Abstract
Amusium pleuronectes, commonly known as the Asian moon scallop, is widely distributed in Indo-Pacific coasts. In this study, the complete mitogenome sequence of A. pleuronectes (18,044 bp) is reported, which represents the first mitogenome from the Amusium genus. This mitogenome contains 13 protein-coding genes, 2 rRNAs, and 22 tRNA genes, showing similar mitogenome features for most marine bivalves. Phylogenetic analysis reveals that within the family Pectinidae, the genus Amusium is closely related to the genus Argopecten. The mitogenome of A. pleuronectes provides a valuable resource for further advancing the understanding of bivalve phylogeny and evolution.
Keywords: Amusium pleuronectes, mitochondrial genome, phylogenetic analysis
Amusium pleuronectes, commonly known as the Asian moon scallop, is widely distributed in Indo-Pacific coastal areas (Minchin 2003) and characterized by beautiful two-colored shells. Except for high economic value, the Asian moon scallop is often used as a biological monitor to evaluate the impact of human activities on the marine environment (Siriprom and Limsuwan 2009). However, genetic information for this species remains scarce. Recently, complete mitogenome has been considered as an effective tool for phylogeny and phylogeography studies (Curole and Kocher 1999; Saccone et al. 1999). In this study, we report the sequencing and assembly of the complete mitogenome of A. pleuronectes, which is the first mitogenome of the genus Amusium.
The example of A. pleuronectes was derived from Tung Ping Chau Bay near Tung Chung village (Guangdong province, China, 22.4907°N, 114.5801°E) at a depth of 7–8 m. All soft tissues were preserved in liquid nitrogen after sampling. Total DNA was extracted using the phenol/chloroform/isoamyl alcohol method (Sambrook et al. 1989). The DNA sample was deposited at the Key Laboratory of Marine Genetics and Breeding (Ministry of Education), Ocean University of China (Specimen code: OUC-MGB-2018-AP-08). Whole genomic sequencing of the individual sample was performed using the Illumina NovaSeq 6000 sequencing platform. The mitogenome of A. pleuronectes was de novo assembled using NOVOPlasty (Dierckxsens et al. 2017), gene information of which was retrieved by the MITOS software (Bernt et al. 2013).
The complete circular mitogenome of A. pleuronectes was 18,044 bp in length. Thirteen protein-coding genes (PCGs), 2 ribosomal RNA genes, and 21 transfer RNA genes were annotated. The 13 conserved PCGs were cytochrome oxidase subunit (I, II, and III), NADH dehydrogenase subunit (1, 2, 3, 4, 5, 6, and 4 L), and ATP synthase subunit (6, and 8), which was similar to the reported mitochondrial genomes of most marine bivalve mollusks (Liu et al. 2019; Ma et al. 2019; Liu et al. 2020). The base composition of A. pleuronectes was A = 22.22%, T = 38.60%, G = 25.55%, and C = 13.63%. The overall AT content (60.82%) was higher than the GC content (39.18%). The small subunit ribosomal RNA (12S rRNA) and large subunit ribosomal RNA (16S rRNA) were annotated with sizes of 968 bp and 1,420 bp, respectively. The length of 22 tRNAs ranged from 63 bp to 72 bp. The whole mitogenome sequence has been deposited in GenBank under the accession number of MT419374.
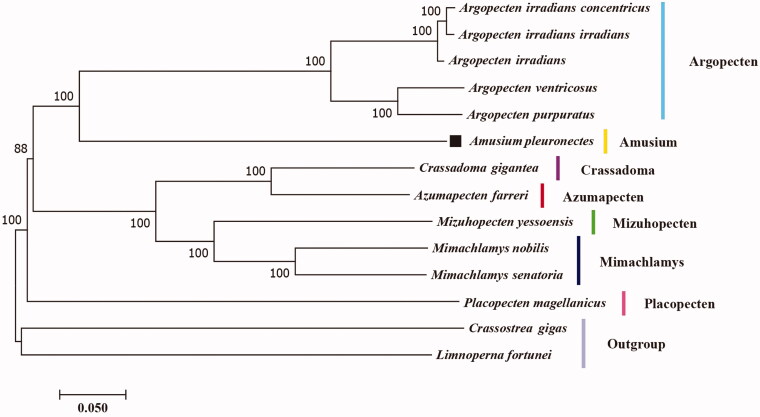
Previous studies of the phylogeny of the family Pectinidae were mostly based on the sequence information of few genes (Serb 2016; Smedley et al. 2019). In this study, a mitogenome-level phylogenetic tree was constructed based on all the available scallop mitogenomes, including A. pleuronectes and 11 other scallop species (from six genera of Pectinidae), as well as two outgroup species (Figure 1). The phylogenetic tree was constructed using the neighbor-joining (NJ) algorithm with 1000 bootstrap trials. The phylogeny tree indicated that the genus Amusium represented by A. pleuronectes was closely related to the genus Argopecten as reported by previous studies (Serb 2016; Smedley et al. 2019). The A. pleuronectes mitogenome provides a valuable resource for further advancing our understanding of bivalve phylogeny and evolution.
Figure 1.
The phylogenetic tree of A. pleuronectes and 13 other bivalves based on complete mitochondrial genomes. The accession numbers of downloaded sequences are as follows: Argopecten irradians concentricus (KT161259.1), Argopecten irradians irradians (NC_012977.1), Argopecten irradians (NC_009687.1), Argopecten venteicosus (KT161261.1), Argopecten purpuratus (NC_027943.1), Crassadoma gigantea (MH016739.1), Azumapecten farreri (NC_012138.1), Mizuhopecten yessoensis (NC_009081.1), Mimachlamys nobilis (NC_011608.1), Mimachlamys senatoria (NC_022416.1), Placopecten magellanicus (NC_007234.1), Crassostrea gigas (NC_001276.1), and Limnoperna fortunei (NC_028706.1).
Funding Statement
We acknowledge the grant support from National Natural Science Foundation of China [U1706203] and Fundamental Research Funds for the Central Universities [201841001].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GeneBank at [https://www.ncbi.nlm.nih.gov/nuccore/MT419374.1/], reference number [Accession number: MT419374].
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Curole JP, Kocher TD.. 1999. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol (Amst). 14(10):394–398. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Li Y, Liu J, Zhang Y, Bao Z, Wang S.. 2019. The complete mitochondrial genome and phylogenetic analysis of Spondylus violaceus. Mitochondrial DNA B. 4(2):2908–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zeng Q, Wang H, Teng M, Guo X, Bao Z, Wang S.. 2020. The complete mitochondrial genome and phylogenetic analysis of the dwarf surf clam Mulinia lateralis. Mitochondrial DNA Part B. 5(1):140–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zhang Y, Zhang Y, Xiao S, Han C, Chen S, Yu Z.. 2019. The complete mitochondrial genome of the giant clam, Tridacna maxima (Tridacnidae Tridacna). Mitochondrial DNA Part B. 4(1):1051–1052. [Google Scholar]
- Minchin D. 2003. Introductions: some biological and ecological characteristics of scallops. Aquat Living Resour. 16(6):521–532. [Google Scholar]
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A.. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 238(1):195–209. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T.. 1989. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Lab Press. [Google Scholar]
- Serb JM. 2016. Reconciling morphological and molecular approaches in developing a phylogeny for the Pectinidae (Mollusca: Bivalvia). Dev Aquacult Fish Sci. 40:1–29. [Google Scholar]
- Siriprom W and Limsuwan P.. 2009. A biomonitoring study: trace metals in Amusium pleuronectes shell from the coastal area of Chon Buri province. Kasetsart J Nat Sci. 43(5):141–145. [Google Scholar]
- Smedley GD, Audino JA, Grula C, Porath-Krause A, Pairett AN, Alejandrino A, Lacey L, Masters F, Duncan PF, Strong EE, et al. 2019. Molecular phylogeny of the Pectinoidea (Bivalvia) indicates Propeamussiidae to be a non-monophyletic family with one clade sister to the scallops (Pectinidae). Mol Phylogenet Evol. 137:293–299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GeneBank at [https://www.ncbi.nlm.nih.gov/nuccore/MT419374.1/], reference number [Accession number: MT419374].