert clinical diagnosis. Arthritis Care Res 2013;65:1299–306. [DOI] [PubMed] [Google Scholar]
- 2. Lindström U, Exarchou S, Sigurdardottir V. et al. Validity of ankylosing spondylitis and undifferentiated spondyloarthritis diagnoses in the Swedish National Patient Register. Scand J Rheumatol 2015;44:369–76. [DOI] [PubMed] [Google Scholar]
- 3. Dubreuil M, Peloquin C, Zhang Y. et al. Validity of ankylosing spondylitis diagnoses in The Health Improvement Network: ankylosing spondylitis diagnostic validity. Pharmacoepidemiol Drug Saf 2016;25:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O'Malley KJ, Cook KF, Price MD. et al. Measuring diagnoses: ICD code accuracy. Health Serv Res 2005;40:1620–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liao KP, Kurreeman F, Li G. et al. Associations of autoantibodies, autoimmune risk alleles, and clinical diagnoses from the electronic medical records in rheumatoid arthritis cases and non-rheumatoid arthritis controls. Arthritis Rheum 2013;65:571–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liao KP, Cai T, Gainer V. et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res 2010;62:1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ananthakrishnan AN, Cagan A, Cai T. et al. Identification of nonresponse to treatment using narrative data in an electronic health record inflammatory bowel disease cohort. Inflamm Bowel Dis 2016;22:151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liao KP, Ananthakrishnan AN, Kumar V. et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS One 2015;10:e0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Imran TF, Posner D, Honerlaw J. et al. A phenotyping algorithm to identify acute ischemic stroke accurately from a national biobank: the Million Veteran Program. Clin Epidemiol 2018;10:1509–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liao KP, Cai T, Savova GK. et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 2015;350:h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu S, Chakrabortty A, Liao KP. et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc 2017;24:e143–e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liao KP, Zhang Y, Cai T. et al. Methods for high-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP). Nat Protoc 2019. [DOI] [PMC free article] [PubMed]
- 13. Zhao SS, Ermann J, Xu C. et al. Comparison of comorbidities and treatment between ankylosing spondylitis and non-radiographic axial spondyloarthritis in the United States. Rheumatology 2019;58:2025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.United States National Library of Medicine. United Medical Language System (UMLS). 2018. https://www.nlm.nih.gov/research/umls/ (5 November 2018, date last accessed).
- 15. Yu S, Cai T. NILE: Fast Natural Language Processing for Electronic Health Records. arXiv:1311.6063. 2019. https://arxiv.org/abs/1311.6063 (16 July 2019, date last accessed).
- 16. van der Linden S, Valkenburg HA, Cats A.. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum 1984;27:361–8. [DOI] [PubMed] [Google Scholar]
- 17. Rudwaleit M, van der Heijde D, Landewé R. et al. The development of Assessment of SpondyloArthritis international Society classification criteria for axial spondyloarthritis (part II): validation and final selection. Ann Rheum Dis 2009;68:777–83. [DOI] [PubMed] [Google Scholar]
- 18. Liao KP, Sun J, Cai TA. et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J Am Med Inform Assoc 2019. doi: 10.1093/jamia/ocz066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walsh JA, Pei S, Penmetsa GK. et al. Cohort identification of axial spondyloarthritis in a large healthcare dataset: current and future methods. BMC Musculoskelet Disord 2018;19:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh JA, Holmgren AR, Krug H, Noorbaloochi S.. Accuracy of the diagnoses of spondylarthritides in veterans affairs medical center databases. Arthritis Rheum 2007;57:648–55. [DOI] [PubMed] [Google Scholar]
- 21. Curtis JR, Harrold LR, Asgari MM. et al. Diagnostic prevalence of ankylosing spondylitis using computerized health care data, 1996 to 2009: underrecognition in a US health care setting. Perm J 2016;20:4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deodhar A, Mittal M, Reilly P. et al. Ankylosing spondylitis diagnosis in US patients with back pain: identifying providers involved and factors associated with rheumatology referral delay. Clin Rheumatol 2016;35:1769–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dean LE, Macfarlane GJ, Jones GT.. Differences in the prevalence of ankylosing spondylitis in primary and secondary care: only one-third of patients are managed in rheumatology. Rheumatology 2016;55:1820–5. [DOI] [PubMed] [Google Scholar]
- 24. Walsh JA, Shao Y, Leng J. et al. Identifying axial spondyloarthritis in electronic medical records of US veterans: axial SpA identification methods. Arthritis Care Res 2017;69:1414–20. [DOI] [PubMed] [Google Scholar]
- 25. Hejblum BP, Cui J, Lahey LJ. et al. Association between anti-citrullinated fibrinogen antibodies and coronary artery disease in rheumatoid arthritis. Arthritis Care Res 2018;70:1113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ananthakrishnan AN, Cagan A, Cai T. et al. Common genetic variants influence circulating vitamin D levels in inflammatory bowel diseases. Inflamm Bowel Dis 2015;21:2507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai T, Zhang Y, Ho Y-L. et al. Association of interleukin 6 receptor variant with cardiovascular disease effects of interleukin 6 receptor blocking therapy: a phenome-wide association study. JAMA Cardiol 2018;3:849–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carroll RJ, Thompson WK, Eyler AE. et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012;19:e162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.