Abstract
Background
Arctic-like (AL) lineages of rabies viruses (RABVs) remains endemic in some Arctic and Asia countries. However, their evolutionary dynamics are largely unappreciated.
Objectives
We attempted to estimate the evolutionary history, geographic origin and spread of the Arctic-related RABVs.
Methods
Full length or partial sequences of the N and G genes were used to infer the evolutionary aspects of AL RABVs by Bayesian evolutionary analysis.
Results
The most recent common ancestor (tMRCA) of the current Arctic and AL RABVs emerged in the 1830s and evolved independently after diversification. Population demographic analysis indicated that the viruses experienced gradual growth followed by a sudden decrease in its population size from the mid-1980s to approximately 2000. Genetic flow patterns among the regions reveal a high geographic correlation in AL RABVs transmission. Discrete phylogeography suggests that the geographic origin of the AL RABVs was in east Russia in approximately the 1830s. The ancestral AL RABV then diversified and immigrated to the countries in Northeast Asia, while the viruses in South Asia were dispersed to the neighboring regions from India. The N and G genes of RABVs in both clades sustained high levels of purifying selection, and the positive selection sites were mainly found on the C-terminus of the G gene.
Conclusions
The current AL RABVs circulating in South and North Asia evolved and dispersed independently.
Keywords: Rabies virus, molecular evolution, Bayesian analysis, phylogeography
INTRODUCTION
Rabies is a neglected zoonotic disease caused by rabies virus (RABV), the prototype species of the genus Lyssavirus of the family Rhabdoviridae [1,2]. The disease is characterized by fatal progressive encephalitis in a wide range of hosts, including humans, domestic animals and wildlife, and remains endemic in most Asian and African countries, which makes it an important public health threat. Generally, RABVs can be divided into seven main lineages based on the molecular phylogeny of the nucleoprotein sequences [3]. The viruses tend to be geographically restricted within lineages, and the name of each lineage is the reflection of its geographic distribution, such as India, Asia, American indigenous, Africa 1, Africa 2, Cosmopolitan and Arctic/AL.
Arctic and Arctic-like (AL) RABVs are phylogenetically closely related viruses mainly circulating among wild and domestic animals in the Arctic regions and in a number of Asian countries. The distribution, phylogeny and evolutionary history of Arctic and AL RABVs were analyzed in recent years. Generally, fox species, such as the Arctic fox (Alopex lagopus) and red fox (Vulpes vulpes), are the primary reservoir of Arctic RABVs; the distribution of this viral lineage covers the majority of the regions around Arctic Circle, such as Russia, Alaska, Canada, and Greenland, while AL RABV isolates mainly circulate among raccoon dogs and domestic dogs spreading over a wider geographic distribution and covering Northeast Asia, West China, South Asia and the Middle East region [3,4,5,6,7,8,9]. The nucleotide substitution rate of the Arctic-related lineage inferred from a database of 212 viral sequences of the N gene fragment was estimated using the Bayesian methods to be 3.817 × 10−4/site/year, and the most recent common ancestor (tMRCA) of the entire lineage has been estimated to have exist existed in approximately the 1820s [5]. Viral distribution, phylogeny and phylogeography suggest that RABVs in the Arctic-related lineage originated from the dog RABVs of the India subcontinent lineage [10]. Isolates circulating around the North Asia and Arctic regions have been speculated to originate from a common ancestor in Central Asia, and transported in the northern direction via human activity or wildlife migration [4,10,11,12].
In recent years, India, Nepal, Bangladesh, South Korea and China witnessed numerous outbreaks of AL RABVs infections in wild and domestic animals [6,11,13,14,15,16,17,18]. Dozens of newly isolated viruses were documented. Arctic-related lineage RABVs mainly circulate among the reservoirs and other spilled-over infected animals; however, concerns of human exposures have increased as more isolates were detected recently. Thus, it is urgent to expand our understanding of the circulation dynamics and evolution of Arctic-related RABVs and especially those AL RABVs that have a complex shared circulation with RABVs of other lineages and additional types of the hosts on the Asian continent. Thus, in this study, we mainly conducted Bayesian analysis of evolutionary history, phylogeography and host range states of Arctic-related RABVs using the updated sequence databases. A better understanding of the evolutionary dynamics of the virus of this lineage is necessary and valuable for future rabies control strategies.
MATERIALS AND METHODS
Sequence datasets
We collected fifty-two brain samples from rabid animals in Northern China from 2006 to 2014. The affected animals included dog (Canis lupus familiaris), raccoon dog (Nyctereutes procyonoides), red fox (Vulpes vulpes), swift fox (Vulpes velox), cow (Bos taurus), sheep (Ovis aries), camel (Camelus bactrianus). They were mostly found in Inner Mongolia, Jilin and Heilongjiang Provinces in north China. For domestic animals and dogs, they were directly submitted by farmers and veterinarians. For raccoon dogs, which were raised for fur production, were submitted by farmers. Clinical signs of suspicious rabies like disease were described. As for two foxes, they were found dead and submitted to sample collectors (Supplementary Table 1). We collected the brain samples and disposed their bodies non-hazardously in case of animal or human exposure. They were not abused in any form. Seven of the brains were confirmed to contain AL RABVs based on the viral N and/or G gene detection. The full N gene coding sequences (1350) of these isolates combined with 93 other publicly available sequences from the GenBank constituted dataset 1; the sequences of Arctic and AL RABV isolates were derived from this dataset and used for evolutionary analysis. Dataset 2 comprised the whole length G gene sequences (1572 nucleotides) from 56 Arctic-related isolates. Dataset 3 included 155 of the N gene sequences (676 nucleotides) from the AL isolates. The full information of these sequences, including isolate names, collection times, locations, and host states, was retrieved from the NCBI nucleotide database. This information is listed in the supplementary file (Supplementary Table 2).
Bayesian evolutionary analysis
For the evolutionary analysis, we reconstructed a maximum clade credibility (MCC) tree for the dataset 1 using the Bayesian Markov chain Monte Carlo (MCMC) analysis implemented in the BEAST package v1.8.0 [19] (http://beast.bio.ed.ac.uk/). The inconsistences in the tree topologies between the previous studies [4,5,6] and our work were resolved by re-evaluation of the nucleotide substitution, molecular clock and demographic models. Initially, the general time reversible (GTR) substitution model with gamma distribution rate (G) and invariant sites (Г4) was selected based on the log likelihood (lnL) and Akaike's information criterion (AIC) scores inferred by MrModeltest v2.3. Then, the combinations of clock models (strict, uncorrelated exponential distribution [UCED] and uncorrelated log-normal distribution [UCLD]) and demographic models (constant size [CS], Bayesian skyline plot [BSP] and exponential growth [EG]) were tested and the best models were selected based on the Bayes factor (BF) test and Akaike's information criterion for MCMC (AICM) values. The best-fit model for the dataset 1 was determined to be GTR+G+Γ4/UCED/BSP. Under these models, the MCMC chains were run for 100,000,000 iterations and were assessed for the good mixing by adequate effective sample size (ESS) by Tracer v1.6 software (http://beast.bio.ed.ac.uk/tracer), ignoring 10% of the chains as burn-in. Evolutionary history was summarized in an annotated MCC tree using TreeAnnotator v 1.8.0 that was visualized in FigTree v 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). Posterior probability values were used to evaluate the statistical support of each node on the tree.
Demographic history
For demographic history estimation of Arctic-related RABVs over time, Bayesian skyline analyses were implemented in the Tracer software using the log file of the MCMC sampling procedures for dataset 1 as described above. The population-size changes were visualized using a skyline plot of changes of genetic diversity versus time.
Selective pressure
Datasets 1 and 2, corresponding to the 100 N and 55 G gene coding sequences, respectively, were used to evaluate the natural selection pressure on the Arctic-related RABVs; prior to this procedure, the intragenic recombination was analyzed by screening of the sequences for recombination breakpoints with SBP and GARD methods implemented in the HyPhy package via the Datamonkey server (http://www.datamonkey.org) [20]. For estimation of (dN-dS) site-by-site selection at every codon in both datasets, single-likelihood ancestor counting (SLAC), fixed effects likelihood (FEL), random effects likelihood (REL) and fast unconstrained Bayesian approximation (FUBAR) methods were selected and used; the codons that showed evidence of positive or negative selection were reported using the default significance levels. In addition, the Mixed Effects Model of Evolution (MEME) module was used to detect the individual sites that are the subjects of episodic diversifying selection [21]. Moreover, synonymous and nonsynonymous substitution rates based on the codon-aligned nucleotide sequences of datasets 1 and 2 were calculated using an online tool SNAP v2.1.1 available at http://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html.
Phylogeny-trait association
The associations between phylogeny and traits (geography) of AL RABVs were investigated using the Bayesian Tip Significance testing (BaTS) method [22]. Using the posterior distribution of phylogenies generated by the BEAST package, phylogeny-geography correlations were analyzed by calculating statistical significance of the null hypothesis that traits are randomly associated with phylogeny tips. Parsimony score (PS), association index (AI) and maximum monophyletic clade (MC) size were reported and the significance of phylogeny-trait associations were assessed by p values.
Discrete phylogeographic analysis of AL RABVs
For the phylogeographic analysis of 155 AL RABVs in dataset 3, spatial dispersal of RABVs among countries and regions was inferred using a Bayesian Markov chain Monte Carlo approach implemented in the BEAUti/BEAST package v1.8.0 [19] integrated with the BEAGLE library (http://beast.bio.ed.ac.uk/BEAGLE) for high performance statistical phylogenetic inference. In brief, we applied a discrete diffusion model to a symmetric substitution model, which specified a discrete phylogeographic analysis, using a standard continuous-time Markov chain (CTMC). We also enabled the MCMC to infer a social network with Bayesian Stochastic Search Variable Selection (BSSVS) procedure by identifying the most parsimonious description of the phylogenetic diffusion process [23]. After adequate chain mixing using the BEAST software, an MCC tree was summarized, visualized and annotated as describe above. Spatial dynamics supported by the Bayes factor test were analyzed using SPREAD v1.0.6 [24] and visualized in the virtual globe software.
RESULTS
Evolutionary history of Arctic-related RABVs
Evolutionary analysis of Arctic-related RABVs was performed by using the full-length N gene from the dataset 1 using the coalescent-based Bayesian MCMC method. The GTR+G+Г4 was suggested as the best-fit nucleotide substitution model (Supplementary Table 3) based on log likelihood and AIC scores. UCED and BSP combination provided the lowest Bayes factors and AICM scores and a narrow margin of 95% HPD and was thus considered the best fit model for the sequences in the dataset 1 (Table 1). Under these model settings, evolutionary history of the Arctic-related RABVs was inferred in a time-scaled MCC tree. The mean nucleotide substitution rate/site/year of the full-length N gene was predicted to be 3.52 × 10-4 with the 95% highest posterior density (HPD) interval distributed from 2.13 × 10-4 to 5.01 × 10-4 (Table 1). Under these substitution rates, the tMRCA of the Arctic-relate lineage (Arctic and AL RABVs) was estimated to be circulated in approximately 1838 with a 95% HPD ranging from 1757 to 1904. Four major clusters (virus groups) of the Arctic-related clade were identified: Arctic, Arctic like-1 (AL-1), Arctic like-2 (AL-2) and Arctic like-3 (AL-3); in addition, ten subgroup clusters were identified within these groups (Fig. 1). The virus in the Arctic clade has diverged into three groups: A-1, A-2 and A-3; the viruses in this clade evolved from a common progenitor that originated in 1940s (tMRCA 1940, 95% HPD 1893-1968). Viruses in the groups A-1 to A-3 were dated back to 1960s; the tMRCAs dated to 1965 (95% HPD 1942-1983), 1968 (95% HPD 1952-1979) and 1965 (95% HPD 1947-1978), respectively. In the AL lineage, the viruses diverged into the AL-1/AL-3 groups and AL-2 group in the 1880s (tMRCA 1881, 95% HPD 1815-1945) and evolved independently in South and West Asia (AL-1 and AL-3) and Northeast Asia (AL-2). In general, a common progenitor of the viruses in the South and West Asia were dated back to 1927 (95% HPD 1861-1965). Viruses in the AL-1 group (tMRCA 1950, 95% HPD 1918-1976) have variable geographic coverage and include samples from India, Pakistan, Nepal, Bhutan, Iraq and Afghanistan, while the viruses in the AL-3 group (tMRCA 1961, 95% HPD 1921-1986) mainly circulates in Nepal. Interestingly, AL-2a is a distinct branch within the AL-2 group encompassing two newly reported RABV isolates from Qinghai and Tibet in West China [18]. This subgroup began to diverge as early as in 1938; after long time of evolution, the viruses emerged in the late 20th century (tMRCA 1997, 95% HPD 1969-2008). Two RABV isolates sampled from North China, DG11248 and TJ11-RD, are segregated into the AL-2c clade which dates back to 1966 (95% HPD 1951-1977). RABVs sampled from South Korea are clustered exclusively within AL-2d, which emerged in 1970s (tMRCA 1974, 95% HPD 1954-1990) (Fig. 1).
Table 1. Bayesian Markov chain Monte Carlo estimates of nucleoprotein coding sequences of 100 Arctic-related rabies viruses.
| Model, substitution/clock/demographic | Bayes factor | AICM | Nucleotide substitution rate* | tMRCA |
|---|---|---|---|---|
| GTR+G+Γ4/UCED/BSP† | −6550.62 | 13238.12 | 3.52 (2.13–5.01) × 10−4 | 1838 (1757–1904) |
| GTR+G+Γ4/UCED/CS | −6554.55 | 13248.08 | 3.17 (2.03–4.40) × 10−4 | 1782 (1550–1905) |
| GTR+G+Γ4/UCED/EG | −6556.009 | 13243.43 | 3.53 (2.35–4.69) × 10−4 | 1827 (1681–1916) |
| GTR+G+Γ4/UCLD/BSP | −6575.49 | 13302.80 | 2.65 (1.88–3.63) × 10−4 | 1803 (1719–1879) |
| GTR+G+Γ4/UCLD/CS | −6576.44 | 13304.55 | 2.90 (2.07–3.85) × 10−4 | 1812 (1720–1877) |
| GTR+G+Γ4/UCLD/EG | −6573.44 | 13313.23 | 2.91 (2.00–3.80) × 10−4 | 1817 (1731–1884) |
| GTR+G+Γ4/strict/BSP | −6659.70 | 15788.33 | 2.60 (1.15–3.20) × 10−4 | 1808 (1722–1873) |
| GTR+G+Γ4/strict/CS | −6592.48 | 13272.98 | 2.51 (1.87–3.23) × 10−4 | 1793 (1704–1866) |
| GTR+G+Γ4/strict/EG | −6591.03 | 13277.56 | 2.54 (1.86–3.28) × 10−4 | 1845 (1753–1913) |
AICM, Akaike's information criterion through Markov chain Monte Carlo; tMRCA, the most recent common ancestor; GTR+G+Γ4, general time reversible substitution model with gamma distribution rate and invariant sites (Г4); UCED, uncorrelated exponential distribution; UCLD, uncorrelated log-normal distribution; BSP, Bayesian skyline plot; CS, constant size; EG, exponential growth; strict, strict clock model.
*Nucleotide substitution rate and tMRCA values are given in the form of median with 95% highest posterior density (HPD) intervals; †Best-fit clock and demographic model is indicated in bold.
Fig. 1. Time-scaled maximum clade credibility phylogeny of Arctic related rabies viruses using 100 complete coding sequences of the N gene from the dataset 1. The horizontal time scale shows the estimated divergence year and the vertical bars define the clustered groups within the lineage. Divergence dates for the major branches together with their 95% highest posterior density ranges are indicated above and below the tree nodes. Posterior probabilities are shown as their values corresponding to the size of the circles and node colors according to the legend on the left. Selected values in the major branches are also shown as numbers.
Population demography of Arctic-related RABVs
It was estimated that the effective population size of Arctic-related (root height), Arctic and AL lineages did not show substantial genetic diversity until the mid-1970s; only a slight growth in population size is apparent. From the mid-1970s to mid-1980s, the population increased substantially faster than before that and the population size reached its peak approximately in 1985. However, the effective number of infections was dramatically decreased in the subsequent fifteen years. The trend of population size decrease stopped at the beginning of the 21st century and began to recover from approximately 2007 to the most recent data point in 2014 (Fig. 2). Similar demographic dynamics can be observed in the case of Arctic-related (root height), Arctic and AL RABVs (data not shown).
Fig. 2. Demographic history of the Arctic-related rabies viruses evaluated by Bayesian skyline plots. Dynamics of the N gene genetic diversity was estimated by the Bayesian skyline reconstruction using the stepwise (constant) variant. The analysis was performed using the trees log file generated by the Markov chain Monte Carlo analysis as described. Dynamics of the mean genetic diversity are shown as a black line and the estimated 95% highest posterior density intervals are presented as the solid blue areas. The maximum clade credibility tree with branches and nodes colored according to the posterior values is superimposed under the demographic history plot, which covers the time interval from 1900 to 2014. Height and width of the plot are proportional to the effective population sizes and time scales.
Selection pressure through evolutionary process
Selective pressure on Arctic-related RABVs through their evolutionary history were estimated using the SLAC, FEL, REL, FUBAR and MEME methods. Generally, the open reading frames of both genes had a high degree of negative (or purifying) selection; the average dS/dN ratios of pairwise comparison of all N and G genes were 111.080 and 18.527 as calculated by the SNAP software. A total of 59 and 18 negatively selected sites were identified by at least three methods including SLAC, FEL, REL and FUBAR. In addition, 6 positively selected codons were found in the G gene coding sequence; however, there was no evidence of positively selected codons within the N gene sequences. Moreover, 3 sites in the N gene and 4 sites within the G gene under the episodic diversifying selection were inferred by the MEME model (Table 2, Supplementary Tables 4, 5, 6, 7).
Table 2. Amino acid sites under natural selection in nucleoprotein and glycoprotein of Arctic-related rabies viruses in dataset 1 and 2.
| Gene | Number of sequences | Mean dN/dS | Positively selective site | Negatively selective site* | Episodic diversifying selection site |
|---|---|---|---|---|---|
| N | 100 | 111.080 | None | 8, 16, 20, 21, 27, 33, 41, 45, 57, 59, 92, 98, 103, 105, 108, 111, 114, 123, 129, 134, 138, 146, 149, 154, 158, 162, 170, 172, 173, 202, 203, 207, 213, 220, 227, 228, 229, 234, 214, 256, 269, 283, 301, 323, 337, 341, 342, 349, 354, 380, 386, 406, 412, 414, 435, 445, 446, 447 | 4, 52, 391 |
| G | 56 | 18.527 | 15, 202, 473, 483, 491, 521 | 4, 6, 8, 71, 102, 149, 231, 239, 317, 332, 343, 360, 387, 436, 440, 455, 480, 509 | 301, 491, 519, 521 |
*The sites found under negative selection by at least three methods are shown, detailed information can be found in Supplementary Tables 3, 4, 5, 6.
Quantifying the phylogeographic structures of AL RABVs
The associations of the phylogeny and geographic trait of AL RABVs were shown in Table 3, the observed values of AI and PS indices from the estimated MCC trees were significantly less that the null values (p = 0), which indicates rejection of the null hypothesis of no association between the phylogeny and geographic trait. The correlation of the phylogeny with the geographic structure suggests that AL RABVs are predominantly clustered by their geographic traits. The phylogeny-trait association for each location was also calculated by maximum monophyletic clade size (MC) statistics. Only a half of the total number of the locations (eight out of sixteen) were supported by the MC statistic, which implies that viruses sampled from these regions were clustered by their geographical locations, while the viruses from the rest of the regions might have witnessed a gene flow from the other regions (Table 3).
Table 3. Estimated geographic structure of 155 Arctic-like rabies viruses.
| Statistic | Sequence number | Observed value (95% CI) | Null value (95% CI) | p value |
|---|---|---|---|---|
| AI | - | 2.10 (1.62–2.60) | 13.63 (12.73–14.40) | 0.00* |
| PS | - | 25.19 (24.00–26.00) | 91.64 (87.94–95.32) | 0.00* |
| MC (Afghanistan) | 6 | 1.35 (1.00–2.00) | 1.08 (1.00–1.86) | 1.00 |
| MC (India) | 49 | 13.55 (10.00–21.00) | 2.62 (2.14–3.34) | 0.01* |
| MC (South Korea) | 42 | 42.00 (42.00–42.00) | 2.37 (1.96–3.14) | 0.01* |
| MC (Nepal) | 20 | 17.54 (16.00–18.00) | 1.59 (1.07–2.17) | 0.01* |
| MC (Pakistan) | 11 | 3.93 (3.00–5.00) | 1.17 (1.00–1.99) | 0.01* |
| MC (Russia) | 4 | 2.00 (2.00–2.00) | 1.02 (1.00–1.07) | 0.01* |
| MC (Alaska) | 1 | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 |
| MC (Bangladesh) | 7 | 4.69 (4.00–6.00) | 1.07 (1.00–1.34) | 0.01* |
| MC (Bhutan) | 1 | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 |
| MC (West China) | 2 | 2.00 (2.00–2.00) | 1.00 (1.00–1.00) | 0.01* |
| MC (North China) | 7 | 6.00 (6.00–6.00) | 1.10 (1.00–1.99) | 0.01* |
| MC (Italy) | 1 | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 |
| MC (Iraq) | 1 | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 |
| MC (Japan) | 1 | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 |
| MC (Mongolia) | 1 | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 |
| MC (Iran) | 1 | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 |
CI, confidence interval; AI, association index; PS, parsimony score; MC, maximum monophyletic clade.
*Statistically significant where p value less than 0.05.
Spatiotemporal dynamics of AL RABVs
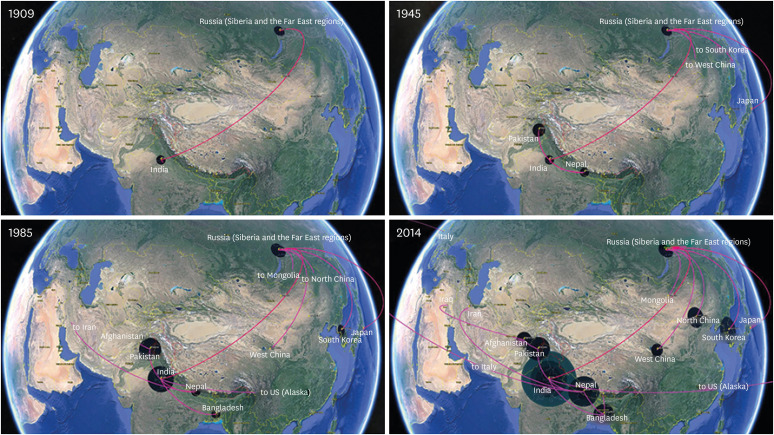
The geographical origin and spatiotemporal dynamics were inferred using a discrete phylogeographic model with Bayesian Stochastic Search Variable Selection (BSSVS) procedure to identify phylogenetic diffusion of the viruses. The root state posterior probabilities for all locations ranged from 0.05 for Iraq to 0.163 for Russia (Fig. 3). Specifically, Siberia and the Far East region in Russia received the highest ancestor support with the posterior probability of 0.163 followed by Japan with the value of 0.129 indicating that the geographic origin of AL RABVs was in Northeast Asia. However, South Asian countries of India and Nepal accounted for the posterior probability distribution of 0.116 and 0.096 in root, respectively (Fig. 3). The geographic location in the location-annotated MCC tree root indicates that tMRCA of the 155 AL RABVs in the dataset 3 emerged year in approximately 1834 (95% HPD 1783-1876) in Russia. This RABVs ancestor in the Northeast Asia subsequently diversified with other ancestral viruses and spread southward to India in the South Asia. Simultaneously, the ancestral RABV in the north dispersed into the adjacent countries, such as Japan, Korea, Mongolia and China, by the transboundary human activities or wildlife migration. In the late 19th and early 20th centuries, the viruses began to diversify and spread to the neighbor regions from Russia in the Northeast Asia and from India in South Asia, which resulted in circulation of three major ancestral clades AL-1, AL-2 and AL-3. Viruses in the AL-1 and AL-3 clades began to diverge in 1909 (95% HPD 1978-1937); AL-3 emerged mainly in Nepal in approximately 1937 (95% HPD 1911-1964) while the viruses in AL-1 mainly circulated and translocated between India, Pakistan, Bangladesh, Bhutan, Afghanistan, etc. RABVs in AL-2 had a common ancestor in Russia. The dispersal of the viruses among the regions began in the 1910s (1911 95% HDP 1884-1936). By the time of the most recent sampling date, the viruses covered a geographical distribution of Japan, Korea, Mongolia, and North and West China (Fig. 3). Notably, the year of the isolate Komatsugawa, which was sampled in 1940s in Japan [25], was estimated to be 1945.2 (95% HPD 1941-1949) by the MCMC method integration over a sampling time interval of 1940 to 1949 assuming a uniform prior distribution. The spatiotemporal dynamics of the migration events are summarized in the keyhole markup language (kml) file which enables the visualization of the spread over time in Google Earth. For convenient visualization of the temporal dynamics of spatial diffusion of 155 RABVs, four time points corresponding to the years of 1909, 1945, 1985 and 2014 were snapshotted and are shown in Figure 4. Additionally, the migration rates were inferred by the Bayes factor (BF) tests; the rates yielding a BF value > 5.0 were considered plus the supported diffusion rates, which are summarized in Figure 5 and Table 4. Accordingly, the RABV diffusion between Afghanistan and Pakistan and between India and Pakistan were predicted with the maximum BF support of 59813.20 and 1166.29, respectively. The migration between Afghanistan and Iraq, North China and Russia (Siberia and the Far East), Bangladesh and India, and Italy and Nepal had moderate BF support ranging from 100 to 1000. Viral diffusion between Japan and Russia, US (Alaska) and India, Bangladesh and Nepal, Mongolia and Russia, Bangladesh and Bhutan, India and Nepal, Japan and South Korea, and Russia and South China were not well supported by the BF tests (5 < BF values < 100). Notably, certain long-distance viral migration, such as between Nepal and Italy and India and US (Alaska), were supported by the BF tests.
Fig. 3. Maximum clade credibility phylogeny of 155 partial N gene sequences of Arctic-like RABVs presented using discrete phylogeography. Colors of the branches represent the most likely location states of the descendant nodes. The color codes for 16 countries or regions are defined in the legend of the left; the posterior probability distributions in the root are shown in the insert chart. Posterior of each node is colored according to its value as described above. Full annotated tree is shown in Supplementary Fig. 1.
Fig. 4. Visualization of spatiotemporal dynamics of 155 Arctic-like RABVs in Google earth. Keyhole markup language file were generated by the SPREAD software using the tree file obtained by discrete phylogeography as described above. Viral dispersal patterns among the countries and regions are indicated as snapshots of four selected time points including 1909, 1945, 1985 and 2014. Geographic coordinates were selected according to the representative regions with the highest number of RABVs sampled or by the capital cities otherwise for convenience.
RABV, rabies viruses.
Fig. 5. Spatial dynamics of Arctic-like RABVs supported by the Bayes factor test. The migration rates between the locations were inferred by the discrete BF tests using the SPREAD software. Rates yielding a BF value >5.0 and supported diffusion events are considered. The Arctic-like RABV-affected countries are colored in light blue within the map. Migrations with different BF value intervals are indicated by the colors and thickness as shown in the legend (n = 155).
RUS, Russia; ALK, Alaska; MGL, Mongolia; nCHN, North China; JPN, Japan; KOR, South Korea; wCHN, West China; BTN, Bhutan; NEP, Nepal; BAN, Bangladesh; IND, India; PAK, Pakistan; AFG, Afghanistan; IRI, Iran; IRQ, Iraq; ITA, Italy; RABV, rabies viruses; BF, Bayes factor.
Table 4. The migrations well-supported by Bayes factor tests*.
| Region between | Bayes factor by BSSRV | |
|---|---|---|
| Afghanistan | Pakistan | 59813.20 |
| India | Pakistan | 1166.29 |
| Afghanistan | Iraq | 468.11 |
| North China | Russia | 436.46 |
| Bangladesh | India | 329.42 |
| Italy† | Nepal | 103.72 |
| Japan | Russia | 27.88 |
| US (Alaska) | India | 22.72 |
| Bangladesh | Nepal | 14.21 |
| Mongolia | Russia | 12.56 |
| Bangladesh | Bhutan | 8.83 |
| India | Nepal | 6.29 |
| Japan | South Korea | 5.61 |
| Russia | West China | 5.02 |
*Bayes factor cutoff value is 5.0; †this case is a dead-end host of organ transplant mediated infection, not an Arctic-like rabies viruses dispersal.
DISCUSSION
Arctic-related RABVs represent a group of distinct viruses of the seven RABV lineages that mainly circulate in the northern-climate countries. The so-called AL RABVs were identified recently to extend the distribution of the viruses in South and West Asia; host spectrum of the viruses were broadened to indicate that dogs and domestic animals are the predominant hosts [5,6,7,11,13,18,26,27,28]. The evolutionary history of this lineage of the viruses has been characterized sporadically by several groups; however, comprehensive analysis of the origin, spread and population dynamics for the viruses of this lineage is largely unappreciated. Insufficient availability of viral sequences and the sampling information may have hindered the characterization of evolutionary dynamics of Arctic-related RABVs. In the recent years, dozens of the N gene sequences of this lineage have been documented; thus, we collected essentially all accessible full-length and partial sequences with traceable date, location and host information, which enabled us to infer the evolutionary history, population dynamics, geographic origin and spreading routes for the newly defined RABVs and to comprehensively estimate the evolutionary dynamics of the entire lineage. We have found that the tMRCA of the current-Arctic related RABVs emerged in the 1830s. This ancestor was subsequently diversified resulting in the independently evolved Arctic and AL RABVs. The N and G genes of both viral lineages were subjected to a high degree of negative (or purifying) selection with sporadic episodic diversifying selection and positive selection sites on the G gene codons. Population demographic analysis indicates that the viruses experienced a gradual growth followed by a sudden decrease in population size. Discrete phylogeography suggests that the geographic origin of AL RABVs is in Siberia and the Far East region in Russia approximately in 1830s; the AL RABVs ancestor then diversified and immigrated to India causing the widespread of the viruses in the North and South Asia. Dogs, domestic animals and raccoon dogs account for the overwhelming majority of the ancestor host distribution.
The nucleotide substitution rate of Arctic-related RABVs during evolutionary process was calculated to be 3.52 × 10-4/site/year (95% HPD 2.13 × 10-4 to 5.01 × 10-4) based on the full length N gene dataset 1; this value is slightly lower although very close to the previous estimates [5,6]. If differences in sample sizes and sequence lengths are not taken into account, the newly added sequences, such as isolates CQH1202D and CXZ1201D sampled from West China, may have accounted for a slight decrease in the substitution rates considering the long evolutionary history of these two isolates. Under this substitution rate, the tMRCA estimates that the Arctic RABVs emerged in 1838 (95% HPD 1757 to 1904) approximately ten years later than assessed by the previous estimates (Nadin-Davis, Sheen, and Wandeler 2012, Pant et al. 2013). Two tree topologies of the Arctic related RABVs were inferred by different research groups [4,5,6], A discrepancy in the main clades in the study by Nadin-Davis et al. includes clear separation of the Arctic and AL clades from the ancestor and independent evolution; however, phylogeny from other studies suggests that the AL 2 clade initially diverged from the root with subsequent separation of the Arctic clade from the AL 1/3 clade. It was suggested that times to tMRCAs differ between the clades. Therefore, we have carefully compared different nucleotide substitutions, molecular clocks and demographic models and selected the best-fit model combination of GTR+G+Γ4/UCED/BSP. Our results of the MCMC analysis tend to advocate the findings of Nadin-Davis and colleagues that Arctic RABVs share a common ancestor with AL RABVs rather than with the AL 1/3 clade.
Reconstructing demographic history of the virus enables us to trace the transmission and spread dynamics of the disease thus providing useful information on the host and anthropogenic factors that have influenced the evolutionary processes [29]; the results of the Bayesian skyline plots indicate that Arctic-related RABVs underwent a 15-year population decrease from the mid-1980s to approximately 2000. The genetic diversity of the viruses is predominantly shaped by the host-pathogen interactions and especially by the immune-driven selection [30]; thus, we assume that a vaccine induced an acquired immunity of the reservoirs and imposed immune pressure on the RABV isolates accounting for a decline in genetic diversities during this period. In addition, a sudden decrease in the number of reservoirs may account for this drop in population. Climatic change, such as global warming, environmental pollution by anthropogenic activities, or dog control programs, may lead to a drop in the number of the hosts. It should be noted that the time point for this population decline corresponds to the initial stage of rabies control campaigns in India and China [31,32], which confirms the effectiveness of the integrated rabies control tactics, such as stray dog control program, dog vaccination campaigns and appropriate postexposure prophylaxis (PEP). However, attention should be paid to the fact that the effective population size of the virus began to rise in the recent years since 2007 as indicated by the plot, which implies that the Arctic-related RABV isolates may have evolved and are now adapted to the immune pressures conferred by current vaccines; therefore, high coverage vaccination for dogs and wildlife is necessary.
The average dS/dN ratios for the N and G genes suggest a strong negative (purifying) selective pressure on both genes. The degree of selection of the N gene, however, is substantially higher than that of the G gene. Correspondingly, higher number of negatively selected sites were identified, which implies the importance of the nucleoprotein during the evolutionary process. High negative selection enables the removal of the deleterious variations from the population maintaining its functions in transcription and replication of the viral genome. In contrast, glycoproteins located on the surfaces of the viruses and infected cells suffer higher selective pressures from the host and especially from the host immune system. Theoretically, the G gene tends to have higher positive selection than the N gene. In agreement with this assumption, six G gene codons are positively selected; four of these codons are located within the cytoplasmic domain at the carboxyl terminus of the glycoprotein. Moreover, three of the four episodic diversifying selection sites were inferred within this region. Relatively higher Darwinian selection in the C-terminus of the G gene leads to consequently higher variant frequency. Indeed, high sequence divergence is present in this region and correlates with neurotropism and pathogenicity as discussed previously [33]. No positively selected codons were found within the four antigenic sites indicating that these selections may not contribute to the evolution of the viral antigenicity.
Previous studies have proposed that the Arctic-related RABVs originate from dog RABVs of the Indian subcontinent lineage in South Asia; this viral ancestor then travelled north and dispersed among the reservoirs in the Arctic regions [4,10,11,12]. However, the ancestral locations and diffusion process estimation of AL RABVs by discrete phylogeography tells another story. Northeast Asia was inferred as the geographic origin of the AL RABVs and the tMRCA emerged approximately in 1834 (95% HPD 1783-1876) (Fig. 3). This result does not conflict with the previous findings as we did not include the RABV sequences from the Indian subcontinent lineage in our phylogeographic analysis. We only conducted the analysis on the geographic origin and migratory events of AL RABVs and thus, information on the origin and dispersal of the entire Arctic-related lineage cannot be obtained. Moreover, the migration between East Russia and India is not supported by the BF test and the translocation event between the two regions remains ambiguous.
Spatiotemporal diffusion process by SPREAD allowed us to reconstruct every history event within the kml visualization tool (Fig. 4). In approximately 1909, a single AL RABVs ancestor emerged in India followed by dispersal to the neighboring countries Nepal and Pakistan. At the same time, ancestor viruses in Russia spread to Japan (beginning from 1929 and reaching in 1945) and South Korea (1923-1963), then to West China (1910-1985), and finally to North China (1996-2004) and Mongolia (1951-2007). The dispersal of AL RABVs from South Asia to West Asia was observed via two transmission routes: one route was the India-Nepal-Iran route, and the other route involved transmission along the India-Pakistan-Afghanistan-Iraq route. After the 1990s, the South Asian countries, especially India, have witnessed a large number of AL RABVs cases; the viruses rapidly spread to Nepal, Bangladesh and Bhutan. Dispersal between India and Nepal, India and Bangladesh, and Bangladesh and Nepal, and between Bangladesh and Bhutan was also inferred with BF support. Interestingly, no viral exchange was observed between The South Asia countries and West China (Qinghai and Tibet provinces). We suggest that the natural barrier of high-altitude landscape between the two regions, Himalayas, halted the spread of the viruses. However, the long-distance dispersal between between India and Alaska was inferred by the BF tests; considering the low sample size (n = 2) and BF factor, this dispersal was not well supported, or it might be the result of human activities since animals in island country often circulated in relatively close ecosystem. Additionally, we included the N gene sequences of Komatsugawa, a Japanese dog isolate sampled in 1940s, in our dataset and the inferred sampling year is 1945. The inclusion of this sequence enabled us to trace the source of ancestral AL RABVs circulating within the South Korea to demonstrate that it was the result of incursions from Japan in agreement with the previous assumptions [5,34]. The Bayesian tip significance test provided significant PS and AI support for the existence of the phylogeographical structure (p = 0.00 for both tests) for AL RABVs indicating that most viruses were clustered by their geographical distributions; however, the MC statistic suggests that the isolates from Afghanistan (n = 6) received nonsignificant geographic structure support, which may be due to the fact that Afghanistan may have acted as an intermediate distribution source; frequent gene flow with other regions may have occurred considering the complexity of RABV epidemiology in Afghanistan [8]. To confirm this hypothesis, the phylogeographic analysis with additional sequences of this lineage in the neighboring countries is needed. Moreover, single-isolate countries were nonsignificant for phylogeny-geographic correlation analysis probably due to the insufficient number of samples for statistical tests.
Footnotes
Funding: This work was supported by the National Key Research and Development Program of China (2016YFD0501001) and a grant from the Science and Technology Bureau of Yantai City, China (2019XDHZ094).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Zhu H, Wen Y.
- Data curation: Yu X, Jiang L.
- Funding acquisition: Zhang X.
- Investigation: Yu X, Zhu H, Bo Y, Li Y.
- Resources: Wen Y.
- Software: Zhu H.
- Validation: Chen G.
- Visualization: Zhang J.
- Writing - original draft: Yu X.
- Writing - review & editing: Zhu H.
SUPPLEMENTARY MATERIALS
Animals and their registered information used in this study
Sequence information of the dataset 1 to 3 used in this study
Scores of 24 nucleotide substitution models inferred by MrModeltest v2.3
Amino acid sites under negative selection in nucleoprotein of Arctic-related RABVs
Amino acid sites under negative selection in nucleoprotein of Arctic-related RABVs
Amino acid sites under negative selection in nucleoprotein of Arctic-related rabies viruses
Amino acid sites under negative selection in nucleoprotein of Arctic-related rabies viruses
Full annotated maximum clade credibility tree for 155 partial N gene sequences of Arctic-like rabies viruses using the discrete phylogeography. Colors of the branches represent the most likely location state of the descendant nodes. The color codes for the 16 countries or regions are defined in the legend on the left. Divergence dates for the major branches and their 95% highest posterior density ranges are indicated above and below the tree nodes. Posterior of each node is colored according to the values as described above. Selected values in the major branches are shown as numbers.
References
- 1.King AM. Family-Rhabdoviridae. In: Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy. 1st ed. San Diego: Elsevier; 2012. pp. 686–713. [Google Scholar]
- 2.Bourhy H, Dautry-Varsat A, Hotez PJ, Salomon J. Rabies, still neglected after 125 years of vaccination. PLoS Negl Trop Dis. 2010;4(11):e839. doi: 10.1371/journal.pntd.0000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nadin-Davis SA, Real LA. Molecular phylogenetics of the lyssaviruses-insights from a coalescent approach. Adv Virus Res. 2011;79:203–238. doi: 10.1016/B978-0-12-387040-7.00011-1. [DOI] [PubMed] [Google Scholar]
- 4.Kuzmin IV, Hughes GJ, Botvinkin AD, Gribencha SG, Rupprecht CE. Arctic and Arctic-like rabies viruses: distribution, phylogeny and evolutionary history. Epidemiol Infect. 2008;136(4):509–519. doi: 10.1017/S095026880700903X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadin-Davis SA, Sheen M, Wandeler AI. Recent emergence of the Arctic rabies virus lineage. Virus Res. 2012;163(1):352–362. doi: 10.1016/j.virusres.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 6.Pant GR, Lavenir R, Wong FY, Certoma A, Larrous F, Bhatta DR, et al. Recent emergence and spread of an Arctic-related phylogenetic lineage of rabies virus in Nepal. PLoS Negl Trop Dis. 2013;7(11):e2560. doi: 10.1371/journal.pntd.0002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuzmin IV, Botvinkin AD, McElhinney LM, Smith JS, Orciari LA, Hughes GJ, et al. Molecular epidemiology of terrestrial rabies in the former Soviet Union. J Wildl Dis. 2004;40(4):617–631. doi: 10.7589/0090-3558-40.4.617. [DOI] [PubMed] [Google Scholar]
- 8.Horton DL, McElhinney LM, Freuling CM, Marston DA, Banyard AC, Goharrriz H, et al. Complex epidemiology of a zoonotic disease in a culturally diverse region: phylogeography of rabies virus in the Middle East. PLoS Negl Trop Dis. 2015;9(3):e0003569. doi: 10.1371/journal.pntd.0003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Real LA, Russell C, Waller L, Smith D, Childs J. Spatial dynamics and molecular ecology of North American rabies. J Hered. 2005;96(3):253–260. doi: 10.1093/jhered/esi031. [DOI] [PubMed] [Google Scholar]
- 10.Bourhy H, Reynes JM, Dunham EJ, Dacheux L, Larrous F, Huong VT, et al. The origin and phylogeography of dog rabies virus. J Gen Virol. 2008;89(Pt 11):2673–2681. doi: 10.1099/vir.0.2008/003913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadin-Davis SA, Turner G, Paul JP, Madhusudana SN, Wandeler AI. Emergence of Arctic-like rabies lineage in India. Emerg Infect Dis. 2007;13(1):111–116. doi: 10.3201/eid1301.060702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assenberg R, Delmas O, Morin B, Graham SC, De Lamballerie X, Laubert C, et al. Genomics and structure/function studies of Rhabdoviridae proteins involved in replication and transcription. Antiviral Res. 2010;87(2):149–161. doi: 10.1016/j.antiviral.2010.02.322. [DOI] [PubMed] [Google Scholar]
- 13.Jamil KM, Ahmed K, Hossain M, Matsumoto T, Ali MA, Hossain S, et al. Arctic-like rabies virus, Bangladesh. Emerg Infect Dis. 2012;18(12):2021–2024. doi: 10.3201/eid1812.120061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao XQ, Yan XJ, Luo GL, Zhang HL, Chai XL, Wang FX, et al. Genetic evidence for domestic raccoon dog rabies caused by Arctic-like rabies virus in Inner Mongolia, China. Epidemiol Infect. 2011;139(4):629–635. doi: 10.1017/S0950268810001263. [DOI] [PubMed] [Google Scholar]
- 15.Oem JK, Kim SH, Kim YH, Lee MH, Lee KK. Reemergence of rabies in the southern Han river region, Korea. J Wildl Dis. 2014;50(3):681–688. doi: 10.7589/2013-07-177. [DOI] [PubMed] [Google Scholar]
- 16.Oem JK, Kim SH, Kim YH, Lee MH, Lee KK. Complete genome sequences of three rabies viruses isolated from rabid raccoon dogs and a cow in Korea. Virus Genes. 2013;47(3):563–568. doi: 10.1007/s11262-013-0923-1. [DOI] [PubMed] [Google Scholar]
- 17.Oh SY, Kim SA, Kim JY, Yoo HS, Lee KK, Shin NS. Detection of antibodies against the rabies virus in Korean raccoon dogs (Nyctereutes procyonoides koreensis) J Zoo Wildl Med. 2012;43(1):174–176. doi: 10.1638/2011-0063.1. [DOI] [PubMed] [Google Scholar]
- 18.Tao XY, Guo ZY, Li H, Jiao WT, Shen XX, Zhu WY, et al. Rabies cases in the west of China have two distinct origins. PLoS Negl Trop Dis. 2015;9(10):e0004140. doi: 10.1371/journal.pntd.0004140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 21.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8(7):e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol. 2008;8(3):239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLOS Comput Biol. 2009;5(9):e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bielejec F, Rambaut A, Suchard MA, Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics. 2011;27(20):2910–2912. doi: 10.1093/bioinformatics/btr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arai YT. [Phylogenetic analysis of two rabies viruses, Takamen and Komatsugawa strains isolated in Japan in the 1940's] Kansenshogaku Zasshi. 2004;78(9):815–822. doi: 10.11150/kansenshogakuzasshi1970.78.815. [DOI] [PubMed] [Google Scholar]
- 26.Hyun BH, Lee KK, Kim IJ, Lee KW, Park HJ, Lee OS, et al. Molecular epidemiology of rabies virus isolates from South Korea. Virus Res. 2005;114(1-2):113–125. doi: 10.1016/j.virusres.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Tenzin WS, Wacharapluesadee S, Denduangboripant J, Dhand NK, Dorji R, Tshering D, et al. Rabies virus strains circulating in Bhutan: implications for control. Epidemiol Infect. 2011;139(10):1457–1462. doi: 10.1017/S0950268810002682. [DOI] [PubMed] [Google Scholar]
- 28.Tenzin SB, Sharma B, Dhand NK, Timsina N, Ward MP. Reemergence of rabies in Chhukha district, Bhutan, 2008. Emerg Infect Dis. 2010;16(12):1925–1930. doi: 10.3201/eid1612.100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho SY, Shapiro B. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol Ecol Resour. 2011;11(3):423–434. doi: 10.1111/j.1755-0998.2011.02988.x. [DOI] [PubMed] [Google Scholar]
- 30.Sironi M, Cagliani R, Forni D, Clerici M. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat Rev Genet. 2015;16(4):224–236. doi: 10.1038/nrg3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menezes R. Rabies in India. CMAJ. 2008;178(5):564–566. doi: 10.1503/cmaj.071488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao XY, Tang Q, Rayner S, Guo ZY, Li H, Lang SL, et al. Molecular phylodynamic analysis indicates lineage displacement occurred in Chinese rabies epidemics between 1949 to 2010. PLoS Negl Trop Dis. 2013;7(7):e2294. doi: 10.1371/journal.pntd.0002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu H, Chen X, Shao X, Ba H, Wang F, Wang H, et al. Characterization of a virulent dog-originated rabies virus affecting more than twenty fallow deer (Dama dama) in Inner Mongolia, China. Infect Genet Evol. 2015;31(4):127–134. doi: 10.1016/j.meegid.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 34.Yang DK, Shin EK, Oh YI, Kang HK, Lee KW, Cho SD, et al. Molecular epidemiology of rabies virus circulating in South Korea, 1998–2010. J Vet Med Sci. 2011;73(8):1077–1082. doi: 10.1292/jvms.10-0440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animals and their registered information used in this study
Sequence information of the dataset 1 to 3 used in this study
Scores of 24 nucleotide substitution models inferred by MrModeltest v2.3
Amino acid sites under negative selection in nucleoprotein of Arctic-related RABVs
Amino acid sites under negative selection in nucleoprotein of Arctic-related RABVs
Amino acid sites under negative selection in nucleoprotein of Arctic-related rabies viruses
Amino acid sites under negative selection in nucleoprotein of Arctic-related rabies viruses
Full annotated maximum clade credibility tree for 155 partial N gene sequences of Arctic-like rabies viruses using the discrete phylogeography. Colors of the branches represent the most likely location state of the descendant nodes. The color codes for the 16 countries or regions are defined in the legend on the left. Divergence dates for the major branches and their 95% highest posterior density ranges are indicated above and below the tree nodes. Posterior of each node is colored according to the values as described above. Selected values in the major branches are shown as numbers.