Abstract
Accurate characterization of the human immunodeficiency virus (HIV) reservoir is imperative to develop an effective cure. HIV was measured in antiretroviral therapy-suppressed individuals using the intact proviral DNA assay (IPDA), along with assays for total or integrated HIV DNA, and inducible HIV RNA or p24. Intact provirus correlated with total and integrated HIV.
Keywords: HIV latency, IPDA, TILDA, p24, integrated HIV
Human immunodeficiency virus type 1 (HIV-1) persists within latently infected cells in the form of integrated proviruses despite active antiretroviral therapy (ART) [1, 2]. The evaluation of eradication strategies is dependent on the development of rapid and scalable assays that can measure it accurately [3]. In ART-treated individuals, measurements of the HIV reservoir are limited in that the vast majority of proviruses are defective and that not all intact genomes are inducible. It is unclear whether ex vivo induction of HIV RNA or proteins reflects activation of defective or intact HIV proviruses [4–6]. Recently, the intact proviral DNA assay (IPDA) was introduced as a strategy to measure intact, defective, and total HIV genomes [7]. In ART-suppressed chronically infected individuals with HIV, we assessed the association between IPDA and 5 non-IPDA HIV measures including measures of integrated and transcriptionally active provirus following ex vivo activation.
METHODS
Participants
Sixty-four individuals participated in the study. Cryopreserved peripheral blood mononuclear cells (PBMC) from leukapheresis and whole blood were used. Due to sample limitations, all assays were not performed for all participants (Supplementary Table 1). All participants were chronically on ART (<50 HIV-1 copies/mL for ≥1 year) with ≥450 CD4+ T cells/mm3 at the time of sample collection. Written informed consent was obtained from all participants, and human experimentation was conducted in accordance with the guidelines of the US Department of Health and Human Services and those of the authors’ institutions.
IPDA HIV Measures
Total, intact, 3′ deleted and/or hypermutated (3′ defective) and 5′ deleted (5′ defective) proviral DNA were assessed by IPDA performed by Accelevir Diagnostics as previously described [7] in CD4+ T cells isolated from leukapheresis-derived PBMC. Genomic DNA was isolated and analyzed by IPDA. Briefly, each IPDA consisted of 2 multiplex droplet digital polymerase chain reactions (ddPCR) performed in parallel: (a) the HIV-1 proviral discrimination reaction, which distinguished intact from defective proviruses, and (b) the copy reference/shearing reaction, which quantified DNA shearing and input diploid cell equivalents. In each sample, the DNA shearing index (DSI: dual fluorescent/single fluorescent droplets) was calculated for both reactions to determine the frequency of intraamplicon shearing. Results were expressed as count of HIV proviruses per 106 CD4+ T cells.
Total HIV DNA Measures
Human CD4+ T cells were isolated from leukapheresis-derived PBMC by negative magnetic selection using the EasySep Human CD4+ T Cell Isolation Kit (StemCell Technology, Cambridge, MA, USA), and used for DNA extraction using the Monarch DNA Purification Kit (New England BioLabs, Ipswich, MA, USA). HIV-1 DNA proviruses were quantified by multiplex quantitative PCR using the HIV-1 Total DNA Quantitative Kit (Ultrabio Technologies, Bothell, WA, USA). Results were expressed as copies of total HIV DNA per 106 CD4+ T cells.
Integrated HIV DNA Measures
Integrated HIV DNA was assessed by (a) Alu-Gag–PCR performed on CD4+ T cells isolated from whole blood-derived PBMC [8] and (b) pulse-field gel electrophoresis (PFGE) and ddPCR [9] performed in leukapheresis-derived PBMC enriched for CD4+ T cells to measure integrated HIV Gag and Pol. All results were expressed as copies of integrated HIV DNA per 106 CD4+ T cells.
Measures of In Vitro HIV Re-activation
In vitro HIV reactivation was measured using CD4+ T cells isolated from leukapheresis-derived PBMC and stimulated for 24 hours with phorbol 12-myristate 13-acetate (PMA) and ionomycin or dimethyl sulfoxide (DMSO). First, the frequency of cells producing tat/rev-RNA was measured by tat/rev-induced limiting dilution assay (TILDA). Briefly, CD4+ T cells were added to a 384 well plate at 4 dilutions, stimulated (24 replicates per stimulation), and tat/rev amplicons were detected in individual wells by reverse transcription (RT) PCR followed by real-time PCR, as previously described [10]. Results were expressed as infectious units per million cells (IUPM). Second, HIV p24 expression was measured by single molecule array (SIMOA) [11] in supernatants collected from the cultures described above. HIV p24 was detected using the Quanterix method, with a limit of detection of 4 fg/mL. Inducible measures for TILDA and SIMOA were used following subtraction of the constitutive measures [ie, (PMA and ionomycin)—DMSO].
Statistical Analysis
Data were described as medians and interquartile ranges. Spearman rank correlation test was used to determine variables associations as measured in the same patient sample. As a descriptive study, unadjusted P-values < .05 were considered statistically significant. All statistics were performed with JMP Pro15 (SAS Institute, Cary, NC, USA).
RESULTS
Demographics
Demographic, clinical characteristics of study participants with type of sample and assays performed in each are summarized in Supplementary Tables 1 and 2.
Associations Between Non-IPDA or IPDA HIV Measures
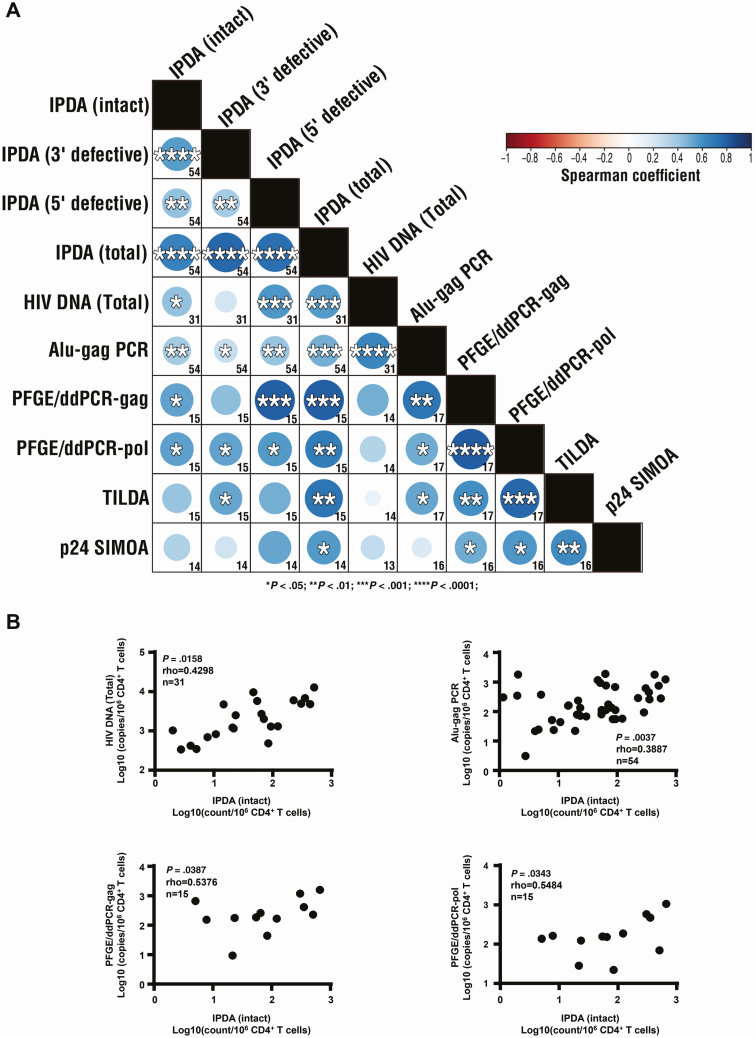
Results showed expected associations between non-IPDA or IPDA HIV measures as shown in Figure 1A (see also Supplementary Table 3 and Supplementary Figures 1–3).
Figure 1.
Associations of HIV burden measures using the (IPDA and alternative assays. A, Matrix of between-assay associations. B, Correlations of intact HIV proviruses assessed by IPDA with total HIV DNA, and integrated HIV DNA measures (assessed by Alu-gag PCR and pulse-field gel electrophoresis and droplet digital PCR [PFGE/ddPCR] [HIV Gag, HIV Pol]). In panel A, numbers in each square indicate the number of participants used for each association; blue circles indicate associations with significant positive correlations with P< .05, <.01, <.001, and <.0001 listed with 1, 2, 3, or 4 asterisks inside the circle. Color of circles represent Spearman rho with negative correlations represented by shades of red and positive correlations represented by shades of blue with darker shades representing stronger rho. Panel B shows available data together with n, rho, and P values. Spearman rank correlation test was used for variables associations in patients with available data on correlated variables. P-values < .05 were considered statistically significant. Abbreviations: HIV, human immunodeficiency virus; IPDA, intact proviral DNA assay; PCR, polymerase chain reaction; SIMOA, single molecule array; TILDA, tat/rev-induced limiting dilution assay.
Associations of IPDA With Non-IPDA HIV Measures
Correlation analysis was performed to assess whether total, integrated or inducible HIV assessed by non-IPDA assays best represented intact or defective proviruses as measured by IPDA. Results are summarized in Figure 1A and Supplementary Table 3, with significant correlations illustrated in Supplementary Figure 4 and nonsignificant correlations illustrated in Supplementary Figure 2. Briefly, IPDA-total proviruses were positively associated with the non-IPDA-total and -integrated HIV DNA measures (Alu-Gag PCR; PFGE/ddPCR HIV [Gag, Pol]), as well as with the non-IPDA-inducible HIV measures (inducible HIV RNA, HIV p24). IPDA-intact proviruses were positively associated with the non-IPDA-total and -integrated HIV DNA measures (Alu-Gag PCR; PFGE/ddPCR HIV [Gag, Pol]). In addition, a positive association was found between IPDA-3′ defective proviruses and non-IPDA-integrated HIV DNA measures (Alu-Gag PCR, PFGE/ddPCR HIV Pol) or -inducible HIV RNA, as well as between IPDA-5′ defective proviruses and non-IPDA-total and -integrated HIV DNA measures (Alu-Gag PCR; PFGE/ddPCR HIV [Gag, Pol]). Furthermore, IPDA-total defective proviruses associations with non-IPDA HIV measures confirmed these findings showing a positive correlation with non-IPDA-total, -integrated HIV DNA, and inducible HIV RNA and HIV p24 (Supplementary Figure 5).
DISCUSSION
In steady-state ART suppression, we report for the first time the relationship of HIV total, integrated, or inducible HIV measures with IPDA-assessed HIV measures. Our data support (1) an association of non-IPDA-total and -integrated HIV DNA measures with IPDA-total, -intact, or -defective proviruses, and (2) an association of inducible HIV measures with IPDA-total or -defective proviruses. We also confirm the presence of a positive association between inducible HIV measures indicating concordant associations between levels of HIV transcription and translation.
In agreement with previous studies [10, 12], we report a positive association between integrated and inducible HIV measures. This, together with our data showing a positive association between inducible HIV measures and IPDA-3′ defective proviruses, suggests that induced viral transcription and translation assays may lead to an overestimation of the size of replication-competent provirus due to the predominance of defective over intact provirus [1, 4, 5]. Recent data showing that defective HIV proviruses can support HIV antigen expression, further support this finding [6]. In contrast to the observed positive associations of inducible HIV RNA (TILDA assay) with total or integrated HIV DNA by Procopio et al [10], our study did not detect inducible HIV RNA as associated with IPDA-intact proviruses. The latter is consistent with induced transcription as a measure of all transcriptionally competent sites and not only intact provirus. In support of this interpretation, IPDA-total but not -intact measures was positively associated with inducible HIV RNA or p24 SIMOA measures. Interestingly, total and integrated HIV measures correlated both with IPDA-intact and IPDA-defective proviruses, suggesting that these measures may provide a global estimate of both intact and defective HIV burden.
A strength of our study is that it was performed in total and not in resting CD4+ T cells. However, a limited sample size together with sample limitations preventing to complete all assays on all samples should be considered as a potential reason for a lack of detection of correlations with greater variance (ie, requiring higher sample size). Our data also do not address duration of infection or time from start of ART in each subject because these data were not available. It should also be taken into consideration when interpreting the results of our study that sequence mismatches between the IPDA primer/probes and the study participants proviruses may have contributed to false negative correlations between IPDA and non-IPDA HIV measures. Future studies could (a) validate our data in a larger cohort and in samples from tissues, (b) explore whether the observed associations between integrated HIV DNA and intact proviruses are maintained in the context of a potential curative intervention, (c) independently validate our IPDA assay results by full genome sequencing, and (d) explore whether translation positive (inducible protein) proviruses that are defective contribute to immune activation and disease.
Taken together, our data support that when testing curative strategies, measures from IPDA or integrated provirus assays could be informative when combined with inducible HIV assays to better estimate changes in intact or integrated proviruses with the potential for antigen expression, of relevance to eradication, and potential for immune stimulation, respectively.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the study participants and their providers.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant number U01AI110434) to L. J. M., the BEAT-HIV Delaney Collaboratory (grant number UM1AI126620), cofunded by the National Institute of Allergy and Infectious Diseases, the National Institute of Mental Health, the National Institute of Neurological Disorders and Stroke, and the National Institute on Drug Abuse; Merck Sharp & Dohme Corp., a subsidiary of Merck & Co, Inc, Kenilworth, NJ, USA; Philadelphia Field Initiative Group HIV Trials; the CLAWS Foundation; AIDS funds from the Commonwealth of Pennsylvania and from the Commonwealth Universal Research Enhancement Program; The Pennsylvania Department of Health; The Philadelphia Foundation (Robert I. Jacobs Fund); Ken Nimblett and the Summerhill Trust; the Penn Center for AIDS Research (grant number P30 AI 045008); and The Wistar Cancer Center Support Grant (grant number P30 CA10815). L. J. M. is supported by the Herbert Kean, MD, Family Professorship. D. R. is supported by the San Diego Center for AIDS Research (grant number AI306214), the Department of Veterans Affairs, and the James B. Pendleton Charitable Trust. M. A-M. is supported by the National Institute of Health (grant numbers R01 AG062383, R21 AI143385, R21 AI129636, and R21 NS106970) and The Foundation for AIDS Research impact grant (grant number 109840-65-RGRL).
Potential conflicts of interest. During the conduct of the study B. J. H., C. B-T., D. J. H., S. L-G., and P. D. Z. reported personal fees from Merck & Co, Inc. outside the submitted work, L. J. M. reported his financial relationship with the Society of Leukocyte Biology, as well as grants and personal fees from the National Institutes of Health, N. C. reported grants from EMD Serono, and J. R. K. reported grants and personal fees from Gilead Sciences. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278:1295–300. [DOI] [PubMed] [Google Scholar]
- 2. Wong JK, Hezareh M, Günthard HF, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278:1291–5. [DOI] [PubMed] [Google Scholar]
- 3. Eriksson S, Graf EH, Dahl V, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 2013; 9:e1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruner KM, Murray AJ, Pollack RA, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016; 22:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ho YC, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Imamichi H, Smith M, Adelsberger JW, et al. Defective HIV-1 proviruses produce viral proteins. Proc Natl Acad Sci U S A 2020; 117:3704–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bruner KM, Wang Z, Simonetti FR, et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019; 566:120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mexas AM, Graf EH, Pace MJ, et al. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS 2012; 26:2295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lada SM, Huang K, VanBelzen DJ, Montaner LJ, O’Doherty U, Richman DD. Quantitation of integrated HIV provirus by pulsed-field gel electrophoresis and droplet digital PCR. J Clin Microbiol 2018; 56:e01158–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Procopio FA, Fromentin R, Kulpa DA, et al. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2015; 2:874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu G, Swanson M, Talla A, et al. HDAC inhibition induces HIV-1 protein and enables immune-based clearance following latency reversal. JCI Insight 2017; 2:e92901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baxter AE, Niessl J, Fromentin R, et al. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe 2016; 20:368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.