Abstract
The Indian leafwing butterfly Kallima paralekta (Horsfield, 1829) (Nymphalidae) is an Asian forest-dwelling, leaf-mimic. Genome skimming by Illumina sequencing permitted assembly of a complete circular mitogenome of 15,200 bp from K. paralekta consisting of 79.5% AT nucleotides, 22 tRNAs, 13 protein-coding genes, two rRNAs and a control region in the typical butterfly gene order. Kallima paralekta COX1 features an atypical CGA start codon, while ATP6, COX1, COX2, ND4, ND4L, and ND5 exhibit incomplete stop codons completed by 3’ A residues added to the mRNA. Phylogenetic reconstruction places K. paraleckta within the monophyletic genus Kallima, sister to Mallika in the subfamily Nymphalinae. These data support the monophyly of tribe Kallimini and contribute to the evolutionary systematics of the Nymphalidae.
Keywords: Illumina sequencing, leaf-mimicry, Lepidoptera, masquerade, mitogenomics
The Living Prairie Mitogenomics Consortium is an undergraduate structured inquiry exercise (Marcus et al. 2010) assembling arthropod mitogenomes for improved DNA-based species identification and phylogenetics (Living Prairie Mitogenomics Consortium 2017, 2018, 2019, 2020; Marcus 2018). Student participants analyzed sequence data (further curated by the instructor) for presentation here.
Alfred Russell Wallace (1867) described the close resemblance between the appearance of Indian leafwing butterfly Kallima paralekta (Nymphalidae) and dead leaves with respect to shape, size, background coloration, markings, and perching behavior as ‘the most wonderful and undoubted case of protective resemblance in a butterfly which we have ever seen’. Now often referred to as masquerade mimicry (Skelhorn 2015), the origin and evolution of this protective resemblance has been an important case study in the discussion of how organisms produce complex phenotypes (Suzuki et al. 2014). Convergent evolution of leaf masquerade mimicry in nymphalid butterfly lineages has also been a challenge for taxonomists interested in Kallima (Shirôzu and Nakanishi 1984; Larsen 1991) and phenotypically similar species in genera such as Kallimoides (Payment et al. 2020a), Mallika (Alexiuk et al. 2020b), Doleschallia (Hamilton et al. 2020), and Junonia (Wahlberg et al. 2005).
To facilitate future phylogenetic and taxonomic work, here we report the complete mitochondrial genome sequence of K. paralekta from specimen Kpar2017.1, collected in Pattaya, Thailand (GPS 12.927608 N, 100.877083 E) in 2017, that has been pinned, spread, and deposited in the Wallis Roughley Museum of Entomology, University of Manitoba (voucher WRME0507735).
DNA was prepared from a specimen leg using a DNeasy Blood and Tissue kit (Qiagen, Düsseldorf, Germany) with slight modifications to the standard protocol as described in McCullagh and Marcus (2015). DNA was sheared by sonication and a fragment library was prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, Massachusetts) as previously described (Peters and Marcus 2017), before sequencing by Illumina NovaSeq6000 (San Diego, California) (Marcus 2018). Mitogenome assembly of K. paralekta (Genbank accession MW192438) was performed by mapping the resulting sequence library of 23,143,566 paired 150 bp reads (Genbank SRA PRJNA667707) to a K. inachus reference mitogenome (HM243591) using 5 iterations of the medium sensitivity settings of Geneious Prime 2020.2. Annotation was in reference to K inachus and Junonia stygia (Nymphalidae, MN623383 (Living Prairie Mitogenomics Consortium 2020)). The K. paralekta nuclear rRNA repeat (MW192439) was also assembled and annotated using J. stygia (MF680448 (Living Prairie Mitogenomics Consortium 2020)), Anartia jatrophae (MT742579 (Payment et al. 2020b)), and Araschnia levana (MT750296 (Alexiuk et al. 2020a)) reference sequences.
The K. paralekta circular 15,200 bp mitogenome assembly was composed of 40,332 paired reads with nucleotide composition: 39.7% A, 12.7% C, 7.8% G, and 39.8% T. The gene order and composition of the K. paraleckta mitogenome is identical to all known butterfly mitogenomes (Cao et al. 2012; McCullagh and Marcus 2015; McCullagh et al. 2020).
Ten K. paralekta mitochondrial protein-coding genes begin with typical ATG or ATT start codons, with the remaining genes beginning with ATC (ND6), GTG (COX2), or CGA (COX1) start codons (Liao et al. 2010). The mitogenome contains three protein-coding genes (COX1, COX2, ND4L) with single-nucleotide (T) stop codons, and three protein-coding genes (ATP6, ND4, ND5) with two-nucleotide (TA) stop codons completed by post-transcriptional addition of 3′ A residues. The locations and structures of tRNAs were determined using ARWEN v.1.2 (Laslett and Canback 2008). The tRNAs have typical cloverleaf secondary structures except for trnS (AGN) where the dihydrouridine arm is replaced by a loop, while the mitochondrial rRNAs and control region are typical for Lepidoptera (McCullagh and Marcus 2015).
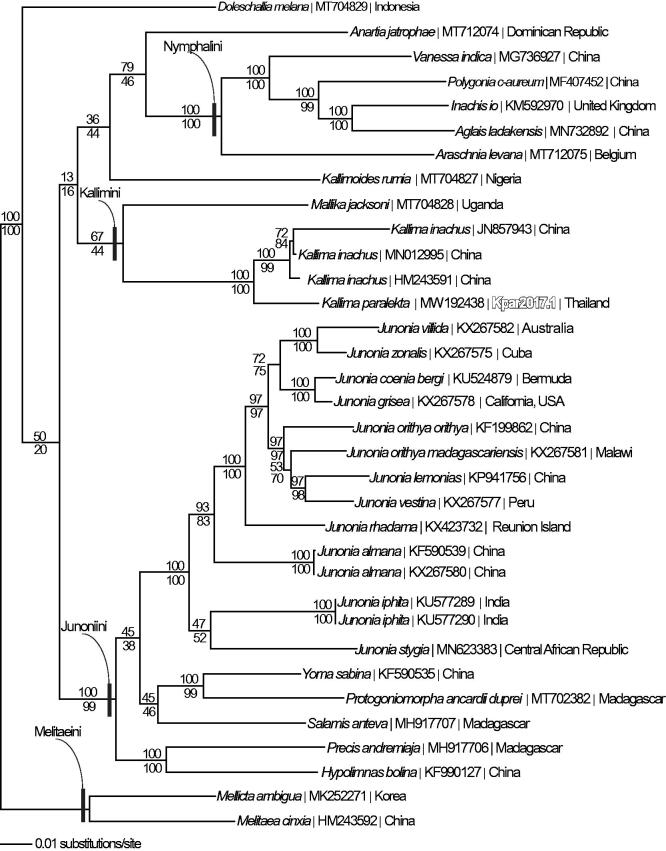
We reconstructed a phylogeny using the complete K. paralekta mitogenome, three K. inachus mitogenomes (Qin et al. 2012; Liu et al. 2020), and 30 additional mitogenomes from family Nymphalidae, including Melitaea cinxia (HM243592) and Mellicta ambigua (MK252271) as outgroup species. Mitogenome sequences were aligned in CLUSTAL Omega (Sievers et al. 2011) and analyzed by parsimony and maximum likelihood (model selected by jModeltest 2.1.7 (Darriba et al. 2012) and likelihood ratio test (Huelsenbeck and Rannala 1997)) in PAUP* 4.0b8/4.0d78 (Swofford 2002) (Figure 1). Phylogenetic analysis places K. paralekta within a monophyletic genus Kallima, which in turn is sister to another leaf-mimicking genus, Mallika. The leaf-mimics in genera Doleschallia, Kallimoides, and Junonia are only distantly related to Kallima and to each other, as has been suggested in some other phylogenetic analyses (Wahlberg et al. 2005; Alexiuk et al. 2020b).
Figure 1.
Maximum likelihood phylogeny (GTR + G model, G = 0.2300, likelihood score 116120.69888) of Kallima paralekta and 33 additional mitogenomes from subfamily Nymphalinae based on 1 million random addition heuristic search replicates (with tree bisection and reconnection). One million maximum parsimony heuristic search replicates produced an identical tree topology (parsimony score 20514 steps). Numbers above each node are maximum likelihood bootstrap values and numbers below each node are maximum parsimony bootstrap values (each from 1 million random fast addition search replicates).
Acknowledgements
We thank Melanie Lalonde and Josephine Payment for assistance with DNA extraction and bioinformatics pipeline development. We thank Genome Quebec for assistance with library preparation and sequencing.
Funding Statement
This work received support from NSERC under Grant [RGPIN-2016-06012] and from the University of Manitoba under the University Research Grants Program.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference numbers PRJNA667707, MW192438, and MW192439.
References
- Alexiuk MR, Marcus JM, Lalonde MML.. 2020a. The complete mitochondrial genome and phylogenetic analysis of the European map butterfly Araschnia levana (Insecta: Lepidoptera: Nymphalidae. Mitochondrial DNA Part B. 5(3):3264–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiuk MR, Marcus JM, Lalonde MML.. 2020b. The complete mitochondrial genome of the Jackson’s Leaf butterfly Mallika jacksoni (Insecta: Lepidoptera: Nymphalidae. Mitochondrial DNA Part B. 5(3):3316–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YQ, Ma C, Chen JY, Yang DR.. 2012. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics. 13:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton RV, Marcus JM, Lalonde MML.. 2020. The complete mitochondrial genome of the black dead leaf butterfly Doleschallia melana (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 5(3):3306–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Rannala B.. 1997. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 276(5310):227–232. [DOI] [PubMed] [Google Scholar]
- Larsen TB. 1991. The butterflies of kenya and their natural history. Oxford: Oxford University Press. [Google Scholar]
- Laslett D, Canback B.. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175. [DOI] [PubMed] [Google Scholar]
- Liao F, Wang L, Wu S, Li YP, Zhao L, Huang GM, Niu CJ, Liu YQ, Li MG.. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci. 6(2):172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chang Z, Chen L, He J, Dong Z, Yang J, Lu S, Zhao R, Wan W, Ma G, et al. 2020. Genome size variation in butterflies (Insecta, Lepidotera, Papilionoidea): a thorough phylogenetic comparison. Syst Entomol. 45(3):571–582. [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2017. The complete mitochondrial genome of the lesser aspen webworm moth Meroptera pravella (Insecta: Lepidoptera: Pyralidae). Mitochondrial DNA Part B. 2(1):344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2018. The complete mitochondrial genome of the giant casemaker caddisfly Phryganea cinerea (Insecta: Trichoptera: Phryganeidae). Mitochondrial DNA Part B. 3(1):375–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2019. The complete mitochondrial genome of the North American pale summer sedge caddisfly Limnephilus hyalinus (Insecta: Trichoptera: Limnephilidae). Mitochondrial DNA Part B. 4(1):413–415. [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2020. The complete mitochondrial genome of the brown pansy butterfly, Junonia stygia (Aurivillius, 1894), (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 5(1):41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JM. 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genet. 5(1):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JM, Hughes TM, McElroy DM, Wyatt RE.. 2010. Engaging first year undergraduates in hands-on research experiences: the upper Green River barcode of life project. J Coll Sci Teach. 39:39–45. [Google Scholar]
- McCullagh BS, Marcus JM.. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia-Pacific Ent. 18(4):749–755. [Google Scholar]
- McCullagh BS, Alexiuk MR, Payment JE, Hamilton RV, Lalonde MML, Marcus JM.. 2020. It’s a moth! It’s a butterfly! It’s the complete mitochondrial genome of the American moth-butterfly Macrosoma conifera (Warren, 1897) (Insecta: Lepidoptera: Hedylidae). Mitochondrial DNA Part B. 5(3):3633–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payment JE, Marcus JM, Lalonde MML.. 2020a. The complete mitochondrial genome of the African leaf butterfly Kallimoides rumia (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 5(3):3415–3417.33458190 [Google Scholar]
- Payment JE, Marcus JM, Lalonde MML.. 2020b. Phylogenetic analysis of the complete mitochondrial genome of the white peacock butterfly Anartia jatrophae saturata (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 5(3):3708–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MJ, Marcus JM.. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst Entomol. 42(1):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XM, Guan QX, Zeng DL, Qin F, Li HM.. 2012. Complete mitochondrial genome of Kallima inachus (Lepidoptera: Nymphalidae: Nymphalinae): comparison of K. inachus and Argynnis hyperbius. Mitochondrial DNA. 23(4):318–320. [DOI] [PubMed] [Google Scholar]
- Shirôzu T, Nakanishi A.. 1984. A revision of the genus Kallima DOUBLEDAY (Lepidoptera, Nymphalidae): I. Generic classification. Tyô to Ga (Lepidoptera Sci). 34:97–110. [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJKarplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelhorn J. 2015. Masquerade. Curr Biol. 25(15):R643–R644. [DOI] [PubMed] [Google Scholar]
- Suzuki TK, Tomita S, Sezutsu H.. 2014. Gradual and contingent evolutionary emergence of leaf mimicry in butterfly wing patterns. BMC Evol Biol. 14:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and Other Methods). Version 4. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Wahlberg N, Brower AVZ, Nylin S.. 2005. Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae). Biol J Linn Soc. 86(2):227–251. [Google Scholar]
- Wallace AR. 1867. Mimicry, and other protective resemblances among animals. Westminster Rev. 32:1–43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference numbers PRJNA667707, MW192438, and MW192439.