Abstract
Background
Bats have been considered natural reservoirs for several pathogenic human coronaviruses (CoVs) in the last two decades. Recently, a bat CoV was detected in the Republic of Korea; its entire genome was sequenced and reported to be genetically similar to that of the severe acute respiratory syndrome CoV (SARS-CoV).
Objectives
The objective of this study was to compare the genetic sequences of SARS-CoV, SARS-CoV-2, and the two Korean bat CoV strains 16BO133 and B15-21, to estimate the likelihood of an interaction between the Korean bat CoVs and the human angiotensin-converting enzyme 2 (ACE2) receptor.
Methods
The phylogenetic analysis was conducted with the maximum-likelihood (ML) method using MEGA 7 software. The Korean bat CoVs receptor binding domain (RBD) of the spike protein was analyzed by comparative homology modeling using the SWISS-MODEL server. The binding energies of the complexes were calculated using PRODIGY and MM/GBGA.
Results
Phylogenetic analyses of the entire RNA-dependent RNA polymerase, spike regions, and the complete genome revealed that the Korean CoVs, along with SARS-CoV and SARS-CoV-2, belong to the subgenus Sarbecovirus, within BetaCoVs. However, the two Korean CoVs were distinct from SARS-CoV-2. Specifically, the spike gene of the Korean CoVs, which is involved in host infection, differed from that of SARS-CoV-2, showing only 66.8%–67.0% nucleotide homology and presented deletions within the RBD, particularly within regions critical for cross-species transmission and that mediate interaction with ACE2. Binding free energy calculation revealed that the binding affinity of Korean bat CoV RBD to hACE2 was drastically lower than that of SARS-CoV and SARS-CoV-2.
Conclusions
These results suggest that Korean bat CoVs are unlikely to bind to the human ACE2 receptor.
Keywords: Bat, coronavirus, SARS-CoV, SARS-CoV-2, spike protein
INTRODUCTION
Bats are strongly regarded as natural reservoirs for newly emerging zoonotic disease-causing viruses, including severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and Ebola virus (EV) [1]. SARS-CoV first emerged in 2002, infecting nearly 8,000 individuals, resulting in more than 750 deaths globally [2]. MERS-CoV was first identified in the Middle East in 2012 and subsequently spread to other countries globally. In the Republic of Korea (ROK), MERS-CoV first emerged in May 2015, infecting 186 individuals and causing 38 deaths [3]. Additionally, the bat-originated EV disease was approximated to have a 50% fatality rate [4].
The newly emerged coronavirus disease 2019 (COVID-19) is caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has been postulated to have probably originated from bats. COVID-19 was first reported in Wuhan city, China, in November 2019, and has rapidly spread across the world [5]. Common symptoms associated with COVID-19 are fever, cough, myalgia, fatigue, and most notably, pneumonia with an abnormal chest computed tomography scan [6]. As of December 3, 2020, a total of 36,332 COVID-19 cases have been documented, including 536 patients who died in the ROK (http://ncov.mohw.go.kr/).
Coronaviruses (CoVs) are enveloped viruses with the largest linear RNA genome, comprised of a single-stranded positive-sense RNA, which is approximately 27–32 kb in length [7]. CoVs are grouped under the following four genera: Alpha-, Beta-, Gamma-, and Delta-coronaviruses (https://talk.ictvonline.org/taxonomy/). With the exception of the genus Gammacoronavirus, the genome of the CoVs encodes 16 non-structural proteins (NSPs) and at least four main structural proteins, namely the spike glycoprotein (S), the envelope (E), the membrane (M), and the nucleocapsid (N) proteins [8]. The S protein, which is essential for infection and virion assembly, is organized into two domains: the N-terminal (S1) and the C-terminal (S2) domain. Viral entry into target cells is mediated by the previously identified interaction between the receptor binding domains (RBDs) within the S1 domain of SARS-CoV and SARS-CoV-2 and the angiotensin-converting enzyme 2 (ACE2) receptor of the host cells [9]. In particular, it is the receptor binding motif (RBM) within the RBD that directly binds to the ACE2 receptor [10]. The ACE2 receptor is known to exist in various mammals, including mice, rats, raccoons, cats, bats, and also humans. The association of the coronaviral S protein with the ACE2 receptor is determined by interactions of amino acids located in critical positions [11]. Recently, SARS-like CoVs have also been found in Korean bats [12,13]. Therefore, in this study, we estimated the likelihood of interaction between the Korean bat CoVs and the human ACE2 receptor through comparative genetic analysis of the Korean bat CoVs, SARS-CoV, and SARS-CoV-2.
MATERIALS AND METHODS
Retrieval of sequence data
To compare the Korean bat CoVs with SARS-CoV and SARS-CoV-2, we retrieved 32 bat and human CoV sequences from the National Center for Biotechnology Information (NCBI) and Global Initiative on Sharing Avian Influenza Data (GISAID; https://www.gisaid.org/) databases. The sequences of two Korean bat CoVs were comprised of the entire sequence of Bat CoV 16BO133 (KY938558) and the spike gene of Bat-CoV B15-21 (KU528591). Furthermore, sequences of SARS-related CoV and SARS-related CoV-2, originating from both animals and humans, were collected, including the SARS-CoV-Tor2 (JX163928), BatCoV RaTG13 (EPI_ISL_420131), SARS-CoV-2 IVDC-HB-01 (EPI_ISL_402119) from China, and KCDC03 (EPI_ISL_407193) from the ROK. The remaining 26 CoV sequences were also retrieved from the NCBI database (Supplementary Table 1).
Construction and analyses of phylogenetic trees
Phylogenetic analysis was carried out based on the complete sequences of the RNA-dependent RNA polymerase (RdRp), the spike gene, and the entire CoV genome. Multiple alignments of the CoV sequences were generated using the Clustal W algorithm in BioEdit v.7.0.9.0 [14]. Nucleotide sequences were used to construct phylogenetic trees by the maximum-likelihood (ML) method based on the General Time Reversible (GTR) model, using the MEGA 7 software as described in a previous report [15,16,17]; bootstrap values were calculated from 1,000 replicates. A discrete Gamma distribution with invariant sites was used to model evolutionary rate differences among sites. All positions containing gaps were eliminated. The tree is drawn to scale, with branch lengths measured as numbers of substitutions per site.
Homology modeling and structural analysis
To further investigate the structural similarity between the spike protein RBD of Korean bat CoVs, SARS-CoV, and SARS-CoV-2, the model of Korean bat CoVs spike protein RBD bound to the hACE2 receptor was obtained by comparative homology modeling using the SWISS-MODEL server [18]. Because the RBD sequences of the two Korean bat CoVs are identical, only one RBD structure using residues 317–491 of the spike protein of the two Korean bat CoVs was generated. The hACE2 sequence was retrieved from the UNIPROT database (Uniprot sequence Q9BYF1). The obtained complex model was checked and validated using PROCHECK [19] and MolProbity [20]. Moreover, the binding energies of the complexes were calculated using PRODIGY [21,22] and MM/GBSA [23,24]. The complex structure was visualized and figures containing structural descriptions were generated using PyMOL (version 1.20, https://pymol.org).
RESULTS
Phylogenetic analysis
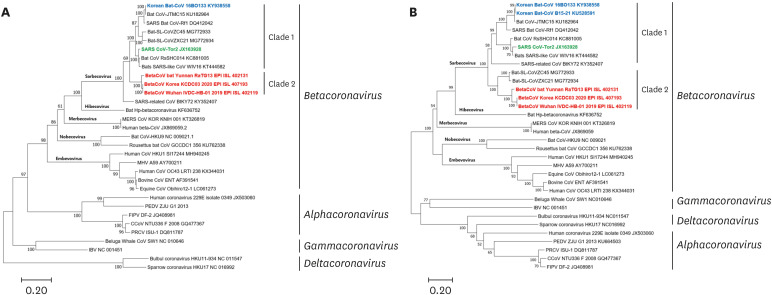
Phylogenetic analysis of the complete RdRp sequence revealed that the Korean bat CoV 16BO133 is included in the subgenus Sarbecovirus (previous lineage B), within the genus Betacoronavirus (Fig. 1A). Korean bat CoV 16BO133 was closely related to the BatCoV JTMC-15 strain that was discovered in Chinese bats (99.6%) (Supplementary Table 2). Conversely, the Korean bat CoV 16BO133 shares 92.4% and 87.5% nucleotide homology with SARS-CoV and SARS-CoV-2, respectively.
Fig. 1. (A) Phylogenetic analysis of complete RdRp sequences of Korean bat coronaviruses and the reference strains. (B) Phylogenetic analysis of the entire spike gene of the two Korean coronaviruses and the reference strains. The phylogenetic tree was constructed using the maximum-likelihood method in MEGA7 (bootstrap value = 1,000 replications). Korean coronaviruses are shown in blue, SARS-CoV in green, BatCoV RaTG13 and SARS-CoV-2 in red.
CoV, coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus.
Moreover, the phylogenetic analysis of the complete spike gene of the Korean bat CoVs 16BO133 and B15-21 strains showed that they have the closest genetic relationship with the JTCMB-15 strain, with 99.1% and 99.0% sequence similarity, respectively (Fig. 1B). The spike gene of the two Korean CoV strains was also shown to have a nucleotide similarity of 72.0-72.1% with SARS-CoV and of 66.8-67.0% with SARS-CoV-2. In addition, the spike genes of the two Korean bat CoV strains have been confirmed to have 99.5% homology at the nucleotide level (Supplementary Table 2). In the spike genes of the Korean bat CoVs, the amino acid similarities of the RBDs of the S1 region showed 64.5% and 65.5% homology with SARS-CoV and SARS-CoV-2, respectively. However, the S2 region showed high sequence similarities of 92.3-93.5% and 89.2-89.4% with SARS-CoV and SARS-CoV-2, respectively (Supplementary Table 3).
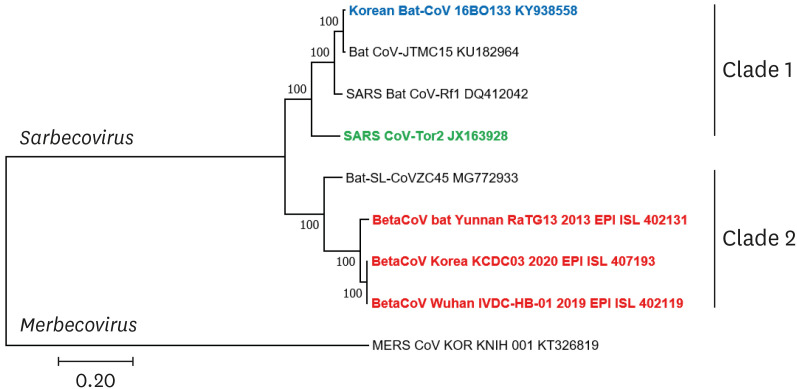
The ML tree using the entire sequence of the CoVs showed that the Korean 16BO133 CoV strain was genetically closest to the JTMC-15 strain, with 98.3% nucleotide similarity. Additionally, the 16BO133 strain shared 82.8% and 76.0% nucleotide similarity with SARS-CoV and SARS-CoV-2, respectively (Supplementary Table 2). Notably, the phylogenetic topology of the complete sequence revealed that the Korean 16BO133 strain was clustered with SARS-CoV; conversely, SARS-CoV-2 belonged to a different cluster (Fig. 2).
Fig. 2. Phylogenetic analysis of the complete gene of Korean bat coronaviruses with the reference strains. The phylogenetic tree was constructed using the maximum-likelihood method in MEGA7 (bootstrap value = 1,000 replications). Korean coronaviruses are shown in blue, SARS-CoV in green, BatCoV RaTG13 and SARS-CoV-2 in red.
CoV, coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus.
Comparative analysis of the sequences of the Spike gene
The length of the complete spike gene of both Korean bat CoV strains was 3,711 bp (1,237 aa), and differed by only six amino acids. Conversely, the lengths of the spike genes of SARS-CoV, SARS-CoV-2, and BatCoV RaTG13 were found to be 3,768 bp (1, 256 aa), 3,822 bp (1,274 aa), and 3,810 bp (1,270 aa), respectively. The spike genes of the Korean 16BO133 CoV strain differed from those of SARS-CoV and SARS-CoV-2 by 303 and 361 amino acids, respectively, whereas the amino acid sequences of the Korean B15-21 strain differed by 311 and 372 residues when compared with SARS-CoV and SARS-CoV-2, respectively.
The amino acid length of the RBD region in the Korean 16BO133 and B15-21 CoV strains was identical with 219 aa, while the lengths of SARS-CoV and SARS-CoV-2 were 237 aa and 238 aa, respectively. Specifically, the length of the RBM region in both the Korean bat CoVs was 53 aa; however, the length of the RBM regions of SARS-CoV and SARS-CoV-2 were 71 aa and 72 aa, respectively.
The amino acid sequences of the spike genes of the two Korean bat CoVs were compared with those of SARS-CoV and the recently emerged SARS-CoV-2. Our sequence alignment showed that the RBD, which is involved in the interaction with the ACE2 receptor, is located at positions 424, 448, 458, 459, 462, 464, 495, 496, 498, 502, 503, 507, 509, 510, 511, 514, and additionally at amino acid positions 492–494 for SARS-CoV, and amino acid positions 455, 484, 491–495 for SARS-CoV-2 (Fig. 3). Notably, large deletions were identified in the RBM region, specifically between positions 454–457 and 482–495, which correspond to the following sequences: STGNY and FSPDGKPCTPPAL in SARS-CoV and VGGNY and YQAGSTPCNGVEGF in SARS-CoV-2 (Fig. 3). In particular, P492, P493, and A494 of SARS-CoV and G455, A484, G491, V492, E493, and G493 of SARS-CoV-2 were included in the deletion regions of two Korean bat CoVs. In addition, the five critical residues within the RBM that were identified to play a key role in cross-species transmission include S464, deletion in 495, S502, T503, and V510 (Fig. 3).
Fig. 3. Comparison between the receptor binding domain sequence of the JTMC-15, Korean bat CoVs, SARS-CoV, and SARS-CoV-2. Receptor binding motif residues are shown in blue and the five critical residues in red. ACE2-interacting residues are written in bold and shaded.
CoV, coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus.
Comparative homology model of the RBD/hACE2 complex
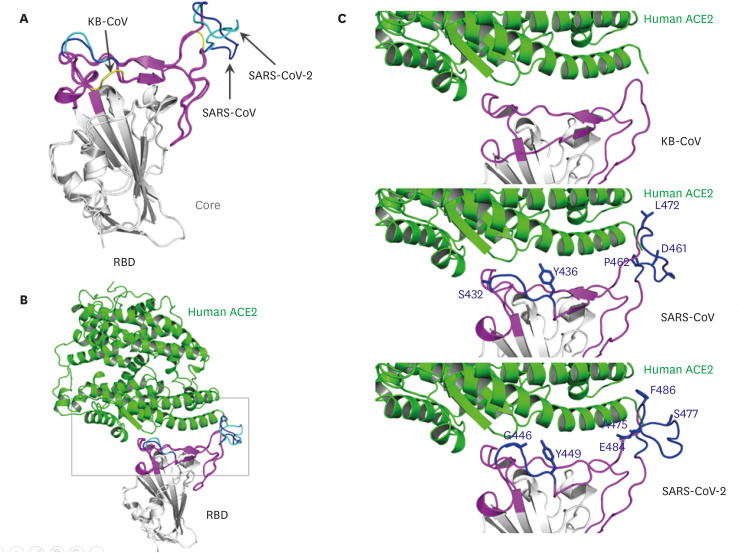
To analyze the interaction between RBD and hACE2 receptors in silico, models of the three-dimensional complexes of each of the two Korean bat CoVs RBDs bound to hACE2 receptor were constructed, based on the SARS-CoV-2 RBD-ACE2 complex crystal structure (PDB ID: 6m0j) by 93% sequence identity (Fig. 4). The root-mean-square deviation (RMSD) of Cα atoms between the template and homology model was 0.121Å (Fig. 4B). As shown in Fig. 4A the core regions of the three RBDs from two Korean bat CoVs, SARS-CoV, and SARS-CoV-2 were superimposable, but the RBM showed drastic differences between Korean bat CoVs, SARS-CoV, and SARS-CoV-2. In the two missing loops of Korean bat CoVs colored in yellow, residues interact with the hACE2 receptor (Fig. 4A and C) [10,25,26]. In addition, the binding energy estimated using PRODIGY was −9.1 kcal/mol, −16.6 kcal/mol, and −17.0 kcal/mol; using MM/GBSA it was −9.06 kcal/mol, −56.98 kcal/mol, and −59.38 kcal/mol for Korean bat RBD, SARS-CoV RBD, and SARS-CoV-2 RBD, respectively (Supplementary Fig. 1).
Fig. 4. Homology complex model of KB RBD and human ACE2. (A) Superimposed RBDs from KB-CoV, SARS-CoV, and SARS-CoV-2. The core of RBD is shown in silver; RBM is in magenta; deleted RBM residues of KB-CoV (yellow) in SARS-CoV and SARS-CoV-2 are in blue and cyan, respectively (B) Superimposed complex model of hACE2 with three RBDs. The boxed are is shown in C. (C) Interface between hACE2 and RMBs from KB (top), SARS-CoV (middle), and SARS-CoV-2 (bottom). For clarity, interacting residues from hACE2 were not shown.
KB, Korean bat; CoV, coronavirus; RBD, receptor binding domain; RBM, receptor binding motif; SARS-CoV, severe acute respiratory syndrome coronavirus.
DISCUSSION
The first novel CoV in bats was identified in 2005 and was soon followed by the detection of SARS-like-CoV in bats [27,28]. In the ROK, SARS-like CoVs were reported for the first time in bat feces collected in 2015 [12]. Subsequently, a few additional SARS-like CoVs have been found in the ROK; moreover, the entire genome and spike gene of Korean bat CoVs were successfully sequenced [13,29]. In this study, the genome sequences of two Korean bat CoVs (entire genome and spike gene sequences, respectively) were compared with those of SARS-CoV and of the newly emerged SARS-CoV-2 (putative bat-derived CoV).
Our phylogenetic analyses of the sequences of the complete RdRp, spike protein, and the entire genome showed that the Korean bat CoVs, strains 16BO133 and B15-21, as well as SARS-CoV and SARS-CoV-2 belong to the subgenus Sarbecovirus (previous lineage B) within the genus Betacoronavirus. Based on the phylogenetic analyses, the two Korean bat CoVs clustered with SARS-CoV; however, SARS-CoV-2 clustered into a distinct clade. These evolutionary data suggest that Korean bat CoVs are genetically more similar to the SARS-CoV than the newly emerged SARS-CoV-2; additionally, the two Korean bat CoVs can be considered to have originated from SARS-CoV.
The amino acid sequences of the spike glycoprotein of bat CoV strains 16BO133 and B15-21 were compared with the corresponding amino acid sequences of other beta-CoVs, including SARS-CoV (SARS-CoV-Tor2), BatCoV RaTG13, and SARS-CoV-2 (IVDC-HB-01). The spike genes of the Korean 16BO133 and B15-21 CoV strains shared low sequence similarities with those of SARS-CoV (72.0% and 72.1%) and SARS-CoV-2 (66.8% and 67.1%, respectively). Additionally, the amino acid sequence of the RBDs of the two Korean bat CoVs was found to have a low sequence similarity with those of SARS-CoV (64.5%) and SARS-CoV-2 (65.5%). However, in a previous study, SARS-like bat CoV (SL-CoV-WIV1), which can directly bind to the human ACE2 receptor, was demonstrated to have a high amino acid similarity (92.1% and 94.9%) with the spike protein and RBD region of SARS-CoV, respectively [30].
The amino acid sequences in the RBD of the two Korean bat CoVs were identical. However, they were identified to have large deletions and substitutions when compared with the sequences of the RBD region of SARS-CoV and SARS-CoV-2. Notably, these deleted regions were identified to be located within the RBM, which is known to play an important role in infections through direct interaction with the ACE2 receptor [10]. Moreover, the two missing residues are included in two loops of RBM that enhance spike expression and cell entry. According to a recent study, when two amino acid residues within these loops were artificially removed, the viruses loss ability of cell entry [31]. Notably, the large regions deleted in Korean bat CoVs include residues interacting with hACE2 receptors. Even in protein modeling, the RBM of the Korean bat CoVs showed significant differences in structure from SARS-CoV and SARS-CoV-2.
Within the RBD, among the 19 and 22 amino acid residues of SARS-CoV and SARS-CoV-2, respectively, only three amino acids were identical in the Korean bat CoVs, SARS-CoV, and SARS-CoV-2. Even the positions corresponding to those of P492, P493, and A494 of SARS-CoV and G455, A484, G491, V492, E493, and G493 of SARS-CoV-2 were deleted in Korean bat CoVs, suggesting low affinity with human ACE receptors. Conversely, five critical amino acids comprising the RBM, which play a key role in cross-species transmission, were found to have either been deleted or mutated in the Korean bat CoVs, including Y/L464S, L/F495deletion, N/Q502S, D/S503T, and T/N510V [30]. Additionally, only one amino acid among the following five critical residues of the SARS-like bat CoV (SL-CoV-WIV1), which binds to human ACE2, was found to be similar to the Korean bat CoVs, although they were similar to the SARS-CoV and SARS-CoV-2: S464S, F495deletion, N502S, D503T, and N510V [32].
Furthermore, the binding free energy estimated from the two methods used indicated that the binding affinity of Korean bat CoV RBD to hACE2 was drastically lower than that of SARS-CoV and SARS-CoV-2. The reduction in binding affinity is most likely due to the missing residues in the loop of RBM. As shown in Fig. 4C, there are several residues that contribute to the interaction with the hACE2 receptor in SARS-CoV and SARS-CoV-2 RBMs [10,25,26]: S432, Y436, D461, P462, and L472 of SARS-CoV, and G446, Y449, A475, S477, E484, and F486 of SARS-CoV-2. The contribution of missing residues in the RBM to the binding energy is plotted based on the MM/GBSA calculation. In this plot, the missing residues contribute to the decrease in the binding energy, indicating an increase in the binding affinity. Hence, based on the comparative genetic analysis within the RBD and homology model of the RBD-hACE2 complex, we suggest that the Korean bat CoVs are unlikely to bind to the human ACE2 receptor. However, further investigations are required to confirm these results.
SARS-CoV and SARS-CoV-2 are believed to have originated from the Rhinolophus bat, namely R. ferrumequinum and R. affinis, respectively [1,5]. The Korean bat CoV strains 16BO133 and B15-21 also originated from R. ferrumequinum. In the ROK, four bat families are known to exist, including Rhinolophedae, Molossidae, Miniopteridae, and Verspertilionidae [33]. In particular, the Rhinolophus bat is mostly phylogenetically related to SARS-CoV [1]. Only one species, R. ferrumequinum within this genus, is found to exist in the ROK [33]. Notably, R. affinis is thought to be a putative natural reservoir of SARS-CoV-2 [5], but it does not exist in the ROK. However, since R. ferrumequinum exists in the ROK, continuous surveillance of CoVs in bats is necessary. Moreover, according to previous studies, pangolins (Manis javanica) are regarded as possible intermediate hosts in the transmission of SARS-CoV-2 from bats to humans [34]. Additionally, various other animals, including ferrets, cats, and dogs are known to be susceptible to SARS-CoV-2 infections [35,36]. Hence, continuous surveillance of wildlife is essential for the identification of intermediate or natural hosts for newly emerging infectious diseases, such as SARS-CoV-2.
In this study, we performed a comparative analysis to predict the potential human infectivity of Korean bat CoVs. The results indicate that the Korean bat CoVs have a low sequence similarity with SARS-CoV and SARS-CoV-2. In particular, there were considerable differences within the RBD of the spike gene, which is known to interact with the human ACE2 receptor. In conclusion, we postulate that Korean bat CoVs are less likely to bind to the human ACE2 receptor than SARS-CoV and SARS-CoV-2.
Footnotes
Funding: This research was supported by the “Research Base Construction Fund Support Program,” funded by the Jeonbuk National University in 2020. This research was also supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2019R1A6A1A03033084). This study was partially supported by the National Research Foundation of Korea (Grant Number: NRF-2018R1D1A1B07041764).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Oem JK.
- Data curation: Na EJ.
- Formal analysis: Na EJ.
- Funding acquisition: Oem JK.
- Investigation: Oem JK.
- Methodology: Lee SY.
- Project administration: Lee SY.
- Resources: Oem JK.
- Software: Na EJ.
- Supervision: Oem JK.
- Validation: Lee SY.
- Visualization: Na EJ.
- Writing - original draft: Na EJ.
- Writing - review & editing: Lee SY.
SUPPLEMENTARY MATERIALS
Reference sequences of coronavirus strains
Nucleotide similarity of the partial RdRp, complete spike gene and entire genome
Amino acid similarity of the S1 and S2 regions of spike gene
Binding energy contribution per residues in RBM. The binding energy obtained MM/GBSA was decomposed per residue. On the top, the RBM residues is shown in magenta, whereas those missing in the Korean bat CoVs are shown in cyan.
References
- 1.Shi Z. Bat and virus. Protein Cell. 2010;1:109–114. doi: 10.1007/s13238-010-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sampathkumar P, Temesgen Z, Smith TF, Thompson RL. SARS: epidemiology, clinical presentation, management, and infection control measures. Mayo Clin Proc. 2003;78:882–890. doi: 10.4065/78.7.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim KH, Tandi TE, Choi JW, Moon JM, Kim MS. Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak in South Korea, 2015: epidemiology, characteristics and public health implications. J Hosp Infect. 2017;95:207–213. doi: 10.1016/j.jhin.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiappelli F, Bakhordarian A, Thames AD, Du AM, Jan AL, Nahcivan M, et al. Ebola: translational science considerations. J Transl Med. 2015;13:11. doi: 10.1186/s12967-014-0362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brian DA, Baric RS. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 11.Xu L, Zhang Y, Liu Y, Chen Z, Deng H, Ma Z, et al. Angiotensin-converting enzyme 2 (ACE2) from raccoon dog can serve as an efficient receptor for the spike protein of severe acute respiratory syndrome coronavirus. J Gen Virol. 2009;90:2695–2703. doi: 10.1099/vir.0.013490-0. [DOI] [PubMed] [Google Scholar]
- 12.Kim HK, Yoon SW, Kim DJ, Koo BS, Noh JY, Kim JH, et al. Detection of severe acute respiratory syndrome-like, Middle East respiratory syndrome-like bat coronaviruses and group H rotavirus in faeces of Korean bats. Transbound Emerg Dis. 2016;63:365–372. doi: 10.1111/tbed.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Jo SD, Son K, An I, Jeong J, Wang SJ, et al. Genetic characteristics of coronaviruses from Korean bats in 2016. Microb Ecol. 2018;75:174–182. doi: 10.1007/s00248-017-1033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall TA. BioEdit: a user friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 1999;41:95–98. [Google Scholar]
- 15.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, He WT, Wang L, Lai A, Ji X, Zhai X, et al. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med. 2020;26:483–495. doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Yang Y, Ren L. Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect Genet Evol. 2020;82:104285. doi: 10.1016/j.meegid.2020.104285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 20.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375-83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vangone A, Bonvin AM. Contacts-based prediction of binding affinity in protein-protein complexes. eLife. 2015;4:e07454. doi: 10.7554/eLife.07454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue LC, Rodrigues JP, Kastritis PL, Bonvin AM, Vangone A. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 23.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H, Li Y, Tian S, Xu L, Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys Chem Chem Phys. 2014;16:16719–16729. doi: 10.1039/c4cp01388c. [DOI] [PubMed] [Google Scholar]
- 25.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 26.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poon LL, Chu DK, Chan KH, Wong OK, Ellis TM, Leung YH, et al. Identification of a novel coronavirus in bats. J Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y, Son K, Kim YS, Lee SY, Jheong W, Oem JK. Complete genome analysis of a SARS-like bat coronavirus identified in the Republic of Korea. Virus Genes. 2019;55:545–549. doi: 10.1007/s11262-019-01668-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:94. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jo YS, Baccus JT, Koprowski JL. Mammals of Korea: a review of their taxonomy, distribution and conservation status. Zootaxa. 2018;4522:1–216. doi: 10.11646/zootaxa.4522.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30:1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu P, Chen W, Chen JP. Viral metagenomics revealed sendai virus and coronavirus infection of Malayan Pangolins (Manis javanica) Viruses. 2019;11:11. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reference sequences of coronavirus strains
Nucleotide similarity of the partial RdRp, complete spike gene and entire genome
Amino acid similarity of the S1 and S2 regions of spike gene
Binding energy contribution per residues in RBM. The binding energy obtained MM/GBSA was decomposed per residue. On the top, the RBM residues is shown in magenta, whereas those missing in the Korean bat CoVs are shown in cyan.