Abstract
Orcokinin neuropeptides are conserved among ecdysozoans, but their functions are incompletely understood. Here, we report a role for orcokinin neuropeptides in the regulation of sleep in the nematode Caenorhabditis elegans. The C. elegans orcokinin peptides, which are encoded by the nlp-14 and nlp-15 genes, are necessary and sufficient for quiescent behaviors during developmentally-timed sleep (DTS) as well as during stress-induced sleep (SIS). The five orcokinin neuropeptides encoded by nlp-14 have distinct but overlapping functions in the regulation of movement and defecation quiescence during SIS. We suggest that orcokinins may regulate behavioral components of sleep-like states in nematodes and other ecdysozoans.
Introduction
Ecdysozoa is comprised of the most diverse group of animals on earth. This clade includes arthropods and nematodes, as well as other smaller phyla, which are united by having a molting cycle (Aguinaldo, Turbeville et al. 1997). Molts occur periodically during growth and are accompanied by elaborate and specific molting behaviors. In the nematode C. elegans, the molting cycle is similar to the circadian cycle in other animals (Hendriks, Gaidatzis et al. 2014). Molt timing is regulated by LIN-42, a worm homolog of the circadian protein PERIOD, and behavior during the molt resembles sleep controlled by circadian timing in other animals (Raizen, Zimmerman et al. 2008).
Orcokinin neuropeptides are strikingly conserved across Ecdysozoa. They have been described in nematodes (Nathoo, Moeller et al. 2001), in several arthropods including cockroaches (Hofer, Dircksen et al. 2005, Hofer and Homberg 2006), kissing bugs (Wulff, Sierra et al. 2017), fruit flies (Chen, Choi et al. 2015), crayfish (Yasuda-Kamatani and Yasuda 2000), lobsters (Dickinson, Stemmler et al. 2009), and in tardigrades (Koziol 2018). Orocokinins are related to pedal peptides (Kim, Go et al. 2018), identified in mollusks (Lloyd and Connolly 1989) and to smooth muscle relaxant peptides (SMPs), identified in echinoderms (Rowe and Elphick 2012, Kim, Kim et al. 2016), suggesting that an ancestor to ecdysozoan orcokinins was present in early bilaterians (Jekely 2013, Semmens and Elphick 2017).
Orcokinins regulate insect ecdysis and circadian activity. In kissing bugs, disruption of orcokinin signaling causes molting defects (Wulff, Sierra et al. 2017), perhaps partially due to a role for these peptides in the biosynthesis of the ecdysteroids (Zitnan, Ross et al. 1999, Yamanaka, Roller et al. 2011). In cockroaches, orcokinins peptides injected into the brain induce a phase shift in circadian sleep/wake behavior (Hofer and Homberg 2006). A role in the regulation of both molting and circadian rhythms suggests the intriguing hypothesis that an ancestral role for orcokinin is in the regulation of behavioral rhythms, specifically sleep/wake behavior. We pursued this hypothesis in C. elegans, given the similarity of its molting cycle to circadian rhythms in other organisms.
C. elegans orcokinins are encoded by the genes nlp-14 and nlp-15 (Nathoo, Moeller et al. 2001). NLP-14 peptides modulate cholinergic signaling during male mating (Sherlekar, Janssen et al. 2013) and mediate decision-making during nociceptive behaviors (Hapiak, Summers et al. 2013). nlp-14 transcripts are upregulated during the 2-hour period prior to ecdysis (George-Raizen, Shockley et al. 2014), when the animals display sleep-like quiescent behavior (Raizen, Zimmerman et al. 2008). Single-cell transcriptomic data suggests that nlp-14 is expressed in the sleep-promoting ALA neuron (Nath, Chow et al. 2016, Taylor, Santpere et al. 2019), while its paralog nlp-15 is expressed in both the ALA neuron and in another sleep promoting neuron called RIS (Taylor, Santpere et al. 2019). Thus, we sought to test the hypothesis that orcokinin neuropeptides regulate sleep.
C. elegans sleep is regulated by neuropeptides. Developmentally timed sleep (DTS) occurs during larval transitions, coincident with the molt (Singh and Sulston 1978, Raizen, Zimmerman et al. 2008). Movement quiescence during DTS is controlled primarily by the RIS neuron which releases FLP-11 neuropeptides (Turek, Lewandrowski et al. 2013, Turek, Besseling et al. 2016) with a minor role for NLP-22 peptides released from the RIA neurons (Nelson, Trojanowski et al. 2013). Arousal during DTS is mediated by pigment dispersing factor neuropeptides (PDF), which also mediate arousal in insects (Renn, Park et al. 1999, Choi, Chatzigeorgiou et al. 2013). SIS is controlled by both the ALA and RIS interneurons, via the release of a collection of neuropeptides (Nelson, Lee et al. 2014, Lenz, Xiong et al. 2015, Turek, Besseling et al. 2016). Based on their spatial and temporal expression patterns, and on their roles in regulating behavioral rhythms in other ecdysozoans, we hypothesized that NLP-14 and NLP-15 play a role in sleep regulation in C. elegans.
We combined the analysis of loss-of-function with over-expression to characterize the function of the C. elegans orcokinins and find that NLP-14 and NLP-15 are required for movement and defecation quiescence that occur during sleep; NLP-14 peptides play a larger role. This work expands our knowledge of the function of orcokinins and suggests a previously unappreciated role in sleep regulation.
Methods
Worm maintenance and strains
Animals were maintained at 20°Celsius on agar plates containing nematode growth medium and fed the OP50 derivative bacterial strain DA837 (Davis, Somerville et al. 1995). The following strains were used in this study:
N2 (Bristol - wild type)
KG532 = kin-2(ce179) X
VC1063 = nlp-15(ok1512) I
PS5009= pha-1(e2132ts); syEx723[hsp-16.2p::lin-3C;myo-2p:gfp; pha-1(+)]
SJU6 = stjEx3[hsp-16.2p::nlp-14; myo-2p::mCherry]
SJU27 = stjIs2[hsp-16.2p::nlp-14; myo-2p::mCherry]
SJU44 = stjEx32[hsp-16.2p::nlp-14(1-3); myo-2p::gfp]
SJU47 = stjEx36[hsp-16.2p::nlp-14(1-2); myo-2p::gfp]
SJU95 = stjEx76[ida-1p::mCherry; nlp-14p::gfp]
SJU96 = stjEx77[ida-1p::mCherry; nlp-14p::gfp]
SJU102 = kin-2(ce179) X; stjIs2
SJU109 = sjtEx123[hsp-16.2p::nlp-14(1); myo-2p::mCherry]
SJU110 = sjtEx123[hsp-16.2p::nlp-14(1); myo-2p::mCherry]
SJU121 = stjEx91[hsp-16.2p::nlp-15; myo-3p::mCherry]
SJU122 = stjEx92[hsp-16.2p::nlp-15; myo-3::mCherry]
SJU154 = nlp-14(tm1880) X
SJU178 = nlp-14(stj10) X
SJU207= nlp-14(tm1880) X; stjEx146[ida-1p::nlp-14; myo-3p::mCherry] (Line#1)
SJU208= nlp-14(tm1880) X; stjEx147[ida-1p::nlp-14; myo-3p::mCherry] (Line#2)
SJU209= nlp-14(tm1880) X; stjEx148[ida-1p::nlp-14;myo-3p::mCherry] (Line#3)
SJU232= stjEx163[hsp-16.2p::nlp-14(3); myo-2p::mCherry]
SJU233= nlp-14(stj18) X
SJU241= stjIs160[hsp-16.2p::nlp-14; myo-2p::mCherry]
SJU244= stjEx167[hsp-16.2p::nlp-14(3); myo-2p::mCherry]
SJU245= stjEx168[hsp-16.2p::nlp-14(3); myo-2p::mCherry]
SJU246= stjEx169[hsp-16.2p::nlp-14(3); myo-2p::mCherry]
SJU247= stjEx170[hsp-16.2p::nlp-14(3); myo-2p::mCherry]
SJU254= stjEx171[hsp-16.2p::nlp-14(1-4); myo-2p::mCherry]
SJU255= stjEx172[hsp-16.2p::nlp-14(1-4); myo-2p::mCherry]
SJU256= stjEx173[hsp-16.2p::nlp-14(1-4); myo-2p::mCherry]
SJU257= stjEx174[hsp-16.2p::nlp-14(1-4); myo-2p::mCherry]
SJU258= stjEx175[hsp-16.2p::nlp-14(1-4); myo-2p::mCherry]
SJU260= nlp-14(stj19) X
SJU262= nlp-15(ok1512) I; nlp-14(stj18) X
SJU272= nlp-14(stj18) X; pha-1(e2132ts) III; syEx723[hsp16.2p::lin-3C; myo-2p::gfp; pha-1(+)]
SJU273= nlp-14(tm1880) X; pha-1(e2132ts) III; syEx723[hsp16.2p::lin-3C; myo-2p::gfp; pha-1(+)]
SJU281= nlp-14(stj13) X; nlp-15(stj25) I
SJU282= stjEx184[nlp-15p::gfp;ida-1p::mCherry; myo-3p::mCherry]
SJU312= nlp-15(stj25) I
Molecular biology and transgenesis
DNA for transgenesis was constructed using overlap extension-polymerase chain reaction (OE-PCR) (Nelson and Fitch 2011). The promoter of the gene hsp-16.2 and the coding sequences of nlp-14 and nlp-15 were amplified from genomic DNA by PCR. The amplicons were fused together by OE-PCR. To over-express subsets of NLP-14 peptides, the hsp-16.2 promoter and a portion of the nlp-14 gene coding for the N-terminal signal peptide and NLP-14(1), (1-2), (1-3) or (1-4) peptide(s), followed by a stop codon, were amplified from genomic DNA. Next, the operon sequence from the genes gpd-2 and gpd-3 and the coding sequence for the red fluorescent protein RFP were amplified from the plasmid pLR304. The three amplicons were fused by OE-PCR. To over-express the NLP-14(3) peptide, the plasmid pSJU8 was commercially engineered to contain sequence for the hsp-16.2 promoter, the coding sequence for the N-terminal signal peptide and NLP-14(3) peptide of the nlp-14 gene and the 3’untranslated region of the gene unc-54 (GeneScript ©). The nlp-14, nlp-15 and ida-1 fluorescent reporters were constructed by amplifying 5’ regulatory DNA for each gene from genomic DNA and the green fluorescent protein (gfp) or mCherry coding sequence from the plasmids pPD95.75 or pCFJ90 (Addgene). The promoter and gfp amplicons were fused for each gene using OE-PCR. To express nlp-14 in the ALA neuron, 5’ regulatory DNA of the gene ida-1 was fused by OE-PCR to the nlp-14 coding sequence. Transgenesis was performed by microinjection, as described (Stinchcomb, Shaw et al. 1985). The strains SJU27, SJU141 and SJU142 were integrated using UV irradiation, as described (Mello and Fire 1995).
Reverse transcription-PCR (RT-PCR) of nlp-14(tm1880) was accomplished by isolating total RNA using an RNeasy mini kit (Qiagen ©), followed by cDNA synthesis using SuperScript™ One-Step RT-PCR System (ThermoFisher ©). Oligonucleotides used are in Table S1. Extrachromosomal arrays and DNA concentrations are listed in Table S2.
Construction of mutants
SJU178, SJU233, SJU262, SJU260, SJU262 and SJU281 were constructed by CRISPR/Cas9 gene editing. Using a published protocol (Arribere, Bell et al. 2014), insertions were made in the nlp-14 or nlp-15 gene at defined sites. Simultaneously, an edit of the dpy-10 gene was made which resulted in an easily identifiable dumpy (Dpy) or roller (Rol) phenotype, to allow for screening. Specifically, a mixture of guide RNA (gRNA) duplexed with Alt-R® CRISPR-Cas9 tracrRNA (IDT ©), Alt-R ® S.p. Cas9 Nuclease V3 (IDT) and oligonucleotide repair templates were injected into day-1 adult wild-type or SJU233 animals. Dpy or Rol progeny of the injected animals were transferred to individual plates and maintained to the next generation. The genomic DNA of 10-15 progeny was used as templates for PCR to amplify a portion of the nlp-14 or nlp-15 gene. The amplicon was treated with NheI restriction enzyme and analyzed by agarose gel electrophoresis. 15-20 non-Dpy non-Rol animals from plates with the desired edit were transferred individually to fresh plates and grown to the next generation. These worms were again screened by PCR combined with restriction digest, and the alleles were confirmed by sequencing (Genewiz ©). The sequences of reagents are listed in Table S3.
The strain SJU154 was generated by crossing the strain FX18180(tm1880) X, which was obtained from the National BioResource Project (PI, Shohei Mitani), to male N2 animals, and then crossing resultant males back to tm1880. This procedure was repeated 3 times to reduce the number of unlinked mutations on the five autosomal chromosomes.
WorMotel behavioral assays
Movement quiescence was quantified during both developmentally timed sleep (DTS) and stress induced sleep (SIS), using the WorMotel, as previously described (Churgin, Jung et al. 2017). For DTS, we monitored active L4 animals (pre-lethargus) of each genotype for 12-hours. Because of day-to-day and chip-to-chip variability in sleep, we statistically compared strains housed in different wells of the same WorMotel. A combination of 24 wild-type, mutant, and/or transgenic active L4 animals were picked onto the agar surfaces of individual wells of the WorMotel polydimethylsiloxane (PDMS) chip. Images were captured every 10-seconds for 12 hours, and quiescence was quantified and DTS was manually measured based on a definable peak of quiescence, as previously described (Raizen, Zimmerman et al. 2008). For SIS, a combination of 24 wild-type, mutant, and/or transgenic day-1 adults were picked individually onto the agar surfaces of a welled PDMS microchip. The chip was placed into a UV-cross linker (Ultraviolet, 254 UVP) and exposed to 1500 J/m2 of UV light to induce SIS (DeBardeleben, Lopes et al. 2017). For over-expression experiments, day-1 adults were heat-shocked on standard growth plates, by submerging them, wrapped in para-film, in a 33°C water bath for 30 minutes. The heat-shocked animals were individually transferred to the agar surfaces of a welled PDMS microchip. For SIS and over-expression, images were captured every 10-seconds for 8 or 4 hours, respectively, and total minutes of quiescence was determined.
Body bending analysis following over-expression
Day-1 adults were heat-shocked by submerging standard growth plates, wrapped in para-film, in a 33°C water bath for 30 minutes. Body bends were counted manually using a stereomicroscope for 60-seconds, 2 hours after heat exposure. A body bend was defined as the movement of the body just posterior to the pharynx to the opposite position from the previous maximal bend.
Defecation analysis during SIS and following overexpression
The defecation cycle was measured manually for 5-6 minutes by visual inspection using a stereomicroscope, using described criteria (Thomas 1990). For over-expression, day-1 adults, on standard growth plates, were submerged, wrapped in para-film, in a 33°C water bath for 30 minutes and analyzed for 5 minutes between 2 and 2.5 hours after heat exposure. For SIS, day-1 adults were exposed to 1500 J/m2 of UV light in a UV-cross linker (Ultraviolet, 254 UVP) on growth plates and the defecation cycle was measured for 5-6 minutes, 85-95 minutes post-UV. For temporal SIS analyses, a single animal was examined for 5-minutes every 30 minutes for 4-hours, post-UV.
Microscopy
Fluorescence microscopy was conducted using an Olympus BX63 wide-field fluorescence microscope equipped with a Hamamatsu FLASH 4.0V3 digital camera and CellSens Dimension Version 2 software. Day-1 adult transgenic animals were immobilized on glass slides containing a 5% agar pad supplemented with 25mM sodium azide.
Alignments
Peptide sequences were obtained from the National Center for Biotechnology Information (NCBI) and aligned using online T-coffee software (Notredame, Higgins et al. 2000). Peptide alignments were annotated using Boxshade (https://embnet.vital-it.ch/software/BOX_doc.html).
Results
NLP-14 and NLP-15 are orcokinin homologs expressed in the ALA and RIS neurons
Neuropeptides coded by the genes nlp-14 and nlp-15 are classified as orcokinins based on sequence similarity to insect and crustacean peptides (Nathoo, Moeller et al. 2001). All five NLP-14 peptides are conserved at the C-terminus, while the NLP-15 peptides show greater conservation at the N-terminus (Figure 1A). nlp-14 expression has been demonstrated in the ventral cord and some sensory and interneurons (Nathoo, Moeller et al. 2001), as well as in male-specific neurons (Sherlekar, Janssen et al. 2013). Single-cell gene expression studies revealed enrichment of both nlp-14 and nlp-15 transcripts in the ALA neuron and enrichment of nlp-15 (but not nlp-14) in the RIS neuron (Nath, Chow et al. 2016, Taylor, Santpere et al. 2019). ALA and RIS are central sleep-promoting neurons (Turek, Lewandrowski et al. 2013, Hill, Mansfield et al. 2014, Konietzka, Fritz et al. 2020). In support of the single-cell transcriptomic data, we found that a GFP transcriptional reporter for nlp-14 colocalizes with an mCherry transcriptional reporter for ida-1, which is strongly expressed in ALA (Zahn, Macmorris et al. 2001) (Figure 1B). Similarly, a GFP transcriptional reporter for nlp-15 showed expression in both ALA and RIS neurons (Figure 1C). The combination of expression during the molt, a C. elegans sleep state, with expression in sleep-regulating neurons led us to hypothesize that nlp-14 and nlp-15 regulate sleep.
Figure 1: NLP-14 and NLP-15 neuropeptides are related to arthropod orcokinins and are expressed in sleep-promoting cells.
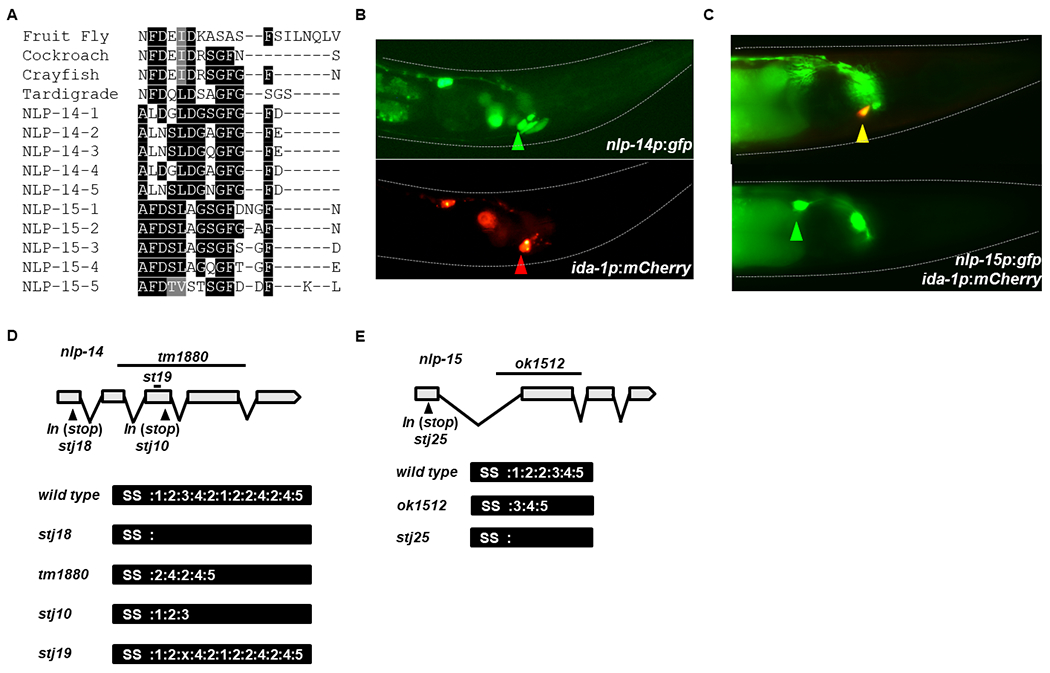
A) Peptide alignment of orcokinins from: Drosophila melanogaster (fruit fly), Blatella germanica (cockroach), Orconectes limosus (crayfish), Hypsibius dujardini (Tardigrade) and C. elegans (Nematode). B) Representative images of an animal expressing gfp from the promoter of nlp-14 (top image) and mCherry from the promoter of ida-1 (bottom image). ALA expression is denoted by a green and red arrowhead. Anterior – Right; Dorsal – Bottom, Ventral – Top. C) Representative images of an animal expressing gfp from the promoter of nlp-15 and the same image superimposed with mCherry from the promoter of ida-1. Expression in the RIS (gfp only) and ALA (gfp and mCherry) are denoted with green and yellow arrowheads, respectively. Anterior – Right; Dorsal – Bottom, Ventral – Top. D) Gene and protein structure for NLP-14, highlighting the location of deletion and insertion alleles. E) Gene and protein diagram for NLP-15, highlighting the location of a deletion and insertion allele. SS denotes signal sequence.
To test this hypothesis, we obtained mutant strains for nlp-14 and nlp-15, which carry the deletion alleles, tm1880 and ok1512, respectively. tm1880 is predicted to cause an in-frame deletion that preserved the signal sequence as well as peptides 2, 4, and 5 of nlp-14. We confirmed this in silico prediction using RT-PCR (Figure 1D). We refer to tm1880 from hereon as nlp-14(2, 4, 5). ok1512 too is predicted to cause an in-frame deletion, which preserves the signal sequence as well as peptides 3-5 of nlp-15 (Figure 1E). We refer to ok1512 as nlp-15(3-5).
To construct complete loss-of-function mutants as well as other mutants, we used CRISPR/Cas9 gene editing technology (Paix, Folkmann et al. 2017). The nlp-14(stj18) strain contains a stop codon in the first exon, after the signal sequence but prior to the sequence encoding all NLP-14 peptides (Figure 1D). Similarly, the nlp-15(stj25) strain contains an insertion of a stop codon 5’ of all five NLP-15 peptides (Figure 1E). Hence, stj18 and stj25 are predicted to make null alleles of nlp-14 and nlp-15, respectively, so we will refer to them as nlp-14(null) and nlp-15(null). The nlp-14(stj10) strain carries an insertion of a stop codon at the 3’-end of the sequence encoding peptide 3 so we will refer to it as nlp-14(1-3). The nlp-14(stj19) strain contains an in-frame deletion that removes only peptide 3, so we will refer to it from hereon as nlp-14(1, 2, 4, 5).
The C. elegans orcokinins are required for DTS
DTS occurs prior to ecdysis in C. elegans (Singh and Sulston 1978, Raizen, Zimmerman et al. 2008). Orcokinins regulate ecdysis and ecdysteroid biosynthesis in insects (Yamanaka, Roller et al. 2011, Wulff, Sierra et al. 2017). We hypothesized that DTS and/or the molt would be disrupted in nlp-14 and/or nlp-15 mutants. Using the WorMotel (Churgin, Jung et al. 2017), we found that DTS was unaltered in nlp-14(null), nlp-15(3-5) or nlp-15(null) single mutant animals (Figure 2A–C). Because we suspected there might be functional redundancy between the two genes, we tested animals that were mutant for both nlp-14(null) and nlp-15(3-5) and nlp-14(null) and nlp-15(null). DTS was modestly reduced in both double mutants (Figure 2D,E). We found no molting defects by carefully scanning double mutant strains via stereomicroscopy or by inspecting select animals at 1000X using wide-field differential interference contrast (DIC) microscopy. Based on these results, we conclude that NLP-14 and NLP-15 are required for movement quiescence during DTS but play a minor role in this behavior perhaps due to compensatory action of other neuropeptides (Nelson, Trojanowski et al. 2013, Turek, Besseling et al. 2016). They are not required for the successful completion of the molt.
Figure 2: The C. elegans orcokinins play a small role during DTS.

A) Movement quiescence during L4 lethargus in wild-type and nlp-14(stj18) animals (N ≥ 32). B) Movement quiescence during L4 lethargus in wild-type and nlp-15(ok1512) animals (N ≥ 20). C) Movement quiescence during L4 lethargus in wild-type and nlp-15(stj25) animals (N ≥ 46). D) Movement quiescence during L4 lethargus in wild-type and nlp-14(stj18); nlp-15(ok1512) animals (N ≥ 33, **p < 0.01). E) Movement quiescence during L4 lethargus in wild-type and nlp-14(stj18); nlp-15(stj25) animals (N ≥ 58, ***p < 0.001). Statistical significance was calculated using Student’s t-test. All error bars represent mean +/− SEM.
NLP-14 peptides are required for movement quiescence during SIS
Since nlp-14 and nlp-15 are expressed in the RIS and/or ALA neurons, which are central regulators of SIS (Hill, Mansfield et al. 2014, Konietzka, Fritz et al. 2020), we tested their necessity for movement quiescence. SIS was induced by UV irradiation (DeBardeleben, Lopes et al. 2017) and animals were monitored on a WorMotel (Churgin, Jung et al. 2017). The nlp-14(null), nlp-14(2, 4, 5) and nlp-14(1-3) animals all displayed reductions in movement quiescence (Figure 3A–C,H,I; Figure S1), while nlp-14(1,2,4,5), nlp-15(3-5) and nlp-15(null) animals did not (Figure 3D,E; Figure S1). nlp-14(null); nlp-15(3-5) and nlp-14(null); nlp-15(null) double mutants displayed reduced total movement quiescence similar to that observed in nlp-14(null) single mutants; i.e. nlp-15 mutations did not enhance the phenotype caused by nlp-14 mutations (Figure 3F,G,J; Figure S1). However, the nlp-15 mutations increased the variance of the nlp-14(null) phenotype (p=0.03, nlp-14(null);nlp-15(null); p=0.003, nlp-14(null);nlp-15(3-5); Levene’s test). Based on these results, we conclude that one or more of the NLP-14 peptides (but not peptide 3) are required for movement quiescence during SIS and that NLP-15 peptides are likely dispensable for SIS total quiescence, but modulate the nlp-14 phenotype in some way.
Figure 3: NLP-14 neuropeptides are required for movement quiescence during SIS.

A) Movement quiescence during UV-induced SIS in wild-type and nlp-14(stj18) animals (N ≥ 33, ***p < 0.001). B) Movement quiescence during UV-induced SIS in wild-type and nlp-14(tm1880) animals (N ≥ 26, ***p < 0.001). C) Movement quiescence during UV-induced SIS in wild-type and nlp-14(stj10) animals (N ≥ 30, *p<0.05). D) Movement quiescence during UV-induced SIS in wild-type and nlp-15(ok1512) animals (N ≥ 17). E) Movement quiescence during UV-induced SIS in wild-type and nlp-15(stj25) animals (N ≥ 33). F) Movement quiescence during UV-induced SIS in wild-type and nlp-14(stj18); nlp-15(stj25) animals (N ≥ 68, ***p < 0.001). G) Movement quiescence during UV-induced SIS in wild-type and nlp-14(stj18); nlp-15(ok1512) animals (N ≥ 24, ***p < 0.001). A-G) Statistical significance was calculated using Student’s t-test. H) Average quiescence in 10-minute windows over 8-hours during UV-induced SIS of wild-type and nlp-14(stj18) animals (N ≥ 33, * P<0.01 at 70, 90-270 minutes). I) Average quiescence in 10-minute windows over 8-hours during UV-induced SIS of wild-type and nlp-14(tm1880) animals (N ≥ 26, * P<0.01 at 40,50 80-190 minutes). J) Average quiescence in 10-minute windows over 8-hours during UV-induced SIS of wild-type and nlp-14(stj18); nlp-15(stj25) animals (N ≥ 68, * P<0.01 at 80-230 minutes). H-J) Statistical significance was calculated using two-way ANOVA followed by Sidak’s multiple comparisons test. All error bars represent mean +/− SEM.
The timing of movement quiescence is regulated by NLP-14 and NLP-15 peptides
We noted that the nlp-14(null), nlp-14(2,4,5) and nlp-14(1-3) mutants all displayed movement quiescence earlier than wild-type controls (Figure 3H,I). In the first hour post UV-stress, each mutant displayed significantly more quiescence than wild-type animals (Figure S2). nlp-14(2, 4, 5) mutants had the most severe defect. We expected that nlp-15 mutants may enhance this phenotype but, to our surprise, the early quiescence phenotype of nlp-14 mutants was suppressed rather than enhanced by nlp-15 mutations. nlp-14(null); nlp-15(3-5) and nlp-14(null); nlp-15(null) double mutants did not show these timing defects (Figure 3J; Figure S2). These data suggest that removal of nlp-14 alters the timing of SIS through nlp-15-dependent mechanisms.
NLP-14 peptides are required for defecation quiescence during SIS
Insect and crustacean orcokinins regulate rhythmic intestinal muscle contractions (Stangier, Hilbich et al. 1992, Hofer and Homberg 2006, Chen, Choi et al. 2015). In C. elegans, the rhythmic defecation cycle is inhibited during SIS. Defecation is precisely timed by NLP-40 neuropeptides released from the posterior intestines (Wang, Girskis et al. 2013), and during sleep it is partially inhibited by peptides released from the ALA (Nath, Chow et al. 2016). Defecation consists of three behaviors, which occur every 50-60 seconds in the following order: a posterior body contraction (pBoc), an anterior body contraction (aBoc) and an expulsion (Exp) (Thomas 1990). The defecation rates of unstressed wild-type and nlp-14(2, 4, 5) animals were similar (Figure 4A), however, in the 10-minute period beginning 85 minutes after UV exposure, the number of expulsions was significantly increased in the nlp-14(2, 4, 5) and nlp-14(null) animals (Figure 4B,D). We also observed that nlp-14(2, 4, 5) animals performed more pBoc and aBoc events without an Exp (Figure 4C). In contrast, nlp-14(1-3) mutant animals did not display defects in defecation quiescence (Figure 4E), suggesting that NLP-14 peptides 4 and 5 are not needed for this behavior. Based on defects observed in the various mutants, we conclude that the NLP-14 peptides 1 and/or peptide 3 are required for the quiescence of defecation during SIS.
Figure 4: NLP-14 neuropeptides are indispensable for defecation quiescence during SIS.

A) Expulsions (x) performed in 6-minutes, of wild-type and nlp-14(tm1880) animals, who had not been exposed to any external stress. B) Wild-type, nlp-14(tm1880) and nlp-14(tm1880); ida-1p::nlp-14 animals 85-95 minutes following UV-stress (N ≥ 20, ***P<0.001). C) p- (Posterior body contraction or pBoc) and a- (Anterior body contraction or aBoc) events or p-, a- and x-events in 6-minutes of wild-type and nlp-14(tm1880) animals, 85-95 minutes post-UV stress (N ≥ 26, ***p<0.001). D) x-events performed in 6-minutes, of wild-type and nlp-14(stj18) animals (N ≥ 26, **p<0.01) and E) wild-type and nlp-14(stj10) animals (N=30). F) Defecation quiescence profile of nlp-14(tm1880), G) nlp-14(stj18), and H) nlp-14(stj10) animals (N=10, *p<0.05 210 minutes). A,D,E) Statistical significance was calculated using Student’s t-test. B-C) Statistical significance was calculated using one-way ANOVA followed by Tukey’s multiple comparisons test. H) Statistical significance was calculated using two-way ANOVA followed by Sidak’s multiple comparisons test. All error bars represent mean +/− SEM.
We performed a temporal analysis of defecation quiescence by counting expulsions for five minutes every 30 minutes after UV irradiation (Table S4). As expected, wild-type animals slowed their defecation rate throughout the first 4 hours of UV-induced SIS. Also as expected, nlp-14(2, 4, 5) and nlp-14(null) animals displayed more frequent events, but there was high variation between animals (Figure 4F,G, Table S4). Both the nlp-14(1-3) and nlp-14(1,2,4,5) animals showed defecation temporal profiles similar to wild type (Figure 4H; Table S4). However, the nlp-14(1-3) mutant animals showed reduced expulsions throughout SIS, which is significantly lower than wild-type animals at later time points. The nlp-15(3-5) single mutants showed no defect in defecation quiescence (Table S4); surprisingly, both nlp-14(null); nlp-15(3-5) and nlp-14(null); nlp-15(null) double mutants also were similar to wild-type animals during these 4 hours (Table S4). Taken together, these data suggest that NLP-14 peptides 1 and 3 are required for defecation quiescence. However, similar to what was observed with the timing defects of nlp-14(null) and nlp-14(2, 4, 5) animals, the defecation defects may be dependent upon the presence of nlp-15.
NLP-14 peptides are secreted from the ALA during SIS
Since nlp-14 is required for quiescence during SIS and is expressed in the ALA neuron (Nath, Chow et al. 2016, Taylor, Santpere et al. 2019), we predicted that quiescence induced by strong activation of this neuron would be blunted in nlp-14 mutants. Epidermal growth factor (EGF) induces sleep in mammals (Kushikata, Fang et al. 1998, Kramer, Yang et al. 2001), Drosophila (Foltenyi, Greenspan et al. 2007) and C. elegans (Van Buskirk and Sternberg 2007), and acts during SIS by stimulating neuropeptide release from both the ALA and RIS neurons (Nelson, Lee et al. 2014, Nath, Chow et al. 2016, Konietzka, Fritz et al. 2020). Overexpression of lin-3, coding for EGF, induces movement quiescence (i.e. EGF-induced sleep) (Van Buskirk and Sternberg 2007). Using the WorMotel, we found that lin-3 overexpression caused prolonged movement quiescence in otherwise wild-type animals (Figure 5A,B), but this quiescence was significantly attenuated in nlp-14(null), nlp-14(2, 4, 5) and nlp-14(1-3) mutant animals (Figure 5A,B). These results suggest that one or more NLP-14 peptides are required for EGF-induced sleep.
Figure 5: NLP-14 peptides function downstream of EGF from the ALA neuron.

A) Total minutes of quiescence in 4-hours, following overexpression of lin-3, in the following genetic backgrounds: Wild type, nlp-14(stj18), nlp-14(tm1880), nlp-14(stj10), and nlp-14(stj19) (N ≥ 31, *p<0.05, ***p<0.001). B) Average quiescence in 10-minute windows over 4-hours following lin-3 over-expression in wild-type and nlp-14(stj18) animals (N =48, p<0.05 100-120 minutes, p<0.001 20-80 minutes). Statistical significance was calculated using two-way ANOVA followed by Sidak’s multiple comparisons test. C) x-, D) p- and E) a-events performed in 5 minutes, 2-hours after the overexpression of lin-3 in wild-type, nlp-14(stj18) and nlp-14(tm1880) animals (N ≥ 10, *p<0.05, **p<0.01). F) Movement quiescence during UV-induced SIS in wild-type, nlp-14(tm1880) and nlp-14(tm1880); ida-1p::nlp-14 animals (N ≥ 11, *p<0.05, ***p<0.001). Numbers below nlp-14(tm1880); ida-1p::nlp-14 indicate distinct transgenic lines. A,C-F) Statistical significance was calculated using one-way ANOVA followed by Tukey’s multiple comparisons test. All error bars represent mean +/− SEM.
In addition to quiescence of body and feeding movements, lin-3 over-expression also induced quiescence of defecation in wild-type animals, where not a single animal performed a pBoc, aBoc, or Exp (Figure 5C–E). EGF-induced defecation quiescence was variably attenuated (i.e. defecation events were observed) in nlp-14(null) and in nlp-14(2, 4, 5) mutants (Figure 5C–E). We conclude that NLP-14 peptides are functioning downstream of EGF signaling to promote quiescence of both movement and defecation.
To test whether activity of NLP-14 peptides in ALA is sufficient to restore quiescent behavior, we made transgenic animals in which nlp-14 expression was controlled by the ida-1 promoter; ida-1 is expressed strongly in ALA, but is also expressed in a few other neurosecretory neurons (Zahn, Macmorris et al. 2001). In three independent transgenic lines, movement quiescence during SIS was significantly increased relative to nlp-14(2,4,5) mutants (Figure 5F). Also, the timing defects we observed in nlp-14(2, 4, 5) animals were corrected by expressing nlp-14 from the ida-1 promoter (Figure S2). Additionally, defecation quiescence during SIS was much more prevalent in these ida-1p::nlp-14 transgenic animals (Figure 4B) and, in fact, it was reduced below that of wild-type control levels (Table S4). Based on expression pattern, the requirement of nlp-14 for EGF-induced sleep and rescue from the ida-1 promoter, our data suggest that the NLP-14 peptides are released from the ALA neuron to regulate both movement and defecation quiescence during SIS. However, our data does not rule out the possibility that NLP-14 peptides are released from other cells as well.
Overexpression of nlp-14 induces movement and defecation quiescence
We predicted that overexpression of nlp-14 would induce quiescence of movement and defecation in active animals, like that observed for other somnogenic neuropeptides such as flp-11, flp-13, flp-24, nlp-8, and nlp-22 (Nelson, Trojanowski et al. 2013, Nelson, Lee et al. 2014, Nath, Chow et al. 2016, Turek, Besseling et al. 2016). We constructed multiple transgenic lines in which nlp-14 expression is controlled by a heat-inducible promoter. To induce strong pan-somatic expression of the gene, we subjected hsp-16p::nlp-14 animals to a 30-minute heat pulse and then waited 2 hours before analysis of behavior. At this 2-hour time point, any direct effect of heat on behavior, which is minor at temperatures less than 35 degrees, had fully dissipated. Wild-type control animals were exposed to the same conditions.
Overexpression of nlp-14 strongly suppressed body movement, which we measured by counting body bends (Figure 6A; Table S5) and by using machine vision, the WorMotel (Figure 6B,C). Overexpression of nlp-14 also caused a significant reduction in defecation events (Figure 6D; Table S6). Thus NLP-14 peptides are both required for and capable of inducing quiescence of movement and defecation. In addition to the movement and defecation phenotypes we were focused on, we incidentally noted that many hsp-16.2p::nlp-14 transgenic animals, even before induced over-expression, displayed a kinked body posture phenotype, where their body resembled a question mark (Video S1).
Figure 6: NLP-14 peptides are capable of inducing sleep-like behaviors.

A) Body bends performed in 1 minute, 2-2.5-hours after a 30-minute, 33 degree Celsius heat shock, by wild-type and hsp-16.2p::nlp-14 (strain SJU27) animals (N = 32, ***p<0.001). B) Movement quiescence during 4-hours following a 30-minute, 33 degree Celsius heat shock, by wild-type and hsp-16.2p::nlp-14 (strain SJU27) animals (N ≥ 19, ***p<0.001). C) Average quiescence in 10-minute windows over 4-hours following a 30-minute, 33 degree Celsius heat shock, wild-type and hsp-16.2p::nlp-14 (strain SJU27) animals (N ≥ 19, ***P<0.001 at all time points displayed). D) x-events performed in 6-minutes, 2-2.5-hours after a 30-minute, 33 degree Celsius heat shock, by wild-type and hsp-16.2p::nlp-14 (strain SJU27) animals (N = 16, ***p<0.001). E) Movement quiescence during 4-hours following a 30-minute, 33 degree Celsius heat shock, hsp-16.2p::nlp-14 (strain SJU27) and kin-2(ce179); hsp-16.2p::nlp-14 animals (N =12, *p<0.001). F) x-events performed in 5-minutes by wild-type and kin-2(ce179) animals without exposure to heat shock and by hsp-16.2p::nlp-14 (strain SJU27) and kin-2(ce179); hsp-16.2p::nlp-14 animals 2-2.5-hours after a 30-minute, 33 degree Celsius heat shock (N = 15). G) Body bends performed in 1-minute, 2-2.5-hours after a 30-minute, 33 degree Celsius heat shock, by wild type and animals over-expressing nlp-14(1), (1-2), (1-3), (1-4) or (3) (N ≥15, ***p<0.001). H) x-events performed in 5-minutes, 2-2.5-hours after a 30-minute, 33 degree Celsius heat shock, by wild type and animals overexpressing nlp-14(1), (1-2), (1-3), (1-4) or (3) (N ≥ 15, *p<0.05, ***p<0.001). A,B,D,E) Statistical significance was calculated using Student’s t-test. C) Statistical significance was calculated using two-way ANOVA followed by Sidak’s multiple comparisons test. F-H) Statistical significance was calculated using one-way ANOVA followed by Tukey’s multiple comparisons test. All error bars represent mean +/− SEM.
Many neuropeptides signal through GPCRs, which increase or decrease signaling of second messenger pathways. Movement quiescence is antagonized by signaling through the cyclic adenosine monophosphate/Protein Kinase A pathway (Cianciulli, Yoslov et al. 2019). In C. elegans, PKA activity can be experimentally increased by genetic impairment in the gene kin-2, which encodes a regulatory subunit of PKA (Charlie, Thomure et al. 2006). We found that the increased PKA activity of kin-2(ce179) mutants (Charlie, Thomure et al. 2006), stimulated movement but not defecation following nlp-14 overexpression (Figure 6E,F). These data suggest that NLP-14 peptides inhibit cAMP/PKA in cells regulating movement quiescence.
Based on our analysis of different nlp-14 loss-of-function alleles, suggesting that removal of subsets of NLP-14 peptides affected behavioral quiescence in unique ways, we over expressed different combinations of the peptides. Overexpression of NLP-14-1 or NLP-14-3 induced quiescence of movement but not defecation, while overexpression of NLP-14-1 and NLP-14-2 strongly induced quiescence of movement and weakly of defecation. Overexpression of NLP-14-(1-3) or NLP-14-(1-4) strongly induced quiescence of both movement and defecation (Figure 6G,H). These data, together with the loss-of-function analyses, suggest that all five NLP-14 peptides regulate movement quiescence; defecation quiescence, however, is prominently regulated by peptides 1 and 3, while the other peptides may play more subtle modulating roles.
Orcokinin receptors are unknown for all ecdysozoans
Orcokinin receptors have not been identified in any animal, despite screening attempts using heterologous expression systems (Yamanaka, Hua et al. 2010, Yamanaka, Roller et al. 2011). In C. elegans, the receptor NPR-10 has been proposed as an NLP-14 receptor, based on genetic interactions and anatomical connectivity (Hapiak, Summers et al. 2013). We did not detect changes in movement quiescence during SIS in the presumptive npr-10(ok1442) null mutants. ok1442 mutants have a 788bp deletion that is predicted to result in a frameshift and premature stop in exon 5 and therefore a truncated protein composed of only five transmembrane domains (Figure S1). Our data suggest that NPR-10 is not the receptor for NLP-14 during sleep regulation or that other receptors function redundantly together with NPR-10. To date, no orcokinin receptor for any animals has been convincingly identified.
Discussion
Orcokinin neuropeptides are conserved in Ecdysozoa, which consists of organisms that undergo molting (Aguinaldo, Turbeville et al. 1997). Millions of years of evolution separate these animals, yet orcokinin peptide sequences are highly similar (Yasuda-Kamatani and Yasuda 2000, Nathoo, Moeller et al. 2001, Hofer, Dircksen et al. 2005, Hofer and Homberg 2006, Dickinson, Stemmler et al. 2009, Chen, Choi et al. 2015, Wulff, Sierra et al. 2017, Koziol 2018). Functional studies have demonstrated that they regulate insect circadian rhythms and molting (Hofer and Homberg 2006, Wulff, Sierra et al. 2017), rhythmic smooth muscle contractions of insects and crustaceans (Stangier, Hilbich et al. 1992, Li, Pulver et al. 2002, Skiebe, Dreger et al. 2002) and decision making behaviors and male mating in C. elegans (Hapiak, Summers et al. 2013, Sherlekar, Janssen et al. 2013). Here we described a novel function for the orcokinin encoded by nlp-14 and nlp-15 during the regulation of sleep.
Using a combination of loss-of-function and overexpression studies we find that NLP-14 and NLP-15 peptides regulate two sleep states, developmentally timed sleep (DTS), which resembles sleep in animals that are strongly circadian (Trojanowski and Raizen 2016), and stress-induced sleep (SIS), a behavior required for recovery following exposure to damaging stress (Hill, Mansfield et al. 2014). The five NLP-14 peptides play a larger role in the regulation of SIS than DTS. They promote movement and defecation quiescence, the latter of which is largely regulated by NLP-14 peptides 1 and 3. We propose that these sleep-regulatory roles are more conserved in Ecdysozoa.
Developmentally timed sleep
Both nlp-14 and nlp-15 are expressed in the sleep regulating neurons ALA and RIS (Van Buskirk and Sternberg 2007, Turek, Lewandrowski et al. 2013). Individually, they are dispensable for DTS but removal of both genes reduces movement quiescence without causing molting defects. This is in contrast to studies done with the kissing bug Rhodnius prolixus where disruption of orcokinin by RNA interference (RNAi) caused molting defects (Wulff, Sierra et al. 2017). We also did not observe molting difficulties when nlp-14 was over-expressed, suggesting that either the role of these peptides is strictly behavioral or that there is degeneracy in the control of molting (Choi, Chatzigeorgiou et al. 2013, Nelson, Trojanowski et al. 2013, Turek, Besseling et al. 2016).
Stress induced sleep
In contrast to its relatively minor roles in DTS, NLP-14 is more important during stress-induced sleep (SIS), where the removal of all or subsets of peptides causes strong defects in movement and defecation quiescence. Numerous neuropeptides regulate movement quiescence, including FLP-11, secreted from the RIS (Konietzka, Fritz et al. 2020) and FLP-13, FLP-24, and NLP-8 (Nelson, Janssen et al. 2015, Nath, Chow et al. 2016), released from ALA. These molecules signal through many GPCRs (Nelson, Janssen et al. 2015, Iannacone, Beets et al. 2017), reducing cAMP/PKA signaling in different cells (Cianciulli, Yoslov et al. 2019). The orcokinins can be added to this expanding list of somnogenic neuropeptides. This observation in C. elegans that multiple peptides can induce quiescence when over-expressed is consistent with studies in fish, which have identified several somnogenic neuropeptides using an over-expression approach (Chiu, Rihel et al. 2016, Lee, Andreev et al. 2017). Therefore, this complexity to sleep regulation appears to be phylogenetically conserved and demonstrates the importance of sleep to all animals.
Our data, however, suggest that the orcokinins in C. elegans are not acting strictly as somnogens. Removal of nlp-14 shifts the timing of SIS, such that it occurs earlier. We propose that NLP-14 peptides may be functioning to promote aversive behaviors associated with nocioception, a previously described role (Hapiak, Summers et al. 2013), and act as a somnogen only at later stages, to facilitate recovery from the stressful exposure.
Surprisingly, this early increased quiescence in nlp-14 mutants is dependent on the presence of nlp-15. An interpretation of this could be that NLP-15 and NLP-14 peptides are antagonizing one another during the injurious response to UV, promoting both quiescence and arousal, respectively. At early time points after UV exposure, NLP-14 peptides may promote behavioral arousal, perhaps to allow for an escape response, whereas NLP-15 may promote quiescence at all time points.
At later time points, both promote sleep. After exposures to injurious conditions such as UV light or high heat, animals must balance the benefits of aversion and escape with those of recovery, which are linked to sleep. More work needs to be done to test this idea.
The defecation motor program is both stimulated (Wang, Girskis et al. 2013) and inhibited by neuropeptides (Nath, Chow et al. 2016). When awake, NLP-40 peptides are released from the posterior intestines following rhythmic calcium fluxes, bind their receptor AEX-2, stimulating cAMP/PKA and calcium signaling in the AVL and DVB neurons. This stimulates GABA release, which excites the enteric muscles and initiates an expulsion (Wang, Girskis et al. 2013). During SIS, the ALA neuron releases NLP-8 peptides to inhibit defecation (Nath, Chow et al. 2016). We find that ALA and/or RIS also release NLP-14 peptides. Our data indicates that NLP-14’s effects on defecation are cAMP/PKA-independent. PKA functions in the AVL and DVB motor neurons during defecation to increase their activity leading to enteric muscle contractions that drive the expulsion events (Wang and Sieburth 2013). Based on this, our data suggest that NLP-14 peptides are functioning either directly on the enteric muscles or downstream of PKA in AVL and DVB. The notion that orcokinins act directly on GI motility would be consistent with observations of crustacean orcokinins, which directly regulate smooth muscle of the gut (Li, Pulver et al. 2002) and deuterostome starfish myorelaxant peptides (SMPs), which promote the relaxation of stomach muscle (Lin, Egertova et al. 2018). There is a sequence similarity between the SMPs and NLP-14 peptides (Kim, Kim et al. 2016). Therefore the role of orcokinin/SMPs in smooth muscle regulation may be conserved.
A conserved sleep-regulating role for orcokinins neuropeptides
Are these sleep functions of NLP-14 and NLP-15 more broadly conserved in Ecdysozoa? Though prior studies have not reported the requirement of orcokinins for sleep, some observations point towards a sleep-regulating role in insects too. Elegant work by Hofer and Homber showed that orcokinin injections result in a circadian phase shift, measured by wheel-running activity in cockroaches. Interestingly, while the authors do not emphasize this point, their actographic data indicate strong inhibition of activity 24-48 hours after the orcokinin injection (Hofer and Homberg 2006). Hence, they observed both a change in sleep timing and in sleep/activity in response to orcokinins injections, much as we observe a change in timing and in sleep following nlp-14 overexpression in C. elegans.
DTS in C. elegans occurs coincident with the molt (Raizen, Zimmerman et al. 2008). Insect larvae can sleep between molts (Szuperak, Churgin et al. 2018) and also become quiescent during the molt, a behavior called molt-sleep (Reinecke, Buckner et al. 1980). Gene-expression analysis suggests that molt-sleep is regulated by neuropeptide signaling (MacWilliam, Arensburger et al. 2015). Removal of orcokinins in the kissing bug causes molting failure and death (Wulff, Sierra et al. 2017). It is possible that inhibition of either the behavioral or physiological aspects of molt-sleep is the cause of this lethality. To test for a conserved sleep-regulating role during the molt, it will be important to measure sleep following orcokinin manipulation during inter-molt and molt-sleep in insects and sleep in crustaceans.
In contrast to effects during DTS, the NLP-14 peptides play a more important role during SIS regulation. Heat-induced recovery sleep occurs in Drosophila melanogaster and is regulated by the same family of neuropeptides controlling SIS in C. elegans (Lenz, Xiong et al. 2015). In an effort to test the generalizability of our findings, we propose that an initial approach would be to test the necessity of orcokinins in Drosophila. It would be particularly interesting to test for an orcokinin role in crayfish, which display slow-wave brain activity, similar to mammals (Ramon, Hernandez-Falcon et al. 2004, Ramon, Mendoza-Angeles et al. 2012). Considering that sickness and injury increase sleep in mammals (Imeri and Opp 2009), SIS may exist in crustaceans as well.
What about tardigrades? These amazingly hardy animals can survive some of the harshest conditions, like desiccation and extreme heat and osmotic pressure. They do so by entering a state of extended quiescence referred to as cryptobiosis (Crowe 1975). This can promote survival for years, but is reversible, at which point their bodies can be remarkably repaired (Wright, Westh et al. 1992). Cryptobiosis may represent an extreme version of SIS. Is this protective behavioral state regulated by orcokinins? If so, orcokinins may be an evolutionarily ancient mechanism controlling protective behavioral quiescence in Ecdysozoa.
Supplementary Material
Acknowledgements
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank the National BioResource Project (PI, Shohei Mitani) for strains. We would also like to thank the SJU Summer Scholars Program, the John P. McNulty Fellows Program and the Peter and Dorothy Kowey fellowship for student funding. MDN was supported by the National Institute of General Medical Sciences of the National Institutes of Health grant R15GM122058 and the National Science Foundation grants IOS-CAREER-1845020 and DBI-MRI-1919847.
Works Cited
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA and Lake JA (1997). “Evidence for a clade of nematodes, arthropods and other moulting animals.” Nature 387(6632): 489–493. [DOI] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS and Fire AZ (2014). “Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans.” Genetics 198(3): 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlie NK, Thomure AM, Schade MA and Miller KG (2006). “The Dunce cAMP phosphodiesterase PDE-4 negatively regulates G alpha(s)-dependent and G alpha(s)-independent cAMP pools in the Caenorhabditis elegans synaptic signaling network.” Genetics 173(1): 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Choi MS, Mizoguchi A, Veenstra JA, Kang K, Kim YJ and Kwon JY (2015). “Isoform-specific expression of the neuropeptide orcokinin in Drosophila melanogaster.” Peptides 68: 50–57. [DOI] [PubMed] [Google Scholar]
- Chiu CN, Rihel J, Lee DA, Singh C, Mosser EA, Chen S, Sapin V, Pham U, Engle J, Niles BJ, Montz CJ, Chakravarthy S, Zimmerman S, Salehi-Ashtiani K, Vidal M, Schier AF and Prober DA (2016). “A Zebrafish Genetic Screen Identifies Neuromedin U as a Regulator of Sleep/Wake States.” Neuron 89(4): 842–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR and Kaplan JM (2013). “Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans.” Neuron 78(5): 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churgin MA, Jung SK, Yu CC, Chen X, Raizen DM and Fang-Yen C (2017). “Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging.” Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciulli A, Yoslov L, Buscemi K, Sullivan N, Vance RT, Janton F, Szurgot MR, Buerkert T, Li E and Nelson MD (2019). “Interneurons Regulate Locomotion Quiescence via Cyclic Adenosine Monophosphate Signaling During Stress-Induced Sleep in Caenorhabditis elegans.” Genetics 213(1): 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe J (1975). “ The physiology of cryptobiosis in tardigrades.” International symposium on tardigrades 32: 37–59. [Google Scholar]
- Davis MW, Somerville D, Lee RY, Lockery S, Avery L and Fambrough DM (1995). “Mutations in the Caenorhabditis elegans Na,K-ATPase alpha-subunit gene, eat-6, disrupt excitable cell function.” J Neurosci 15(12): 8408–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBardeleben HK, Lopes LE, Nessel MP and Raizen DM (2017). “Stress-Induced Sleep After Exposure to Ultraviolet Light Is Promoted by p53 in Caenorhabditis elegans.” Genetics 207(2): 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PS, Stemmler EA, Barton EE, Cashman CR, Gardner NP, Rus S, Brennan HR, McClintock TS and Christie AE (2009). “Molecular, mass spectral, and physiological analyses of orcokinins and orcokinin precursor-related peptides in the lobster Homarus americanus and the crayfish Procambarus clarkii.” Peptides 30(2): 297–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ and Newport JW (2007). “Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila.” Nat Neurosci 10(9): 1160–1167. [DOI] [PubMed] [Google Scholar]
- George-Raizen JB, Shockley KR, Trojanowski NF, Lamb AL and Raizen DM (2014). “Dynamically-expressed prion-like proteins form a cuticle in the pharynx of Caenorhabditis elegans.” Biol Open 3(11): 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapiak V, Summers P, Ortega A, Law WJ, Stein A and Komuniecki R (2013). “Neuropeptides amplify and focus the monoaminergic inhibition of nociception in Caenorhabditis elegans.” J Neurosci 33(35): 14107–14116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks GJ, Gaidatzis D, Aeschimann F and Grosshans H (2014). “Extensive oscillatory gene expression during C. elegans larval development.” Mol Cell 53(3): 380–392. [DOI] [PubMed] [Google Scholar]
- Hill AJ, Mansfield R, Lopez JM, Raizen DM and Van Buskirk C (2014). “Cellular stress induces a protective sleep-like state in C. elegans.” Curr Biol 24(20): 2399–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer S, Dircksen H, Tollback P and Homberg U (2005). “Novel insect orcokinins: characterization and neuronal distribution in the brains of selected dicondylian insects.” J Comp Neurol 490(1): 57–71. [DOI] [PubMed] [Google Scholar]
- Hofer S and Homberg U (2006). “Evidence for a role of orcokinin-related peptides in the circadian clock controlling locomotor activity of the cockroach Leucophaea maderae.” J Exp Biol 209(Pt 14): 2794–2803. [DOI] [PubMed] [Google Scholar]
- Hofer S and Homberg U (2006). “Orcokinin immunoreactivity in the accessory medulla of the cockroach Leucophaea maderae.” Cell Tissue Res 325(3): 589–600. [DOI] [PubMed] [Google Scholar]
- Iannacone MJ, Beets I, Lopes LE, Churgin MA, Fang-Yen C, Nelson MD, Schoofs L and Raizen DM (2017). “The RFamide receptor DMSR-1 regulates stress-induced sleep in C. elegans.” Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imeri L and Opp MR (2009). “How (and why) the immune system makes us sleep.” Nat Rev Neurosci 10(3): 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekely G (2013). “Global view of the evolution and diversity of metazoan neuropeptide signaling.” Proc Natl Acad Sci U S A 110(21): 8702–8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Go HJ, Oh HY, Elphick MR and Park NG (2018). “Identification of evolutionarily conserved residues required for the bioactivity of a pedal peptide/orcokinin-type neuropeptide.” Peptides 103: 10–18. [DOI] [PubMed] [Google Scholar]
- Kim CH, Kim EJ, Go HJ, Oh HY, Lin M, Elphick MR and Park NG (2016). “Identification of a novel starfish neuropeptide that acts as a muscle relaxant.” J Neurochem 137(1): 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konietzka J, Fritz M, Spiri S, McWhirter R, Leha A, Palumbos S, Costa WS, Oranth A, Gottschalk A, Miller DM 3rd, Hajnal A and Bringmann H (2020). “Epidermal Growth Factor Signaling Promotes Sleep through a Combined Series and Parallel Neural Circuit.” Curr Biol 30(1): 1–16 e13. [DOI] [PubMed] [Google Scholar]
- Koziol U (2018). “Precursors of neuropeptides and peptide hormones in the genomes of tardigrades.” Gen Comp Endocrinol 267: 116–127. [DOI] [PubMed] [Google Scholar]
- Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC and Weitz CJ (2001). “Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling.” Science 294(5551): 2511–2515. [DOI] [PubMed] [Google Scholar]
- Kushikata T, Fang J, Chen Z, Wang Y and Krueger JM (1998). “Epidermal growth factor enhances spontaneous sleep in rabbits.” Am J Physiol 275(2): R509–514. [DOI] [PubMed] [Google Scholar]
- Lee DA, Andreev A, Truong TV, Chen A, Hill AJ, Oikonomou G, Pham U, Hong YK, Tran S, Glass L, Sapin V, Engle J, Fraser SE and Prober DA (2017). “Genetic and neuronal regulation of sleep by neuropeptide VF.” Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz O, Xiong J, Nelson MD, Raizen DM and Williams JA (2015). “FMRFamide signaling promotes stress-induced sleep in Drosophila.” Brain Behav Immun 47: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Pulver SR, Kelley WP, Thirumalai V, Sweedler JV and Marder E (2002). “Orcokinin peptides in developing and adult crustacean stomatogastric nervous systems and pericardial organs.” J Comp Neurol 444(3): 227–244. [DOI] [PubMed] [Google Scholar]
- Lin M, Egertova M, Zampronio CG, Jones AM and Elphick MR (2018). “Functional characterization of a second pedal peptide/orcokinin-type neuropeptide signaling system in the starfish Asterias rubens.” J Comp Neurol 526(5): 858–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd PE and Connolly CM (1989). “Sequence of pedal peptide: a novel neuropeptide from the central nervous system of Aplysia.” J Neurosci 9(1): 312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacWilliam D, Arensburger P, Higa J, Cui X and Adams ME (2015). “Behavioral and genomic characterization of molt-sleep in the tobacco hornworm, Manduca sexta.” Insect Biochem Mol Biol 62: 154–167. [DOI] [PubMed] [Google Scholar]
- Mello C and Fire A (1995). “DNA transformation.” Methods Cell Biol 48: 451–482. [PubMed] [Google Scholar]
- Nath RD, Chow ES, Wang H, Schwarz EM and Sternberg PW (2016). “C. elegans Stress-Induced Sleep Emerges from the Collective Action of Multiple Neuropeptides.” Curr Biol 26(18): 2446–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathoo AN, Moeller RA, Westlund BA and Hart AC (2001). “Identification of neuropeptide-like protein gene families in Caenorhabditiselegans and other species.” Proc Natl Acad Sci U S A 98(24): 14000–14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MD and Fitch DH (2011). “Overlap extension PCR: an efficient method for transgene construction.” Methods Mol Biol 772: 459–470. [DOI] [PubMed] [Google Scholar]
- Nelson MD, Janssen T, York N, Lee KH, Schoofs L and Raizen DM (2015). “FRPR-4 Is a G-Protein Coupled Neuropeptide Receptor That Regulates Behavioral Quiescence and Posture in Caenorhabditis elegans.” PLoS One 10(11): e0142938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MD, Lee KH, Churgin MA, Hill AJ, Van Buskirk C, Fang-Yen C and Raizen DM (2014). “FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans.” Curr Biol 24(20): 2406–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MD, Trojanowski NF, George-Raizen JB, Smith CJ, Yu CC, Fang-Yen C and Raizen DM (2013). “The neuropeptide NLP-22 regulates a sleep-like state in Caenorhabditis elegans.” Nat Commun 4: 2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG and Heringa J (2000). “T-Coffee: A novel method for fast and accurate multiple sequence alignment.” J Mol Biol 302(1): 205–217. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A and Seydoux G (2017). “Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans.” Methods 121-122: 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV and Pack AI (2008). “Lethargus is a Caenorhabditis elegans sleep-like state.” Nature 451(7178): 569–572. [DOI] [PubMed] [Google Scholar]
- Ramon F, Hernandez-Falcon J, Nguyen B and Bullock TH (2004). “Slow wave sleep in crayfish.” Proc Natl Acad Sci U S A 101(32): 11857–11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon F, Mendoza-Angeles K and Hernandez-Falcon J (2012). “Sleep in invertebrates: crayfish.” Front Biosci (Schol Ed) 4: 1190–1200. [DOI] [PubMed] [Google Scholar]
- Reinecke J, Buckner JS and Grugel SR (1980). “Life cycle of laboratory-reaered tobacco hornworms, Manduca sexta, a study of development and behavior, using time-lapse cinematography.” The Biological Bulletin 158(1): 129. [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC and Taghert PH (1999). “A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila.” Cell 99(7): 791–802. [DOI] [PubMed] [Google Scholar]
- Rowe ML and Elphick MR (2012). “The neuropeptide transcriptome of a model echinoderm, the sea urchin Strongylocentrotus purpuratus.” Gen Comp Endocrinol 179(3): 331–344. [DOI] [PubMed] [Google Scholar]
- Semmens DC and Elphick MR (2017). “The evolution of neuropeptide signalling: insights from echinoderms.” Brief Funct Genomics 16(5): 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlekar AL, Janssen A, Siehr MS, Koo PK, Caflisch L, Boggess M and Lints R (2013). “The C. elegans male exercises directional control during mating through cholinergic regulation of sex-shared command interneurons.” PLoS One 8(4): e60597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN and Sulston JE (1978). “Some Observations On Moulting in Caenorhabditis Elegans.” Nematologica 24(1): 63–71. [Google Scholar]
- Skiebe P, Dreger M, Meseke M, Evers JF and Hucho F (2002). “Identification of orcokinins in single neurons in the stomatogastric nervous system of the crayfish, Cherax destructor.” J Comp Neurol 444(3): 245–259. [DOI] [PubMed] [Google Scholar]
- Stangier J, Hilbich C, Burdzik S and Keller R (1992). “Orcokinin: a novel myotropic peptide from the nervous system of the crayfish, Orconectes limosus.” Peptides 13(5): 859–864. [DOI] [PubMed] [Google Scholar]
- Stinchcomb DT, Shaw JE, Carr SH and Hirsh D (1985). “Extrachromosomal DNA transformation of Caenorhabditis elegans.” Mol Cell Biol 5(12): 3484–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuperak M, Churgin MA, Borja AJ, Raizen DM, Fang-Yen C and Kayser MS (2018). “A sleep state in Drosophila larvae required for neural stem cell proliferation.” Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SR, Santpere G, Reilly M, Glenwinkel L, Poff A, McWhirter R, Xu C, Weinreb A, Basavaraju M, Cook SJ, Barrett A, Abrams A, Vidal B, Cros C, Rafi I, Sestan N, Hammarlund M, Hobert O and Miller DM (2019). “Expression profiling of the mature C. elegans nervous system by single-cell RNA-Sequencing.” bioRxiv: 737577. [Google Scholar]
- Thomas JH (1990). “Genetic analysis of defecation in Caenorhabditis elegans.” Genetics 124(4): 855–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski NF and Raizen DM (2016). “Call it Worm Sleep.” Trends Neurosci 39(2): 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek M, Besseling J, Spies JP, Konig S and Bringmann H (2016). “Sleep-active neuron specification and sleep induction require FLP-11 neuropeptides to systemically induce sleep.” Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek M, Lewandrowski I and Bringmann H (2013). “An AP2 transcription factor is required for a sleep-active neuron to induce sleep-like quiescence in C. elegans.” Curr Biol 23(22): 2215–2223. [DOI] [PubMed] [Google Scholar]
- Van Buskirk C and Sternberg PW (2007). “Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans.” Nat Neurosci 10(10): 1300–1307. [DOI] [PubMed] [Google Scholar]
- Wang H, Girskis K, Janssen T, Chan JP, Dasgupta K, Knowles JA, Schoofs L and Sieburth D (2013). “Neuropeptide secreted from a pacemaker activates neurons to control a rhythmic behavior.” Curr Biol 23(9): 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H and Sieburth D (2013). “PKA controls calcium influx into motor neurons during a rhythmic behavior.” PLoS Genet 9(9): e1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JC, Westh P and Ramlov H (1992). “Cryptobiosis in Tardigrada.” Biological Reviews 67: 1–29. [Google Scholar]
- Wulff JP, Sierra I, Sterkel M, Holtof M, Van Wielendaele P, Francini F, Broeck JV and Ons S (2017). “Orcokinin neuropeptides regulate ecdysis in the hemimetabolous insect Rhodnius prolixus.” Insect Biochem Mol Biol 81: 91–102. [DOI] [PubMed] [Google Scholar]
- Yamanaka N, Hua YJ, Roller L, Spalovska-Valachova I, Mizoguchi A, Kataoka H and Tanaka Y (2010). “Bombyx prothoracicostatic peptides activate the sex peptide receptor to regulate ecdysteroid biosynthesis.” Proc Natl Acad Sci U S A 107(5): 2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N, Roller L, Zitnan D, Satake H, Mizoguchi A, Kataoka H and Tanaka Y (2011). “Bombyx orcokinins are brain-gut peptides involved in the neuronal regulation of ecdysteroidogenesis.” J Comp Neurol 519(2): 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda-Kamatani Y and Yasuda A (2000). “Identification of orcokinin gene-related peptides in the brain of the crayfish Procambarus clarkii by the combination of MALDI-TOF and on-line capillary HPLC/Q-Tof mass spectrometries and molecular cloning.” Gen Comp Endocrinol 118(1): 161–172. [DOI] [PubMed] [Google Scholar]
- Zahn TR, Macmorris MA, Dong W, Day R and Hutton JC (2001). “IDA-1, a Caenorhabditis elegans homolog of the diabetic autoantigens IA-2 and phogrin, is expressed in peptidergic neurons in the worm.” J Comp Neurol 429(1): 127–143. [DOI] [PubMed] [Google Scholar]
- Zitnan D, Ross LS, Zitnanova I, Hermesman JL, Gill SS and Adams ME (1999). “Steroid induction of a peptide hormone gene leads to orchestration of a defined behavioral sequence.” Neuron 23(3): 523–535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


