ABSTRACT
A cohort of the first 50 COVID-19 patients in East Tyrol, a region in the southwest of Austria, were monitored in home quarantine. Specific viral ribonucleic acid was detected in throat swabs and stool samples. Analysis indicated a median virus shedding duration of 13 days; however, statistical outliers highlight the importance of consequent testing. This underlines the need of negative throat swabs prior to removing quarantine. We monitored the disease's characteristics via an in-house score called Corona Severity Index, in order to predict an aggravation of the disease. Special attention was paid to early symptoms, such as headache, which appeared to be significantly more common in younger patients (p=0.019). Anosmia and ageusia showed a predominance in female patients (p=0.028). Investigation revealed seven relapses and viral shedding fluctuation in four cases. A follow-up examination shed light on seroconversion which could be observed in 35 of 40 participants. This further clarifies the necessity of establishing discharge standards and follow-up management for COVID-19 patients.
KEYWORDS: COVID-19; home quarantine; virus shedding; clinical characteristics,; recurrences
Introduction
In early 2020, COVID-19 caused by SARS-CoV-2 spread all over the world after its initial outbreak in Wuhan, China. As of 10 September 2020, there have been 28,181,141 confirmed cases, 910,686 fatal outcomes have been registered and 20,214,594 patients have recovered from COVID-19.1 The disease causes symptoms located predominantly in the lung and the upper respiratory tract. Additionally, the gastrointestinal tract is affected in some patients. The course of the disease reaches from asymptomatic presentation to acute respiratory distress syndrome; patients may thus require mechanical ventilation and the disease can have a fatal outcome.2–4 The incubation period ranges from 1–14 days, with the period of 5 days being most common.5 According to prior evaluations, viral ribonucleic acid (RNA) was detectable for about 3 weeks despite resolution of symptoms. Stool samples, and nasopharyngeal and throat swabs were tested positive for specific RNA, whereas urine samples have shown equivocal results up to now.6,7
In this prospective study we investigated clinical characteristics and correlations, the way and duration of virus shedding as well as further aspects of home quarantine in a rural environment in East Tyrol, Austria. All patients had been in home quarantine due to a laboratory-confirmed SARS-CoV-2 infection.
Methods
Enrolment and data collection
Between 19 March 2020 and 09 April 2020, we monitored the first 50 patients of East Tyrol, Austria, with laboratory confirmed COVID-19 in home quarantine in the context of primary care (in the case of two hospitalised patients undergoing mechanical ventilation, we had to act outside of this time range in order to get two consecutive negative test results and, thus, we considered the first test results after extubation). Informed consent was obtained from all participants. In a 2-day interval, patients drew throat swabs as well as stool and urine specimens themselves. They were visited at home by medical staff every second day, whereby clinical data was also collected. All those samples were investigated for specific viral load in the laboratory using quantitative real-time polymerase chain reaction test (qRT-PCR). Clinical data were obtained with a standardised data collection form.
In order to end quarantine, two negative throat swabs were necessary, where the final one was drawn by the researchers.
Following the time range of initial data collection, we conducted a 2-week follow-up examination on 40 of the 50 patients to evaluate possible recurrences and seroconversion. Therefore, weekly throat swabs as well as stool and blood samples were collected.
Laboratory investigation
The extraction of nucleic acid was performed using an automated IndiMag 48 and an IndiMag Pathogen kit in accordance with the manufacturer's instructions (INDICAL BIOSCIENCE, Leipzig, Germany).
Extracts were tested for SARS-CoV-2 by qRT-PCR using a BioRad CFX96 system (Roche, Basel, Switzerland) with a LightMix Modular Assay kit in accordance with the modified Charité guidelines.8 10 μL of extracted RNA were added into 15 μL reaction mixture (Mastermix). Each 15 μL Mastermix contained 12.5 μL buffer solution, 0.25 μL enzyme mix, 1.75 μL distilled water and 0.5 μL primer probe wHCoV (E-gene, occasionally N-gene and RdRP gene). Reactions were incubated at 55°C for 5 minutes and 95°C for 5 minutes in order to conduct reverse transcription of viral RNA, sample denaturation and enzyme activation. These steps were followed by PCR-amplification including 45 cycles at 95°C for 5 seconds, 60°C for 15 seconds and 72°C for 15 seconds. Cooling was implemented at 40°C for 30 seconds.
Results were interpreted based on the quantification cycle (Cq) value according to the MIQE guidelines.9 A Cq value lower than or equal to 37 was assigned to confirmation by RdRP and N-gene. One of the two genes had to be positive in order to be defined as a test result. A Cq value higher than 37 was defined as a negative test result.
Seroconversion was investigated using a commercial COVID-19 immunuglobulin G (IgG) ELISA kit (Epitope Diagnostics, San Diego, USA) 2 weeks after onset of symptoms.
Clinical evaluation and severity classification
Clinical data were obtained every second day during a face-to-face visit. We defined a standardised data collection form named Corona Severity Index (CSI), which is a modified clinical prediction scale for measuring the probability of morbidity and mortality among patients with community-acquired pneumonia.10 Clinical data were collected via the CSI and additional monitoring if necessary. Within the CSI, we evaluated age, gender, comorbidities and physical examination values, such as altered mental status, respiratory rate, heart rate, body temperature and arterial blood oxygenation (supplementary material S1).
The parameters mentioned were rated and summarised resulting in a definite number of points. Based on this score, we evaluated if drug administration or medical intervention was of advantage.
An impression of the psychological status could be gained during conversation and personal contact and questioning. Difficulties with local authorities could be assessed by interaction with both patients and authorities.
Statistics
Data are given as frequencies for counted data and as mean with standard deviation or median with range for continuous data. Groups were compared by means of the chi-squared or the Mann–Whitney U test, as appropriate, whereas times to event data were analysed by means of the Kaplan–Meier estimator and groups were compared using the log-rank test. All p values are two sided and a value lower than 0.05 was considered statistically significant. SPSS version 26.0 (IBM, Armonk, USA) was used for statistical analysis.
Results
Epidemiological and clinical characteristics
In our study, we included the first 50 patients that tested positive for COVID-19 in East Tyrol. All of them stayed in home quarantine. Median observation time was 14 days with a minimum of 4 days and a maximum of 33 days. Two of the patients (4%) had a severe course of the disease and were hospitalised with the need of mechanical ventilation in an intensive care unit setting. In both cases, the CSI score increased and therefore proved to be a suitable instrument of prediction in combination with the regular monitoring. This allowed a fast intervention. Both patients were able to be discharged, and no fatal outcomes were registered. Demographic and clinical information are summarised in Table 1.
Table 1.
Demographic and clinical data
| Age, years, median (range) | 48 (12–75) |
|---|---|
| <18 years old, n (%) | 3 (6) |
| 18–59 years old, n (%) | 39 (78) |
| ≥60 years old, n (%) | 8 (16) |
| Male, n (%) | 23 (46) |
| Comorbidity, n (%) | |
| Neoplastic, liver or renal disease | 0 (0) |
| Cerebrovascular disease | 1 (2) |
| Heart disease | 4 (8) |
| Diabetes | 1 (2) |
| Hypertension | 4 (8) |
| Immunosuppression | 1 (2) |
| Current smoker, n (%) | 4 (8) |
| Interval between symptom onset and diagnosis, days, median (range) | 3 (0–17) |
| Symptoms and signs, n (%) | |
| Body temperature ≥37.5°C | 26 (52) |
| Diarrhoea | 18 (36) |
| Cough | 32 (64) |
| Dyspnoea | 10 (20) |
| Sore throat | 13 (26) |
| Nausea | 10 (20) |
| Myalgia | 18 (36) |
| Headache | 33 (66) |
| Nasal congestion | 8 (16) |
| Anosmia and ageusia | 32 (64) |
| Pruritus | 2 (4) |
The mean age of the patients was 46±15.7 years (range 13–75) the median age was 48 years. Twenty-three (46%) patients were male and 27 (54%) female. Out of these 50 patients, eight (16%) had one or more coexisting medical conditions, four (8%) of them were current smokers and one (2%) was immunosuppressed.
We compared appearance of selected symptoms under consideration of age groups, divided by the median age of 48 years and gender. In this evaluation, 6/50 (12%) patients were completely asymptomatic. Thirty-three out of 49 (67%) patients complained about headache in the course of the disease (one patient's data was not evaluable due to hospitalisation). This symptom appeared to be significantly more common in younger patients (22 vs 11; p=0.019).
Anosmia and ageusia were reported by 32/50 (64%) patients, showing a significantly higher rate in women (21 vs 11; p=0.028).
Twenty-seven out of 50 (54%) patients showed signs of gastrointestinal disease, such as nausea or diarrhoea; however, no significance was detectable regarding age and gender distribution. Time range between symptom onset in symptomatic patients and diagnosis, which was defined as the first positive throat swab, showed a median of 3 days (95% confidence interval (CI) 2.3–3.7) and a mean duration of 3.9 days (95% CI 0.6–2.8). In some of the patients, symptom onset was after the first positive throat swab, as they were tested in the course of contact tracing. Considering age distribution, the evaluation suggested a nonsignificant (p=0.08) difference in time range. The median time between symptom onset and diagnosis was 2 days in younger (mean 3 days) and 3 days (mean 5 days) in elderly patients.
Recurrences were defined by a positive throat swab after at least two consecutive negative ones. This occurred in 7/50 cases (14%; 95% CI 6.5%–25.5%) during evaluation time; three were male and four were female. The 2-week follow-up revealed additional recurrences in two patients, which were identified by positive throat swabs. Additionally, a fluctuation of virus shedding can be assumed due to one sporadic negative throat swab within a positive testing period, which appeared in four cases.
Psychological assessment revealed that 10/50 patients (20%; 95% CI 10.8%–32.6%) had difficulties in coping with the disease and quarantine. Five complained about social stigmatisation and the effect of the disease on their social environment. Five suffered distress from regular visits by the police. Two of them had to seek psychological treatment because of home quarantine experience and circumstances.
During cooperation with patients and local authorities, discrepancies became evident. End of quarantine management was not performed concordantly in all patients, as some administrative decisions claimed two negative throat swabs at the end of quarantine and others did not.
Laboratory parameters
Virus shedding was detected by qRT-PCR and evident in throat swabs and stool samples, but not in the 52 urine samples as none tested positive. The median sample collection was five throat swabs (range 2–8) and four stool samples (range 0–5).
The duration of virus shedding was defined as the time between the onset of symptoms (or the first positive test result in initially or continuously asymptomatic patients) and the second of two consecutive negative test results from throat swabs. The median virus shedding duration lasted for 13 days (95% CI 11.9–14.1) with a minimum of 4 days in an asymptomatic patient and a maximum of 30 days. This value differed according to gender, with a median of 13 days, women showed significantly shorter virus shedding duration than men who had a median virus shedding duration of 15 days (p=0.013; Fig 1).
Fig 1.
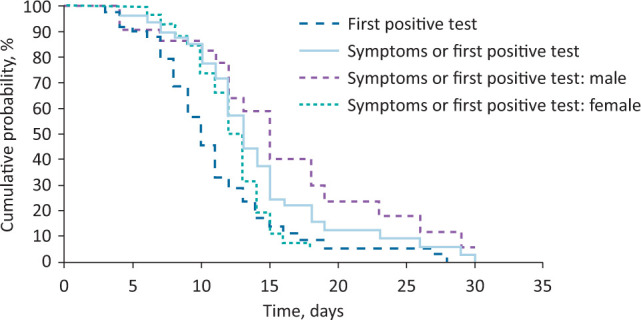
Duration from symptom onset or first positive throat swab to the second of two consecutive negative throat swabs.
The median duration of a positive throat swab and two consecutive negative test results was 10 days (95% CI 8.8–11.2). While women showed a faster elimination of the virus, with a median of 10 days, men stayed tested positive longer, with a median duration of 12 days (p=0.006; Fig 1).
There is a median delay of 9 days (95% CI 6.9–11.0) until stool can be tested positive. Furthermore, it was observed that virus shedding in stool was prolonged compared to throat swabs and that it transcended the occurrence of symptoms.
Stool continued to be positive for a median duration of 21 days (95% CI not calculable) and an average duration of 13.9 days (95% CI 10.1–17.6).
Throat swabs and stool samples combined (maximum duration) revealed a total median positive duration of 21 days (95% CI 13.0–29.0).
Virus shedding in stool occurred frequently, 32/46 (69%) patients showed viral RNA in faeces (four patients had to be excluded from stool sample analysis because sampling was not possible). Out of the 32 (70%) patients with virus shedding in stool, 19 (59%) showed gastrointestinal symptoms, whereas only six (43%) of the 14 patients without virus shedding had a gastrointestinal course of disease (p=0.301).
IgG-antibodies were analysed in order to confirm seroconversion 14 days post-infection. During the 2-week follow-up examination, this could be detected in 35/40 participants (87.5%; 95% CI 74.8%–95.1%). No significant correlation could be detected between seroconversion and course of disease. However, viral load in the first positive throat swab appeared to be low (Cq >30) in four of five patients whose seroconversion was not traceable.
Discussion
Our data reveal differences in gender distribution; however, in contrast to other studies with hospitalised patients, we had a predomination of women in our study population.11,12 The results indicate that there is a tendency to a less severe course of disease in women. Our investigation supports this hypothesis since both hospitalised patients were male. Fever and dropping values of oxygen saturation resulting in a higher CSI score indicated an aggravation of disease in these two cases. Patients with increasing score should thus be monitored closely in order to detect the need for hospitalisation or oxygen support.
The symptom profile of milder cases is diverse, which points out the need of consequent testing for SARS-CoV-2 in people with flu-like symptoms. Headaches appeared to be a very common symptom according to our evaluation, especially around disease onset. In other studies, this symptom occurred infrequently and was not defined as one of the most characteristic symptoms of this disease.11,13 The most common initial symptoms were cough, fever, myalgia and sore throat.11 Based on our observation, especially headache as a symptom should be considered for further diagnostics regarding COVID-19.
Anosmia and ageusia seem to be pathognomonic for COVID-19, as the frequency of these symptoms is very high in patients. This symptom complex mostly appears slightly delayed and can persist longer than other medical conditions and positive testing. Even patients with comparatively mild symptoms seemed to be affected. The expression of the ACE2 receptor in neurons suggests that SARS-CoV-2 may have higher neuro-invasive potential compared to previous coronaviruses.14 We could also see that women especially are very sensitive to this kind of impairment. This may suggest a gender difference in ACE2-receptors responsible for the docking of SARS-CoV-2 as sex hormones influence ACE2 receptor expression.15
Gastrointestinal symptoms are reported to be common in patients with COVID-19.16,17 Our results revealed no significant coherence between gastrointestinal symptoms and virus shedding in stool. The lacking correlation requires consequent testing for appearance of viral RNA in faeces, irrespective of gastrointestinal symptoms.
Furthermore, the observation of stool samples staying positive longer than throat swabs can be used for retrospective studies. In order to define infectivity of stool, complete gene sequencing should be considered, and cultivation would be necessary.
Data for virus excretion in faeces underlies certain limitations. The commencement of sample collection was delayed, which makes it difficult to determine the exact onset of virus secretion in this medium. Secondly, the primary endpoint was defined by negative throat swabs. Consequently, stool samples were not collected until negative in all patients.
Studies on virus shedding in urine are controversial.6,13,18,19 In our study, virus RNA could not be detected in urine samples. Considering this finding, urine is unlikely to be a transmission route and most likely it has no infectious potential.
Virus shedding duration appears to be shorter in women than in men. Accordingly, women are more likely to eliminate the virus faster. In addition, we established that after 14 days, which is the actual duration of home quarantine, 20% of the patients were still testing positive in throat swabs. Only after approximately 25 days, 95% of the study population test negative. In order to detect patients with prolonged virus shedding, it is necessary to conduct testing at the end of home quarantine. Ten days after symptom onset or first positive testing, 50% of the patients had two consecutive negative test results. This point of time may be a good starting point for regular throat swabs justifying the end of quarantine.
Our regular testing interval demonstrated that recurrences are more frequent than expected. It is difficult to distinguish between a manifest recurrence and a fluctuation in virus shedding. These findings suggest that the virus can change its reproduction site in the course of disease. As a consequence, a negative throat swab does not necessarily prove the complete absence of the virus. Thus, a recurrence would be an indication for persistence in possibly still symptomatic patients. Yet, recurrence might as well be the result of an autogenous reinfection via virus shedding in stool. In order to confirm this faecal-oral route, proof of infectivity in stool would be necessary.20
Further limitations could be seen in asymptomatic patients since the duration of virus shedding is occasionally misinterpreted. In fact, these patients are tested with a delay in time, resulting in shorter virus excretion period. In this context, it must be questioned if asymptomatic patients, particularly those with a noticeably short positive testing period, might be merely colonised instead of manifestly diseased. Consequently, it is important to pay attention to inflammatory signs, which confirm the manifestation of a disease in comparison with colonisation. Asymptomatic people implicate a great risk of infection for other people via contact. Therefore, intensive and consistent testing of exposed individuals is necessary in order to avoid an aggravation of the pandemic.
A limiting aspect of this research might be the sample size which is comparatively small. In order to provide more meaningful and detailed information, an extended sample size would be of advantage.
During the follow-up examination, seroconversion could be detected. However, only 40 out of 50 patients were willing to participate in the 2-week follow-up examination. Course of disease or duration of virus shedding did not show any correlation with seroconversion. Even asymptomatic patients showed partly high values of IgG-antibodies. Nevertheless, there is a minor tendency that severe course of disease implies efficient seroconversion. Subsequent analysis considering the course of time of the viral load is necessary to investigate a possible correlation. In order to identify a more precise point of time of seroconversion, sampling has to be started as early as possible after onset of disease.
In contrast to hospitalised patients, the special challenge in this case was the collaboration of medical professionals with local authorities. Contradictory information and individually different decisions on quarantine duration were confusing for the parties concerned. Some patients obligatorily had to confirm two consecutively negative throat swabs in order to end quarantine. For others, this was not necessary, which interfered with medical security standards.
Home quarantine and associated isolation was experienced as a psychological burden by many participants. Patients reported feeling like criminals as a result of frequent interference by the executive authorities, and they feared stigmatisation of society. These aspects should be taken into consideration for future decisions regarding disease management.
Conclusion
Patients in home quarantine should be monitored regularly for aggravation of disease. This guarantees an early detection of potential aggravation and enables fast intervention.
Discharge should be based on clinical and laboratory findings. Authorities should insist on two consecutively negative throat swabs prior to removing quarantine. Even after end of quarantine patients should be monitored for recurrence of disease or long-term sequelae.
Faecal shedding of the virus should be investigated as it lasts longer than those in throat swabs and may be a source of viral spreading.
Besides flu-like symptoms of the upper respiratory tract, headache, anosmia and ageusia should raise awareness for COVID-19.
Summary
| What is known? |
|
| What is the question? |
|
| What was found? |
|
| What is the implication for practice now? |
|
Supplementary material
Additional supplementary material may be found in the online version of this article at www.rcpjournals.org/clinmedicine:
S1 – Example of the in-house developed Corona Severity Index.
Acknowledgements
The authors would like to thank Erna Inwinkl for her support regarding sample collection.
References
- 1.Worldometer COVID-19 coronavirus pandemic. Worldometer. www.worldometers.info/coronavirus [Accessed 10 September 2020]. [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology 2020;5:536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M, Kleine-Weber H, Krüger N, et al. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv 2020. 01.31.929042. [Google Scholar]
- 4.Zhou M, Zhang X, Qu J. Coronavirus disease 2019 (COVID-19): a clinical update. Front Med 2020;14:126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 2020;172:577–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9. [DOI] [PubMed] [Google Scholar]
- 7.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corman V, Bleicker T, Brünink S, et al. Diagnostic detection of 2019-nCoV by real-time RT-PCR: Protocol and preliminary evaluation as of Jan 17, 2020. World Health Organization, 2020. www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf?sfvrsn=a9ef618c_2 [Google Scholar]
- 9.Bustin SA, Benes V, Garson JA, et al. The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 2009;55:611–22. [DOI] [PubMed] [Google Scholar]
- 10.Fine MJ, Auble TE, Yealy DM, et al. A prediction rule to identify low-risk patients with community-acquired pneumonia. N Engl J Med 1997;336:243–50. [DOI] [PubMed] [Google Scholar]
- 11.Garg S, Kim L, Whitaker M, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019 — COVID-NET, 14 States, March 1–30, 2020. Weekly 2020;69:458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T, Dai Z, Mo P, et al. Clinical characteristics and outcomes of older patients with coronavirus disease 2019 (COVID-19) in Wuhan, China (2019): a single-centered, retrospective study. J Gerontol A Biol Sci Med Sci 2020;75:1788–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kujawski SA, Wong KK, Collins JP, et al. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med 2020;26:861–8. [DOI] [PubMed] [Google Scholar]
- 14.Natoli S, Oliveira V, Calabresi P, et al. Does SARS-Cov-2 invade the brain? Translational lessons from animal models. Eur J Neurol 2020;27:1764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.La Vignera S, Cannarella R, Condorelli RA, et al. Sex-specific SARS-CoV-2 mortality: among hormone-modulated ace2 expression, risk of venous thromboembolism and hypovitaminosis D. Int J Mol Sci 2020;21:2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmulson M, Dávalos MF, Berumen J. Beware: Gastrointestinal symptoms can be a manifestation of COVID-19. Rev Gastroenterol Mex 2020;85:282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klopfenstein T, Kardiane-Oussou NJ, Royer PY, et al. Diarrhea: An underestimated symptom in Coronavirus disease 2019. Clin Res Hepatol Gastroenterol 2020;44:282–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020;323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng L, Liu J, Xu W, et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J Med Virol 2020;92:1676–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling Y, Xu SB, Lin YX, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl) 2020;133:1039–43. [DOI] [PMC free article] [PubMed] [Google Scholar]


