ABSTRACT
Disinfectants play an essential role in controlling the dissemination of bacteria in health care settings, but it may also contribute to the selection of antibiotic resistance bacteria. This study looked at Klebsiella pneumoniae isolates collected from three hospitals in Lima, Peru, in order to evaluate: their susceptibility to chlorhexidine [CHG] and isopropanol [ISP]), and their association with antimicrobial susceptibility. We analyzed 59 K. pneumoniae isolates and assessed their CHG and ISP susceptibility by minimum inhibitory concentrations (MICs). Additionally, we performed a regression analysis to assess the association between disinfectant tolerance and antibiotic resistance (measured by the disc diffusion method), colistin resistance (by microdilution), carbapenemases presence (by polymerase chain reaction [PCR]), and clonal relationships (by pulsed-field gel electrophoresis [PFGE]). Eleven K. pneumoniae strains were isolated from fomites, and 48 strains from clinical samples. The MIC range of these isolates was 8–128 µg/ml for CHG and 16–256 mg/ml for ISP. We found that resistance to trimethoprim/sulfamethoxazole (TMP/SMX) was the main factor associated with CHG log2 MIC (ß = 0.65; 95%CI: 0.03, 1.27; R2 = 0.07). In the case of ISP, the log2(MIC) was associated with the institution of origin, showing lower ISP log2(MIC) in fomites compared to clinical samples(ß = −0.77; 95%CI: −1.54, −0.01; R2 = 0.08). Resistance to CHG and ISP among K. pneumoniae isolates found in Peruvian hospitals seems to be elevated and highly variable. Further studies are needed to confirm our results and implement actionable interventions if necessary.
KEYWORDS: Disinfectants, chlorhexidine, alcohol-based disinfectants, kKebsiella pneumoniae, cross-resistance, antibiotic resistance
Introduction
Biocides are routinely used in healthcare settings and play an essential role in infection control [1], one that has been highlighted during the fight against SARS-CoV-2. Two of the most frequently used types of biocides in hospitals are chlorhexidine digluconate (CHG) and isopropyl alcohol (ISP). Chlorhexidine is a bisbiguanide antiseptic used against a wide range of microorganisms. It functions by forming a bridge between phospholipids with subsequent displacement of cations (Mg2+ and Ca2+) [2], resulting in membrane disruption via a reduction in the capacity of the bacterial membrane to osmoregulate and changes in enzymes associated with metabolic membrane capability [2]. Chlorhexidine is often used for procedures such as handwashing, preoperative preparation, and surface disinfection because of its bacteriostatic and bactericidal properties [3,4], as well as its ability to rapidly denature the proteins of microorganisms [5].
In recent decades, infections associated with multidrug-resistant (MDR) pathogens have increased, being Klebsiella pneumoniae one of the most resistant species. This is of concern because K. pneumoniae has also been associated with high mortality rates and an increased use of last-resort antibiotics, such as carbapenems, colistin, and their combinations [6]. These MDR pathogens are responsible for increasing mortality rates secondary to hospital-acquired infections, readmissions, and emergency department visits after discharge [7].
There is an ongoing debate about whether there is an association between an increased tolerance of this bacteria to disinfectants and its resistance to antibiotics. There is evidence suggesting that hygiene promotion, which includes biocides use, helps prevent bacteria transmission across health care settings, although further research is needed to determine the consequence of biocides uses on the appearance or selection of antibiotic resistance bacteria [8,9]. This study aimed to test the levels of tolerance of K. pneumoniae strains to CHG and ISP and determine their association with antimicrobial resistance in Peruvian health care settings before COVID-19.
Materials and methods
Bacterial strains
The Klebsiella spp. strains evaluated were obtained from bacterial collections at the Molecular Microbiology Laboratory of the Universidad Científica del Sur in Lima, Peru. The K. pneumoniae isolates were from 3 different Institutions in Lima: the Hospital Nacional 2 de Mayo (HN2M) (fomites isolates collected from clinical mobile phones and uniforms), as well as Instituto Nacional Materno Perinatal (INMP) and Hospital Nacional Guillermo Almenara Irigoyen (HNGAI), both of which provided clinical isolates (bacteremia, secretions). The strains were collected in 2018, and the identification was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS).
This study was approved by the Ethical Committee of the Universidad Cientifica del Sur in Lima, Peru.
Preparation of disinfectant (chlorhexidine and isopropanol) solutions
Stock solutions of CHG (CHG; 40 mg/mL) were prepared in sterile distilled water at a concentration of 768 µg/ml prior to further dilution (256, 128, 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125 µg/ml) in Mueller Hinton broth and distributed into 96-well plates. Stock solutions of ISP (ISO; EMSURE®) were prepared at 768 mg/L in sterile distilled water and were also diluted (256, 128, 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125 mg/ml) in Mueller Hinton broth and placed in 96-well plates. We used the strains ATCC 13883 Klebsiella pneumoniae as a control. The disinfectants were added separately to these strains and used as a positive control. For negative control, only the culture media was used to monitor the sterility.
Disinfectant susceptibility testing
Minimum inhibitory concentrations (MICs) were determined using the Mueller Hinton broth microdilution method established by the Clinical Laboratory Standards Institute (CLSI) [10]. Briefly, bacterial strains were resuspended in phosphate-buffered saline (PBS) 0.5x to a 0.5 McFarland concentration. Lecture was done after 18–20 hours of incubation at 37°C. The MIC was defined as the lowest concentration of the disinfectant agents that inhibits the visible growth of the isolate tested observed with the unaided eye. In the absence of an established cutoff, we defined MIC50 as the MIC necessary to inhibit the growth of 50% of the study isolates, while MIC90 was considered the MIC necessary to inhibit the growth of 90% of the study isolates.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by a conventional disc diffusion method. The antimicrobials included were meropenem, imipenem, piperacillin/tazobactam, chloramphenicol, trimethoprim/sulfamethoxazole, nitrofurantoin, levofloxacin, ciprofloxacin, aztreonam, cefotaxime, amoxicillin plus clavulanic acid, gentamicin, and amikacin. The category of resistance was interpreted according to CLSI guidelines [10]. The Escherichia coli strain ATCC 25922 was used as a control. Screening for colistin was evaluated by microdilution as recommended by the CLSI-EUCAST Polymyxin Breakpoints Working Group in order to describe MICs [11].
Presence of carbapenemases
The presence of carbapenemases was evaluated in isolates with diminished carbapenem susceptibility. Thus, these strains were screened by a multiplex polymerase chain reaction (PCR) test for the blaKPC, blaOXA-48, blaVIM, blaIMP, and blaNDM genes [12].
Clonality studies
The isolates’ clonal relatedness was determined by pulsed-field gel electrophoresis (PFGE), as described previously [13]. Band patterns were analyzed using InfoQuestTM FP v.5.4 software. Bacteria with a similarity > 80% were considered to be clonally related.
Statistical analysis
Descriptive analysis of disinfectant susceptibility was performed using the MICs 50, MIC 90, and CHG and ISP range separately. We conducted a bivariate analysis using the Kruskal Wallis test for means log2(MIC) comparisons. We estimated the magnitude of association modeling the log2(MIC) using generalized linear models, a Gaussian family, link identity, and robust variances. During this process, we included the following variables as factors potentially associated with CHG and ISP log2(MIC), separately: institution and source of isolate origin, carbapenemases positivity, clonality, and resistance to common and last-resort antibiotics. In all cases, we estimated 95% confidence intervals (95% CI), and the STATA MP v14.0 statistical package (Stata Corp LP, College Station, Texas) was used for the statistical analyses. We used a p-value of 0.05 to measure the statistical significance and a p-value of 0,10 for exploratory analysis.
Results
Bacterial strains
A total of 59 K. pneumoniae isolates were included in the study. Among these, 11 (18.6%) were isolated from HN2M, 7 (64%) were isolated from mobile phones, and 4 (36%) were isolated from the clothing of health care workers. 23 (38.9%) were clinical isolates from blood samples of bacteremia from the HNGAI, and 25 (42.3%) were from blood and secretions of patients from the INMP. All the isolates belonged to a collection obtained in 2018 and stored at −80°C in the Universidad Científica del Sur Microbiology Laboratory.
Antibiotic susceptibility
The K. pneumoniae collection that was studied presented a high rate of resistance to ciprofloxacin, amoxicillin plus clavulanic acid, ceftriaxone, and ceftazidime (86.4, 72.4, 74.6, and 75.8% respectively). Resistance rates of nearly 60% were found to aztreonam, trimethoprim/sulfamethoxazole, and nitrofurantoin (60.7%, 60.3%, and 66.1%, respectively), 51.9% to piperacillin/tazobactam, 51.7% to both imipenem and chloramphenicol, 42.1% to levofloxacin, 48.2% to meropenem, and 45.6% to gentamicin. Finally, low resistance levels to amikacin (36.2%) and colistin (8.5%) were reported.
Disinfectant susceptibility
When analyzing CHG resistance, we observed high variability of MICs (coefficient of variation [CV] = 0.81), ranging from 8 to 128 µg/ml (Figure 1). The MIC50 of all the isolates included in the study was 32 µg/ml. In the bivariate analysis, we found marginal MIC differences between CHG isolates testing positive versus negative to carbapenemases and resistant to levofloxacin. In contrast, statistically significant differences were found between those testing positive versus negative to resistance to TMP/SMX (Tables 1 and 2). On the other hand, when analyzing ISP resistance, we also observed a wide range of MICs against ISP (CV = 0.80), 16–256 mg/ml (Figure 1). The ISP MIC50 of all the isolates was 64 mg/ml. In the bivariate analysis, we found significant differences among sample sources (p = 0.04), and marginal differences among institutions (p = 0.08). Specifically, we observed a lower MIC50 and MIC90 among HN2M isolates (32 and 64 mg/ml, respectively) compared to those obtained from HNGAI (64 and 128 mg/ml, respectively) and INMP (64 and 128 mg/ml, respectively). Likewise, we observed a lower MIC50 and MIC90 among isolates from fomites from health workers’ mobile phones/clothing (32 and 64 mg/ml, respectively) compared to those from blood and secretions (64 and 128 mg/ml, respectively) (Table 1).
Figure 1.
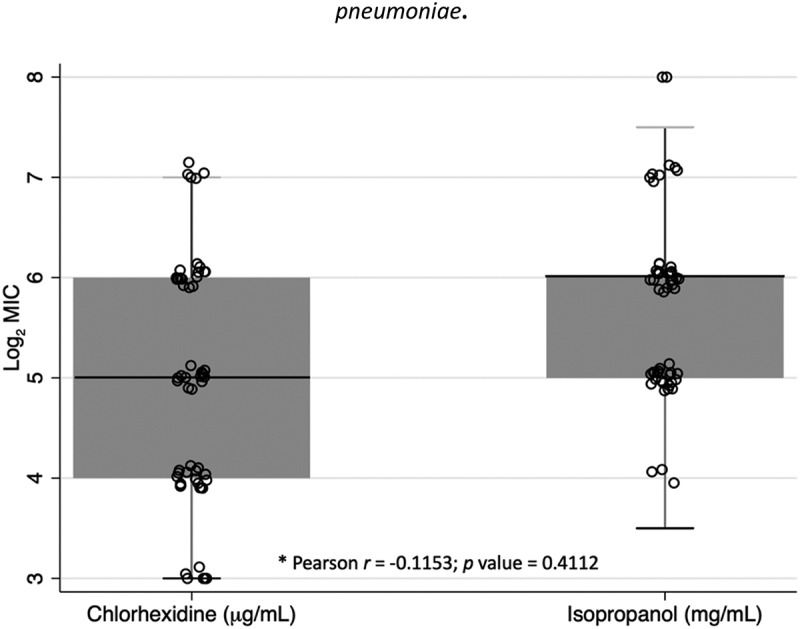
Log2(MIC) of chlorhexidine and isopropanol in strains of Klebsiella pneumoniae.
Table 1.
MICs of chlorhexidine and isopropanol by the study sample’s characteristics
| Characteristic | Chlorhexidine MIC (µg/mL) |
p-value |
Isopropanol MIC (mg/mL) |
p-value |
||||
|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |||
| All | 32 | 64 | 8–128 | 64 | 128 | 16–256 | ||
| Institution | ||||||||
| INMP | 16 | 64 | 8–128 | 0.123 | 64 | 128 | 32–256 | 0.084† |
| HNGAI | 64 | 128 | 8–128 | 64 | 128 | 16–256 | ||
| HN2M | 32 | 64 | 16–64 | 32 | 64 | 32–64 | ||
| Sample source | ||||||||
| Fomite | 32 | 64 | 16–64 | 0.541 | 32 | 64 | 32–64 | 0.038‡ |
| Blood/Secretions | 32 | 128 | 8–128 | 64 | 128 | 16–256 | ||
| Clonality | ||||||||
| Negative | 16 | 64 | 8–128 | 0.192 | 64 | 128 | 16–64 | 0.239 |
| Positive | 64 | 16–256 | 32 | 32–64 | ||||
| Carbapenemases | ||||||||
| Absence | 16 | 64 | 8–128 | 0.071† | 64 | 128 | 32–256 | 0.455 |
| Presence | 64 | 128 | 8–128 | 64 | 128 | 16–256 | ||
MIC, Minimal inhibitory concentration; INMP, Instituto Nacional Materno Perinatal; HNGAI, Hospital Nacional Guillermo Almenara Irigoyen; HN2M, Hospital Nacional Dos de Mayo; †, p value <0.10; ‡, p value <0.05
Table 2.
MICs of chlorhexidine and isopropanol by antibiotics
| Antimicrobial agents |
Chlorhexidine MIC (µg/mL) |
p-value |
Isopropanol MIC (mg/mL) |
p-value |
||||
|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |||
| Colistin | ||||||||
| Sensitive | 32 | 64 | 8–128 | 0.329 | 64 | 128 | 16–256 | 0.609 |
| Resistant | 32 | 32 | 8–32 | 80 | 128 | 32–128 | ||
| Imipenem | ||||||||
| Sensitive | 32 | 64 | 8–128 | 0.105 | 64 | 128 | 32–128 | 0.675 |
| Resistant | 32 | 128 | 8–128 | 64 | 128 | 16–256 | ||
| Meropenem | ||||||||
| Sensitive | 32 | 64 | 8–128 | 0.158 | 64 | 128 | 32–256 | 0.647 |
| Resistant | 32 | 128 | 8–128 | 64 | 128 | 16–256 | ||
| PIP/TZB | ||||||||
| Sensitive | 32 | 64 | 8–128 | 0.165 | 64 | 128 | 32–128 | 0.815 |
| Resistant | 32 | 128 | 8–128 | 64 | 128 | 16–128 | ||
| Chloramphenicol | ||||||||
| Sensitive | 32 | 128 | 8–128 | 0.183 | 32 | 128 | 16–256 | 0.101 |
| Resistant | 16 | 64 | 32–256 | 64 | 128 | 32–256 | ||
| TMP/SMX | ||||||||
| Sensitive | 16 | 64 | 8–128 | 0.039‡ | 64 | 128 | 32–128 | 0.708 |
| Resistant | 32 | 128 | 8–128 | 64 | 128 | 16–256 | ||
| Nitrofurantoin | ||||||||
| Sensitive | 32 | 64 | 8–64 | 0.875 | 48 | 128 | 16–128 | 0.610 |
| Resistant | 32 | 128 | 8–128 | 64 | 128 | 16–256 | ||
| Aztreonam | ||||||||
| Sensitive | 32 | 64 | 8–128 | 0.598 | 32 | 128 | 32–256 | 0.510 |
| Resistant | 32 | 64 | 8–128 | 64 | 128 | 16–256 | ||
| Ciprofloxacin | ||||||||
| Sensitive | 64 | 128 | 8–128 | 0.138 | 48 | 256 | 32–256 | 0.697 |
| Resistant | 32 | 64 | 8–128 | 64 | 128 | 16–256 | ||
| Levofloxacin | ||||||||
| Sensitive | 32 | 128 | 8–128 | 0.059† | 64 | 128 | 16–256 | 0.596 |
| Resistant | 16 | 64 | 8–128 | 64 | 128 | 32–128 | ||
| Cefotaxime | ||||||||
| Sensitive | 32 | 128 | 8–128 | 0.979 | 64 | 128 | 32–256 | 0.991 |
| Resistant | 32 | 64 | 8–128 | 64 | 128 | 16–256 | ||
| Ceftazidime | ||||||||
| Sensitive | 32 | 64 | 8–64 | 0.881 | 64 | 64 | 32–128 | 0.781 |
| Resistant | 32 | 128 | 8–128 | 64 | 128 | 16–256 | ||
| AMC | ||||||||
| Sensitive | 16 | 64 | 8–128 | 0.266 | 64 | 256 | 32–256 | 0.199 |
| Resistant | 32 | 64 | 8–128 | 64 | 128 | 16–128 | ||
| Gentamicin | ||||||||
| Sensitive | 32 | 64 | 8–128 | 0.115 | 64 | 128 | 32–256 | 0.586 |
| Resistant | 64 | 128 | 8–128 | 64 | 128 | 16–128 | ||
| Amikacin | ||||||||
| Sensitive | - | NA | - | NA | ||||
| Resistant | 32 | 64 | 8–128 | 64 | 128 | 16–256 | ||
MIC, Minimal Inhibitory Concentration; Me, Median; IQR, Interquartile range; Pip/tzb, piperacillin/tazobactam; TMP/SMX, trimethoprim/sulfamethoxazole; AMC, amoxicillin/clavulanate; †, p value <0.10; ‡, p value <0.05
Presence of carbapenemases
The isolates that presented a resistance phenotype to carbapenems (either imipenem or meropenem) were tested by PCR, detecting the blaNDM gene in 23 of them (38.9%). We did not find other carbapenemases genes, such as blaKPC, blaoxa-48, blaIMP, or blaVIM.
Clonality analysis
When testing PFGE patterns, we observed that all the isolates belonged to different PFGE types, except for 20 strains classified in the same group. Despite these results, we did not observe significant or marginal differences in the MICs of CHG and ISP compared with the rest of the isolates.
Association of antibiotic resistance with disinfectant susceptibility
In the bivariate regression analysis, a significant direct association was found between the CHGlog2(MIC) and resistance to TMP/SMX (p = 0.041). Also, marginal direct associations were observed with positivity to carbapenemases (p = 0.072) and resistance to imipenem (p = 0.094) and levofloxacin (p = 0.061) (Table 3). However, in the multivariate regression analysis, the only resistance to TMP/SMX was observed as the main predictor of the CHG log2(MIC) (ß = 0.65; 95%CI: 0.03, 1.27; R2 = 0.1127). On the other hand, when modeling the ISP log2(MIC), we observed a significant association with the institution of origin of the sample, with HN2M samples showing a lower ISP log2(MIC) compared to isolates from the INMP (p = 0.048). No association was observed between the ISP log2(MIC) and any antibiotic resistance tested in the study.
Table 3.
Regression analysis for log2(MIC) of chlorhexidine and isopropanol
| Associated Factor | Chlorhexidine MIC (Log2µg/mL) ß (95% CI) |
p-value | Isopropanol MIC (Log2 mg/mL) ß (95% CI) |
p-value |
|---|---|---|---|---|
| Institution | ||||
| INMP | Ref. | Ref | ||
| HNGAI | 0.66 (−0.01, 1.32) | 0.054† | −0.23 (−0.76, 0.29) | 0.375 |
| HN2M | 0.53 (−0.31, 1.37) | 0.210 | −0.77 (−1.54, −0.01) | 0.048‡ |
| Fomites source | −0.22 (−1.01, 0.58) | 0.588 | −0.22 (−1.01, 0.58) | 0.588 |
| PFGE Type 4 | 0.51 (−0.26, 1.28) | 0.191 | −0.11 (−0.63, 0.41) | 0.680 |
| Carbapenemases | 0.66 (−0.06, 1.38) | 0.072† | −0.23 (−0.79, 0.32) | 0.400 |
| Colistin | −0.56 (−1.68, 0.56) | 0.321 | 0.29 (−0.67, 1.25) | 0.543 |
| Imipenem | 0.52 (−0.09, 1.13) | 0.094† | −0.10 (−0.41, 0.61) | 0.703 |
| Meropenem | 0.44 (−0.17, 1.06) | 0.158 | −0.15 (−0.66, 0.36) | 0.547 |
| PIP/TZB | 0.45 (−0.16, 1.07) | 0.147 | −0.04 (−0.55, 0.48) | 0.889 |
| Chloramphenicol | −0.46 (−1.14, 0.23) | 0.186 | 0.43 (−0.12, 0.98) | 0.122 |
| TMP/SMX | 0.65 (0.03, 1.27) | 0.041‡ | −0.07 (−0.59, 0.44) | 0.779 |
| Nitrofurantoin | 0.09 (−0.55, 0.75) | 0.764 | 0.16 (−0.37, 0.69) | 0.547 |
| Aztreonam | −0.51 (−0.51, 0.81) | 0.645 | 0.09 (−0.46, 0.63) | 0.741 |
| Ciprofloxacin | −0.68 (−1.56, 0.21) | 0.132 | −0.32 (−1.11, 0.47) | 0.422 |
| Levofloxacin | −0.60 (−1.23, 0.03) | 0.061† | 0.11 (−0.42, 0.64) | 0.678 |
| Cefotaxime | −0.02 (−0.74, 0.69) | 0.946 | −0.07 (−0.66, 0.52) | 0.814 |
| Ceftazidime | 0.07 (−0.66, 0.81) | 0.840 | −0.03 (−0.63, 0.58) | 0.934 |
| AMC | 0.38 (−0.30, 1.05) | 0.270 | −0.49 (−1.03, 0.06) | 0.078 |
| Gentamicin | 0.48 (−0.15, 1.11) | 0.129 | −0.25 (−0.77, 0.27) | 0.339 |
MIC, Minimal Inhibitory Concentration; INMP, Instituto Nacional Materno Perinatal; HNGAI, Hospital Nacional Guillermo Almenara Irigoyen; HN2M, Hospital Nacional Dos de Mayo; PFGE, pulsed field gel electrophoresis; PIP/TZB, piperacillin/tazobactam; TMP/SMX, trimethoprim/sulfamethoxazole; AMC, amoxicillin/clavulanate; ß, linear regression coefficient; 95% CI, 95% confidence interval; †, p value <0.10; ‡, p value <0.05
Discussion
The main findings of our study include elevated and highly variable levels of resistance to CHG and ISP among clinically-related K. pneumoniae isolates from Peru. Additionally, we observed an association between CHG resistance and TMP/SMX resistance plus a significant variability of ISP resistance according to institution source. These results are important because both CHG and ISP are two of the main disinfectants used in health care facilities and because their use has increased substantially during COVID-19 pandemic.
Our study findings coincide with previous studies that reported a lower susceptibility of bacteria to CHG and ISP [14], which are commonly used to control the spread of K. pneumoniae and other bacteria and viruses in most health care settings around the world. Disinfectants are crucial to correct infection control and are extensively used in hospitals, veterinary medicine, the food industry, farms, and homes [14], but their use could associate antimicrobial resistance increase [8]. Surveillance studies are needed to evaluate the microorganisms’ tolerance to disinfectants at an institutional level to establish adequate disinfection programs.
Although there are no cutoffs by international organizations such as CLSI or EUCAST to guide the interpretation of susceptibility to these substances, epidemiological cutoff values have been proposed to indicate CHG resistance in Klebsiella (64 µg/ml) [15]. In our study, we found a wide range of MICs of CHG in Klebsiella isolates, with MIC50 of the HNGAI samples being the highest (64 µg/ml), and according to the cutoff, they were resistant to this disinfectant. In short, our results showed 21 (35,6%) samples with MIC≥64 µg/ml, which, according to the epidemiological cutoff, is considered resistant. Although not statistically significant, we observed higher CHG resistance levels in the HNGAI sample compared to those from INMP, as well as among fomites (HN2M). This finding could be related to greater use of CHG in intensive care units in which patients receive daily CHG bathing, exerting selection pressure on microorganisms previously related to bacteria more tolerant to CHG [16].
We found a significant association between TMP/SMX resistance and higher levels of CHG MIC values. This association has been described in bacterial species such as Pantoea spp. and Salmonella spp., but to our knowledge, not in K. pneumoniae [17]. In addition, the present data demonstrate that other antibiotics have shown this association. Thus, the presence of carbapenemases is directly associated with higher CHG MIC values, and the resistance to levofloxacin has a negative association since higher CHG MIC values are related to the presence of strains susceptible to levofloxacin. Our results correlate with a previous study performed in China that suggested decreased sensitivity against commonly used disinfectants, including CHG, in carbapenem-resistant strains. Furthermore, Chen et al.[18] reported the presence of carbapenem-resistant K. pneumoniae with decreased susceptibility to CHG (MIC 32 µg/ml, which is the limit of the ecological cutoff of 64 µg/ml). It is important to mention that only six phylogenetically related strains were included in the study and all carbapenemases that were found were blaNDM [19]. Our data showed a negative association between fluoroquinolones and CHG resistance, while a positive association has been previously described in Pseudomonas aeruginosa [19]. Thus, in vitro exposure to CHG has been related to the selection of quinolone-resistant P. aeruginosa due to over-expression of the efflux pumps MexAB-OprM, MexCD-OprJ, and MexXY-OprM [20], none of which are related to K. pneumoniae.
We found no relationship between colistin resistance and increased tolerance to CHG compared to the study by Wand et al. (2017), which found that the MICs of CHG were the same in both colistin-susceptible and colistin-resistant K. pneumoniae strains [6]. Likewise, it has been described that the adaptation to CHG led to resistance against colistin in K. pneumoniae and, instead, the adaptation to colistin does not lead to resistance against CHG [6]. In K. pneumoniae, in vitro exposure to CHG increased the colistin resistance likely by induction of point mutations in pmrBA and phoPQ related to colistin resistance and/or smvR [21]. SmvA is an efflux pump belonging to the major facilitator superfamily (MFS), which has been suggested to play a major role in CHG resistance in K. pneumoniae strains [6].
Increasing CHG resistance has been related to the over-expression of efflux pumps involved eliminating CHG [22]. Notably, different MDR efflux systems can export both antibiotics and biocides, contributing to the co-selection of resistance to different agents. There is a gene family related to diminished CHG susceptibility called qac (quaternary ammonium compound) (e.g., qacA/B and qacC, among others) [23]. Of note, some qac genes have been shown to contribute to the increasing resistance to specific antibiotics; for instance, QacA and QacBIII contribute to the final norfloxacin and ciprofloxacin MIC levels [23]. These qac genes encode for Qac efflux proteins belonging to the MFS and SMR efflux pump families. While the role of these proteins is well known in Gram-positive strains, previous findings have led to controversy about the role and frequency of the cepA, qacA, and qacE genes in Gram-negative strains, with more or less importance being given depending on the study [18, 21, 24, 25].
Moreover, it has been described that the qacEΔ1 gene is located in an integron class 1 structure upstream of the sul1 gene that confers sulfonamide resistance and is often flanked by the dihydrofolate reductase gene dfrA1 related to trimethoprim resistance [18]. These gene cassette structures are related to the CHG tolerance, and TMP/SMX resistance found in our study (p < 0.05), with both genes likely being disseminated together in the same mobile structure within the same plasmid. This relationship could be associated with the described propriety of CHG that could stimulate the horizontal transfer of antibiotic resistance [26], leading to the potential to co- and cross-select antibiotic resistance mechanisms [27]. Future studies are needed to determine the gene content in integrons inside plasmids and relate their frequency with the MIC of the biocides and diminished antimicrobial susceptibility strains.
Regarding ISP-based disinfectants, the present study found lower resistance in isolates from fomites compared to clinical isolates. This may be because strains found in potential fomites (mobile devices, health workers’ clothing, etc.) are less exposed to these disinfectants than hospital surfaces.
It is worth noting that there are studies that reported cases of disinfectant-induced antibiotic resistance in bacterial strains [8,17]. This may have contributed to the antibiotic resistance in our strains if previously exposed to disinfectants in hospital settings.
No relation was found between antibiotic resistance and alcohol tolerance, which supports a previous report evidencing that no cross-resistance to antibiotics after low-level exposure to alcohol-based disinfectants has so far been described [17]. This might occur because the mechanisms of action of alcohol-based disinfectants are not related to mechanisms causing antibiotic resistance, such as inhibition and uncoupling of messenger ribonucleic acid (RNA) and protein synthesis through specific effects on RNA polymerase and ribosomes [28], or interference with cellular metabolism, disruptions of cytoplasmic integrity and cell lysis [29].
Our results showed a high proportion of CHG and ISP resistant strains. However, the clinical importance of these findings is not yet evident, since the concentrations of disinfectants used in clinical settings are, at present, greater than the highest MICs found in the study. Globally, very few studies have discussed and analyzed MIC rates of biocides together with antibiotics and their molecular mechanisms [31–33]. Antimicrobial resistance genotypes vary among different countries, regions, and hospital settings, and the same could occur with disinfectant-tolerant bacteria and their mechanisms [16]. Therefore, it is important to highlight that these results provide a baseline for measuring disinfectant susceptibility to assist in future surveillance studies in Peru.
30This study evaluated pre-pandemic levels of CHG and ISP resistance. Considering the wide-scale use of biocides for personal and environmental disinfection during the COVID-19 pandemic, the current levels of resistance may be a global public health threat. Future surveillance studies are essential to understand the effect of disinfectant use and observe the possible emergence of microorganisms that can survive exposure to disinfectants.
Acknowledgments
We thank the members of the Microbiology Laboratory team of the three hospitals involved for their kind collaboration. The authors thank Donna Pringle for editorial assistance.
Funding Statement
This work was supported by the Universidad Cientifica del Sur, ‘Semilla 2019-01 grant’.
Disclosure statement
The authors reported no potential competing interests.
References
- [1].Rutala WA. APIC guideline for selection and use of disinfectants. Am J Infect Control. 1996. August;24(4):313–342. [DOI] [PubMed] [Google Scholar]
- [2].Hugo WB, Longworth AR.. The effect of chlorhexidine on the electrophoretic mobility, cytoplasmic constituents, dehydrogenase activity and cell walls of Escherichia coli and Staphylococcus aureus. J Pharm Pharmacol. 1966. September;18(9):569–578. [DOI] [PubMed] [Google Scholar]
- [3].Denny J, Munro CL.. Chlorhexidine bathing effects on health-care-associated infections. Biol Res Nurs. 2017. March;19(2):123–136. [DOI] [PubMed] [Google Scholar]
- [4].Lakhani N, Vandana KL. Department of periodontics, college of dental sciences, Davangere, Karnataka, India. Chlorhexidine – An insight. Int J Adv Res. 2016. July 31;4(7):1321–1328. [Google Scholar]
- [5].Boyce JM. Alcohols as surface disinfectants in healthcare settings. Infect Control Hosp Epidemiol. 2018. March;39(3):323–328. [DOI] [PubMed] [Google Scholar]
- [6].Wand ME, Bock LJ, Bonney LC, et al. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob Agents Chemother. 2017. January;61(1):e01162–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Barrasa-Villar JI, Aibar-Remón C, Prieto-Andrés P, et al. Impact on morbidity, mortality, and length of stay of hospital-acquired infections by resistant microorganisms. Clin Infect Dis. 2017. August 15;65(4):644–652. [DOI] [PubMed] [Google Scholar]
- [8].Caselli E, Brusaferro S, Coccagna M, et al. Reducing healthcare-associated infections incidence by a probiotic-based sanitation system: A multicentre, prospective, intervention study. PLoS One. 2018;13(7):e0199616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sanchez P, Moreno E, Martinez JL. The biocide triclosan selects Stenotrophomonas maltophilia mutants that overproduce the SmeDEF multidrug efflux pump. Antimicrob Agents Chemother. 2005;49(2):781–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].M100-S11 . Performance standards for antimicrobial susceptibility testing. Clin Microbiol Newsl. 2001;23(6):49. [Google Scholar]
- [11].Turlej-Rogacka A, Xavier BB, Janssens L, et al. Evaluation of colistin stability in agar and comparison of four methods for MIC testing of colistin. Eur J Clin Microbiol Infect Dis. 2018;37(2):345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bogaerts P, Rezende de Castro Ŗ, de Mendonc R, et al. Validation of carbapenemase and extended-spectrum b-lactamase multiplex endpoint PCR assays according to ISO 15189. J Antimicrob Chemother. 2013;68:1576–1582. [DOI] [PubMed] [Google Scholar]
- [13].Durmaz R, Otlu B, Koksal F, et al. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis. 2009;62(5):372–377. [PubMed] [Google Scholar]
- [14].Fraise AP. Choosing disinfectants. J Hosp Infect. 1999;43(4):255–264. [DOI] [PubMed] [Google Scholar]
- [15].Morrissey I, Oggioni MR, Knight D, et al. Evaluation of epidemiological cut-off values indicates that biocide resistant subpopulations are uncommon in natural isolates of clinically-relevant microorganisms. Manganelli R, editor. PLoS ONE. 2014. 23;9(1):e86669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Suwantarat N, Carroll KC, Tekle T, et al. High prevalence of reduced chlorhexidine susceptibility in organisms causing central line-associated bloodstream infections. Infect Control Hosp Epidemiol. 2014;35(9):1183–1186. [DOI] [PubMed] [Google Scholar]
- [17].Kampf G. Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics (Basel). 2018;7(4):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen Y, Liao K, Huang Y, et al. Determining the susceptibility of carbapenem resistant Klebsiella pneumoniae and Escherichia coli strains against common disinfectants at a tertiary hospital in China. BMC Infect Dis. 2020;20(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stein C, Vincze S, Kipp F, et al. Carbapenem-resistant Klebsiella pneumoniae with low chlorhexidine susceptibility. Lancet Infect Dis. 2019;19(9):932–933. [DOI] [PubMed] [Google Scholar]
- [20].Kawamura M, Sato T, Fujimura S. In vitro exposure of chlorhexidine induce quinolone resistance in Pseudomonas aeruginosa. J Infect Public Health. 2020;13(2):317–318. [Google Scholar]
- [21].Zhang Y, Zhao Y, Xu C, et al. Chlorhexidine exposure of clinical Klebsiella pneumoniae strains leads to acquired resistance to this disinfectant and to colistin. Int J Antimicrob Agents. 2019;53(6):864–867. [DOI] [PubMed] [Google Scholar]
- [22].Rossi L, Bertoncelli A, Tomi ED, et al. Susceptibility to chlorhexidine of Klebsiella pneumoniae strains: KPC producers and potential role of efflux pumps. 29th ECCMID (P2631) 2019.
- [23].Nakaminami H, Noguchi N, Sasatsu M. Fluoroquinolone efflux by the plasmid-mediated multidrug efflux pump QacB variant QacBIII in Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54(10):4107–4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Guo W, Shan K, Xu B, et al. Determining the resistance of carbapenem-resistant Klebsiella pneumoniae to common disinfectants and elucidating the underlying resistance mechanisms. Pathog Glob Health. 2015;109(4):184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vijayakumar R, Sandle T, Al-Aboody MS, et al. Distribution of biocide resistant genes and biocides susceptibility in multidrug-resistant Klebsiella pneumoniae, Pseudomonas aeruginosa and Acinetobacter baumannii — A first report from the Kingdom of Saudi Arabia. J Infect Public Health. 2018;11(6):812–816. [DOI] [PubMed] [Google Scholar]
- [26].Jutkina J, Marathe NP, Flach C-F, et al. Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci Total Environ. 2018;616–617:172–178. [DOI] [PubMed] [Google Scholar]
- [27].Webber MA, Whitehead RN, Mount M, et al. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemother. 2015;70(8):2241–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Haft RJF, Keating DH, Schwaegler T, et al. Correcting direct effects of ethanol on translation and transcription machinery confers ethanol tolerance in bacteria. Proc Natl Acad Sci. 2014. 24;111(25):E2576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Block SS. Disinfection, sterilization, and preservation. Philadelphia, PA: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- [30].Abuzaid A, Hamouda A, Amyes SGB. Klebsiella pneumoniae susceptibility to biocides and its association with cepA, qacΔE and qacE efflux pump genes and antibiotic resistance. J Hosp Infect. 2012. June;81(2):87–91. [DOI] [PubMed] [Google Scholar]
- [31].Vijayakumar R, Aboody M, AlFonaisan M, et al. Determination of minimum inhibitory concentrations of common biocides to multidrug-resistant gram-negative bacteria. Appl Med Res. 2016;2(3):56. [Google Scholar]
- [32].Lambert RJW, Joynson J, Forbes B. The relationships and susceptibilities of some industrial, laboratory and clinical isolates of Pseudomonas aeruginosa to some antibiotics and biocides. J Appl Microbiol. 2001;91(6):972–984. [DOI] [PubMed] [Google Scholar]


