ABSTRACT
Background
Breast milk is a complex biofluid that provides nutrients and bioactive agents, including bacteria, for the development of the infant gut microbiota. However, the impact of maternal diet and other factors, such as mode of delivery and antibiotic exposure, on the breast milk microbiota has yet to be understood.
Objectives
This study aimed to examine the association between maternal diet and breast milk microbiota and to ascertain the potential role of mode of delivery and antibiotic exposure.
Methods
In a cross-sectional study of the MAMI cohort, breast milk microbiota profiling was assessed in 120 samples from healthy mothers by 16S rRNA gene sequencing. Maternal dietary information was recorded through an FFQ, and clinical characteristics, including mode of delivery, antibiotic exposure, and exclusive breastfeeding, were collected.
Results
Maternal diet was grouped into 2 clusters: Cluster I (high intake of plant protein, fiber, and carbohydrates), and Cluster II (high intake of animal protein and lipids). Breast milk microbiota was shaped by maternal dietary clusters. Staphylococcus and Bifidobacterium were associated with carbohydrate intake whereas the Streptococcus genus was associated with intakes of the n–3 PUFAs [EPA and docosapentaenoic acid (22:5ω-3)]. Mode of delivery and antibiotic exposure influenced breast milk microbiota in a diet cluster–dependent manner. Differences between/among the maternal dietary clusters were found in the milk microbiota of the cesarean-section (C-section)/antibiotic group, whereas no differences were observed in vaginal births. Lower abundances of Lactobacillus, Bacteroides, and Sediminibacterium genera were observed in Cluster II/C-section/antibiotic exposure compared with the other groups.
Conclusions
Maternal diet shapes the composition and diversity of breast milk microbiota, with the most important contributions coming from dietary fiber and both plant and animal protein intakes. The relation between the maternal diet and the milk microbiota needs further research because it has a key impact on infant microbiota development and contributes to infant health outcomes in the short and long term.
This trial was registered at clinicaltrials.gov as NCT03552939.
Keywords: maternal diet, breast milk, microbiota, plant protein, animal protein
See corresponding commentary on page 278.
Introduction
Breast milk is the best food for infants during early life (1, 2). It represents the most relevant postnatal link between mothers and newborns and it drives the microbial, metabolic, and immunological programming of infants’ health (3, 4). Breastfed infants harbor higher abundances of Bifidobacterium and Lactobacillus in the gut than formula-fed infants (5), contributing to a distinct immune system maturation, protection against infections, and also to reduction in the risk of disease, such as allergies, obesity, and other disorders later in life (3, 6, 7).
Breast milk provides an optimal source of nutrients and beyond; it contains bioactive compounds, including immunoglobulins, lysozymes, growth factors, oligosaccharides, and beneficial microorganisms (4, 8–10). Breast milk microbiota composition is diverse and commonly characterized by Staphylococcus and Streptococcus spp. followed by other genera, such as Veillonella, Propionibacterium, lactic acid bacteria, and Bifidobacterium spp. (11). Furthermore, there is a large inter- and intraindividual variability in breast milk composition, and the contribution of these variations to infant development is not yet completely understood. Maternal and infant factors, including lactational stage, maternal pregestational BMI, weight gain over the pregnancy, mode of delivery, geographical location, antibiotics, parity, infant gender, and also the method of breastfeeding all shape the breast milk microbiota composition (12–16).
Limited data are available on the impact of maternal diet on breast milk microbiota. We hypothesized that breast milk microbiota composition and diversity would be shaped by perinatal factors in a diet-dependent manner. Thus, this study aimed to assess whether maternal diet and specific nutrients during pregnancy would shape breast milk microbiota, and to ascertain the potential influence of other perinatal factors, including mode of delivery and antibiotic exposure, on breast milk microbiota composition according to maternal diet.
Methods
Study design and volunteers
A total of 120 healthy mothers from the observational study MAMI (MAternal MIcrobes) cohort were included in this cross-sectional study according to the full availability of biological samples and clinical and dietary data (Supplemental Figure 1). The MAMI cohort is a prospective mother-infant cohort in the Spanish Mediterranean area, as described elsewhere (17).
Nutritional, anthropometrical, and clinical parameters were collected including maternal antibiotic exposure, pregestational BMI, weight gain during pregnancy, mode of delivery, and for newborns gestational age, infant feeding, and weight and length at birth, 7, and 15 d. BMI (kg/m2) was stratified according to the Sociedad Española para el Estudio de la Obesidad criteria: normal weight (18.5–25.0); overweight (25.1–29.9); and obese (≥30.0) (18). None of the participating volunteers were diagnosed with any disease; none were receiving drug treatment or pro- and prebiotic administration, with the exception of antibiotic exposure during pregnancy and/or at delivery.
All participants received oral and written information about the study and written consent was obtained. The study was approved by the hospital ethics committees (Hospital Clínico Universitario de Valencia and Hospital Universitario y Politécnico La Fe). The study was registered on clinicaltrials.gov with registration number NCT03552939.
Maternal nutritional assessment
The maternal diet was estimated with the use of an FFQ (19). The FFQ questionnaire included 140 items, covering usual foods and recipes, as well as the frequency of consumption (daily, weekly, monthly, or during the end of pregnancy and birth time), the number of times the participant consumed a particular food item, the median portion (in household measures—grams and milliliters), and the size of each participant's portion. All the food surveys were conducted by trained nutritionists. FFQ information was analyzed using data from the Food Composition Tables developed by the Centro de Enseñanza Superior de Nutrición Humana y Dietética (20). The intake of soluble and insoluble fiber types was determined by using the Marlett food composition tables (21). Polyphenol content was obtained from the Phenol-Explorer database (22). Data were standardized by 2500 kcal/d energy intake to control for overconsumption or underconsumption and to reduce the potential measurement error in the dietary assessment (23, 24). The FFQ data have been validated elsewhere for collecting information on diet during pregnancy (19), and we have also validated our results’ data in a randomly selected subsample (n = 20) through the use of a 3-d recall food record questionnaire for the intake of dietary nutrients (19) (Supplemental Table 1).
Maternal dietary profile determination
Maternal diet clustering was performed as described elsewhere (25). Briefly, Jensen–Shannon distance and partitioning around medoid clustering were used. The optimal number of clusters was calculated by the Calinski–Harabasz index (CHI). The clusters were generated using the phyloseq (26), cluster (27), MASS (28), clusterSim (29), and ade4 (30) R packages (R Foundation). Clustering was carried out by using specific dietary information, such as intake of animal protein, plant protein, soluble and insoluble fiber, SFAs, total MUFAs, total PUFAs and trans fatty acids, and carbohydrates.
Influence of mode of birth and antibiotics on breast milk microbiota
In our cohort, intrapartum antibiotic exposure was mainly associated with cesarean section (C-section) births, although in our study, some vaginal deliveries were also exposed to antibiotics (n = 3, representing 3.84%). Due to the low number of samples exposed to antibiotics during vaginal delivery, we removed them from the analysis to make homogeneous groups: 1) vaginal birth and no antibiotic exposure; and 2) C-section birth and antibiotic exposure.
To identify the impact of mode of birth/antibiotics on milk microbiota, depending on maternal diet, 4 groups were distinguished, according to the mode of delivery and maternal diet (Cluster I_C-section, Cluster I_VAG, Cluster II_C-section, and Cluster II_VAG) (Supplemental Figure 1).
Breast milk samples
Breast milk samples were collected during 7–15 d after birth, following a standardized protocol. In brief, breast skin was cleansed with soap or 0.5% chlorhexidine solution and the first drops were discarded. Morning collection was recommended. Then, breast milk was collected from 1 breast with the use of a sterile pump and stored in sterile bottles to normalize the milk collection protocol. Finally, breast milk samples were sent to a biobank and then aliquoted and stored at −80°C until further analysis.
DNA extraction and total bacterial levels
Total DNA was isolated from 1.5–2.0-mL breast milk samples using the Master-Pure DNA Extraction Kit (Epicentre) according to the manufacturer's instructions; modifications included physical and enzymatic treatments as previously detailed (31). DNA purification was performed using a MagSi-NGS Plus kit (AMSBIO) and quantification was assessed by Qubit 2.0 Fluorometer (Life Technologies). Breast milk is a low-microbial biomass sample that would be affected by potential contaminants from the environment and DNA extraction kit reagents (“kitome”). Controls during DNA extraction and PCR amplification were also included and sequenced.
Total bacterial quantification was performed using targeted qPCR in a LightCycler 480 real-time PCR system (Roche Technologies). Each reaction mixture of 10 μL was composed of SYBR Green PCR Master Mix (Roche), 0.25 μL of each of the specific primers 789R (5′-GCGTGGACTACCAGGGTATCT) and 515F (5′-GTGCCAGCMGCCGCGGTAA) (32, 33) at a concentration of 10 μM, and 1 μL of template DNA, with an annealing temperature of 62°C.
16S rRNA amplicon sequencing
Breast milk microbiota composition was determined by V3–V4 variable regions of the 16S rRNA gene sequencing, following Illumina protocols. A Nextera XT Index Kit (Illumina) was used for the multiplexing step, and a Bioanalyzer DNA 1000 chip (Agilent Technologies) for checking the PCR product quality. Libraries were sequenced using a 2 × 300 bppaired-end run (MiSeq Reagent Kit v3) on a MiSeq-Illumina platform (FISABIO sequencing service), according to the manufacturer's instructions (Illumina) (34).
Sequences and data analysis
A DADA2 pipeline was used to achieve quality filtering, sequence joining, and chimera removal (35). Using the SILVA v132 database (36), taxonomic assignment was performed also including the species-level classification. Samples with a relative abundance <0.01% and those present <3 times in ≥20% of the samples were filtered. Furthermore, the decontam package (37) was used to determine the presence of potential contaminant-related sequences, resulting in 48 amplicon sequence variants classified as possible contaminants, which were also filtered. Sequences classified as cyanobacteria and chloroplasts were also removed from the final data set as well as samples with ≤1000 reads. Predictive inferred functional analysis was performed using the Piphillin pipeline (38)
GraphPad Prism v. 5.04 was used for t-test and Mann–Whitney analysis, according to data normality assessed by Kolmogorov–Smirnov and Shapiro–Wilk tests. χ2 tests (2 × 2) were performed to assess the significance of the differences in the characteristics according to categorical variables; these were conducted in SPSS v.25 (IBM).
Spearman correlations between relative abundances of bacterial groups and nutrient intakes were also performed in SPSS v.25. Linear discriminant analysis effect size (LEfSe) was used to identify microbial genera enriched in maternal diet clusters. A redundancy discriminant analysis (RDA) was applied to study the statistical effect of maternal diet and delivery mode on breast milk microbiota. A permutational multivariate analysis of variance (PERMANOVA) based on Bray–Curtis distance, and α diversity by Shannon (diversity) and Chao1 indexes (richness) were performed using Calypso online platform (v8.84) software (39). RStudio was also used to represent the heatmaps between bacterial groups at the genus level and dietary components through the ggplot package (40).
Results
Maternal diet clusters and clinical data
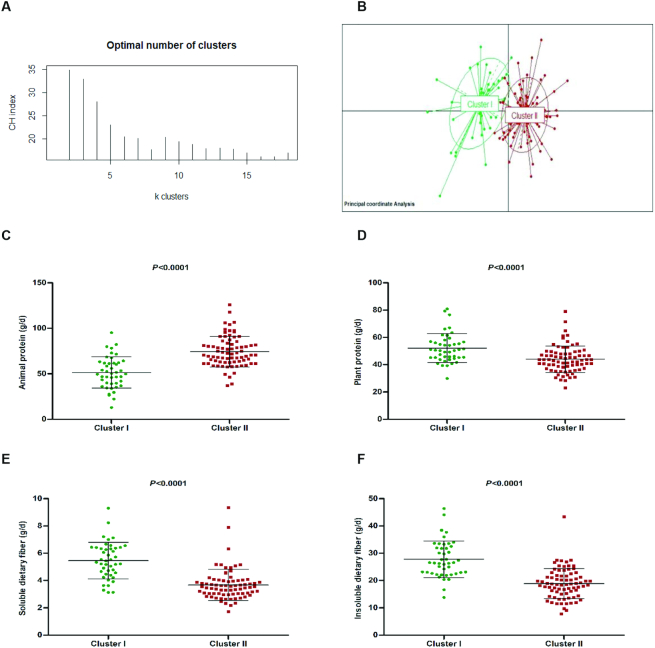
A dietary intake–based clustering using Jensen–Shannon distance revealed two distinct clusters validated by the CHI (Figure 1A, B). Cluster I (n = 44) was characterized by plant protein, fiber, and carbohydrates, whereas Cluster II (n = 76) was characterized by animal protein and lipids (Figure 1C–F). No significant differences were found in the maternal-neonatal clinical characteristics (Table 1). Interestingly, higher pregestational BMI was observed in Cluster II compared with Cluster I (P = 0.016); however, both were classified as normal-weight women (BMI ≤25).
FIGURE 1.
Maternal diet clusters and representative dietary components. The partitioning around medoid method showed that women were grouped (A) into 2 clusters and both distinct clusters (Cluster I = green, n = 44; and Cluster II = red, n = 76) are represented in the Principal Coordinate Analysis (PCoA) (B). The strongest contributors to the clustering were higher intake of animal protein and lower plant protein and dietary fiber. Diet was standardized by total energy intake to 2500 kcal/d (C–F). Middle line represents the media of all values and also, the upper and lower lines represent the standard deviation (SD)..
TABLE 1.
Characteristics of the volunteers included in the analysis (n = 120)
| Total sample (n = 120) | Cluster I (n = 44) | Cluster II (n = 76) | P value1 | |
|---|---|---|---|---|
| Maternal characteristics2 | ||||
| Pregestational BMI, kg/m2 | 23 ± 3.6 | 22 ± 2.8 | 24 ± 3.8 | 0.016 |
| Weight gain during pregnancy, kg | 12 ± 4.6 | 12 ± 3.9 | 12 ± 4.9 | 0.89 |
| Gestational age, wk | 40 ± 1.3 | 40 ± 1.2 | 39 ± 1.3 | 0.24 |
| Intrapartum antibiotic exposure, n (%) | 44 (37) | 17 (39) | 27 (36) | 0.73 |
| Mode of delivery: vaginal birth, n (%) | 78 (65) | 29 (66) | 49 (64) | 0.87 |
| Infant characteristics2 | ||||
| Gender: female, n (%) | 68 (57) | 28 (64) | 40 (53) | 0.24 |
| Weight, kg | ||||
| Birth | 3.3 ± 0.46 | 3.4 ± 0.57 | 3.3 ± 0.40 | 0.40 |
| 7 d | 3.3 ± 0.49 | 3.4 ± 0.57 | 3.3 ± 0.43 | 0.40 |
| 15 d | 3.6 ± 0.51 | 3.6 ± 0.59 | 3.6 ± 0.45 | 0.42 |
| Length, cm | ||||
| Birth | 50 ± 2.2 | 50 ± 2.7 | 50 ± 1.8 | 0.57 |
| 7 d | 51 ± 2.1 | 51 ± 2.3 | 51 ± 2.0 | 0.74 |
| 15 d | 52 ± 2.1 | 52 ± 2.3 | 52 ± 2.0 | 0.87 |
| Exclusive breastfeeding until 6 mo, n (%) | 94 (78) | 33 (75) | 61 (80) | 0.50 |
Student t test and χ2 test were performed to assess the significance between maternal clusters' characteristics in numerical variables and categorical variables, respectively. P < 0.05 was considered statistically significant.
Numerical data are expressed as mean ± SD; categorical data as number of cases and percentage.
Detailed information about nutrient intakes according to the clustering is shown in Table 2. A significantly higher intake of total protein (P < 0.0001), mainly animal protein (P < 0.0001), was observed in women from Cluster II compared with those observed in Cluster I. Furthermore, Cluster II also presented a higher total lipid intake, mainly SFAs (P < 0.0001) as well as total MUFAs (P < 0.0001) and total PUFAs (P = 0.003). But there were no significant differences in the intakes of starch and fatty acids, such as ω-6 conjugated linoleic acid, ω-3 α-linolenic acid, ω-3 docosapentaenoic acid (DPA), and DHA between clusters. However, Cluster I was characterized by significantly higher dietary intakes of plant protein, carbohydrates, and total, insoluble, and soluble dietary fibers (all P < 0.0001).
TABLE 2.
Specific dietary intakes of the healthy mothers involved in the study1
| Dietary intakes2, 3 | Total sample (n = 120) | Cluster I (n = 44) | Cluster II (n = 76) | P value4 |
|---|---|---|---|---|
| Total protein, g/d | 116 ± 19 | 106 ± 15 | 122 ± 19 | <0.0001 |
| Animal protein, g/d | 66 ± 20 | 51 ± 17 | 74 ± 17 | <0.0001 |
| Plant protein, g/d | 47 ± 11 | 52 ± 11 | 44 ± 10 | <0.0001 |
| Lipids, g/d | 114 ± 16 | 102 ± 13 | 120 ± 13 | <0.0001 |
| SFAs | 24 ± 7.2 | 21 ± 4.8 | 26 ± 7.7 | <0.0001 |
| Trans fatty acids | 0.007 ± 0.01 | 0.005 ± 0.01 | 0.008 ± 0.01 | 0.009 |
| MUFAs | 46 ± 11.0 | 41 ± 10.0 | 50 ± 10.0 | <0.0001 |
| PUFAs | 17 ± 4.0 | 16 ± 2.9 | 18 ± 4.3 | 0.003 |
| n–6 CLA | 0.003 ± 0.01 | 0.003 ± 0.01 | 0.003 ± 0.01 | 0.57 |
| n–6 EDA | 0.007 ± 0.01 | 0.005 ± 0.01 | 0.008 ± 0.01 | 0.14 |
| n–3 ALA | 0.14 ± 0.1 | 0.15 ± 0.1 | 0.13 ± 0.1 | 0.34 |
| n–3 EPA | 0.17 ± 0.1 | 0.15 ± 0.1 | 0.19 ± 0.1 | 0.025 |
| n–3 DPA | 0.09 ± 0.1 | 0.08 ± 0.1 | 0.09 ± 0.1 | 0.10 |
| n–3 DHA | 0.41 ± 0.3 | 0.36 ± 0.2 | 0.44 ± 0.3 | 0.10 |
| Carbohydrates, g/d | 249 ± 40 | 285 ± 30 | 228 ± 28 | <0.0001 |
| Total dietary fiber, g/d | 36 ± 11 | 44 ± 11 | 32 ± 7.3 | <0.0001 |
| Insoluble dietary fiber, g/d | 22 ± 7.4 | 28 ± 6.7 | 19 ± 5.5 | <0.0001 |
| Soluble dietary fiber, g/d | 4.3 ± 1.5 | 5.5 ± 1.3 | 3.7 ± 1.1 | <0.0001 |
| Starch, g/d | 29 ± 12 | 30 ± 12 | 29 ± 12 | 0.66 |
| Polyphenols, g/d | 1.8 ± 759 | 2.2 ± 941 | 1.5 ± 497 | <0.0001 |
| Folic acid, μg/d | 143 ± 142 | 197 ± 160 | 112 ± 120 | 0.001 |
| Vitamin A, mg/d | 1.37 ± 533 | 1.44 ± 528 | 1.33 ± 535 | 0.25 |
| Vitamin B-6, mg/d | 3.5 ± 1.4 | 4.1 ± 1.6 | 3.2 ± 1.2 | 0.001 |
| Vitamin B-12, μg/d | 7.8 ± 3.1 | 7.3 ± 3.1 | 8.1 ± 3.0 | 0.13 |
| Vitamin C, mg/d | 156 ± 71 | 192 ± 73 | 134 ± 61 | <0.0001 |
| Vitamin D, μg/d | 103 ± 82 | 89 ± 67 | 112 ± 89 | 0.15 |
| Vitamin E, mg/d | 13 ± 5.0 | 12 ± 3.9 | 13 ± 4.8 | 0.34 |
| Iron, mg/d | 21 ± 4.9 | 22 ± 5.6 | 20 ± 4.3 | 0.08 |
| Calcium, g/d | 1.17 ± 277 | 1.14 ± 272 | 1.20 ± 279 | 0.28 |
| Selenium, μg/d | 161 ± 29 | 154 ± 30 | 165 ± 28 | 0.05 |
| Sodium, g/d | 2.5 ± 587 | 2.5 ± 596 | 2.5 ± 585 | 0.95 |
| Magnesium, mg/d | 566 ± 148 | 589 ± 118 | 553 ± 161 | 0.20 |
| Manganese, mg/d | 6.7 ± 1.7 | 7.6 ± 1.8 | 6.2 ± 1.5 | <0.0001 |
| Zinc, mg/d | 14 ± 2.8 | 15 ± 3.5 | 14 ± 2.4 | 0.17 |
n–3 ALA, ω-3 α-linolenic acid; n–6 CLA, ω-6 conjugated linoleic acid; n–3 DPA, ω-3 docosapentaenoic acid; n–6 EDA, ω-6 eicosadienoic acid.
Numerical data expressed as mean ± SD.
Diets were standardized by total energy intake to 2500 kcal/d.
Student t test was performed to assess the significance of the differences in the dietary intakes between clusters section in numerical variables and categorical variables, respectively. P < 0.05 was considered statistically significant.
Breast milk microbiota is shaped by maternal diet cluster
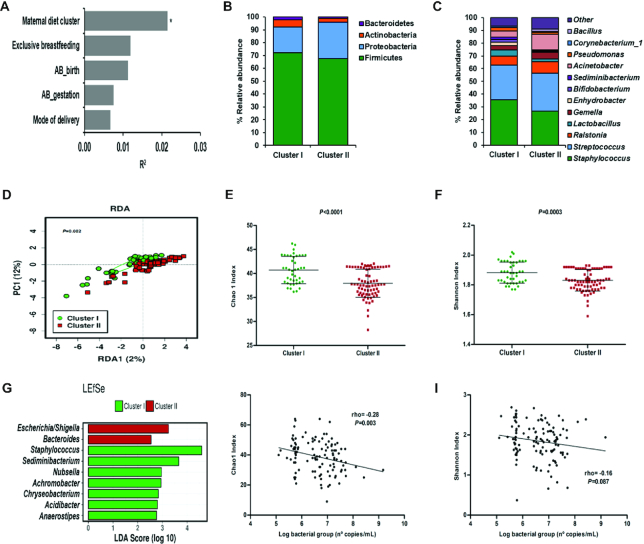
In general, the most abundant genera present in all the breast milk samples included in the study were Staphylococcus and Streptococcus followed by Ralstonia, Gemella, Lactobacillus, and others (Supplemental Figure 3A). We performed a PERMANOVA based on Bray–Curtis distance to identify the main factors contributing to the breast milk microbiota (Figure 2A). The combination of all factors had an impact on the breast milk microbiota (PERMANOVA Bray–Curtis model P = 0.051), whereas maternal diet cluster was the main contributor among the others affecting the milk microbiota variation (P < 0.01). Breast milk microbiota was significantly different depending on maternal dietary profile (PERMANOVA Bray–Curtis distance R2 = 0.021 and P = 0.007). This observation was also confirmed by a multivariate analysis (RDA test, variance = 1.91; P = 0.002, Figure 2D).
FIGURE 2.
Breast milk microbiota is shaped by maternal diet clusters. Maternal factors contributing to milk microbiota variation (PERMANOVA Bray–Curtis model with 5 variables, P = 0.051) (n = 115) (A). The relative abundance of bacteria present in breast milk at phylum (B) and genus level (C) on maternal diet cluster, Cluster I (n = 42) and Cluster II (n = 73). Multivariate redundancy discriminant analysis (RDA) showed significant differences in breast milk microbial communities depending on maternal diet clusters, Cluster I (n = 42) and Cluster II (n = 73) (D). Microbial α diversity indexes according to maternal diet cluster, Cluster I (n = 42) and Cluster II (n = 73): richness (Chao1 index, E) and diversity (Shannon index, F). Richness and diversity values were adjusted for total bacterial load to normalize the values and avoid the impact of bacterial levels. Linear discriminant analysis (LDA) effect size (LEfSe) plot of taxonomic biomarkers was identified in both clusters: Cluster I (n = 42) (green) and Cluster II (n = 73) (red) (G). Specific associations between total bacterial load estimated by qPCR and α-diversity indexes, Chao 1 (H) and Shannon index (I) (n = 115). AB_birth, intrapartum antibiotic exposure; AB_gestation, antibiotic treatment during gestation; PC, Principal component; PERMANOVA, Permutational multivariate analysis of variance.
Distinct bacterial profiles at phylum and genus levels were observed in Clusters I and II (Figure 2B, C). At the phylum level, Cluster I showed higher relative abundance of Bacteroidetes (P < 0.001) and Actinobacteria (P = 0.014) whereas at the genus level, a higher relative abundance of Staphylococcus (P = 0.036) and Lactobacillus (P = 0.022) compared with Cluster II was observed. In addition, higher relative abundances of Bifidobacterium (P = 0.026) and Sediminibacterium (P < 0.001) and lower relative abundances of Bacteroides (P = 0.023) and Escherichia/Shigella (P = 0.023) genera were also observed in Cluster I compared with Cluster II. Furthermore, LEfSe [linear discriminant analysis (LDA) >2.5] analysis showed that Cluster II was characterized by Bacteroides and Escherichia/Shigella genera whereas Staphylococcus spp. were characteristic of Cluster I (Figure 2G).
Regarding α diversity, significantly higher bacterial richness (measured by the Chao1 index; P < 0.0001) and diversity (measured by Shannon index; P = 0.0003) were observed in Cluster I compared with Cluster II (Figure 2E, F). Despite differences in α diversity, the total bacterial counts in milk samples were similar between clusters [median (IQR) log10 bacterial gene copies per milliliter: Cluster I = 6.52 (5.76–7.20) compared with Cluster II = 6.70 (5.77–7.20)]. Additionally, we found that higher bacterial richness but not diversity was associated with lower total bacterial load (ρ = −0.28; P = 0.003) (Figure 2H, I).
We also found significant differences in breast milk microbiota inferred function, depending on the maternal cluster (Supplemental Table 2).
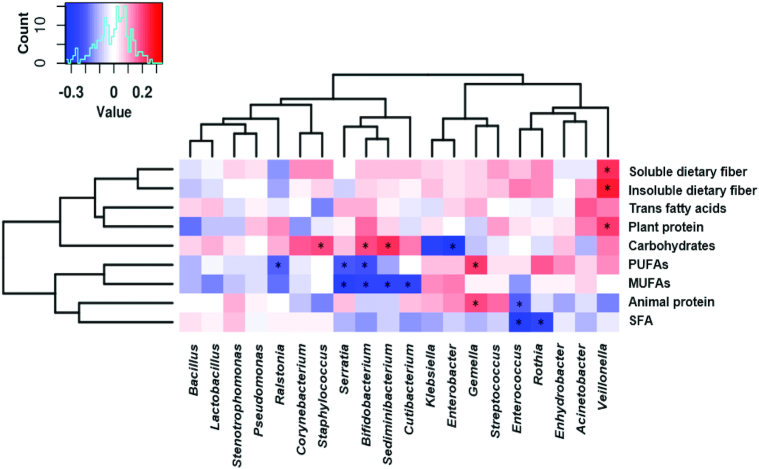
Association between breast milk microbiota and specific dietary nutrients
Significant associations between dietary nutrients and specific breast milk microbial genera were identified (Figure 3, Supplemental Figure 3). Staphylococcus was associated with higher intake of carbohydrates (ρ = 0.19; P = 0.038) and with lower total protein intake (ρ = −0.21; P = 0.026) whereas Streptococcus genus was directly associated with higher intake of total protein (ρ = 0.22; P = 0.018) as well as higher EPA (ρ = 0.19; P = 0.040) and DPA (ρ = 0.19; P = 0.044), selenium (ρ = 0.26; P = 0.005), and zinc (ρ = 0.18; P = 0.049).
FIGURE 3.
Correlations between breast milk microbial top 20 genera and the main dietary nutrients. Heatmaps of Spearman rank correlations between specific dietary nutrient intakes and the top 20 breast milk bacterial genus abundances (n = 115). Significant correlations (P < 0.05) are marked by an asterisk (*). Blue squares represent negative correlations, whereas red squares show positive correlations.
Furthermore, Bifidobacterium abundance was associated with a higher intake of carbohydrates (ρ = 0.20; P = 0.031) and polyphenols (ρ = 0.19; P = 0.042), and also with lower total lipid intake (ρ = −0.23; P = 0.012), mainly with lower total MUFA (ρ = −0.26; P = 0.006) and total PUFA (ρ = −0.22; P = 0.016) intakes. Interestingly, whereas Enterococcus genus was negatively associated with total animal protein (ρ = −0.20; P = 0.036) intake and saturated fat (ρ = −0.30; P = 0.001), the Gemella genus was positive correlated with animal protein (ρ = 0.20; P = 0.029), and also with n–3 PUFAs such as n–3 DPA (ρ = 0.26; P = 0.005), n–3 DHA (ρ = 0.26; P = 0.004), and n–3 EPA (ρ = 0.26; P = 0.006) intake. Additionally, although total lipid intake presented a positive correlation with Klebsiella and Enterobacter, both bacterial features showed a negative association with carbohydrates. Calcium intake was associated with lower relative abundance of Veillonella genus (ρ = −0.19; P = 0.043) and higher relative abundance of Stenotrophomonas (ρ = 0.20; P = 0.032). Significantly higher Veillonella abundance was associated with higher total fiber (ρ = 0.20; P = 0.034) and plant protein (ρ = 0.22; P = 0.019) and insoluble dietary fiber intakes (ρ = 0.27; P = 0.004). In addition, higher abundance of Enterococcus was associated with higher vitamin A (ρ = 0.20; P = 0.037), and lower vitamin D (ρ = −0.19; P = 0.043).
Higher microbial richness measured by the Chao1 index was associated with lower MUFA intake (ρ = −0.19; P = 0.048). However, higher microbial diversity measured by the Shannon index was associated with higher maternal total fiber intake (ρ = 0.18; P = 0.055).
Impact of mode of birth and antibiotics on breast milk microbiota is modulated by maternal diet
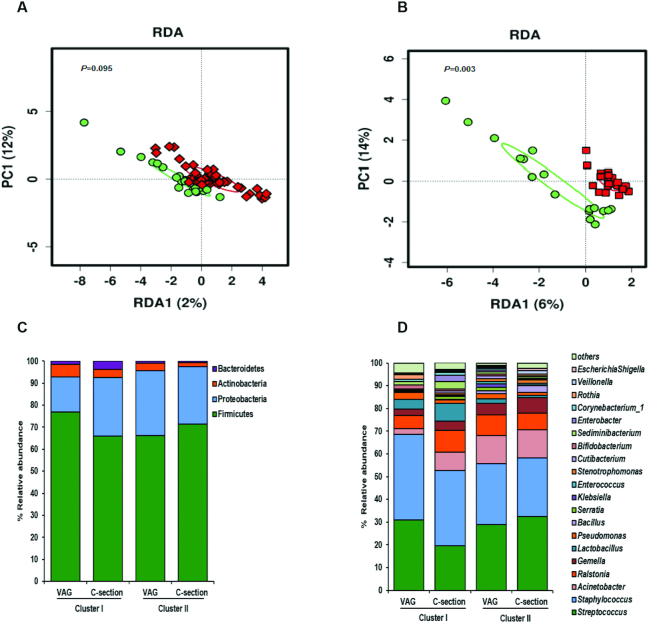
Significant differences were observed in the whole microbial community between dietary clusters in mothers under antibiotic exposure and C-section (RDA test variance = 4.9, P = 0.003; and PERMANOVA Bray–Curtis R2 = 0.046, P = 0.037), whereas no differences were detected in vaginal deliveries (RDA test variance = 1.57, P = 0.095; PERMANOVA Bray–Curtis R2 = 0.021, P = 0.121) (Figure 4A, B).
FIGURE 4.
Impact of mode of delivery and antibiotic exposure on breast milk microbiota according to maternal diet clusters. Multivariate redundancy discriminant analysis (RDA) showed significant differences in breast milk microbial communities between maternal diet clusters depending on antibiotic exposure and mode of delivery: vaginal/nonantibiotics (A) and C-section/antibiotics (B). Cluster I (n = 40) (green) and Cluster II (n = 72) (red). Breast milk microbiota profiles at phylum level (C) and at genus level (D) according to maternal diet cluster and mode of delivery and antibiotic exposure. C-section, cesarean section; PC1, principal component 1; VAG, vaginal delivery.
Distinct bacterial profiles at the phylum and family level were observed (Figure 4C, D). Lower Actinobacteria (P = 0.043) and lower Bacteroidetes (P = 0.005) phylum members were found in C-section/antibiotics Cluster II compared with the other groups. Significantly lower abundances of Lactobacillus (P = 0.042), Bacteroides (P = 0.026), and Sediminibacterium genera (P = 0.003) were observed in C-section/Cluster II compared with the other groups. Interestingly, Rothia genus was significantly higher (P = 0.041) in vaginal Cluster I compared with the other groups.
Significant differences were observed in microbial communities grouped according to antibiotic exposure/mode of delivery and maternal dietary clusters (RDA test variance = 3.58; P = 0.002) as well as in the microbial diversity and richness, depending on antibiotic exposure/mode of birth and diet cluster (Supplemental Figure 3B, D). Higher microbial diversity and richness were observed in women from Cluster I and vaginal/non–antibiotic exposure compared with the other groups. Interestingly, higher microbial richness (Chao1 index) was found in the C-section/antibiotics group in Cluster I than in Cluster II, whereas no differences were found in diversity (measured by Shannon index).
Discussion
Our study showed the influence of maternal diet on the composition and diversity of breast milk microbiota. We observed specific associations between maternal nutrient intake and breast milk microbiota. Maternal diet, mode of delivery, and antibiotic exposure influence the breast milk composition, opening new possibilities to develop targeted dietary interventions during lactation.
In agreement with previous data (16, 41, 42), we reported that Firmicutes and Proteobacteria followed by Actinobacteria were the most abundant phyla in breast milk samples. Furthermore, Streptococcus and Staphylococcus were also identified as the most abundant genera (11, 43). Other studies have also shown that the genera Staphylococcus, Lactobacillus, Enterococcus, and Bifidobacterium in breast milk are shared with mother-to-infant microbiota (5, 44).
Maternal diet, lactation stage, weight status, genetics, and geographical location are known to affect breast milk composition (e.g., protein, fatty acids, immunological profile, oligosaccharides) (12, 45–48). However, the impact of these factors on breast milk microbial communities is still unclear. The vertical transfer of microorganisms through lactation has a relevant role in the correct establishment of the infant gut microbiota (41). Therefore, it is necessary to identify which factors might influence the diversity and composition of breast milk microbiota, with a special emphasis on modifiable factors, such as dietary patterns and lifestyle. For example, specific bacteria from fermented food are present and shared between maternal and neonatal microbiota including breast milk (49); this highlights the key diet–breast milk–microbiota interplay.
In our study, maternal diet intakes were clustered into 2 distinct groups: Cluster I was characterized by a higher intake of dietary fiber, plant protein, carbohydrates, and polyphenols whereas Cluster II was defined by a higher intake of animal protein, total lipids, total PUFAs, total MUFAs, and SFAs. The clusters were each associated with a specific microbial composition and diversity. Cluster I was linked to a higher presence of the Staphylococcus and Bifidobacterium genera in breast milk. Additionally, we also observed that these bacterial genera were positively related to carbohydrates and polyphenols. Furthermore, these microorganisms, mainly Bifidobacterium, are the hallmark of breastfed infant gut microbiota, suggesting a possible link between breast milk and the neonatal gut, as reported previously (44).
A significant impact of specific macronutrient and micronutrient intake on milk microbial communities in healthy lactating women has been reported (50). High maternal protein consumption was associated with high abundance of Gemella in breast milk, whereas high consumption of SFAs and MUFAs was associated with low abundance of Corynebacterium in breast milk (50). In the present study, Gemella was associated with the intake of animal protein and also with PUFAs, such as n–3 DPA, n–3 EPA, and n–3 DHA. Our findings also showed an association between calcium intake and a low relative abundance of Veillonella genus. By contrast, it has been reported that calcium was positively associated with Veillonella (50). Moreover, a recent study reported an association between vitamin C intake and Staphylococcus, and between total PUFA intake and the Bifidobacterium genus (15).
Our findings clearly suggest that the Bifidobacterium genus is associated with a high intake of polyphenols and that the Staphylococcus genus is related to carbohydrates. Although the relation between polyphenols and breast milk microbiota is poorly understood, several studies have suggested that dietary polyphenols have prebiotic properties and antimicrobial activities as well as modulating the composition and functionality of the gut microbiota (51, 52). Moreover, some phenolic compounds can promote the growth of specific bacterial taxa, such as the Lactobacillus and Bifidobacterium genera (53, 54).
Microbial diversity measured by the Shannon index, but not microbial richness, was associated with higher total fiber maternal intake. We also observed that Cluster I milk samples, which are associated with higher fiber and plant protein intakes, presented higher microbial diversity and richness than breast milk samples from Cluster II. Thus, we suggest that lower fiber and plant protein intake is linked to lower microbial diversity and richness. A recent study suggested that the consumption of breast milk with reduced microbial richness during the first month of life is associated with allergy development in children (55). Therefore, maternal diet would shape breast milk microbial diversity and richness with potential impact on infant health outcomes. Furthermore, other studies have reported that breast milk microbiota diversity was higher in women who delivered vaginally than in those who delivered via C-section and had antibiotic exposure (42). Our data showed that antibiotic exposure and/or mode of delivery shaped the breast milk microbiota composition and diversity in a cluster-dependent manner. Higher microbial diversity and richness was observed in breast milk from Cluster I and vaginal/non–antibiotic exposure compared with the other groups. Interestingly, breast milk samples from women who underwent C-section and were exposed to antibiotics had higher microbial richness (Chao1 index) but not diversity (Shannon index) in Cluster I compared with Cluster II. These results would suggest that fiber and plant protein intakes can modulate the potential impact of antibiotics and C-section on the microbial diversity of breast milk. These data could promote the development of potential new dietary interventions targeted at milk microbiota in women exposed to antibiotics during C-section procedures. Thus, our data together with the available evidence highlight the potential for modulation of breast milk microbiota by perinatal factors, which consequently would influence child microbiota development and health outcomes.
C-sections and antibiotic use have been observed to impact both maternal and neonatal gut microbiota (56). These 2 are related factors and cannot be studied separately (42). Although considerable evidence is available regarding the effect of C-section birth and antibiotics on gut microbiota, studies focused on breast milk remain scarce. Contradictory findings regarding the effect of mode of delivery on milk microbiota can be found in the literature. Some studies have shown higher microbial diversity and relative abundance of Bifidobacterium and Lactobacillus spp. in the breast milk samples of women who delivered vaginally compared with those who delivered via C-section (12, 13, 42), whereas other studies reported no differences (57, 58). Hence, the debate is still open, and additional studies are needed (12). Some studies have shown that breast milk from women undergoing a C-section delivery and/or who received anesthesia during delivery tended to be less likely to contain lactobacilli (5). Mode of delivery and intrapartum antibiotic exposure were both associated with overall microbial composition in breast milk samples at 1 mo postpartum (42). Interestingly, differences between elective and emergency C-section birth seem to influence the microbiota composition (12). Milk samples from women who underwent emergency C-section showed similar microbial profiles to breast milk from those who delivered vaginally, whereas breast milk from those who had elective surgery showed microbial profiles close to those of skin and oral microbial communities (12), although other studies reported no differences (58). These results suggest the contributory role of the physiological labor process, stress, and/or hormonal signals in milk microbiota.
Similarly, antibiotics have been proposed to have an impact on milk microbiota composition (5, 42). Low abundance of Lactobacillus and Bifidobacterium in breast milk was found in mothers treated with antibiotics during pregnancy or lactation. Chemotherapy during lactation decreased the relative abundance of Bifidobacterium, Staphylococcus, and Eubacterium spp. in the milk samples (59). Several authors have related the Bifidobacterium and Lactobacillus genera to infant health outcomes, proposing the shaping of breast milk microbiota as a gateway to disease prevention (60). Lower Bifidobacterium abundances have been observed in the breast milk of allergic mothers and in their offspring's gut microbiota compared with nonallergic healthy ones (61). Thus, the metabolism of dietary compounds by both Lactobacillus and Bifidobacterium would influence the child´s development through several unknown mechanisms. We found a depletion of Lactobacillus genus in the breast milk of mothers from the Cluster II/C-section group. However, the breast milk from mothers in Cluster I who had a vaginal birth was enriched in Actinobacteria phylum. Interestingly, similar results were found in the analysis of the effect of maternal diet on neonatal gut microbiota (62). These results suggested that the possible interaction between maternal diet and mode of delivery has an effect on breast milk microbiota.
Limitations
This study was a cross-sectional study with a limited sample size and a power analysis that could have hindered—to a certain extent—the possibility of detecting other significant associations. Thus, large-scale prospective longitudinal studies are recommended in the future. Furthermore, an FFQ was chosen for the dietary records as a complete tool to estimate regular intake. However, FFQs can also introduce errors due to a lack of perception of food proportion sizes. Although we validated our data with a random subset of a 3-d diet recall questionnaire, better dietary collection tools and a wider data set would be needed in future studies. Furthermore, other health-related habits, including exercise, stress handling, and home environment have also not been considered in the analysis. The potential associations of maternal diet and other breast milk compounds (such as human milk oligosaccharides, cytokine profile, and metabolites, among others) as well as the potential impact of the observed associations on infant microbiota need to be considered and warrant further studies. Moreover, the study of breast milk samples from vaginal deliveries exposed to maternal intrapartum antibiotics would provide novel insights into the impact of maternal diet on milk microbiota.
Despite all these limitations, our study expands knowledge about the impact of maternal diet on breast milk composition and diversity, which can have potential effects in infant microbiota colonization and immune system maturation patterns.
Conclusions
In conclusion, maternal diet shapes the composition and diversity of breast milk microbiota. Macronutrients as well as other specific dietary nutrients, such as soluble and insoluble fiber and polyphenols, influence milk microbial communities. The consumption of food rich in soluble and insoluble fiber and plant protein can be related to microbiota with higher Staphylococcus, Bifidobacterium, and Lactobacillus abundance. The intake of food with high animal protein content and fats, such as n–3 PUFAs and MUFAs, showed a negative correlation with Enterococcus and Bifidobacterium and a positive correlation with Gemella, which might play a role in the composition of the developing infant's gut microbiota. Furthermore, we found an association among breast milk microbiota composition, dietary pattern, and antibiotic exposure/mode of delivery. This finding suggests a complex interaction between different maternal and perinatal factors that affects breast milk microbiota. Thus, further analysis is warranted to clarify the relations in this network and design interventions that can modify such relations in case one of the factors is affected.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the families involved in the MAMI study as well as all the members of the MAMI cohort study.
The authors’ responsibilities were as follows—MCC: designed the study; CM-C: recruited the families and collected the biological samples; IG-M, SG: were responsible for the dietary information and analysis; EC-M, MS-R: were responsible for the microbiota data and statistical analysis; EC-M: drafted the first draft; and all authors: read and approved the final manuscript.
Notes
This work was supported by the European Research Council under the European Union's Horizon 2020 research and innovation program (ERC starting grant, no. 639226). EC-M and MS-R are supported by a Predoctoral Fellowship from Generalitat Valenciana—European Social Fund (GRISOLÍA2019 and ASCII2016, respectively).
We would also like to thank the support from LaMaratò-TV3 (DIM-2-ELI, ref. 2018-27/31). We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Author disclosures: The authors report no conflicts of interest.
Data described in the manuscript will be made available upon request.
Supplemental Figures 1–3 and Supplemental Tables 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: CHI, Calinski–Harabasz index; C-section, cesarean section; DPA, docosapentaenoic acid (22:5ω-3); LEfSe, linear discriminant analysis effect size; PERMANOVA, permutational multivariate analysis of variance; RDA, redundancy discriminant analysis.
Contributor Information
Erika Cortes-Macías, Institute of Agrochemistry and Food Technology (IATA-CSIC), National Research Council, Valencia, Spain.
Marta Selma-Royo, Institute of Agrochemistry and Food Technology (IATA-CSIC), National Research Council, Valencia, Spain.
Izaskun García-Mantrana, Institute of Agrochemistry and Food Technology (IATA-CSIC), National Research Council, Valencia, Spain.
Marta Calatayud, Institute of Agrochemistry and Food Technology (IATA-CSIC), National Research Council, Valencia, Spain.
Sonia González, Department of Functional Biology, Faculty of Medicine, University of Oviedo, Oviedo, Spain; Diet, Microbiota and Health Group, Instituto de Investigación Sanitaria del Principado de Asturias (DIMISA, ISPA), Oviedo, Spain.
Cecilia Martínez-Costa, Department of Pediatrics, School of Medicine, University of Valencia, Valencia, Spain; Pediatric Gastroenterology and Nutrition Section, Hospital Clínico Universitario Valencia, INCLIVA, Valencia, Spain.
Maria Carmen Collado, Institute of Agrochemistry and Food Technology (IATA-CSIC), National Research Council, Valencia, Spain.
References
- 1. Walker A Breast milk as the gold standard for protective nutrients. J Pediatr. 2010;156:S3–7. [DOI] [PubMed] [Google Scholar]
- 2. Lönnerdal B Bioactive proteins in human milk: mechanisms of action. J Pediatr. 2010;156:S26–30. [DOI] [PubMed] [Google Scholar]
- 3. Le Huërou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. 2010;23:23–36. [DOI] [PubMed] [Google Scholar]
- 4. Gomez-Gallego C, Garcia-Mantrana I, Salminen S, Collado MC. The human milk microbiome and factors influencing its composition and activity. Semin Fetal Neonatal Med. 2016;21:400–5. [DOI] [PubMed] [Google Scholar]
- 5. Soto A, Martín V, Jiménez E, Mader I, Rodríguez JM, Fernández L. Lactobacilli and Bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors. J Pediatr Gastroenterol Nutr. 2014;59:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Turfkruyer M, Verhasselt V. Breast milk and its impact on maturation of the neonatal immune system. Curr Opin Infect Dis. 2015;28:199–206. [DOI] [PubMed] [Google Scholar]
- 7. Guaraldi F, Salvatori G. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol. [Internet]2012;2.doi:10.3389/fcimb.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hennet T, Borsig L. Breastfed at Tiffany's. Trends Biochem Sci. 2016;41:1–11. [DOI] [PubMed] [Google Scholar]
- 9. Munblit D, Treneva M, Peroni DG, Colicino S, Chow LY, Dissanayeke S, Pampura A, Boner AL, Geddes DT, Boyle RJet al. . Immune components in human milk are associated with early infant immunological health outcomes: a prospective three-country analysis. Nutrients. [Internet]2017;9 doi:10.3390/nu9060532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boix-Amorós A, Puente-Sánchez F, du Toit E, Linderborg KM, Zhang Y, Yang B, Salminen S, Isolauri E, Tamames J, Mira Aet al. . Mycobiome profiles in breast milk from healthy women depend on mode of delivery, geographic location, and interaction with bacteria. Appl Environ Microbiol. 2019;85:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunt KM, Foster JA, Forney LJ, Schütte UME, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire MA. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011;6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–51. [DOI] [PubMed] [Google Scholar]
- 13. Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. 2016;7:54–60. [DOI] [PubMed] [Google Scholar]
- 14. Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, Nicol MP, Isolauri E, Yang B, Collado MCet al. . Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. [Internet]2016;7 10.3389/fmicb.2016.01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Padilha M, Danneskiold-samsøe NB, Brejnrod A, Hoffmann C, Cabral VP, Iaucci J, de M, Sales CH, Fisberg RM, Cortez RV, Brix Set al. . The human milk microbiota is modulated by maternal diet. Microorganisms[Internet] 2019;7 doi:10.3390/microorganisms7110502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, Lix LM, de Souza RJ, Becker AB, Mandhane PJet al. . Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25:324–35. [DOI] [PubMed] [Google Scholar]
- 17. García-Mantrana I, Alcántara C, Selma-Royo M, Boix-Amorós A, Dzidic M, Gimeno-Alcañiz J, Úbeda-Sansano I, Sorribes-Monrabal I, Escuriet R, Gil-Raga Fet al. . MAMI: a birth cohort focused on maternal-infant microbiota during early life. BMC Pediatr. 2019;19:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salas-Salvadó J, Rubio MA, Barbany M, Moreno B, Grupo colaborativo de la SEEDO. Consenso SEEDO 2007 para la evaluación del sobrepeso y la obesidad y el establecimiento de criterios de intervención terapéutica. Medicina Clínica. 2007;128:184–96.. Spanish. [DOI] [PubMed] [Google Scholar]
- 19. Mouratidou T, Ford F, Fraser RB. Validation of a food-frequency questionnaire for use in pregnancy. Public Health Nutr. 2006;9:515–22. [DOI] [PubMed] [Google Scholar]
- 20. Cervera P, Farran A, Zamora-Ros R. Tablas de composición de alimentos del CESNID: Taules de composició d'aliments del CESNID. Rev Esp Salud Pública. 2004;78:407. [Google Scholar]
- 21. Marlett JA, Cheung T-F. Database and quick methods of assessing typical dietary fiber intakes using data for 228 commonly consumed foods. J Am Diet Assoc. 1997;97:1139–51. [DOI] [PubMed] [Google Scholar]
- 22. Neveu V, Perez-Jiménez J, Vos F, Crespy V, Chaffaut L, Mennen L, Knox C, Eisner R, Cruz J, Wishart Det al. . Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database. [Internet]2010. doi:10.1093/database/bap024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tognon G, Nilsson LM, Lissner L, Johansson I, Hallmans G, Lindahl B, Winkvist A. The Mediterranean diet score and mortality are inversely associated in adults living in the subarctic region. J Nutr. 2012;142:1547–53. [DOI] [PubMed] [Google Scholar]
- 24. Voortman T, Kiefte-de Jong JC, Geelen A, Villamor E, Moll HA, de Jongste JC, Raat H, Hofman A, Jaddoe VWV, Franco OHet al. . The development of a diet quality score for preschool children and its validation and determinants in the generation R study. J Nutr. 2015;145:306–14. [DOI] [PubMed] [Google Scholar]
- 25. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JMet al. . Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K, Studer M, Roudier P. Cluster: cluster analysis basics and extensions. R Package version 2.1.0. R Foundation; 2019. [Google Scholar]
- 28. Venables WN, Ripley BD. Modern applied statistics with S. Springer; 2002. [Google Scholar]
- 29. Walesiak M, Dudek A. ClusterSim: searching for optimal clustering procedure for a data set. R Package version 0.48-3. R Foundation; 2019. [Google Scholar]
- 30. Bougeard S, Dray S. Supervised multiblock analysis in R with the ade4. J Stat Soft. 2018;86:1–17. [Google Scholar]
- 31. Boix-Amorós A, Collado MC, Mira A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front Microbiol. 2016;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cruaud P, Vigneron A, Lucchetti-Miganeh C, Ciron PE, Godfroy A, Cambon-Bonavita MA. Influence of DNA extraction method, 16S rRNA targeted hypervariable regions, and sample origin on microbial diversity detected by 454 pyrosequencing in marine chemosynthetic ecosystems. Appl Environ Microbiol. 2014;80:4626–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. García-Mantrana I, Selma-Royo M, González S, Parra-Llorca A, Martínez-Costa C, Collado MC. Distinct maternal microbiota clusters are associated with diet during pregnancy: impact on neonatal microbiota and infant growth during the first 18 months of life. Gut Microbes. 2020;11:962–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. [Internet]2018. doi:10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, DeSantis TZ. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One. 2016;11:e0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion MJ, Berger B, Krause L. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017;33:782–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller Set al. . gplots: various R programming tools for plotting data. R Foundation; 2019; [cited May 2020] [Internet]. Available from: https://cran.r-project.org/web/packages/gplots/index.html. [Google Scholar]
- 41. Murphy K, Curley D, O'Callaghan TF, O'Shea C, Dempsey EM, O'Toole PW, Ross RP, Ryan CA, Stanton C. The composition of human milk and infant faecal microbiota over the first three months of life : a pilot study. Sci Rep. 2017;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hermansson H, Kumar H, Collado MC, Salminen S, Isolauri E, Rautava S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front Nutr. 2019;6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lundgren SN, Madan JC, Karagas MR, Morrison HG, Hoen AG, Christensen BC. Microbial communities in human milk relate to measures of maternal weight. Front Microbiol. 2019;10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martín V, Maldonado-Barragán A, Moles L, Rodriguez-Baños M, del Campo R, Fernández L, Rodríguez JM, Jiménez E. Sharing of bacterial strains between breast milk and infant feces. J Hum Lact. 2012;28:36–44. [DOI] [PubMed] [Google Scholar]
- 45. Collado MC, Laitinen K, Salminen S, Isolauri E. Maternal overweight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr Res. 2012;72:77–85. [DOI] [PubMed] [Google Scholar]
- 46. Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, Foster JA, Sellen DW, Kamau-Mbuthia EW, Kamundia EWet al. . What's normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: the INSPIRE Study. Front Nutr. [Internet]2019;6 doi:10.3389/fnut.2019.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tan SS, Khor GL, Stoutjesdijk E, Ng KWT, Khouw I, Bragt M, Schaafsma A, Dijck-brouwer DAJ, Muskiet FAJ. Case study of temporal changes in maternal dietary intake and the association with breast milk mineral contents. J Food Compos Anal. 2020;89:103468. [Google Scholar]
- 48. Moro GE, Bertino E, Bravi F, Tonetto P, Gatta A, Quitadamo PA, Salvatori G, Profeti C, Di Nicola P, Decarli Aet al. . Adherence to the traditional Mediterranean diet and human milk composition: rationale, design, and subject characteristics of the MEDIDIET study. Front Pediatr. [Internet]2019;7 doi:10.3389/fped.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Albesharat R, Ehrmann MA, Korakli M, Yazaji S, Vogel RF. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst Appl Microbiol. 2011;34:148–55. [DOI] [PubMed] [Google Scholar]
- 50. Williams JE, Carrothers JM, Lackey KA, Beatty NF, York MA, Brooker SL, Shafii B, Price WJ, Settles ML, McGuire MAet al. . Human milk microbial community structure is relatively stable and related to variations in macronutrient and micronutrient intakes in healthy lactating women. J Nutr. 2017;147:1739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Faraldo Corrêa TA, Rogero MM, Mariko Hassimotto AN, Lajolo FM. The two-way polyphenols-microbiota interactions and their effects on obesity and related metabolic diseases. Front Nutr. [Internet]2019;6 doi:10.3389/fnut.2019.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee HC, Jenner AM, Low CS, Lee YK. Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res Microbiol. 2006;157:876–84. [DOI] [PubMed] [Google Scholar]
- 53. Ozdal T, Sela DA, Xiao J, Boyacioglu D, Chen F, Capanoglu E. The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients. 2016;8:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. García-Mantrana I, Calatayud M, Romo-Vaquero M, Espín JC, Selva M V, Collado MC. Urolithin metabotypes can determine the modulation of gut microbiota in healthy individuals by tracking walnuts consumption over three days. Nutrients. 2019;11:2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dzidic M, Mira A, Artacho A, Abrahamsson TR, Jenmalm MC, Collado MC. Allergy development is associated with consumption of breastmilk with a reduced microbial richness in the first month of life. Pediatr Allergy Immunol. 2020;31:250–7. [DOI] [PubMed] [Google Scholar]
- 56. Rutayisire E, Huang K, Liu Y, Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: a systematic review. BMC Gastroenterol. [Internet]2016;16 doi:10.1186/s12876-016-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sakwinska O, Moine D, Delley M, Combremont S, Rezzonico E, Descombes P, Vinyes-Pares G, Zhang Y, Wang P, Thakkar SK. Microbiota in breast milk of Chinese lactating mothers. PLoS One. 2016;11:e0160856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. [Internet]2016;4 doi:10.1186/s40168-015-0145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Urbaniak C, McMillan A, Angelini M, Gloor GB, Sumarah M, Burton JP, Reid G. Effect of chemotherapy on the microbiota and metabolome of human milk, a case report. Microbiome. [Internet]2014;2:24 doi:10.1186/2049-2618-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Van Den Elsen LWJ, Garssen J, Burcelin R, Verhasselt V. Shaping the gut microbiota by breastfeeding: the gateway to allergy prevention?. Front Pediatr. [Internet]2019;7 10.3389/fped.2019.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grönlund M, Gueimonde M, Laitinen K, Kociubinski G, Grönroos T, Salminen S, Isolauri E. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exp Allergy. 2007;37:1764–72. [DOI] [PubMed] [Google Scholar]
- 62. Lundgren SN, Madan JC, Emond JA, Morrison HG, Christensen BC, Karagas MR, Hoen AG. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome. [Internet]2018;6 doi:org/10.1186/s40168-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.