Abstract
Childhood obesity is a strong risk factor for adult obesity, type 2 diabetes, and cardiovascular disease. The mechanisms that link early adiposity with late-onset chronic diseases are poorly characterised. We developed a mouse model of early adiposity through litter size reduction. Mice reared in small litters (SLs) developed obesity, insulin resistance, and hepatic steatosis during adulthood. The liver played a major role in the development of the disease.
Objective
To gain insight into the molecular mechanisms that link early development and childhood obesity with adult hepatic steatosis and insulin resistance.
Methods
We analysed the hepatic transcriptome (Affymetrix) of control and SL mice to uncover potential pathways involved in the long-term programming of disease in our model.
Results
The circadian rhythm was the most significantly deregulated Gene Ontology term in the liver of adult SL mice. Several core clock genes, such as period 1–3 and cryptochrome 1–2, were altered in two-week-old SL mice and remained altered throughout their life course until they reached 4–6 months of age. Defective circadian rhythm was restricted to the periphery since the expression of clock genes in the hypothalamus, the central pacemaker, was normal. The period-cryptochrome genes were primarily entrained by dietary signals. Hence, restricting food availability during the light cycle only uncoupled the central rhythm from the peripheral and completely normalised hepatic triglyceride content in adult SL mice. This effect was accompanied by better re-alignment of the hepatic period genes, suggesting that they might have played a causal role in mediating hepatic steatosis in the adult SL mice. Functional downregulation of Per2 in hepatocytes in vitro confirmed that the period genes regulated lipid-related genes in part through peroxisome proliferator-activated receptor alpha (Ppara).
Conclusions
The hepatic circadian rhythm matures during early development, from birth to postnatal day 30. Hence, nutritional challenges during early life may misalign the hepatic circadian rhythm and secondarily lead to metabolic derangements. Specific time-restricted feeding interventions improve metabolic health in the context of childhood obesity by partially re-aligning the peripheral circadian rhythm.
Keywords: Developmental origins of health and disease (DOHaD), Childhood obesity, Circadian rhythm, Litter size reduction, Time-restricted feeding (TRF), Non-alcoholic fatty liver disease (NAFLD)
Highlights
-
•
Relative neonatal overfeeding rapidly misaligned the hepatic circadian rhythm of 2-week old mice.
-
•
The hepatic circadian clock remained altered throughout adulthood and contributed to alterations in lipid metabolic genes.
-
•
Time-restricted feeding reduced adiposity and normalised hepatic triglyceride content of adult SL mice.
-
•
These improvements were accomplished, in part, through re-alignments of the hepatic circadian rhythm.
1. Introduction
According to the World Health Organisation, “childhood obesity is one of the most serious public health challenges of the twenty-first century” [1]. In 2016, more than 41 million children under the age of five years were overweight or obese. Obesity is a major risk factor for other chronic diseases, including type 2 diabetes (T2D), cardiovascular disease, kidney failure, and some types of cancer [[1], [2], [3], [4], [5], [6], [7]], which shorten the lifespan of individuals [8].
The developmental origins of health and disease (DOHaD) hypothesis suggests that early environmental cues, including nutrition, induce long-term physiological effects by permanently altering the expression of target genes [9]. Childhood overweight/obesity can be considered a part of the DOHaD paradigm because it is strongly associated with late-onset metabolic derangements. The molecular mechanisms that link metabolic adaptations to childhood overweight/obesity with long-term metabolic diseases are poorly characterised. Litter size reduction in mice and rats has been widely used as a model of childhood obesity and neonatal developmental programming of adult disease (reviewed in [10,11]). Briefly, rodents raised in small litters (SLs) exhibit rapid weight gain and late-onset metabolic disorders, including obesity, insulin resistance, impaired glucose tolerance, hepatic steatosis, and renal and cardio-metabolic defects. These long-term metabolic derangements can be due to alterations in either the central nervous system typically leading to hyperphagia [[12], [13], [14], [15]] or in peripheral tissues [[16], [17], [18]].
We developed a mouse model of developmental programming and early adiposity (i.e. childhood obesity) by litter size reduction [19,20] (Figure 1A). Mice reared in SLs showed transient hyperphagia, augmented lipid intake, and increased adiposity as early as 2 weeks of age. Furthermore, the SL mice developed several features of metabolic syndrome later in life, including obesity, hepatic steatosis, insulin resistance, and glucose intolerance [19]. The liver played a major role in driving early onset insulin resistance [19,20]. This is an excellent model of the disease because, similar to human pathophysiology, developmental metabolic derangements associated to early adiposity in mice also lead to late-onset metabolic derangements.
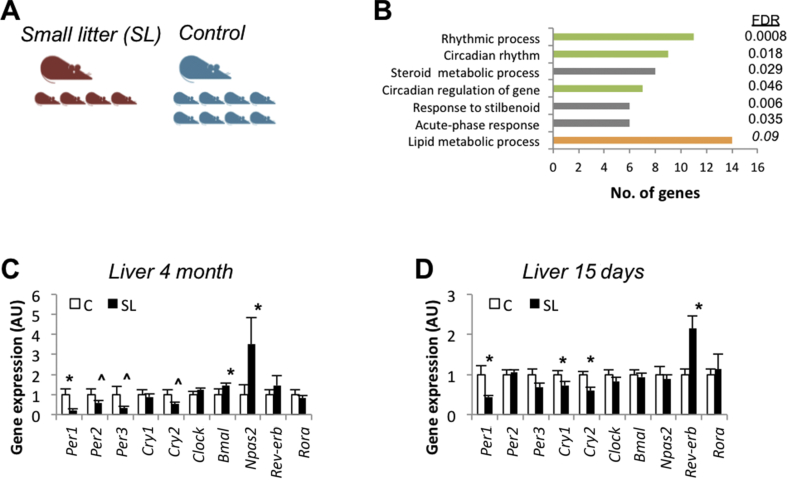
Figure 1.
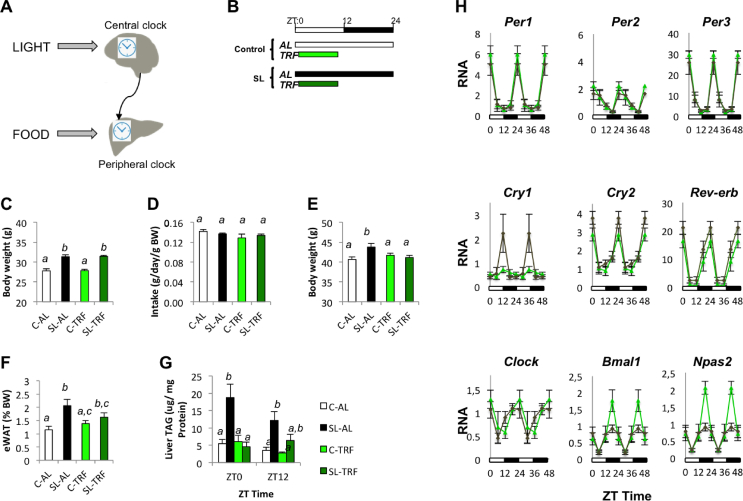
Early overfeeding altered the hepatic circadian clock in young and adult mice. (A) Mouse model of early adiposity by litter size reduction. Litter size was adjusted at birth to eight pups (control group, C) or four pups per dam (small litter group, SL). (B) David functional clustering enrichment analysis from hepatic gene expression (Affymetrix). The analysis included genes showing statistically significant expression between 4-month-old C and SL mice. Clock genes mRNA levels/expression (qPCR) in the liver of 4-month-old C and SL mice (C) and 15-day-old mice (D). Data represent means ± S.E.M. ∗p < 0.05 vs C mice. Student's t test.
The circadian rhythm is an evolutionarily conserved system that helps organisms anticipate predictable environmental changes by generating near 24-h rhythms in behaviour and physiology. At the molecular level, the core components of the clock system are the transcription factors circadian locomotor output cycles kaput (Clock), neuronal PAS domain protein 2 (Npas2), and brain and muscle aryl hydrocarbon receptor nuclear translocator-like protein 1 (Bmal1) (Fig. S1A). Clock heterodimerises with Bmal1 or its brain homolog Npas2 and regulates the transcription of their targets, including the period (Per1, Per2, and Per3) and cryptochrome genes (Cry1 and Cry2). In turn, Per1-3 and Cry1-2 rhythmically accumulate and form a repressor complex that interacts with Clock/Bmal1 to inhibit their transcription [21]. Other genes, including the nuclear receptor subfamily 1 group D member 1 (Nrd1d1, also known as Rev-erb) and RAR-related orphan receptor alpha (Rora), play additional roles in the circadian gene network [22]. The purpose of the circadian circuitry is to produce rhythms in frame with external cues (light/dark [LD] and feeding/fasting cycles) to maintain energy homeostasis. In accordance, it has been shown that alterations in circadian systems may contribute to the development of metabolic syndrome and obesity-related complications in humans [[23], [24], [25], [26]]. Furthermore, genetic ablation of core components of the clock system supports a causal role of the circadian rhythm in mediating metabolic disorders, including hyperglycaemia, hyperinsulinemia, and impaired lipid metabolism [[27], [28], [29]].
In this study, we aimed to shed light on the mechanism/pathways that may link early overfeeding-associated developmental programming with late-onset liver dysfunction in our model. Specifically, we conducted an unbiased transcriptomic analysis of the livers of the control and SL mice. We found that the circadian rhythm ranked as the most significantly deregulated Gene Ontology term (GO: 0007623). Herein, we provide evidence that (1) early overfeeding rapidly misaligns the hepatic circadian clock, (2) which remains altered throughout the life course, and (3) this misalignment precedes hepatic steatosis and insulin resistance in our model in part through a Ppara/Rora-dependent mechanism.
2. Materials and methods
2.1. Animal care and experimental design
Protocols were approved by the Universitat de Barcelona Animal Care and Use Committee. ICR mouse strain (ICR-CD1, Envigo Laboratories, Barcelona, Spain) was chosen for this study based on its fast-somatic growth, especially during the neonatal period. Mouse breeding was conducted as previously described [20]. Briefly, at birth, the litter size was adjusted to eight pups (control group, C) or four pups per dam (small litter group, SL). At weaning, all mice had free access to standard chow (2014 Tekland Global, Harlan Iberica, Barcelona, Spain) and water. In this study, metabolic analysis was conducted in males only because SL females were protected against hepatic steatosis-hepatic insulin resistance [19]. Food intake was recorded from 2-month-old individual mice every 24 h and the weight difference was a measure of daily food intake. Average food intake is presented as g/day/BW at each week over the course of 4 weeks. The mice were maintained under constant temperature (21–23 °C) and humidity (55 ± 10%).
2.2. Monitoring motor activity/locomotor activity rhythms
Motor activity was measured in 2-month-old C and SL mice during 30 consecutive days by using activity meters that used two perpendicular crossed infrared beams situated 3 cm above the floor of the cage. Each beam interruption represented an activity count that was registered and compiled in 15-min intervals, obtaining a motor activity time series. Light dark cycles were recorded simultaneously to the activity in a separate channel connected to a photocell pulse generator. The mice were first maintained on a 12:12-h light/dark (LD) cycle for 10 days and later on constant darkness (DD) for 20 days. Data from the first 3 days were not used for the analysis. The mice had ad libitum access to water and a standard chow diet. Motor activity data were first visually inspected by double-plotted graphs. Temporal series of motor activity and light intensity were analysed using El Temps (v293) (Diez-Noguera, University of Barcelona, Spain, www.el-temps.com), an integrated package for chronobiological analysis. Periodogram and spectral analyses among others were used to analyse the activity rhythms.
2.3. Indirect calorimetry
Indirect calorimetry was carried out using a 16-chamber Phenomaster monitoring system (TSE Systems GmbH, Bad Homburg, Germany). Individually caged mice were acclimated in the measuring room for three days before the onset of the experiments. During the experiments, a 12-h light–dark cycle (lights on at 8:00 am) was kept and the room temperature was maintained at 22 + 2 °C. Food and water were available ad libitum. Oxygen consumption and CO2 production were directly measured over a period of 48–72 h, and the respiratory exchange ratio (RER) and energy expenditure were calculated from these values. Locomotor activity was monitored by counting infrared photocell beam breaks on the X and Y axes.
2.4. Time-restricted feeding (TRF)
We conducted a dietary intervention to decouple the central from peripheral clocks. Specifically, the liver is entrained by light:dark cues through the hypothalamus and by nutritional cues. Importantly, mice are crepuscular and consume >80% of their food during the dark phase of the light–dark cycle. Hence, we forced them to consume all their calories during the light phase. Two-month-old C and SL mice were randomly assigned to ad libitum (AL) or time-restricted feeding (TRF) groups. The mice included in the TRF protocol had access to a chow diet for 8 h per day during the light cycle, from Zeitgeber time 1 (ZT1, 9 am) to ZT9 (5 pm). ZT0 was lights on (8 am) and ZT12 was lights off (8 pm). Food access was controlled via programmable automatic feeders (SmartWaiter, Cibertec, Madrid, Spain). The mice were euthanised 4 weeks after commencing the TRF intervention. The liver was weighted, rapidly frozen in liquid nitrogen, and stored at −80 °C for further analyses (qPCR and TAG content).
2.5. Epididymal fat mass assessment
Fat mass was determined in 4-month-old mice. Epididymal fat depots were dissected and fat mass was calculated as a percentage of wet tissue per whole-body weight.
2.6. Liver TAG analysis
A total of 100 mg of frozen liver sample was taken and homogenised in 500 ul of SDS 0.1% using Next Advance Bullet Blender Storm 24. Then, 350 ul was mixed with 350 ul of methanol. Equal volumes of chloroform were added and the samples were chilled for 30 min in ice. Next, 48 ul of KCl 0.5 M was added and maintained for 30 min on ice. The samples were centrifuged (10 min at 2000 rpm at 4 °C), the SN was discarded, and 300 ul of TAG (lower part) was left to evaporate O/N under the hood. The pellets were then mixed in 50 ul of pure EtOH. Liver TAG was measured using specific kits from Sigma–Aldrich (Madrid, Spain): free glycerol reagent (#F6428-40ML), triglyceride reagent (#T2449-10ML), and glycerol standard solution (#G7793-5ML). All samples were normalised by the protein content using Pierce BCA (Thermo Fisher Scientific, Madrid, Spain).
2.7. Microarray analysis
Microarray hybridisation and analysis were performed as previously described using GeneChip Affymetrix Mouse 430 2.0 whole genome arrays [29]. Briefly, 3 microarrays were hybridised for each group (control and SL). Each array contained the pooled RNA from three independent mice. Expression values were summarised after background correction and normalisation steps using the RMA methodology [30]. Differential expression analysis was conducted using the non-parametric Rank Prod approach [31]. Oligonucleotides presenting changes between groups with q values lower than 0.1 were considered significant. The David tool [32] was used to calculate the functional clustering enrichment statistical analysis of the Gene Ontology terms and Kegg pathways databases considering the list of significant genes. The data were deposited at the GEO (accession number GSE55304).
2.8. Real-time quantitative PCR (qPCR)
Total RNA was isolated from previously frozen livers (TRIzol, Sigma–Aldrich, Madrid, Spain) of 8-h fasted 4-month-old mice or randomly fed 15-day-old mice and used for cDNA synthesis (Promega). Transcript levels were then quantified by qPCR using SYBR Green PCR Master Mix (Promega). Results of the respective genes of interest were normalised to b-actin and subsequently median normalised to 1. The list of primers used is detailed in Supplementary Table 1.
2.9. Cell culture
The murine hepatocarcinoma Hepa1-6 cell line (ATCC CRL-1830) was used to test Per1-3 shRNA and later perform qPCR expression analyses. Cells were maintained in DMEM supplemented with penicillin (100 units/ml), streptomycin (100 ug/ml), l-glutamine (200 mM), and 10% FBS (Sigma–Aldrich, Steinheim, Germany) at 37 °C and 5% CO2. The cells were sub-cultured at 90% confluence.
2.10. Adenoviral infections (shRNA)
Adenoviral vectors were purchased from Vector Biolabs (Malvern, PA, USA). The viral vectors contained short hairpin RNAs to inhibit the specific expression of Per1 (Ad-GFP-U6-mPER1-shRNA) and Per2 (Ad-GFP-U6-mPER2-shRNA). An adenoviral vector containing a scramble shRNA sequence with the co-expression of green fluorescent protein (GFP) was used as a control (Ad-GFP-U6-shRNA). Next, 80% confluent cells were infected with the corresponding vector at a multiplicity of infection of 10 in DMEM without serum for 2 h. These conditions led to 80–90% viral transduction as assessed by GFP fluorescence. After infection, the cells were washed with PBS and maintained for 48 h in growth media supplemented with FBS (10%). At the end of incubation, the cells were washed, RNA was extracted (TRIzol), and the RNA concentration was analysed by qPCR as previously described.
2.11. Circadian rhythm analysis (cosinor method)
Liver RNA samples from the C and SL mice were collected at different ZT times along the 24-h cycle and analysed. The number of mice at each time point in the experiments is provided in the figure legends. The presence of rhythmicity in each time series of data was statistically evaluated by adjusting the data to a 24-h sinusoidal pattern and significance was tested using the cosinor method [34,35] and F test of sinusoidality [35].
2.12. In silico identification of transcription factors
We first analysed the putative binding sites for transcription factors in the promoter regions of lipid-related genes. Briefly, we obtained the nucleotide sequence of 1 kb upstream of the transcription start site of the genes (UCSC Genome Browser; https://genome.ucsc.edu/). We imported the sequence (FASTA format) into the MEME Suite to elucidate the regions containing putative motifs for clock genes (http://meme-suite.org/).
2.13. Statistical analysis
The results are expressed as mean ± SEM. Statistical analysis was performed using a two-tailed t test or one-way ANOVA as indicated (IBM SPSS Statistics 19, Madrid, Spain). A ∗p value < 0.05 and ∗∗∗p value < 0.001 was considered significant. The rhythm variables derived from the cosinor analysis, amplitude (difference between the mesor and peak), and acrophase (clock time of the peak value) were also evaluated. Differences between two rhythms were tested by comparisons of cosinors [35]. Indirect calorimetry experiments were analysed by two-way ANOVA followed by Bonferroni's post hoc test.
3. Results
3.1. Neonatal relative overfeeding induced early and sustained misalignment of hepatic clock genes
We developed a mouse model of early adiposity (childhood obesity) and adult metabolic dysfunction by litter size reduction (Figure 1A) [19]. In this study, we aimed to gain further insight into the molecular mechanisms that link developmental adaptations that are related to early adiposity with adult insulin resistance/glucose intolerance. We previously showed that the liver is the primary mediator of whole-body metabolic dysfunction in adult SL mice. Therefore, we used an unbiased approach and analysed the hepatic gene expression profile in 4-month-old control and SL mice (Affymetrix). Overall, 245 annotated genes were differentially expressed in the liver of the adult SL mice (Table S2), with the circadian rhythm being the Gene Ontology term with the highest significance (David enrichment analysis) (Figure 1B). The genes included in this ontology (Table S3) encompassed several core Clock genes (Per1, Per3, and Npas2) and Clock Output genes (Rev-erb and Dbp) (Fig. S1A).
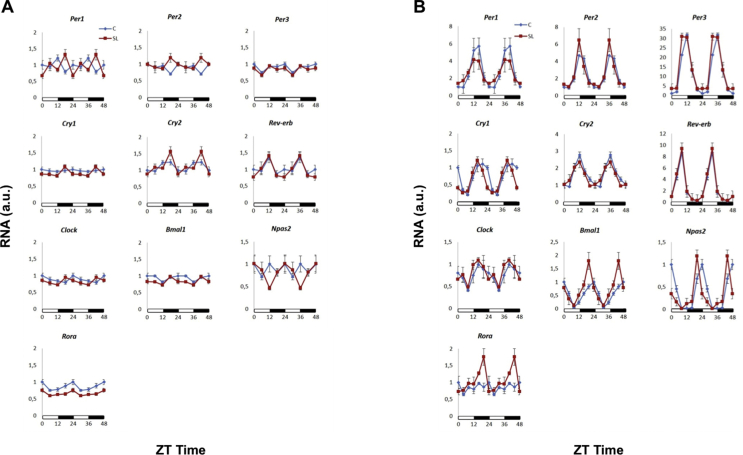
We then confirmed (qPCR) that the expression of some clock genes, namely the period and cryptochrome genes, was altered in the livers of the adult SL mice (Figure 1C). In contrast, the expression of clock genes remained largely normal in other tissues, including the hypothalamus and white adipose tissue (Fig. S1B and C). We previously reported that young SL mice developed hepatic insulin resistance by 2 weeks of age [19]. Hence, we measured the expression of clock genes in the liver of 2-week-old mice (qPCR) and found that Per1, Cry1, Cry2, and Rev-erb were already altered at this developmental stage (Figure 1D). Again, no changes were observed in the hypothalamus of the young SL mice (Fig. S1D). These data were acquired between 9 and 11 am (ZT1-ZT3) under static conditions. We then analysed the expression of clock genes during a 24-h cycle (Figure 2 and Table S4). At postnatal day 15, only a few clock genes showed rhythmicity in both the control and SL mice. Specifically, in the control mice, significant rhythms (p < 0.05) were detected only for Per3, Cry2, Bmal1, and Rora (Figure 2A and Table S4). In contrast, Cry1, Rev-erb, Bmal1, Clock, and Npas2 showed significant rhythmic behaviour in the young SL mice. Thus, by age 15 days, the livers of the SL mice showed an almost opposite behaviour compared to those of the controls. The genes that already cycled in the young C mice did not cycle in the SL mice and vice versa. This difference greatly dissipated by 4 months. All the clock genes showed rhythmicity in both groups (Figure 2B and Table S4). Although the mean acrophases and amplitudes of Per1, Per2, Cry2, Reverb, and Npas2 tended to be lower in the SL mice, they were not statistically significant among the groups.
Figure 2.
mRNA expression levels of clock genes during a 24-h cycle in control (blue) and SL (red) (A) 15-day-old mice and (B) 4-month-old mice. The graphs represent double plots in which the 24-h cycles are plotted twice to provide a better graphical view of the rhythmic behaviour.
These data suggested that SL-derived developmental programming induced rapid and sustained deregulation of clock genes, namely the period and cryptochrome genes. Several studies have demonstrated that the circadian rhythm plays a causal role in mediating metabolic dysfunction, including altered hepatic lipid metabolism [[37], [38], [39]]. Therefore, we subsequently determined whether the clock genes might also play a role in the development of hepatic steatosis and whole-body metabolic dysfunction in our model.
3.2. Clock genes regulated hepatic lipid metabolism in the SL mice
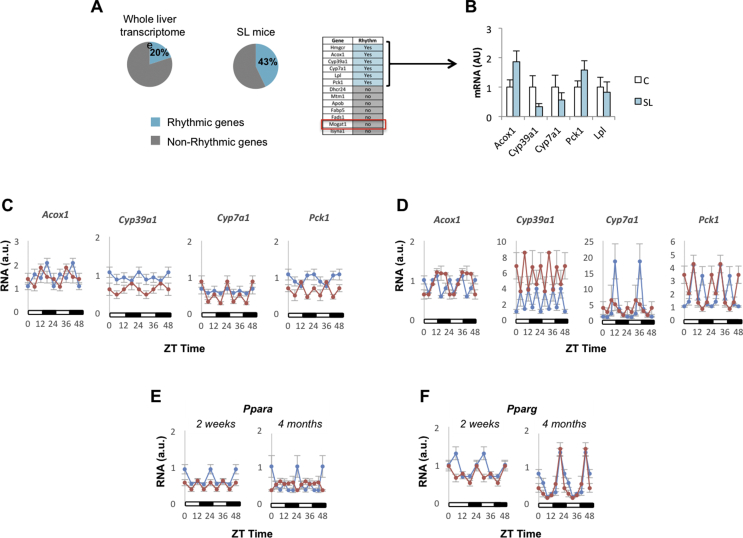
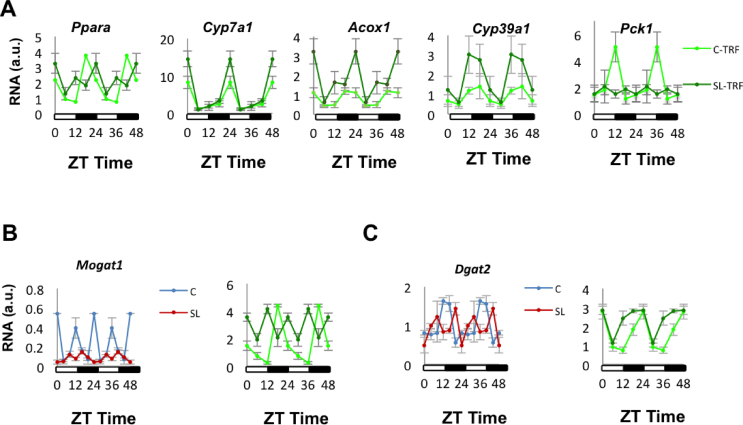
First, we found that the lipid metabolic process was significant in our enrichment study (Figure 1B and Table S4). It has been reported that 15–20% of the liver transcriptome shows some degree of rhythmicity [[38], [39], [40], [41], [42]] (Figure 3A, left panel). We estimated how many genes in the lipid metabolic ontology process exhibited rhythmic behaviour using the CircadiOmics database (http://circadiomics.ics.uci.edu) [42,43]. This database contains more than 230 high-throughput omic datasets corresponding to “over 74 million measurements sampled over 24-h cycles” [43]. Information about the oscillatory trajectory of specific genes, in particular tissues and species, can be visualised using a comprehensive web server. This information includes periodicity statistics (e.g. period, amplitude, phase, P value, and q value) and oscillatory trajectories. We found that 40% of the genes included in the lipid metabolic term showed rhythmic behaviour in the mouse liver (Figure 3A, right panel). This over-representation (enrichment) suggested that the circadian clock might have been involved, at least in part, in regulating hepatic lipid metabolism in our model. We next confirmed (qPCR) that the expression of oscillatory lipid-related genes, including Acox1, Cyp39a1, Cyp7a1, and Pck1, was altered in the liver of the adult SL mice (Figure 3B). Their rhythmic behaviour was misaligned in the young (Figure 3C) and adult SL mice (Figure 3D).
Figure 3.
The lipid metabolism process Gene Ontology term was significantly enriched in genes that showed rhythmic behaviour. (A) Left panel: This graph describes the approximate number of genes that exhibit oscillatory behaviour in the liver. The graph was constructed based on available published information [[39], [40], [41], [42]]. Right panel: The diagram represents the genes from our data set that according to the information included in the CircadiOmics database (http://circadiomics.ics.uci.edu) show hepatic rhythmicity in rodents. (B) Comparison of SL oscillatory lipid-related genes mRNA levels/expression (qPCR) in 4-month-old liver of C and SL mice. (C–D) Oscillatory lipid-related genes mRNA expression levels in 15-day-old (C) and 4-month-old mice (D) Control and SL liver during a 24-h cycle. (E)Ppara and (F)Pparg expression (qPCR) during a 24-h cycle. Data represent means ± S.E.M. ∗p < 0.05 vs C mice; ˆp < 0.1 vs C mice. Student's t test.
How are these lipid-related genes linked to the circadian clock? In silico analysis of their promoter region (encompassing 1 kb upstream of the transcription start site) showed no consensus-binding motifs for the core clock genes (Fig. S2A). Instead, clock output nuclear transcription factors, namely Rev-erb and to a lesser extent Rora, have putative binding sites for most of them. Although Rev-erb could explain the altered expression of the lipid-related genes, it did not appear dramatically deregulated in the adult SL mice. The KEGG pathway enrichment analysis indicated that PPAR signalling (mmu03320:PPAR signalling pathway) might be a potential upstream regulator of lipid-related genes (Fig. S3). Notably, some of them (Pck1, Cyp7a1, Cyp39a1, and Acox1) are actually PPAR targets [41]. PPARs are transcription factors that link the circadian rhythm with the regulation of metabolism [[42], [43], [44], [45]]. Ppara expression was altered in the liver of the 15-day-old SL mice and worsened with age (Figure 3E). In contrast, Pparg was fairly normal in the adult mice (Figure 3F). These data suggested that the misexpression of the lipid-related genes might have been due to the complex interaction of their upstream nuclear receptors, including Ppara, Rev-erb, and Rora.
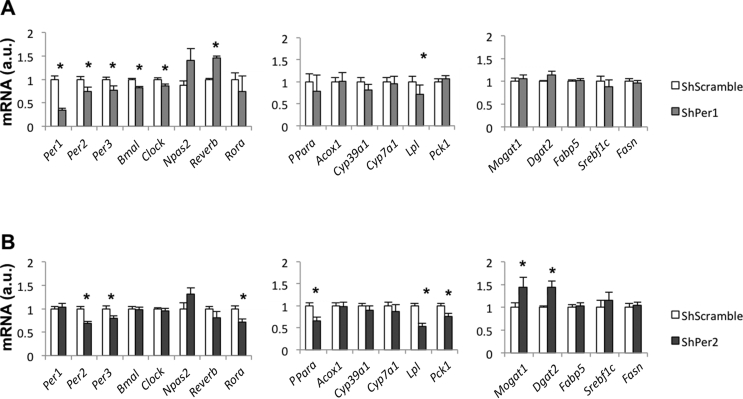
Integrating these data with the previous finding that the period genes were primarily altered in our model (Figure 1), it is tempting to speculate that the clock genes, namely Per1-3, might influence the expression of lipid-related genes, possibly through coordinated action of Rora/Ppara. To prove a causal relationship between the period genes and lipid metabolism, we selectively inhibited the expression of Per1 and Per2 in Hepa1-6 cells using adenoviral vectors carrying specific shRNA constructs. We confirmed that Ad-shPer1 reduced Per1 mRNA content by 60% (Figure 4A). Likewise, Per2 and Per3 mRNA contents were also moderately reduced (20–30%). Per1 downregulation caused a major impact on the expression of the core clock genes: Per2, Per3, Clock, and Bmal1 decreased, whereas Rev-erb was significantly elevated (Figure 4A). In contrast, Per1 downregulation did not influence Ppara and its downstream targets (Figure 4A). However, Ad-shPer2 effectively reduced Per2 mRNA content by 40% (Figure 4B). Unlike Per1 inhibition, Per2 downregulation did not impact other clock genes, but reduced the expression of Rora-Ppara and some Ppara downstream targets, such as Lpl and Pck1 (Figure 4B). We previously reported that, in SL mice, hepatic lipid accumulation was largely attributable to an increase in the expression of key genes involved in free fatty acid esterification, including Mogat1 and Dgat2 (Figure 3A) [19]. In the present study, we found that Ad-shPer2, but not Ad-shPer1, significantly increased the expression of both of these genes (Figure 4B). Our data suggested that the period genes likely influenced the deregulation of the clock-core system and hepatic lipid-related pathways. However, proof of direct regulation (via ChIP-qPCR or similar approaches) is currently lacking. This relationship clearly deserves investigation. We then addressed whether liver-specific changes in the clock-component genes influenced whole-body rhythmicity.
Figure 4.
(A–B) Circadian clock and hepatic lipid-related mRNA levels/expression in Hepa-1c cells after infection with adenoviral vectors carrying specific Per1-shRNA (A) and Per2-shRNA (B) constructs. Data represent means ± S.E.M. ∗p < 0.05 vs Scramble-shRNA Hepa1-6 infected cells. Student's t test.
3.3. Endogenous peripheral locomotor activity was misaligned in the adult SL mice
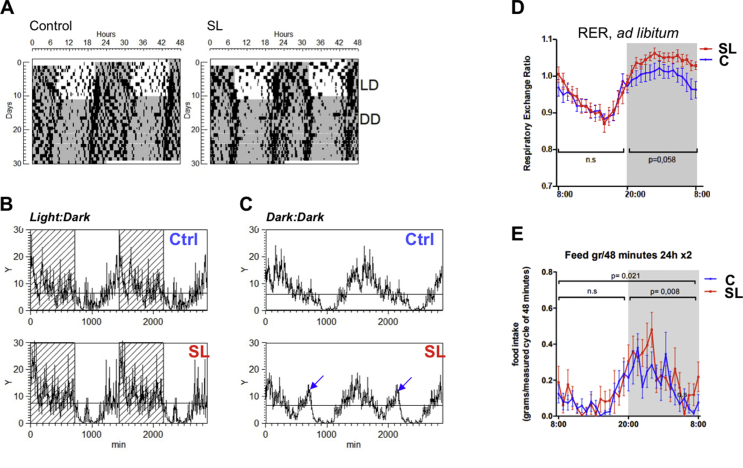
We analysed the locomotor activity of the adult control and SL mice under a standard LD 12-h:12-h cycle (Figure 5A). Under standard LD conditions, the control and SL mice showed no differences in period, amplitude, or mean motor activity (Figure 5B). Likewise, during the LD cycle, fasting or ad libitum feeding did not significantly influence SL locomotor activity (Fig. S4A and B). However, under constant dark conditions (DD), the pattern of the activity rhythm was significantly different among the groups (Figure 5C). Specifically, the SL mice showed a bimodal pattern (12-h rhythm). Of note, the bimodal pattern was quantified using the power of the second harmonic of a spectral analysis. The power of the 12-h rhythm (second harmonic) of the spectral analysis was higher in the SL group than in the control groups only in DD stage but not under LD.
Figure 5.
(A) Locomotor activity measurements of 2-month-old C and SL mice under standard light/dark (LD) 12-h:12-h cycles or constant darkness for 21 consecutive days (DD). (B) Pattern of locomotor activity of C and SL mice under LD conditions. (C) Pattern of locomotor activity of C and SL mice under constant dark conditions (DD). Note the bimodal rhythmicity in SL mice (blue arrows). Measurements are means of 21 consecutive days. (D) Respiratory exchange ratio (RER) representation, which reflects the glucose-to-lipid oxidation ratio of C and SL mice during 12-h light–dark cycle upon ad libitum conditions. (E) Feeding behaviour that represents food intake registered every 48 min through a 24-h cycle. Data represent means ± S.E.M of animals from the same group. (D–E) A two-way ANOVA analysis was used to test the effect of the animal model (C and SL) on RER and feeding behaviour during day and light periods. Significance at p < 0.05 represented in graphs corresponds to animal model variables, whereas no significant interaction effects were found in the analysed parameters.
Although we did not find differences in activity during standard LD conditions, some markers of whole-body energy metabolism showed modifications during a specific period of the day. For example, the respiratory exchange ratio (RER), which reflects the glucose-to-lipid oxidation ratio, was moderately elevated in the ad libitum-fed SL mice during the dark phase (Figure 5D). This effect was accompanied by increased food intake during the dark phase of the 24-h cycle (Figure 5E). Furthermore, oxygen consumption and energy expenditure (EE) were significantly reduced in the ad libitum-fed SL mice during the light cycle (Fig. S4C and D). Upon fasting, both the control and SL mice showed an appropriate decrease in RER, although a minor transient difference was observed during the LD transition among the groups (Fig. S4E). Likewise, oxygen consumption and EE tended to be lower in the fasted SL mice than in the control mice, (Fig. S4F and G). These differences were in concordance with the obese phenotype of SL mice.
Together, the previous data strongly suggested that there was an intrinsic metabolic alteration affecting different phases of the daily light–dark oscillations in the adult SL mice. This desynchrony was primarily attributable to the periphery, since the SL mice maintained normal locomotor activity under standard LD conditions. During the LD, the hypothalamus was able to synchronise whole-body rhythmicity. However, differences in activity under free running conditions (DD) were likely due to intrinsic alterations in peripheral tissues. This hypothesis was supported by the previous expression data (Figure 1), which indicated the liver as the organ that is likely involved in these effects.
3.4. Time-restricted feeding (TRF) re-aligned clock-controlled genes and improved lipid metabolism in the adult SL mice
In mammals, the hepatic clock is entrained by (1) nutritional cues, such as fasting/feeding or high-fat feeding, and (2) light via the central pacemaker (Figure 6A). Hence, we reasoned that restricting food availability during a specific period of time (TRF) might re-programme the hepatic circadian clock and improve the overall physiology of the adult SL mice. We designed a dietary intervention to uncouple the LD cycle from the nutritional cues. Specifically, the mice included in the TRF intervention had access to the diet from 9 am to 5 pm (ZT1-ZT9) (Figure 6B). During this 8-h window, all mice had unrestricted ad libitum access to food.
Figure 6.
(A) Schematic representation of the interplay between the central clock in the hypothalamus and peripheral clocks. The central clock is entrained by the light/dark cues through the retina. The hepatic clock is entrained by hypothalamic-derived signals and nutritional cues. (B) TRF intervention. Two-month-old mice had access to a chow diet ad libitum (AL) or for 8 h per day during the light cycle (TRF) from Zeitgeber time (ZT) 1 (9 am) to ZT time 9 (5 pm) for 4 consecutive weeks. (C) Body weight at the beginning of the dietary intervention. (D) Average of food intake measurements (g/day/g body weight) during the 4 weeks of TRF intervention. (E) Final body weight of C and SL mice subjected to ad libitum or TRF at the end of the intervention. (F) Fat mass (epididymal white adipose tissue, eWAT) at the end of the TRF intervention. (G) Means of hepatic lipid content (TAG levels) at ZT0 and ZT12 in C and SL mice. (H) mRNA expression levels of clock genes during a 24-h cycle in control-TRF (light green) and SL-TRF (dark green) at the end of the 4-week intervention. Data represent means ± S.E.M of all the animals from the same group. Samples were subjected to a two-way ANOVA analysis in which discordant vowels denote statistical significance.
At the beginning of the intervention, the SL mice were heavier than the controls and all of the mice were randomly distributed into 4 groups: C-AL, SL-AL, C-TRF, and SL-TRF (Figure 6C). As described by others [48], the average food intake was similar among the 4 groups (Figure 6D). However, despite this, the SL-TRF mice stayed leaner than the SL-AL mice and adiposity diminished, although this was not statistically significant (Figure 6E,F). More strikingly, the intervention restored the hepatic lipid content to that of the control mice (Figure 6G). These data indicated that TRF was an efficient method to improve hepatic lipid homeostasis.
We postulated that TRF-mediated improvements in hepatic lipid homeostasis were partly mediated through the re-programming of peripheral clocks. First, the intervention clearly shifted the acrophase estimations of Per1, Per2, Per3, Cry2, and Reverb by 6–12 h compared with the rhythms under ad libitum feeding (Fig. S5 and Table S4). Intriguingly, these were the genes that showed reduced amplitudes in the SL-AL mice compared to the controls (Figure 2B). TRF shortened the amplitude differences between the C-TRF and SL-TRF mice, with Reverb significantly surpassing the control-TRF (p < 0.05) (Figure 6H and Table S4). We then explored whether TRF also modified the rhythmicity of Ppara and its downstream targets (Figure 7). We found that the rhythmicity of Ppara and some of its downstream targets, including Cyp7a1, Cyp39a1, Pck1, and Acox1, largely improved in the SL-TRF mice as compared to that in the SL-AL mice (Figure 7A). We also measured the expression of Mogat1 and Dgat2. They appeared extremely altered in the liver of the SL-AL mice (Figure 7B,C). The nutritional intervention partially restored the cycling of both these genes in the liver of the SL-TRF mice (Figure 7B,C). In summary, these data indicated that TRF might have restored hepatic lipid content at least in part by re-aligning clock-mediated Ppara downstream lipid targets.
Figure 7.
(A) mRNA expression levels of oscillatory lipid-related genes. (B)Mogat1 expression during a 24-h cycle in the liver of mice fed ad libitum (left panel) or subjected to TRF (right panel). (C)Dgat2 expression during a 24-h cycle in the liver of mice fed ad libitum (left panel) or subjected to TRF (right panel). Data represent means ± S.E.M of all the animals from the same group.
4. Discussion
We previously developed a mouse model of early adiposity (childhood obesity) through litter size reduction [19,20]. Mice reared in SLs developed metabolic disorders during adulthood, including obesity, hepatic steatosis, insulin resistance, and glucose intolerance [19,20]. The liver played a major role in driving these derangements. Indeed, young SL mice developed hepatic insulin resistance by 2 weeks of age and fatty liver by a month. These metabolic imbalances persisted until adulthood (4–6 months).
In the present study, we sought to gain further insight into the molecular mechanisms involved in the development and long-term maintenance of hepatic dysfunction. To this end, we performed an unbiased transcriptomic analysis using microarrays (Affymetrix) of the livers of the adult control and SL mice. While other techniques, such as RNA-Seq, are more versatile and offer the possibility of exploring wider ranges of RNA abundance in biological samples, microarrays are excellent tools to uncover novel unexpected pathways involved in specific physiological processes. Strikingly, we found that the circadian rhythm Gene Ontology term was significantly deregulated in our model. We confirmed that the expression of several hepatic core clock genes, namely Per1-3 and Cry1-2, was already altered by 2 weeks of age and remained as such until adulthood (4–6 months). Our data indicated that, in SL mice, the circadian clock might have played a causal role in mediating hepatic steatosis by influencing in part the expression of lipid-related genes in a Ppara-dependent manner.
The circadian system is an endogenous regulatory system that helps organisms anticipate predictable environmental changes by generating near 24-h rhythms in behaviour and physiology [[21], [22], [23]]. In humans, alterations of the circadian rhythm have been associated with chronic diseases, including obesity and T2D [25,41,50,51]. Genetic manipulation in rodents confirmed a causal role of the circadian rhythm in mediating metabolic derangements. For example, ablation of Clock/Bmal1, Per1-3, or Rev-erb leads to whole-body insulin resistance and hepatic steatosis [[26], [27], [28],44,52,53]. The mammalian clock system is organised in a hierarchical manner. The master or “central” clock is located in the hypothalamus within the suprachiasmatic nucleus (SCN). The central clock acts as an “orchestra director” that coordinates whole-body temporal physiology by synchronising peripheral clocks that are present in all the body's organs [54]. Interestingly, in the 2-week-old SL mice, the clock genes in the liver were primarily altered and remained as such until 4 months of age. We did not find such alterations in other peripheral tissues or the hypothalamus. These data suggested that SL-associated signals were extremely specific in programming the periphery (namely the liver) while having no major effects on the central pacemaker.
Supporting the role of peripheral clocks in our model, we found that whole-body locomotor activity followed a normal pattern when the adult SL mice were maintained under a normal LD cycle. During the LD, the hypothalamus, which showed a normal expression of clock genes in the young and adult SL mice, sensed the daily light–dark cycle and was able to properly synchronise whole-body rhythmicity. However, under DD conditions, an endogenous desynchrony manifested in the SL mice. This desynchrony was likely due to intrinsic alterations in peripheral tissues because under DD conditions, the central pacemaker has weaker influence on the periphery and endogenous (abnormal) rhythmicity is partially revealed. Intriguingly, although the SL mice maintained normal activity under LD conditions, they still showed mild temporal physiological perturbations. First, the SL mice exhibited increased food intake during the dark phase and reduced oxygen consumption and energy expenditure during the light phase. This behaviour contributed to explain the obese phenotype of the SL mice. Furthermore, the SL mice also showed increased RER during the dark period (which is the active phase in rodents), suggesting a preference for oxidising carbohydrates over fat. We speculate that increased RER in the SL mice might have been partly attributed to combined elevation of the free fatty acid esterification pathway (through increased expression of Mogat1 and Dgat2) and impairment of fatty acid oxidation (through Ppara misexpression). Further experiments are needed to fully elucidate the mechanisms leading to physiological defects. In summary, these data strongly supported that defective whole-body rhythmicity might have been attributable primarily to peripheral clocks, as the liver is a major player.
Why is the periphery, particularly the liver, affected by early overfeeding? This is likely due to the combination of two factors. First, hepatic circadian rhythmicity starts developing at birth and is completed by 30 days of age [55]. Second, it is well known that while the central clock is primarily entrained by LD information through the retina [57], the peripheral clocks, including that of the liver, are additionally entrained by nutritional cues, such as the feeding/fasting cycle and macronutrient composition [37,[58], [59], [60], [61], [62]]. We previously reported that SL mice were exposed to high calorie intake and different macronutrient composition (high TAG milk concentration) during the lactation period [19,20]. Based on these observations, we postulate that the hepatic clock was rapidly misaligned in the young SL mice because they were exposed to complex interactions or early nutritional cues, including high calorie intake, differences in macronutrient composition, and possibly differences in offspring feeding periodicity. For example, it might have been possible that the SL females fed their offspring more frequently than the controls, but at the cost of spending less time during the feeding period. While speculative, this hypothesis deserves future investigation.
It was intriguing that the misalignment observed in the young SL mice was fairly normalised in the adult SL mice. The difference between both stages was probably due to the fact that, as previously described, in young mice, hepatic rhythmic behaviour is still under maturation; it is completed by postnatal day 30 [64]. After ageing, the differences in rhythmic behaviours are minimal, probably because the central pacemaker is able to re-synchronise peripheral clocks. The endogenous defects can be partially revealed through nutritional cues.
At the molecular level, nutritional cues do not equally influence the expression of all of the clock genes. In this regard, the period and cryptochrome genes are susceptible to regulation through nutrition signals [63], whereas Clock/Bmal1/Npas2 are primarily regulated by signals derived from the SCN, which is driven by LD cycles [64]. Indeed, in our model, Per1-3 and Cry1-2 were altered in both the young and adult SL mice. Therefore, our data indicated that early nutrition imbalances primarily influenced the expression of nutrition-sensitive clock genes. Given the importance of feeding–fasting cycles in mediating temporal regulation of metabolism, we investigated whether TRF could improve overall physiology in the adult SL mice [65]. TRF refers to the idea of restricting food availability during specific windows of the day. During periods when food is available, individuals have ad libitum access to the food [49,66,67]. As previously described, the hepatic circadian rhythm is largely entrained by the timing of food consumption [65,68]. Therefore, we conducted a temporal dietary intervention in which food was available for 8 h during the light cycle only, from ZT1 to ZT9. As rodents consume most of their calories (>70%) during the dark cycle, this TRF forced the animals to exclusively consume their food during the light phase. Using this intervention, we could partially uncouple the peripheral (hepatic) oscillators from the SCN [58,67]. Our goal was to maintain the light/dark (hypothalamic) signals as they are normally perceived. Therefore, by changing only the nutritional cues, we could explore the involvement of the periphery (liver) in our model by comparing its response to individuals maintained ad libitum.
Strikingly, this TRF was very effective at reducing body fat mass and liver TAG content. These metabolic improvements were associated with better alignment of the hepatic clock genes, suggesting that desynchronised peripheral clocks might partially underlie metabolic deregulation in our model. Together, our interpretation of the TRF data was that the liver of the SL mice had endogenous rhythmic defects (already suggested in Figure 5; DD) that were somewhat masked by the hypothalamus (Figure 5; LD). TRF supported that the nutritional cues may have directly influenced the hepatic clock gene expression and therefore the liver rhythmic behaviour and metabolism (Table S4). The TRF protocol we outlined clearly created a desynchrony that did not pretend to become a therapeutic intervention. It was just a means to reveal the relative contributions of the peripheral-to-central signals in mediating hepatic behaviour in our models. Likewise, it is very likely that in the long run, this type of desynchrony would potentially lead to further abnormal behaviour and additional metabolic defects. Indeed, we found that Clock, Rora, and Npas2 lost significant rhythmicity in the SL mice. However, it is plausible that devising rational therapeutic TRFs can improve overall SL physiology. In agreement with this, other authors have also reported that different temporal restriction schedules may protect mice from hepatic steatosis by re-setting their hepatic clocks [69,70].
Our TRF data provide indirect support for the circadian clock playing a causal role in hepatic lipid dysregulation in our model. While these data are associative, additional lines of evidence support this hypothesis. First, the lipid metabolic process Gene Ontology term was significantly altered in the liver of the adult SL mice. It is known that approximately 20% of the hepatic transcriptome shows rhythmic behaviour. Strikingly, we confirmed that 40% of the genes included in our gene list exhibit rhythmic behaviour in the liver [60]. This enrichment strongly suggested that, to some extent, lipid metabolic deregulation in the adult SL mice might have been under the control of the clock genes. Interestingly, the rhythmic expression of the lipid-related genes was dramatically misaligned, whereas the clock gene expression was mildly deregulated. This apparent discrepancy could be partly attributed to a cascade effect. For example, small changes in clock gene expression translate into larger deregulation of clock-controlled transcription factor(s) that in turn are further amplified, resulting in striking misalignment of the final targets (lipid-related genes). We performed an in silico analysis of the promoter regions of the chrono-disrupted lipid genes and did not find evidence supporting a direct interaction of clock genes in mediating lipid dysfunction. Instead, their effect should have been exerted through an indirect mechanism. In line with this possibility, one well-known feedback loop connecting clock genes and metabolic regulators in the liver is interactions between Per2 and nuclear receptors Ppara and Pparg [71,72].
This observation led us to propose that chrono-disruption of hepatic lipid metabolism might occur in part via the Per2-Ppara axis targeting lipid genes [42,73]. In agreement with this, we found that Ppara, but not Pparg, was profoundly chrono-disrupted in the young and adult mice. Second, Ppara rhythmicity was partially reset in the SL-TRF mice. We tested this hypothesis through functional assays in vitro. Specifically, we inhibited the expression of Per1 and Per2 in hepatocytes in vitro using adenoviral vectors containing specific shRNAs. Downregulation of Per1 (60%) induced general derangement of clock genes. However, Per2 inhibition (>40%) decreased Ppara expression. Importantly, Per2 inhibition also reduced the expression of Ppara target genes (Lpl and Pck1) and genes involved in free fatty acid esterification (Mogat1 and Dgat2), which were previously associated with the development of steatosis in our model [19]. In summary, these data indicated that early over-nutrition may lead to hepatic steatosis in part through deregulation of clock genes, namely period 1–3, which secondarily influence the expression of lipid genes partially via Ppara. We recognise that, despite the functional loss-of-function assays, the aforementioned interpretation was arrived at through associative experiments. Further experiments with knock-out models, such as the period 1–3 KO or the Ppara KO, would help determine the causal relationship between clock genes (namely period 1–2) and the expression of hepatic lipid-related genes.
Similar observations in closely related models have been described. For example, in utero exposure to an “obesogenic” environment altered the hepatic amplitudes of core clock genes and circadian metabolic genes in offspring [76]. These oscillatory derangements were partially attributed to changes in Ppara expression. The authors of this study suggested that offspring of obese dams were unable to adequately “mount a response to metabolic demands that require mobilisation of lipids (for example, under fasting of high-fat feeding) via an inability to induce Ppara and its downstream targets” [77]. Another model of maternal obesity also led to the development of hepatic steatosis in offspring [78]. Again, non-alcoholic fatty liver was partly attributed to chronic disruption of Clock/Bmal1 and Cry2 transcription, which was characterised by a biphasic rhythmic pattern. These data indicate that nutritional imbalances during developmental stages (including prenatal and early postnatal life) chrono-disrupt the early expression of hepatic core clock genes and downstream targets throughout the life course.
The DOHaD hypothesis proposes that early environmental challenges may have long-lasting effects and influence adult physiology [79]. Our mouse model exposed to high-calorie intake and increased TAG availability during neonatal development clearly fits within this paradigm. A current key question in the field is the mechanisms that drive these long-term effects. It has been proposed that epigenetic mechanisms, including DNA methylation, histone modifications, and/or non-coding RNAs, might play a role in such long-lasting effects [80]. Indeed, we previously showed that specific histone modifications partly explain early and late-onset upregulation of Mogat1 in the liver of both young and adult SL mice [19]. It might be possible that epigenetic mechanisms also underlie the misexpression of the clock genes in our model. In line with this, it has been shown that, in the liver, specific histone modifications, such as H3K4me3 and H3K9ac, follow a rhythmic pattern and contribute to circadian chromatin remodelling [81]. In mice, HDAC3 is recruited to the promoter of liver Rev-erb, regulating its rhythmic behaviour [82]. Deletion of HDAC3 creates circadian rhythm-related modifications in chromatin remodelling that are associated with an increase in hepatic TAG content. In addition or in parallel to histone modifications, changes in DNA methylation may also contribute to the regulation of clock genes in the context of DOHaD [78,81]. For example, cytosine methylation of specific CpG sites in the promoter region of Per1 is established during development [84]. Likewise, the promoter regions of Bmal1 and Per2 were found to be hypermethylated in white blood cells of obese women [85]. These data indicate that the circadian rhythm itself might be under the control of some epigenetic machinery, including histones and DNA methylation [80,83,84,86]. Hence, studies are necessary to confirm whether rhythmic histone codes and/or DNA methylation influenced the expression of the clock genes in our model.
5. Conclusion
Our data indicated that litter size reduction was related to early life liver circadian rhythm misalignment and liver-related metabolic dysfunction. Strikingly, TRF interventions greatly improved whole-body adiposity and completely re-established the hepatic TAG content back to the levels of the control mice, suggesting a causal role of peripheral (hepatic) clock genes. We have provided some evidence that the circadian clock in SL mice might deregulate lipid metabolic processes through Per-Ppara axis-dependent lipid metabolism. This axis contributes to hepatic lipid accumulation in SL mice, which in turn drives late-onset metabolic syndrome development. Our data support a rationale for designing TRFs for the treatment or prevention of metabolic derangements associated with early adiposity. It is likely that TRFs mediate their beneficial effects though partially re-aligning the circadian clock.
Acknowledgements
J.C.J.-C. was supported by grants from FEDER/Ministerio de Ciencia, Innovación y Universidades (SAF2017-84542-R) and Instituto de Salud Carlos III (PI14/00035 and CPII16/00046), co-funded by the European Regional Development Plan, ‘‘A way to make Europe.’’ M.P.-V. was recipient of a postdoctoral fellowship from Mexico’s National Council for Science and Technology (CONACYT), Government of Mexico.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101162.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Table S1. List of primers and sequences included in this study.
Table S2. List of genes from the microarray analysis (Affymetrix). We compared the expression in the liver of 4-month-old control and SL mice. Differential expression analysis was performed using the non-parametric Rank Prod approach. Overall, 245 annotated genes were differentially expressed in the liver of SL mice vs control mice.
Table S3. List of genes belonging to the most significant Gene Ontology term resulting from DAVID functional clustering enrichment analysis: circadian rhythm.
Table S4. Functional annotation of the periodicity statistics associated with the clock genes of the control and SL mice. The information includes the period, acrophase, mensor, and significance of the periodicity.
Table S5. List of genes belonging to the lipid metabolic process Gene Ontology term resulting from DAVID functional clustering enrichment analysis.
References
- 1.Kahn B.B., Flier J.S. Obesity and insulin resistance. Journal of Clinical Investigation. 2000 doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smyth S., Heron A. Diabetes and obesity: the twin epidemics. Nature Medicine. 2006 doi: 10.1038/nm0106-75. [DOI] [PubMed] [Google Scholar]
- 3.Pulgaron E.R., Delamater A.M. Obesity and type 2 diabetes in children: epidemiology and treatment. Current Diabetes Reports. 2014 doi: 10.1007/s11892-014-0508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freedman D.S., Dietz W.H., Srinivasan S.R., Berenson G.S. The relation of overweight to cardiovascular risk factors among children and adolescents: the Bogalusa Heart Study. Pediatrics. 1999 doi: 10.1542/peds.103.6.1175. [DOI] [PubMed] [Google Scholar]
- 5.The N.S., Suchindran C., North K.E., Popkin B.M., Gordon-Larsen P. Association of adolescent obesity with risk of severe obesity in adulthood. JAMA - Journal of the American Medical Association. 2010 doi: 10.1001/jama.2010.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd L.J., Langley-Evans S.C., McMullen S. Childhood obesity and risk of the adult metabolic syndrome: a systematic review. International Journal of Obesity. 2012 doi: 10.1038/ijo.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logue J., Sattar N. Childhood obesity: a ticking time bomb for cardiovascular disease. Clinical Pharmacology & Therapeutics. 2011 doi: 10.1038/clpt.2011.88. [DOI] [PubMed] [Google Scholar]
- 8.Reilly J.J., Kelly J. Long-term impact of overweight and obesity in childhood and adolescence on morbidity and premature mortality in adulthood: systematic review. International Journal of Obesity. 2011 doi: 10.1038/ijo.2010.222. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Gronert M.S., Ozanne S.E. Mechanisms underlying the developmental origins of disease. Reviews in Endocrine & Metabolic Disorders. 2012;13(2):85–92. doi: 10.1007/s11154-012-9210-z. [DOI] [PubMed] [Google Scholar]
- 10.Habbout A., Li N., Rochette L., Vergely C. Postnatal overfeeding in rodents by litter size reduction induces major short- and long-term pathophysiological consequences. Journal of Nutrition. 2013 doi: 10.3945/jn.112.172825. [DOI] [PubMed] [Google Scholar]
- 11.Parra-Vargas M., Ramon-Krauel M., Lerin C., Jimenez-Chillaron J.C. Size does matter: litter size strongly determines adult metabolism in rodents. Cell Metabolism. 2020 doi: 10.1016/j.cmet.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Plagemann A., Harder T., Brunn M., Harder A., Roepke K., Wittrock-Staar M. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. Journal of Physiology. 2009 doi: 10.1113/jphysiol.2009.176156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G., Kohorst J.J., Zhang W., Laritsky E., Kunde-Ramamoorthy G., Baker M.S. Early postnatal nutrition determines adult physical activity and energy expenditure in female mice. Diabetes. 2013 doi: 10.2337/db12-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collden G., Balland E., Parkash J., Caron E., Langlet F., Prevot V. Neonatal overnutrition causes early alterations in the central response to peripheral ghrelin. Molecular Metabolism. 2015 doi: 10.1016/j.molmet.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu S., Eclarinal J., Baker M.S., Li G., Waterland R.A. Developmental programming of energy balance regulation: is physical activity more “programmable” than food intake? Proceedings of the Nutrition Society. 2016 Feb;75(1):73–77. doi: 10.1017/S0029665115004127. [DOI] [PubMed] [Google Scholar]
- 16.Bei F., Jia J., Jia Y.Q., Sun J.H., Liang F., Yu Z.Y. Long-term effect of early postnatal overnutrition on insulin resistance and serum fatty acid profiles in male rats. Lipids in Health and Disease. 2015 doi: 10.1186/s12944-015-0094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conceição E.P.S., Franco J.G., Oliveira E., Resende A.C., Amaral T.A.S., Peixoto-Silva N. Oxidative stress programming in a rat model of postnatal early overnutrition - role of insulin resistance. Journal of Nutritional Biochemistry. 2013 doi: 10.1016/j.jnutbio.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Du Q., Hosoda H., Umekawa T., Kinouchi T., Ito N., Miyazato M. Postnatal weight gain induced by overfeeding pups and maternal high-fat diet during the lactation period modulates glucose metabolism and the production of pancreatic and gastrointestinal peptides. Peptides. 2015 doi: 10.1016/j.peptides.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Ramon-Krauel M., Pentinat T., Bloks V.W., Cebrià J., Ribo S., Pérez-Wienese R. Epigenetic programming at the Mogat1 locus may link neonatal overnutrition with long-term hepatic steatosis and insulin resistance. Federation of American Societies for Experimental Biology Journal. 2018 doi: 10.1096/fj.201700717RR. [DOI] [PubMed] [Google Scholar]
- 20.Pentinat T., Ramon-Krauel M., Cebria J., Diaz R., Jimenez-Chillaron J.C. Transgenerational inheritance of glucose intolerance in a mouse model of neonatal overnutrition. Endocrinology. 2010 doi: 10.1210/en.2010-0684. [DOI] [PubMed] [Google Scholar]
- 21.Bass J., Takahashi J.S. Circadian integration of metabolism and energetics. Science. 2010 doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohawk J.A., Green C.B., Takahashi J.S. Central and peripheral circadian clocks in mammals. Annual Review of Neuroscience. 2012 doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheer F.A.J.L., Hilton M.F., Mantzoros C.S., Shea S.A. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proceedings of the National Academy of Sciences of the United States of America. 2009 doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spiegel K., Tasali E., Leproult R., Van Cauter E. Effects of poor and short sleep on glucose metabolism and obesity risk. Nature Reviews Endocrinology. 2009 doi: 10.1038/nrendo.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maury E., Ramsey K.M., Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circulation Research. 2010 doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudic R.D., McNamara P., Curtis A.M., Boston R.C., Panda S., Hogenesch J.B. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biology. 2004 doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turek F.W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E. Obesity and metabolic syndrome in circadian Clock mutant nice. Science. 2005 doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimba S., Ogawa T., Hitosugi S., Ichihashi Y., Nakadaira Y., Kobayashi M. Deficient of a clock gene, brain and muscle arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PloS One. 2011 doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez D., Pentinat T., Ribó S., Daviaud C., Bloks V.W., Cebrià J. In utero undernutrition in male mice programs liver lipid metabolism in the second-generation offspring involving altered Lxra DNA methylation. Cell Metabolism. 2014 doi: 10.1016/j.cmet.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 30.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U. Biostatistics; Oxford, England: 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. [DOI] [PubMed] [Google Scholar]
- 31.Breitling R., Armengaud P., Amtmann A., Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Letters. 2004 doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 32.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009 doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Cornélissen G., Halberg F., Stebbings J., Halberg E., Carandente F., Hsi B. Chronobiometry with pocket calculators and computer systems. La Ricerca in Clinica e in Laboratorio. 1980 doi: 10.1007/BF02905347. [DOI] [PubMed] [Google Scholar]
- 35.Bingham C., Arbogast B., Cornelissen Guillaume G., Lee J.K., Halberg F. Inferential statistical methods for estimating and comparing cosinor parameters. Chronobiologia. 1982;9(4):397–439. [PubMed] [Google Scholar]
- 37.Mazzoccoli G., Pazienza V., Vinciguerra M. Clock genes and clock-controlled genes in the regulation of metabolic rhythms. Chronobiology International. 2012 doi: 10.3109/07420528.2012.658127. [DOI] [PubMed] [Google Scholar]
- 38.Panda S., Antoch M.P., Miller B.H., Su A.I., Schook A.B., Straume M. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002 doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 39.Reddy A.B., Karp N.A., Maywood E.S., Sage E.A., Deery M., O'Neill J.S. Circadian orchestration of the hepatic proteome. Current Biology. 2006 doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 40.Eckel-Mahan K., Sassone-Corsi P. Metabolism and the circadian clock converge. Physiological Reviews. 2013 doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmers C., Gill S., DiTacchio L., Pulivarthy S.R., Le H.D., Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009 doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ceglia N., Liu Y., Chen S., Agostinelli F., Eckel-Mahan K., Sassone-Corsi P. CircadiOmics: circadian omic web portal. Nucleic Acids Research. 2018 doi: 10.1093/nar/gky441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel V.R., Eckel-Mahan K., Sassone-Corsi P., Baldi P. CircadiOmics: integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nature Methods. 2012 doi: 10.1038/nmeth.2111. [DOI] [PubMed] [Google Scholar]
- 44.Kersten S., Rakhshandehroo M., Knoch B., Müller M. Peroxisome proliferator-activated receptor alpha target genes. PPAR Research. 2010 doi: 10.1155/2010/612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X., Downes M., Yu R.T., Bookout A.L., He W., Straume M. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006 doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 48.Reinke H., Asher G. Crosstalk between metabolism and circadian clocks. Nature Reviews Molecular Cell Biology. 2019 doi: 10.1038/s41580-018-0096-9. [DOI] [PubMed] [Google Scholar]
- 49.Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E.A., Gill S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metabolism. 2012 doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garaulet M., Ordovás J.M., Madrid J.A. The chronobiology, etiology and pathophysiology of obesity. International Journal of Obesity. 2010 doi: 10.1038/ijo.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shetty A., Hsu J.W., Manka P.P., Syn W.K. Role of the circadian clock in the metabolic syndrome and nonalcoholic fatty liver disease. Digestive Diseases and Sciences. 2018 doi: 10.1007/s10620-018-5242-x. [DOI] [PubMed] [Google Scholar]
- 52.Ferrell J.M., Chiang J.Y.L. Circadian rhythms in liver metabolism and disease. Acta Pharmaceutica Sinica B. 2015 doi: 10.1016/j.apsb.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qian J., Scheer F.A.J.L. Circadian system and glucose metabolism: implications for physiology and disease. Trends in Endocrinology and Metabolism. 2016 doi: 10.1016/j.tem.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi D., Chen J., Wang J., Yao J., Huang Y., Zhang G. Circadian clock genes in the metabolism of non-alcoholic fatty liver disease. Frontiers in Physiology. 2019 doi: 10.3389/fphys.2019.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaix A., Lin T., Le H.D., Chang M.W., Panda S. Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metabolism. 2019 doi: 10.1016/j.cmet.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brooks E., Canal M.M. Development of circadian rhythms: role of postnatal light environment. Neuroscience & Biobehavioral Reviews. 2013 doi: 10.1016/j.neubiorev.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Varcoe T.J., Gatford K.L., Kennaway D.J. Maternal circadian rhythms and the programming of adult health and disease. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2018 doi: 10.1152/ajpregu.00248.2017. [DOI] [PubMed] [Google Scholar]
- 59.Damiola F., Le Minli N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & Development. 2000 doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koronowski K.B., Kinouchi K., Welz P.S., Smith J.G., Zinna V.M., Shi J. Defining the independence of the liver circadian clock. Cell. 2019 doi: 10.1016/j.cell.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eckel-Mahan K.L., Patel V.R., De Mateo S., Orozco-Solis R., Ceglia N.J., Sahar S. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013 doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kinouchi K., Magnan C., Ceglia N., Liu Y., Cervantes M., Pastore N. Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Reports. 2018 doi: 10.1016/j.celrep.2018.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pendergast J.S., Branecky K.L., Yang W., Ellacott K.L.J., Niswender K.D., Yamazaki S. High-fat diet acutely affects circadian organisation and eating behavior. European Journal of Neuroscience. 2013 doi: 10.1111/ejn.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sládek M., Jindráková Z., Bendová Z., Sumová A. Postnatal ontogenesis of the circadian clock within the rat liver. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2007 doi: 10.1152/ajpregu.00184.2006. [DOI] [PubMed] [Google Scholar]
- 65.Bae S.A., Androulakis I.P. The synergistic role of light-feeding phase relations on entraining robust circadian rhythms in the periphery. Gene Regulation and Systems Biology. 2017 doi: 10.1177/1177625017702393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Menet J.S., Pescatore S., Rosbash M. CLOCK: BMAL1 is a pioneer-like transcription factor. Genes & Development. 2014 doi: 10.1101/gad.228536.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaix A., Manoogian E.N.C., Melkani G.C., Panda S. Time-restricted eating to prevent and manage chronic metabolic diseases. Annual Review of Nutrition. 2019 doi: 10.1146/annurev-nutr-082018-124320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manoogian E.N.C., Panda S. Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Research Reviews. 2017 doi: 10.1016/j.arr.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Longo V.D., Panda S. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metabolism. 2016 doi: 10.1016/j.cmet.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Asher G., Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015 doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 71.Chaix A., Zarrinpar A., Miu P., Panda S. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metabolism. 2014 doi: 10.1016/j.cmet.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guan D., Xiong Y., Borck P.C., Jang C., Doulias P.T., Papazyan R. Diet-induced circadian enhancer remodeling synchronizes opposing hepatic lipid metabolic processes. Cell. 2018 doi: 10.1016/j.cell.2018.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grimaldi B., Bellet M.M., Katada S., Astarita G., Hirayama J., Amin R.H. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metabolism. 2010 doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Borengasser S.J., Kang P., Faske J., Gomez-Acevedo H., Blackburn M.L., Badger T.M. High fat diet and in utero exposure to maternal obesity disrupts circadian rhythm and leads to metabolic programming of liver in rat offspring. PloS One. 2014 doi: 10.1371/journal.pone.0084209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patel M.S., Nielsen J.H. Taylor & Francis Group; 2017. Fetal and early postnatal programming and its influence on adult health. [Google Scholar]
- 78.Mouralidarane A., Soeda J., Sugden D., Bocianowska A., Carter R., Ray S. Maternal obesity programs offspring non-alcoholic fatty liver disease through disruption of 24-h rhythms in mice. International Journal of Obesity. 2015 doi: 10.1038/ijo.2015.85. [DOI] [PubMed] [Google Scholar]
- 79.Hanson M.A., Gluckman P.D. Early developmental conditioning of later health and disease: physiology or pathophysiology? Physiological Reviews. 2014 doi: 10.1152/physrev.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiménez-Chillarón J.C., Díaz R., Martínez D., Pentinat T., Ramón-Krauel M., Ribó S. The role of nutrition on epigenetic modifications and their implications on health. Biochimie. 2012 doi: 10.1016/j.biochi.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 81.Koike N., Yoo S.H., Huang H.C., Kumar V., Lee C., Kim T.K. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012 doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feng D., Liu T., Sun Z., Bugge A., Mullican S.E., Alenghat T. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011 doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gabory A. Epigenetic mechanisms involved in developmental nutritional programming. World Journal of Diabetes. 2011 doi: 10.4239/wjd.v2.i10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ripperger J.A., Merrow M. Perfect timing: epigenetic regulation of the circadian clock. FEBS Letters. 2011 doi: 10.1016/j.febslet.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 85.Milagro F.I., Gómez-Abellán P., Campión J., Martínez J.A., Ordovás J.M., Garaulet M. CLOCK, PER2 and BMAL1 DNA methylation: association with obesity and metabolic syndrome characteristics and monounsaturated fat intake. Chronobiology International. 2012 doi: 10.3109/07420528.2012.719967. [DOI] [PubMed] [Google Scholar]
- 86.Sahar S., Sassone-Corsi P. 2013. The epigenetic language of circadian clocks. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of primers and sequences included in this study.
Table S2. List of genes from the microarray analysis (Affymetrix). We compared the expression in the liver of 4-month-old control and SL mice. Differential expression analysis was performed using the non-parametric Rank Prod approach. Overall, 245 annotated genes were differentially expressed in the liver of SL mice vs control mice.
Table S3. List of genes belonging to the most significant Gene Ontology term resulting from DAVID functional clustering enrichment analysis: circadian rhythm.
Table S4. Functional annotation of the periodicity statistics associated with the clock genes of the control and SL mice. The information includes the period, acrophase, mensor, and significance of the periodicity.
Table S5. List of genes belonging to the lipid metabolic process Gene Ontology term resulting from DAVID functional clustering enrichment analysis.