Abstract
There is an increasing awareness that the gut microbiome plays a critical role in human health and disease, but mechanistic insights are often lacking. In June 2018, the Health and Environmental Sciences Institute (HESI) held a workshop, “The Gut Microbiome: Markers of Human Health, Drug Efficacy and Xenobiotic Toxicity” (https://hesiglobal.org/event/the-gut-microbiome-workshop) to identify data gaps in determining how gut microbiome alterations may affect human health. Speakers and stakeholders from academia, government, and industry addressed multiple topics including the current science on the gut microbiome, endogenous and exogenous metabolites, biomarkers, and model systems. The workshop presentations and breakout group discussions formed the basis for identifying data gaps and research needs. Two critical issues that emerged were defining the microbial composition and function related to health and developing standards for models, methods and analysis in order to increase the ability to compare and replicate studies. A series of key recommendations were formulated to focus efforts to further understand host-microbiome interactions and the consequences of exposure to xenobiotics as well as identifying biomarkers of microbiome-associated disease and toxicity.
Keywords: microbiome, gut, health, xenobiotics, environmental, biotransformation, biomarkers, animal models
The gastrointestinal (GI) microbiota play an underlying role in health. Alterations in stability and functional capabilities of microbiota are associated with disease although is unclear whether this is a cause or a result of the disease. Although there is emerging research regarding the physiological functions of intestinal microbes, less is known about the consequences of chemically induced alterations to intestinal microbiome composition and toxicological effects to the host. The interactions of xenobiotics with the microbiota as a result of drug therapy or environmental exposures are of increasing interest to public health.
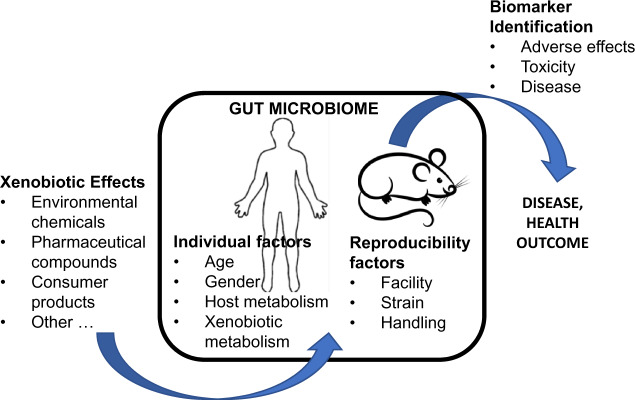
Health and Environmental Sciences Institute (HESI) assembled a cross-disciplinary group of experts (Table 1) to examine gut microbial-host dynamics in order to better understand what is currently known about the effect of chemical exposures on the microbiome and to identify key knowledge gaps related to such exposures (Figure 1).
Table 1.
Workshop Topics and Presenters
| Workshop Topics | Expert Presenters |
|---|---|
| Drugging gut microbial enzymes for treatment of CVD |
|
| Biotransformation |
|
| Biomarkers of adverse effects |
|
| Biomarkers of toxicity and disease |
|
| Human susceptibility |
|
| Animal models |
|
Experts from all sectors addressed state of the science and need for identification of biomarkers to advance understanding and decision making on efficacy and safety of xenobiotics. Presentations can be found on the HESI website here (https://hesiglobal.org/event/the-gut-microbiome-workshop/).
Figure 1.
Key data gaps in the microbiome research field identified at the 2018 HESI workshop. HESI’s June 2018 workshop identified key data gaps in the gut microbiome research field. The results identify needs for easily accessible biomarkers and improved human and animal experimental model studies for testing and validation of the impact of changes in the gut microbiome on human health outcomes.
KEY ISSUE NO. 1: DEFINING GUT MICROBIOMES ASSOCIATED WITH HEALTH
The intestinal microbiome is made up of between 500 and 1000 bacterial species as well as viruses, archaea, and eukaryotic microorganisms, many of which play a role in human health and disease (Backhed et al., 2012; Gilbert et al., 2018; Hanson and Weinstock, 2016; Ogilvie and Jones, 2015; Qin et al., 2010; Tuddenham and Sears, 2015; Virgin, 2014; Ward et al., 2018). There is great variation in species diversity, taxonomic composition, and population density of microbiota in the gut of humans and animal species. For those taxa that contribute to human health little is known about the part that each microbial species is filling, how well they perform that function, their interactions with other microorganisms and with the host, and where in the host they are acting (Backhed et al., 2012; Barratt et al., 2017; Cho and Blaser, 2012; Gilbert et al., 2018). Understanding the functions, features and normal ranges of the microbial communities that support health will be essential in addressing impacts of environmental agents and potentially identifying microbial configurations that result in disease (Lloyd-Price et al., 2016, 2019). The inclusion of functional measurements may lead to different interpretations of microbiota diversity, compared with taxonomic classification alone, leading to greater analytical challenges (Fu et al., 2016; Zhu et al., 2015). However, there is a growing consensus that functional characterizations are necessary for understanding and modeling the physiology of the microbiome (Heintz-Buschart and Wilmes, 2018; Moya and Ferrer, 2016; Zhu et al., 2015).
Before beneficial or adverse effects of chemical exposure can be determined, it is necessary to have a baseline for the bacterial species diversity as well as an understanding of the functional capacities associated with health. Given the considerable variation across human populations, among individuals and even within an individual, this will not be a single microbiome but a group of features, capabilities and characteristics of microbial communities that contribute to health. In defining such microbiomes, temporal variability in these communities, such as short-term perturbations associated with diet, lifestyle traits, antibiotic exposure, or acute illness, as well as more long-term impacts that may lead to deleterious chronic health outcomes must be considered (Kundu et al., 2017). In addition, factors such as geographic location, ethnicity, age, physical activity, genetics and gender, resistance (the ability to withstand stress or perturbations), and resilience (the capacity to return to a healthy state) influence an individual’s microbiome (Backhed et al, 2012). For example, diet is associated with significant differences in microbiota both across populations and with longitudinal studies, yet there may not be apparent adverse impacts to human health (Clemente et al, 2015; Yatsunenko et al, 2012). Such observations suggest that there may be functional redundancy in biochemical pathways in the microbiome (Baumler and Sperandio, 2016). Large cohort studies may be needed in defining normal variations within a microbiome, especially if the studies include longitudinal sampling of the microbiome (Faust et al., 2015; Fettweis et al. 2019; Flores et al., 2014; Lloyd-Price et al., 2019; Proctor et al., 2019; Vandeputte et al., 2017a,b; Yassour et al., 2016; Zhou et al., 2019). Measurement of blood metabolome is useful in predicting gut α-diversity in such studies (Price et al., 2017; Wilmanski et al., 2019).
For most healthy adults, there appears to be a window of microbiome normalcy whose bounds are not typically exceeded, even though there may be minor transitory variation in composition observed with dietary changes or exposures to certain xenobiotics (Kundu et al., 2017; Moya and Ferrer, 2016). However, changes in microbial communities can be associated with adverse health consequences. In one model, this may occur if a person’s microbiome leaves their respective window of normalcy reaching a “tipping point” where their health is affected. Alternatively, rather than a distinct shift there can be stochastic changes in the microbiome leading to dispersion of the microbiome composition as well as increased functions that are related to disease (Armour et al., 2019; Zaneveld et al. 2017).
RESEARCH NEED NO. 1
In order to understand, adverse health effects of the microbiome resulting from disease or xenobiotic exposure, there is a need to better understand the essential components, capabilities, and range of microbial variation linked to a person’s health. This requires comprehension of both the structure and the function of the microbial community. Identification of key conserved functional, metabolic, and biochemical pathways present in healthy individuals may provide a strong starting point.
It is evident that multiple factors contribute to the composition, diversity and function of the gut microbiome with regard to health, drug efficacy, and xenobiotic toxicity. The list includes genetics, diet, age, obesity, concomitant diseases, drugs, and gender to name a few. There is a need to not only understand the impact of a single factor on the dynamics between gut microbiome and the host but also to determine contributions of combinations of these factors. However, defining what constitutes microbial diversity and functionally of both healthy and unhealthy individuals will help to elucidate the relative contributions of host factors.
Large cohort studies are valuable sources of data for defining microbiomes associated with health, but comparison and reproducibility across such studies is hindered by differences in experimental design and data analysis. Variation and bias in sampling procedures, stabilization conditions, shipment and storage practices, methods for analyte extraction and purification, sample preparation and analysis, bioinformatic analysis algorithms as well as the reference databases used are contributing factors (Backhed et al., 2012; Gilbert et al., 2018; Sinha et al., 2017; Stulberg et al., 2016). Standardization of methods and analytical approaches is needed to develop a well-characterized measurement pipeline. However, procedures may need to remain somewhat fluid to account for continuing technological developments and evolving understanding of the biology involved.
KEY ISSUE NO. 2: DEFINING CAUSES AND EFFECTS OF ALTERING THE GUT MICROBIOME
GI microbiota are essential in maintaining health and wellness by digesting food nutrients, producing endogenous metabolites and biotransforming xenobiotics (Dietert and Silbergeld, 2015b). Furthermore, in a healthy individual the gut microbiome, together with the intestinal mucosa, is necessary to maintain gut homeostasis and provide an epithelial barrier between the lumen and the rest of the body (Hiippala et al., 2018; Rogers, 2015).
Loss, disruption or dispersion of a functional microbiome can be associated with acute adverse health effects or chronic diseases. For example, antibiotics can alter a healthy individual’s microbiome resulting in diarrhea, leaving the body susceptible to the development of Clostridium difficile infection (Backhed et al., 2012; Schaffler and Breitruck, 2018; Schubert et al., 2014). Changes in gut microbiota may be linked to the development of inflammatory bowel disease, colorectal cancer, and fatty liver disease (Miyoshi et al., 2017; Sharpton et al., 2019; Wang et al., 2017).
Many of these chronic diseases have inflammatory or altered immune system characteristics. Critical immune functions related to epithelial signaling, inflammatory responses, production of antimicrobial factors, and induction of Immunoglobulin A antibodies are propagated locally in the GI tract and associated secondary lymphoid tissues (Peyer’s Patches; Baumler and Sperandio, 2016; Dietert and Dietert, 2015a; Hiippala et al., 2018). The microbiome contributes to the development and maintenance of the immune system (Agace and McCoy, 2017). Early life is characterized by a period of microbial flux and assembly which is affected by multiple factors such as manner of delivery (vaginal or c-section), nutrition (breast milk or formula) and antibiotic exposure. Although dynamic in nature, in adults the majority of the composition and function of the microbiome of a healthy person is fairly stable, but this decreases with age due to the loss of key species and a progressive gain of pathobiotic bacteria (Buford, 2017).
The systemic influence of the GI microbiota on human health can be altered by exogenous compounds during drug therapy or environmental exposures. Antibiotics are used to remove a pathogen from the host, but an unintended consequence can be alteration of the composition and function of the gut microbiota (Miyoshi et al., 2017). Exposure to nicotine, arsenic, or polybrominated diphenyl ethers change the gut microbiome diversity (Chi et al., 2017, 2018; Li et al., 2018).
Exposure to xenobiotics can have effects in multiple ways. These include those that are transitory, established, or developmentally programmed (Dietert and Dietert, 2015a; Faust et al., 2015; Fofanova et al., 2016; Moya and Ferrer, 2016; Norman et al., 2015; Pascal et al., 2017; Sanz et al., 2015; Stenman et al., 2016). The transitory outcomes, such as changes after eating, are typically easily correctable and the gut microbiome return to the composition and functionality exhibited before the insult. Established outcomes occur when there is the alteration of a keystone species within the microbiota which would require significant host and microbiota changes to recover the original microbiota (eg, after C. difficile infection). The developmentally programmed outcomes are those that occur when the adverse impact is the result of alteration during a critical window of the microbiome development and can lead to later-in-life problems associated with the microbiome (eg, antibiotics during early childhood).
RESEARCH NEED NO. 2
The gut microbiome plays an essential role in maintaining health and modification of the microbial communities can have negative effects in the body. Defining what changes in the microbiota are associated with acute or chronic adverse effects, as well as the magnitude of change necessary, is needed. Adverse effects can range from changes to the intestinal barrier integrity to alterations or dispersion of the normal microbiota of the host population of microbiota leading to a diseased state. Identification of such adverse effects may provide biomarkers of disease progression. To begin to identify causes and effects of changes in the microbial population, 2 approaches are possible. One is to start from a healthy individual’s baseline microbiome then determine deviations associated with a disease state. The second is determining what is a diseased state and ascertaining if there are key nodes, drivers, or contributing factors that move individuals from health to illness.
Despite the improvements in analytical, genomic, and bioinformatic techniques related to structure and function of the microbiome in health and disease, there is still a knowledge gap in the translation of high-throughput data from genotype to phenotype and microbiome composition stability/instability versus core function of the microbial community.
KEY ISSUE NO. 3: ACCOUNTING FOR BIOTRANSFORMATION
GI microbiota have the capacity to metabolize endogenous and exogenous compounds. The metabolites can have positive, neutral, or negative effects on the host. Endogenous microbial metabolites can exert physiological functions. Gut microbe-derived metabolites can signal via receptors at the epithelium interface and communicate with the host. For example, microbial metabolism of tryptophan generates aryl hydrocarbon receptor (AHR) ligands, such as indole, indirubin, and indigo (Hubbard et al., 2015). These ligands activate AHR and promote intestinal homeostasis through regulation of innate cytokine or chemokine gene expression, regulation of enterocyte differentiation, as well as regulation and development of intraepithelial lymphocytes and innate lymphoid cells. Gut microbial metabolism can result in products associated with adverse health and disease progression. The microbiome converts dietary lipid phosphatidylcholine to trimethylamine (TMA), which is then metabolized by hepatic flavin monooxygenases to TMA-N-oxide (TMAO). TMAO is a proatherogenic factor associated with cardiovascular disease (CVD; Brown and Hazen, 2015). Such findings suggest that endogenous microbial metabolites could be used as potential biomarkers of health or disease.
GI microbiota express biotransformation enzymes that metabolize a variety of therapeutic drugs and environmental compounds that can result in changes in efficacy and toxicity (Klaassen and Kui, 2015; Spanogiannopoulos et al., 2016). Gut bacteria can convert a prodrug to an active drug or detoxify the parent compound leading to modified drug availability, as well as changes in pharmacokinetics-pharmacodynamics (PKPDs). Sulfasalazine, a prodrug for ulcerative colitis, is directly metabolized by the gut bacteria at the azo bond to generate 2 bioactive metabolites, sulfapyridine (antimicrobial) and 5-aminosalicylate (anti-inflammatory; Peppercorn and Goldman, 1972). The cardiac drug glycoside digoxin undergoes reduction to an inactive metabolite, dihydrodigoxin, which is significantly decreased if antibiotics are preadministered (Haiser et al., 2013; Lindenbaum et al., 1981). Microbial metabolites of a drug can also increase toxicity. The chemotherapeutic drug irinotecan is metabolized to SN-38 which is both the active and toxic metabolite. SN-38 is formed and glucuronidated by the liver and then transported to the gut where it undergoes deconjugation by gut bacteria. This results in longer gastrointestinal (GI) exposure, reabsorption, and greater bioavailability of SN-38 (Wallace et al., 2010). The gut microbiome is also implicated in the interindividual variability in metabolism of the analgesic and antipyretic acetaminophen. The endogenous microbial metabolite p-cresol and acetaminophen are substrates for sulfotransferases. Individuals with high levels of p-cresol generated by bacteria may have decreased acetaminophen metabolism through competition for hepatic sulfonation, which could result in an elevated risk for acetaminophen-induced hepatotoxicity (Clayton et al., 2009).
Nontherapeutic drugs and environmental xenobiotics can induce compositional changes and functional changes an in gut microbiota. In turn, these compounds may also be biotransformed by the microbiome. Arsenic is metabolized by gut bacteria and also alters the abundance and profile of the gut microbiome (Chi et al., 2018; Gokulan et al., 2018; Lu et al., 2014). Polychlorinated biphenyl or polybrominated diphenyl ether exposed mice show altered bile acid homeostasis resulting in part from changes in gut microbiota bile acid metabolism (Cheng et al, 2018; Li et al, 2018). Gut anaerobes are capable of transforming Hg2+ to highly toxic and permeable methylmercury and significantly contributing to its body burden and poisoning (Edwards and McBride, 1975).
In addition to directly biotransforming chemicals, the gut microbiome can influence the host xenobiotic metabolizing capability. Comparison of hepatic drug metabolizing genes in conventional and germ-free (GF) mice show that 34 genes, including Cyp3a, decrease, whereas 21 genes, including Cyp4a, increase (Selwyn et al., 2015). A cluster of 112 hepatic genes linked to xenobiotic metabolism and retinoid X receptor-inhibiting pathways are differentially expressed in conventional and GF mice. This results in more efficient pentobarbital metabolism and shorter anesthesia time in GF mice (Bjorkholm et al., 2009).
RESEARCH NEED NO. 3
Current experimental approaches to evaluate microbe-mediated biotransformation primarily include: (1) in vitro incubation of individual bacterial strains, mixed cultures in bioreactors, or purified enzymes with compounds, (2) ex vivo incubation of a fecal microbiome community (or other GI regions of interest) with compounds, and (3) in vivo administration of compounds into animals such as rodents. These approaches provide evidence in support of microbial metabolism of many approved drugs (Sousa et al., 2008; Zimmermann et al., 2019). However, more caution is required when extrapolating these results into humans than with traditional PKPD approaches, given the high diversity and interindividual variability of the human gut microbiome. Comprehensive and accurate modeling approaches for analyzing microbial metabolism information and incorporating into host PKPD models are necessary.
As more information on biotransformation of xenobiotics by the microbiome is published, a public database would be valuable. Such a database would be a repository centralizing comprehensive data that identifies compounds, responsible bacteria and/or enzymes, metabolites generated and metabolic pathways. This requires a collaborative effort involving researchers from academia, government and industry, working together with the regulatory agencies to promote a better understanding of microbiome-mediated biotransformation.
KEY ISSUE NO. 4: DETERMINING BIOMARKERS OF DISEASE AND TOXICITY
Biomarkers of disease and toxicity resulting from perturbations of the gut microbiota composition and function, from changes in microbial metabolites, or as a consequence of microbial biotransformation of xenobiotics would be useful. There are a number of characteristics for an ideal biomarker. It should be able to differentiate between disease progression and/or response to treatment. Stool samples or accessible tissues and/or biofluids should be used and analysis be affordable to facilitate acceptance and use for large scale screening. The biomarker needs to work for high-risk populations and distinguish between the disease and other potentially microbiome-mediated effects.
There are some examples of microbiome biomarkers of disease that are promising. Alterations in gut microbiota are associated with hypertension. Exposure of hypertensive rats to the antibiotic minocycline attenuates blood pressure (Yang et al., 2015). In animals and humans systolic blood pressure is correlated with microbiota composition and metabolites, as well as alterations in gut structure (Kim et al., 2018). Zonulin, a bacterial product and a marker of intestinal permeability, is elevated with high systolic blood pressure but is not an ideal biomarker since it is also associated with celiac disease (Fasano et al., 2000; Kim et al., 2018). A more specific indicator may be the negative correlation of butyrate producing bacteria which alter systolic blood pressure and plasma butyrate levels (Kim et al., 2018).
TMAO blood levels are of interest as a biomarker for CVD, insulin resistance, and type 2 diabetes (Miao, 2015). The production of TMAO in mammals primarily occurs via gut bacteria. Studies in humans and animals show circulating levels of TMAO are associated with risk and progression of CVD (Tang et al., 2014). Sensitive assays, particularly those amenable to the clinical setting, are being developed which will facilitate use of TMAO as a biomarker (Garcia et al, 2017). The initial association of TMAO with CVD resulted from using untargeted metabolomics in cardiac patients (Wang et al., 2011). This approach can be expanded to use multiple data streams (clinical tests, metabolomes, proteomes, genome sequence, and microbiome) to develop correlation networks in order to identify bacterial analytes associated with normal physiology or disease (Price et al., 2017). For example, there is an association between phenylacetylglutamine and the Coriobacteriaceae and Mogibacteriaceae families. Phenylacetylglutamine is a microbial metabolite and is a risk factor for CVD in those with chronic kidney disease (Poesen et al., 2016).
Microbial biomarkers may also prove useful for identifying xenobiotic toxicity. Low dose exposure of mice to polychlorinated biphenyls alters microbiota composition leading to a greater abundance of species that generate secondary bile acids causing increases in serum bile acids (Cheng et al., 2018). At higher doses serum bile acids were not affected due to an increase in hepatic efflux transporters (Cheng et al., 2018).
Although in vivo studies are useful for discovering biomarkers, there is also a need for in vitro methods. In mice, tempol acts as an antioxidant by detoxifying reactive oxygen species and modulates the gut microbiome host signaling axis resulting in prevention of weight gain (Cai et al., 2016; Li et al., 2013). Incubation of mouse cecal content with tempol and analysis of samples using flow cytometry as well as high-throughput mass spectrometry and nuclear magnetic resonance-based metabolomics show that tempol disrupted microbiota membrane physiology and metabolic activity consistent with the in vivo data (Cai et al., 2016). Such a multi-prong in vitro approach holds promise for screening xenobiotics for gut microbial toxicity and identifying potential biomarkers.
RESEARCH NEED NO. 4
Case studies provide examples of potential biomarkers that could serve as the basis for noninvasive diagnostic tests to identify disease and track its progression. When an association between a disease and gut microbiota is recognized, additional work will be needed to distinguish between microbial changes that are a cause or a result of the disease. Biomarkers should also be applicable beyond their discovery cohorts and be able to distinguish between disease states of interest and nontarget diseases, as well as having a reasonable degree of specificity.
Discovery and qualification of biomarkers will also be advanced by the adoption of standard protocols which will generate robust and reproducible data that facilitates comparisons of studies. Detailed methods and procedures for analysis of gut microbiota structure and function that will be useful for toxicologic studies are available (Nichols et al., 2018).
KEY ISSUE NO. 5: OPTIMIZING ANIMAL MODELS
Although animal species have limitations in completely replicating humans, they provide an opportunity to manipulate host genetics and the environment to allow insight into the relationship between the microbiota, xenobiotics, toxicity, and disease. Although species differences must be recognized in extrapolating animal studies to humans, such data are valuable.
Rodents are widely used in studying the microbiome. The compositions of gut microbiota vary among genetically distinct strains of mice (Friswell et al., 2010). Differences also occur in laboratory strains compared with genetically similar wild populations (Rosshart et al., 2017). GF mice and gene-knockout mice are also useful models. GF mice can be colonized with a single (mono-associated) agent, a defined bacterial combination such as altered Schardler flora or human fecal samples (De Palma et al., 2017; Franklin and Ericsson, 2017; Orcutt et al., 1987; Ridaura et al., 2013). Interestingly, introducing microbiota from wild mice into laboratory mouse strains increases resistance to influenza infection, decreases inflammation and increases resistance to certain types of cancer (Rosshart et al., 2017). The wild mouse microbiome develops as a result of evolutionary pressures due to continuous exposure to natural toxins and pathogens, thus it may offer a better model for human disease (Rosshart et al., 2017).
Disparities in gut microbiota of the same mouse strain can be attributed to vendor or institutional location (Friswell et al., 2010). Within a facility, the room, cage housing, food, water, and bedding all have an effect (Ericsson et al., 2018; Friswell et al., 2010; Hildebrand et al., 2013; Hugenholtz and de Vos, 2018; Robertson et al., 2019).
Although rodents are the most commonly used models in microbiome studies, zebrafish are also proving to be useful. Zebrafish develop rapidly and externally to the mother, thereby enabling direct embryo exposures. The GI tract is homologous to that in mammals, although there are differences in the immune system. Zebrafish are kept at a lower temperature than mammals which may compromise the ability to study human-relevant microorganisms. Xenobiotic-microbiome interactions can be evaluated in the zebrafish model. For example, exposure of zebrafish larvae to the antimicrobial compound triclosan results in changes in the community structure of the microbiome. There is an increase in triclosan resistant species which results in a greater ability to biotransform triclosan (Weitekemp et al., 2019). Gut microbiota in zebrafish also alter metabolism and mediate the neurodevelopmental toxicity of 17-β-estradiol (Catron et al., 2019).
RESEARCH NEED NO. 5
Study reproducibility in the biosciences is an ongoing concern (Mullane and Williams, 2017). Variations in experimental design, methods, and statistical analysis can contribute to an inability to replicate results and conclusions. Complicating this further is the diversity and function of the gut microbiota (Franklin and Ericsson, 2017; Turner, 2018).
Reproducibility in studies of gut microbiome host dynamics can be increased by recognizing the sources of variation, prioritizing them; then, identifying ways to address them (Ericsson and Franklin, 2015; Franklin and Ericsson, 2017). To limit variation in mouse models, numerous approaches have been developed to standardize gut microbiota profiles including cohousing, cross fostering, and GF derivation, although each has advantages and disadvantages (Ericsson and Franklin, 2015; Franklin and Ericsson, 2017). Maintaining a stable microbiome composition across animals and experiments is a problem that is not easily solved. Banking fecal samples annually and defining the microbiome at the initiation of a study would allow institutional drift to be monitored (Franklin and Ericsson, 2017). In some instances, repeating studies in the second generation addresses the need for exposure to microbiota in early development (Franklin and Ericsson, 2017). For zebrafish, protocols for GF derivation and gnotobiotic husbandry provide useful guidance for standardization (Melancon et al., 2017).
Often the discrepancies in reported results can be attributed to variations in study design. Unfortunately, key details are often missing from publications. Omissions can be reduced by adoption of Animal Research: Reporting of Experiments guidelines (Kilkenny et al., 2012). This checklist of 20 items covers species, strain and number of animals, husbandry, experimental design, methods, and statistical analysis.
Determining the optimal microbiota to facilitate translating results from mice to humans depends on the question being asked. The current model of colonizing GF mice with microbiota derived from inbred, wild murine, or human sources has great potential. This approach is currently limited by the availability of the number of defined combinations and characterized mouse or human material. The development of a wider array of standardized samples would be useful.
Several in vitro models of host-microbiome interactions have been described that complement in vivo studies (Arnold et al., 2016; von Martels et al., 2017). Although there are anaerobic bacteria gut epithelial cell cocultures showing feasibility for experimental use, further refinements and standardization are still necessary (von Martels et al., 2017). It is also recognized that no one model is perfect, the best one to use will depend on the question being addressed.
RECOMMENDATIONS
The gut microbiome plays a pivotal part in health, but understanding its multiple roles still needs to be fully elucidated. HESI’s gut microbiome workshop was convened to identify research areas critical to determining how gut microbiome alterations may influence human health. Data gaps and research needs to address specific issues have been identified based on workshop presentations and breakout group discussions (Table 2).
Table 2.
Key Issues and Research Needs
| Key Issue | Research Needs |
|---|---|
| Defining gut microbiomes associated with health |
|
| Defining causes and effects altering the gut microbiome |
|
| Accounting for biotransformation |
|
| Determining biomarkers of disease and toxicity |
|
| Optimizing animal models |
|
Key research areas and needs, as identified by workshop speakers and participants who participated in breakout group discussions.
There are 2 topics that cut across the key issues. The first is the lack of clarity in what constitutes gut microbiomes in healthy individuals. It is recognized that there are many factors that contribute to variation across and within human populations. Consequently, there is not a single microbiome but rather a suite of microbiomes that are associated with health. As yet, these have not yet been adequately defined. Having information on which species comprise such microbiomes and the normal range of variation is critical in order to define disease and adverse effects. The second is the need to adopt standards for methods, models and data analysis. This will facilitate comparisons across studies and reproducibility of data.
Moving forward the key recommendations are to focus efforts on the following important areas:
Defining the range of gut microbiota composition and function gut microbiomes associated with health and/or disease;
Identifying microbiome changes linked to disease and adverse health effects;
Characterizing the formation and function of microbiota metabolism of endogenous products on health and disease;
Determining the effects of xenobiotics on microbiota composition and function;
Increasing understanding of the impact of microbiota biotransformation of xenobiotics on efficacy and toxicity;
Identifying a suite of biomarkers to monitor health, disease and adverse effects resulting from microbiota-host interactions;
Standardizing variables such as husbandry, study design, sample collection, analysis, and statistical methods
Addressing these issues will provide further insight into the role of the microbiome in human health, disease and toxicity.
ACKNOWLEDGMENTS
Health and Environmental Sciences Institute (HESI) is an independent nonprofit that collaboratively identifies and helps to resolve global and environmental challenges by convening experts through multi-sector forums. HESI’s work aims to move scientific principles into tested solutions that can be broadly applied to benefit health and the environment. The authors gratefully acknowledge the contributions of the speakers and the workshop participants, the discussions of which form the basis for this article. The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the author employers.
FUNDING
This work was provided in-kind by the Health and Environmental Sciences Institute (HESI) Microbiome Subcommittee, which is supported by sponsorships from member companies. HESI’s scientific initiatives are primarily supported by the in-kind contributions (from public and private sector participants) of time, expertise, and experimental effort. These contributions are supplemented by direct funding (that primarily supports program infrastructure and management) provided primarily by HESI’s corporate sponsors.
DECLARATION OF CONFLICTING INTERESTS
The authors of the article volunteered to serve on the HESI Microbiome committee and were involved in the planning and execution of the workshop. The views expressed in this article reflect the discussions from the workshop and do not necessarily reflect the views or policies of their employers.
Disclaimer: This workshop report is for information purposes only and shall not be construed to represent the official position or an obligation on the part of the U.S. Federal Government or any individual organization to provide support for any ideas or recommendations identified in it.
REFERENCES
- Agace W. W., McCoy K. D. (2017). Regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 46, 532–548. [DOI] [PubMed] [Google Scholar]
- Arnold J. W., Roach J., Azcarate-Peril M. A. (2016). Emerging technologies for gut microbiome research. Trends Microbiol. 24, 887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour C. R., Nayfach S., Pollard K. S., Sharpton T. J. (2019). A metagenomic meta-analysis reveals functional signatures of health and disease in the human gut microbiome. mSystems 4, e00332–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F., Fraser C. M., Ringel Y., Sanders M. E., Sartor R. B., Sherman P. M., Versalovic J., Young V., Finlay B. B. (2012). Defining a healthy human gut microbiome: Current concepts, future directions, and clinical applications. Cell Host Microbe 12, 611–622. [DOI] [PubMed] [Google Scholar]
- Barratt M. J., Lebrilla C., Shapiro H. Y., Gordon J. I. (2017). The gut microbiota, food science, and human nutrition: A timely marriage. Cell Host Microbe 22, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumler A. J., Sperandio V. (2016). Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkholm B., Bok C. M., Lundin A., Rafter J., Hibberd M. L., Pettersson S. (2009). Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4, e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. M., Hazen S. L. (2015). The gut microbial endocrine organ: Bacterially derived signals driving cardiometabolic diseases. Annu. Rev. Med. 66, 343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buford T. W. (2017). (Dis)Trust your gut: The gut microbiome in age-related inflammation, health, and disease. Microbiome 5, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Zhang L., Jones R. A., Correll J. B., Hatzakis E., Smith P. B., Gonzalez F. J., Patterson A. D. (2016). Antioxidant drug tempol promotes functional metabolic changes in the gut microbiota. J. Proteome Res. 15, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catron T. R., Swank A., Wehmas L. C., Phelps D., Keely S. P., Brinkman N. E., McCord J., Singh R., Sobus J., Wood C. E., et al. (2019). Microbiota alter metabolism and mediate neurodevelopmental toxicity of 17beta-estradiol. Sci. Rep. 9, 7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. L., Li X., Lehmler H. J., Phillips B., Shen D., Cui J. Y. (2018). Gut microbiota modulates interactions between polychlorinated biphenyls and bile acid homeostasis. Toxicol. Sci. 166, 269–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi L., Gao B., Tu P., Liu C. W., Xue J., Lai Y., Ru H., Lu K. (2018). Individual susceptibility to arsenic-induced diseases: The role of host genetics, nutritional status, and the gut microbiome. Mamm. Genome 29, 63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi L., Mahbub R., Gao B., Bian X., Tu P., Ru H., Lu K. (2017). Nicotine alters the gut microbiome and metabolites of gut-brain interactions in a sex-specific manner. Chem. Res. Toxicol. 30, 2110–2119. [DOI] [PubMed] [Google Scholar]
- Cho I., Blaser M. J. (2012). The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 13, 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton T. A., Baker D., Lindon J. C., Everett J. R., Nicholson J. K. (2009). Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. U.S.A. 106, 14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente J. C., Pehrsson E. C., Blaser M. J., Sandhu K., Gao Z., Wang B., Magris M., Hidalgo G., Contreras M., Noya-Alarcón Ó., et al. (2015). The microbiome of uncontacted Amerindians. Sci. Adv. 1, e1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma G., Lynch M. D., Lu J., Dang V. T., Deng Y., Jury J., Umeh G., Miranda P. M., Pigrau Pastor M., Sidani S., et al. (2017). Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci. Transl. Med. 9, eaaf6397. [DOI] [PubMed] [Google Scholar]
- Dietert R. R., Dietert J. M. (2015. a). The microbiome and sustainable healthcare. Healthcare (Basel) 3, 100–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietert R. R., Silbergeld E. K. (2015. b). Biomarkers for the 21st century: Listening to the microbiome. Toxicol. Sci. 144, 208–216. [DOI] [PubMed] [Google Scholar]
- Edwards T., McBride B. C. (1975). Biosynthesis and degradation of methylmercury in human faeces. Nature 253, 462–464. [DOI] [PubMed] [Google Scholar]
- Ericsson A. C., Franklin C. L. (2015). Manipulating the gut microbiota: Methods and challenges. ILAR J. 56, 205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson A. C., Gagliardi J., Bouhan D., Spollen W. G., Givan S. A., Franklin C. L. (2018). The influence of caging, bedding, and diet on the composition of the microbiota in different regions of the mouse gut. Sci. Rep. 8, 4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano A., Not T., Wang W., Uzzau S., Berti I., Tommasini A., Goldblum S. E. (2000). Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet 355, 1518–1519. [DOI] [PubMed] [Google Scholar]
- Faust K., Lahti L., Gonze D., de Vos W. M., Raes J. (2015). Metagenomics meets time series analysis: Unraveling microbial community dynamics. Curr. Opin. Microbiol. 25, 56–66. [DOI] [PubMed] [Google Scholar]
- Fettweis J. M., Serrano M. G., Brooks J. P., Edwards D. J., Girerd P. H., Parikh H. I., Huang B., Arodz T. J., Edupuganti L., Glascock A. L., et al. (2019). The vaginal microbiome and preterm birth. Nat. Med. 25, 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores G. E., Caporaso J. G., Henley J. B., Rideout J. R., Domogala D., Chase J., Leff J. W., Vazquez-Baeza Y., Gonzalez A., Knight R., et al. (2014). Temporal variability is a personalized feature of the human microbiome. Genome Biol. 15, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofanova T. Y., Petrosino J. F., Kellermayer R. (2016). Microbiome-epigenome interactions and the environmental origins of inflammatory bowel diseases. J. Pediatr. Gastroenterol. Nutr. 62, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin C. L., Ericsson A. C. (2017). Microbiota and reproducibility of rodent models. Lab. Anim. (NY )46, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friswell M. K., Gika H., Stratford I. J., Theodoridis G., Telfer B., Wilson I. D., McBain A. J. (2010). Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One 5, e8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B. C., Randolph T. W., Lim U., Monroe K. R., Cheng I., Wilkens L. R., Le Marchand L., Hullar M. A., Lampe J. W. (2016). Characterization of the gut microbiome in epidemiologic studies: The multiethnic cohort experience. Ann. Epidemiol. 26, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E., Wolak-Dinsmore J., Wang Z., Li X. S., Bennett D. W., Connelly M. A., Otvos J. D., Hazen S. L., Jeyarajah E. J. (2017). NMR quantification of trimethylamine-N-oxide in human serum and plasma in the clinical laboratory setting. Clin. Biochem. 50, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Blaser M. J., Caporaso J. G., Jansson J. K., Lynch S. V., Knight R. (2018). Current understanding of the human microbiome. Nat. Med. 24, 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokulan K., Arnold M. G., Jensen J., Vanlandingham M., Twaddle N. C., Doerge D. R., Cerniglia C. E., Khare S. (2018). Exposure to arsenite in CD-1 mice during juvenile and adult stages: Effects on intestinal microbiota and gut-associated immune status. mBio 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser H. J., Gootenberg D. B., Chatman K., Sirasani G., Balskus E. P., Turnbaugh P. J. (2013). Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B. M., Weinstock G. M. (2016). The importance of the microbiome in epidemiologic research. Ann. Epidemiol. 26, 301–305. [DOI] [PubMed] [Google Scholar]
- Heintz-Buschart A., Wilmes P. (2018). Human gut microbiome: Function matters. Trends Microbiol. 26, 563–574. [DOI] [PubMed] [Google Scholar]
- Hiippala K., Jouhten H., Ronkainen A., Hartikainen A., Kainulainen V., Jalanka J., Satokari R. (2018). The Potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 10, 988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F., Nguyen T. L., Brinkman B., Yunta R. G., Cauwe B., Vandenabeele P., Liston A., Raes J. (2013). Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 14, R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T. D., Murray I. A., Perdew G. H. (2015). Indole and tryptophan metabolism: Endogenous and dietary routes to Ah receptor activation. Drug Metab. Dispos. 43, 1522–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz F., de Vos W. M. (2018). Mouse models for human intestinal microbiota research: A critical evaluation. Cell Mol. Life Sci. 75, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C., Browne W. J., Cuthi I., Emerson M., Altman D. G. (2012). Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. Vet. Clin. Pathol. 41, 27–31. [DOI] [PubMed] [Google Scholar]
- Kim S., Goel R., Kumar A., Qi Y., Lobaton G., Hosaka K., Mohammed M., Handberg E. M., Richards E. M., Pepine C. J., et al. (2018). Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin. Sci. (Lond.) 132, 701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen C. D., Cui J. Y. (2015). Review: Mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab. Dispos. 43, 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P., Blacher E., Elinav E., Pettersson S. (2017). Our gut microbiome: The evolving inner self. Cell 171, 1481–1493. [DOI] [PubMed] [Google Scholar]
- Li, F., Jiang, C., Krausz, K. W., Li, Y., Albert, I., Hao, H., Fabre, K. M., Mitchell, J. B., Patterson, A. D., and Gonzalez, F. J. (2013). Microbiome Remodelling Leads to Inhibition of Intestinal Farnesoid X Receptor Signalling and Decreased Obesity. Nat. Commun. 4, 2384–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Y., Dempsey J. L., Wang D., Lee S., Weigel K. M., Fei Q., Bhatt D. K., Prasad B., Raftery D., Gu H., et al. (2018). PBDEs altered gut microbiome and bile acid homeostasis in male C57BL/6 mice. Drug Metab. Dispos. 46, 1226–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbaum J., Rund D. G., Butler V. P. Jr, Tse-Eng D., Saha J. R. (1981). Inactivation of digoxin by the gut flora: Reversal by antibiotic therapy. N. Engl. J. Med. 305, 789–794. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J., Abu-Ali G., Huttenhower C. (2016). The healthy human microbiome. Genome Med. 8, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J., Arze C., Ananthakrishnan A. N., Schirmer M., Avila-Pacheco J., Poon T. W., Andrews E., Ajami N. J., Bonham K. S., Brislawn C. J., et al. ; IBDMDB Investigators. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel disease. Nature 569, 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Abo R. P., Schlieper K. A., Graffam M. E., Levine S., Wishnok J. S., Swenberg J. A., Tannenbaum S. R., Fox J. G. (2014). Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: An integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 122, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon E., Gomez De La Torre Canny S., Sichel S., Kelly M., Wiles T. J., Rawls J. F., Eisen J. S., Guillemin K. (2017). Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods Cell Biol. 138, 61–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi J., Bobe A. M., Miyoshi S., Huang Y., Hubert N., Delmont T. O., Eren A. M., Leone V., Chang E. B. (2017). Peripartum antibiotics promote gut dysbiosis, loss of immune tolerance, and inflammatory bowel disease in genetically prone offspring. Cell Rep. 20, 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Ling A. V., Manthena P. V., Gearing M. E., Graham M. J., Crooke R. M., Croce K. J., Esquejo R. M., Clish C. B., Vicent D., Biddinger S. B.; Morbid Obesity Study Group. (2015). Flavin-containing monooxygenase 3 as a potential player in diabetes-associated atherosclerosis. Nat. Commun. 6, 6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A., Ferrer M. (2016). Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 24, 402–413. [DOI] [PubMed] [Google Scholar]
- Mullane K., Williams M. (2017). Enhancing reproducibility: Failures from reproducibility initiatives underline core challenges. Biochem. Pharmacol. 138, 7–18. [DOI] [PubMed] [Google Scholar]
- Nichols R. G., Cai J., Murray I. A., Koo I., Smith P. B., Perdew G. H., Patterson A. D. (2018). Structural and functional analysis of the gut microbiome for toxicologists. Curr. Protoc. Toxicol. 78, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J. M., Handley S. A., Baldridge M. T., Droit L., Liu C. Y., Keller B. C., Kambal A., Monaco C. L., Zhao G., Fleshner P., et al. (2015). Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie L. A., Jones B. V. (2015). The human gut virome: A multifaceted majority. Front. Microbiol. 6, 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orcutt R., Gianni F., Judge R. (1987). Development of an ‘Altered Schaedler Flora’ for NCI gnotobiotic rodents. Microecol. Ther. 17, 59. [Google Scholar]
- Pascal V., Pozuelo M., Borruel N., Casellas F., Campos D., Santiago A., Martinez X., Varela E., Sarrabayrouse G., Machiels K., et al. (2017). A microbial signature for Crohn’s disease. Gut 66, 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppercorn M. A., Goldman P. (1972). The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J. Pharmacol. Exp. Ther. 181, 555–562. [PubMed] [Google Scholar]
- Poesen R., Claes K., Evenepoel P., de Loor H., Augustijns P., Kuypers D., Meijers B. (2016). Microbiota-derived phenylacetylglutamine associates with overall mortality and cardiovascular disease in patients with CKD. J. Am. Soc. Nephrol. 27, 3479–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N. D., Magis A. T., Earls J. C., Glusman G., Levy R., Lausted C., McDonald D. T., Kusebauch U., Moss C. L., Zhou Y., et al. (2017). A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 35, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor L. M., Creasy H. H., Fettweis J. M., Lloyd-Price J., Mahurkar A., Zhou W., Buck G., Snyder M. P., Strauss J. F., Weinstock G., et al. (2019). The integrative human microbiome project. Nature 569, 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., et al. ; MetaHIT Consortium. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V. K., Faith J. J., Rey F. E., Cheng J., Duncan A. E., Kau A. L., Griffin N. W., Lombard V., Henrissat B., Bain J. R., et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214–1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S. J., Lemire P., Maughan H., Goethel A., Turpin W., Bedrani L., Guttman D. S., Croitoru K., Girardin S. E., Philpott D. J. (2019). Comparison of co-housing and littermate methods for microbiota standardization in mouse models. Cell Rep. 27, 1910–1919 e2. [DOI] [PubMed] [Google Scholar]
- Rogers G. B. (2015). The human microbiome: Opportunities and challenges for clinical care. Intern. Med. J. 45, 889–898. [DOI] [PubMed] [Google Scholar]
- Rosshart S. P., Vassallo B. G., Angeletti D., Hutchinson D. S., Morgan A. P., Takeda K., Hickman H. D., McCulloch J. A., Badger J. H., Ajami N. J., et al. (2017). Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171, 1015–1028 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz Y., Olivares M., Moya-Perez A., Agostoni C. (2015). Understanding the role of gut microbiome in metabolic disease risk. Pediatr. Res. 77, 236–244. [DOI] [PubMed] [Google Scholar]
- Schaffler H., Breitruck A. (2018). Clostridium difficile - From colonization to infection. Front. Microbiol. 9, 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert A. M., Rogers M. A., Ring C., Mogle J., Petrosino J. P., Young V. B., Aronoff D. M., Schloss P. D. (2014). Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio 5, e01021–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn F. P., Cui J. Y., Klaassen C. D. (2015). RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab. Dispos. 43, 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton S. R., Ajmera V., Loomba R. (2019). Emerging role of the gut microbiome in nonalcoholic fatty liver disease: From composition to function. Clin. Gastroenterol. Hepatol. 17, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R., Abu-Ali G., Vogtmann E., Fodor A. A., Ren B., Amir A., Schwager E., Crabtree J., Ma S., Abnet C. C., et al. ; Microbiome Quality Control Project, Consortium. (2017). Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat. Biotechnol. 35, 1077–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa T., Paterson R., Moore V., Carlsson A., Abrahamsson B., Basit A. W. (2008). The gastrointestinal microbiota as a site for the biotransformation of drugs. Int. J. Pharm. 363, 1–25. [DOI] [PubMed] [Google Scholar]
- Spanogiannopoulos P., Bess E. N., Carmody R. N., Turnbaugh P. J. (2016). The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman L. K., Burcelin R., Lahtinen S. (2016). Establishing a causal link between gut microbes, body weight gain and glucose metabolism in humans - Towards treatment with probiotics. Benef. Microbes 7, 11–22. [DOI] [PubMed] [Google Scholar]
- Stulberg E., Fravel D., Proctor L. M., Murray D. M., LoTempio J., Chrisey L., Garland J., Goodwin K., Graber J., Harris M. C., et al. (2016). An assessment of US microbiome research. Nat. Microbiol. 1, 15015. [DOI] [PubMed] [Google Scholar]
- Tang W. H., Wang Z., Fan Y., Levison B., Hazen J. E., Donahue L. M., Wu Y., Hazen S. L. (2014). Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: Refining the gut hypothesis. J. Am. Coll. Cardiol. 64, 1908–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuddenham S., Sears C. L. (2015). The intestinal microbiome and health. Curr. Opin. Infect. Dis. 28, 464–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. V. (2018). The role of the gut microbiota on animal model reproducibility. Animal Model Exp. Med. 1, 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte D., Kathagen G., D’Hoe K., Vieira-Silva S., Valles-Colomer M., Sabino J., Wang J., Tito R. Y., De Commer L., Darzi Y., et al. (2017. a). Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511. [DOI] [PubMed] [Google Scholar]
- Vandeputte D., Tito R. Y., Vanleeuwen R., Falony G., Raes J. (2017. b). Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 41, S154–S167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin H. W. (2014). The virome in mammalian physiology and disease. Cell 157, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Martels J. Z. H., Sadaghian Sadabad M., Bourgonje A. R., Blokzijl T., Dijkstra G., Faber K. N., Harmsen H. J. M. (2017). The role of gut microbiota in health and disease: In vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe 44, 3–12. [DOI] [PubMed] [Google Scholar]
- Wallace B. D., Wang H., Lane K. T., Scott J. E., Orans J., Koo J. S., Venkatesh M., Jobin C., Yeh L. A., Mani S., et al. (2010). Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Klipfell E., Bennett B. J., Koeth R., Levison B. S., DuGar B., Feldstein A. E., Britt E. B., Fu X., Chung Y.-M.,. et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang Y., Huycke M. M. (2017). Microbiome-driven carcinogenesis in colorectal cancer: Models and mechanisms. Free Radic. Biol. Med. 105, 3–15. [DOI] [PubMed] [Google Scholar]
- Ward T. L., Dominguez-Bello M. G., Heisel T., Al-Ghalith G., Knights D., Gale C. A. (2018). Development of the human mycobiome over the first month of life and across body sites. mSystems 3, e00140–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitekamp C. A., Phelps D., Swank A., McCord J., Sobus J. R., Catron T., Keely S., Brinkman N., Zurlinden T., Wheaton E., et al. (2019). Triclosan-resistant host-associated microbiota perform xenobiotic biotransformations in larval zebrafish. Toxicol. Sci. 172, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmanski T., Rappaport N., Earls J. C., Magis A. T., Manor O., Lovejoy J., Omenn G. S., Hood L., Gibbons S. M., Price N. D. (2019). Blood metabolome predicts gut microbiome α-diversity in humans. Nat. Biotechnol. 37, 1217–1228. [DOI] [PubMed] [Google Scholar]
- Yang T., Santisteban M. M., Rodriguez V., Li E., Ahmari N., Carvajal J. M., Zadeh M., Gong M., Qi Y., Zubcevic J., et al. (2015). Gut dysbiosis is linked to hypertension. Hypertension 65, 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M., Vatanen T., Siljander H., Hamalainen A. M., Harkonen T., Ryhanen S. J., Franzosa E. A., Vlamakis H., Huttenhower C., Gevers D., et al. ; on behalf of the DIABIMMUNE Study Group. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8, 343ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., Magris M., Hidalgo G., Baldassano R. N., Anokhin A. P., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld J. R., McMinds R., Vega Thurber R. (2017). Stress and stability applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2, 1–8. [DOI] [PubMed] [Google Scholar]
- Zhou W., Sailani M. R., Contrepois K., Zhou Y., Ahadi S., Leopold S. R., Zhang M. J., Rao V., Avina M., Mishra T., et al. (2019). Longitudinal multi-omics of host microbe dynamics in prediabetes. Nature 569, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A., Sunagawa S., Mende D. R., Bork P. (2015). Inter-individual differences in the gene content of human gut bacterial species. Genome Biol. 16, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A. L. (2019). Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]