Abstract
Background
This study aimed to isolate potent thermophilic and amylolytic bacteria from a hot spring of Pharaoh’s bath, Sinai, Egypt, and screen its degradative activity. The amylolytic activity was further optimized using a statistical full factorial design followed by response surface methodology.
Results
A thermophilic bacterium was isolated from the hot spring of Pharaoh’s Bath, Sinai, Egypt. The isolate produced amylase, cellulase, and caseinase and was identified by 16S rRNA gene sequencing as Parageobacillus thermoglucosidasius Pharon1 (MG965879). A growth medium containing 1% soluble starch was found to optimize the amylase production. Dinitrosalycalic acid method (DNS) was used to estimate the amount of reducing sugar produced. Statistical full factorial and response surface designs were employed to optimize physical variables affecting the α-amylase production and determine the significant interactions of the studied variables during the fermentation process. According to the results obtained by the response optimizer, the maximum amylase activity reached 76.07 U/mL/ min at 54.1°C, pH 5.6 after 98.5 h incubation under aerobic conditions. Moreover, the produced enzyme was thermostable and retained most of its activity when exposed to a high temperature of 100°C for 120 min. Maximum enzyme activity was attained when the enzyme was incubated at 70°C for 30 min.
Conclusions
This is the first report of the production of thermostable α-amylase by the potent thermophilic Parageobacillus thermoglucosidasius. The enzyme endured extreme conditions of temperature and pH which are important criteria for commercial and industrial applications.
Supplementary Information
The online version contains supplementary material available at 10.1186/s43141-021-00123-4.
Keywords: Thermostable amylase, Thermophilic, Parageobacillus thermoglucosidasius, Hot spring, Full factorial design, Response surface optimization
Background
Amylases are important enzymes that play a pivotal role in biotechnology. They are produced by plants, animals, and microorganisms. Amylases are widely used in baking and bread industry, fermentations, textiles, alcohols, pharmaceuticals, and detergents [1]. Moreover, they are used in the production of corn and chocolate syrup, production of low-calorie beer, purification of apple and pear juice, malt production, and in removing stickiness in the paper industry [2–4].
Microbial amylases have been generally favored over other plant’s and animal’s amylases because they are easily produced at low cost and in a shorter time. Moreover, the production of bacterial amylase is cheaper and faster than amylases produced by other microorganisms. Many industrial enzymatic reactions occurring at high temperatures have advantages in decreasing the contamination risk, increasing the diffusion rate, being resistant to denaturing agents, and proteolytic enzymes [5].
The amylases produced by different Bacillus species differ in their types, range of pH, and temperature [6]. The hot springs are a promising source for isolation of thermophilic bacteria producing thermo-stable α-amylase required in many industrial applications [7, 8]. The demand for thermostable amylases in biotechnology and industrial applications is increasing [9]. However, reports about bacterial strains that could produce thermostable amylase are still limited [10].
Statistical and mathematical techniques for predicting the behavior of process variables and explaining their interactions, and response surface methodology (RSM) have been applied in the optimization of amylase production [11–13].
Methods
Sampling and isolation of thermophilic bacteria
A soil sample from hot springs of Pharaoh’s bath (29° 12′ 24.9′′ N, 32° 57′ 35.4′′ E), Sinai, Egypt, was collected in a sterile container and kept in an icebox for isolation of thermophilic bacteria. The sample was 10-fold diluted in sterile 0.09% saline solution, then 100 μL from each dilution was cultured on nutrient agar plates. The plates were incubated at 70°C for 48 h. After the incubation period, bacterial colonies of distinctive morphology were selected and purified on fresh sterile nutrient agar plates.
Selection of a degradative organism
The isolates were screened to produce amylase, cellulase, and caseinase through sub-culturing on agar plates supplemented with 1% soluble starch [14], carboxy methyl cellulose [15], and 15% skimmed milk [16], in triplicates, respectively. The plates were incubated at 70°C for 24 h. The isolate which produced the 3 hydrolytic enzymes was selected and identified.
Molecular identification
Total DNA of the bacterial isolate was extracted according to the instruction manual using DNA extraction kits (Thermo, Fisher Scientifics, USA) and stored frozen at – 20°C until PCR reaction was carried out. A pair of flanking sequences, 16S-1F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 16S-517R (5′-ATTACCGCGGCTGCTGG-3′), was used for primer binding sites to partially amplify the target gene. Two microliters of the bacterial DNA was used as a template for PCR reaction. PCR was carried on using Premix Taq (MyTaq, Bioline, UK) according to the instruction manual. PCR was performed in genius model FGENO2TD thermal cycler (Techne, England). The PCR conditions were adjusted to 5 min for initial denaturation at 94°C, then 35 cycles of 1 min at 94°C, 1 min at 54°C, and 1 min at 72°C, and finally 10 min at 72°C for amplification of genes. The amplified genes were run on 1% agarose gel with a size marker to determine the size and purity of the products. The amplified bands were cleaned using a PCR product purification kit (Thermo, Fisher Scientifics, USA).
Sequencing of forward directions of partial 16S rRNA gene was performed in Macrogen, Korea. The sequence was identified using the BLAST search program, National Center for Biotechnology Information (NCBI), and National Library of Medicine, USA [17].
Sequence alignments were performed by Clustal W 1.83 XP software, and a phylogenetic tree was constructed using the neighbour-joining method using MEGA 6 software. Then, the sequence was submitted using Bankit tool (NCBI website; www.ncbi.nlm.nih.gov) to obtain the accession number.
Selection of the best amylase production medium
The selected isolate was inoculated on four different starch containing media, M1 medium of the following composition: 2% starch, 1% yeast extract, 0.1% peptone, 0.1% beef extract, 0.05% MgSO4, and 0.04% CaCl2 [18]; M2 medium: 0.5% soluble starch, 0.5% yeast extract, 0.25% (NH4)2SO4, 0.02% MgSO4·7H2O, 0.3% KH2PO4, and 0.025% CaCl2·2H2O [16]; M3 medium: 2% soluble starch, 0.3% yeast extract, 0.3% tryptone, 0.03% sodium dodecyl sulfate, 0.5% polyethylene glycol, 0.02% MgSO4 7H2O, 1.0% K2HPO4, and 1.0% NaCl (pH 7) [19]; and M4 medium :1% soluble starch, 0.2% yeast extract, 0.5% peptone, 0.05% MgSO4, 0.05% NaCl, and 0.015% CaCl2 (pH 7.0) [7]. Then, the flasks were incubated at 60°C for 24 h to determine highest activity.
Detection of amylase activity
The amylase activity was assayed according to Miller [20] using 3,5-dinitrosalicylic acid (DNS) method through incubation of 300 μL of 1% starch in 50 mM sodium phosphate buffer (pH 7.5) with 200 μL cell-free supernatant for 30 min at 60, 70, and 80°C. The reaction was stopped by adding 500 μL DNS reagent and boiling it in a water bath for 10 min. After cooling at room temperature, the amount of glucose released was determined by measuring the absorbance at 550 nm using Unico 7200 Spectrophotometer. Amylase activity was estimated using a calibration curve for glucose over a concentration range of 15 to 100 μmol. One unit of enzyme activity was defined as the amount of enzyme that releases 1 μmol reducing sugars per min [21].
Statistical factorial designs
The two experimental designs namely Full Factorial Design and Central Composite Design (CCD) were adopted to study the effects of four independent variables with their interactions on amylase activity and to determine the optimum variable level for maximum enzyme activity. The Minitab Software v. 18 program was used for designing the involved models and statistical-analyzing the obtained results of the response (α-amylase activity). Experiments were performed in 250 mL flasks containing 100 mL sterile M4 medium inoculated with 100 μL of (106 CFU per mL) of the selected bacterial isolate. All experiments were triplicated and the mean amylase activity was calculated.
Full factorial design
A 24 full factorial design was used to study the statistical significance of the following variables: growth medium pH (A), incubation temperature (B), incubation period (C), and aeration (D) besides their interactions towards the amylase activity. Each factor was studied at two levels (low and high) chosen according to previous studies and literature review (Table 1). A total of 16 sets of experiments were carried out to determine the significant factors that affect the amylase activity.
Table 1.
The investigated variables with their codes and levels for the full factorial design
| Variables | Code | Low level (-1) | High level (+1) |
|---|---|---|---|
| pH | A | 5 | 8 |
| Temperature (°C) | B | 50 | 75 |
| Incubation period (h) | C | 24 | 96 |
| Aeration | D | Aerobic conditions | Anaerobic conditions |
Central composite design (CCD)
The central composite design is one of the designs in Response Surface Methodology (RSM). Variables that showed a significant effect in the previous full factorial model were inserted in a three-factor CCD to study their interactions and to determine the optimum variable level for maximum amylase production.
Determination of amylase thermo-stability
The crude enzyme was incubated at different temperatures of 60, 70, 80, 90, and 100°C for zero, 30, 60, 90, and 120 min to determine the thermal stability using the DNS method. Thermal stability was expressed as percent residual activity, and the initial enzyme activity was taken as 100 % [22].
Statistical analysis
The data obtained from both the full factorial and the RSM on amylase activity optimization were analyzed by analysis of variance (ANOVA) test. The regression of tested variables and their interactions, model significance, and the coefficient of determination (R2) of the generated models were estimated. The results were used to fit a polynomial model equation to represent the behavior of the system and correlate the relation between the studied variables and amylase activity. All experiments were triplicated, and the mean amylase activity was calculated.
Results
Isolation, screening, and identification of degradative bacterial isolate
The isolate growing on a nutrient agar plate, incubated at 70°C and the production ability of amylase, cellulase, and caseinase was selected.
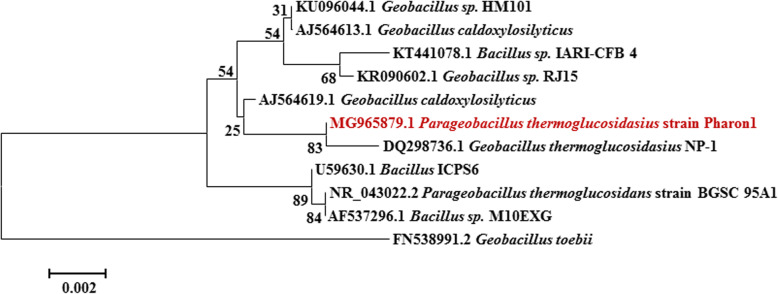
The isolate was identified by partial amplification and sequencing of 16S rRNA gene as P. thermoglucosidasius (99% sequence similarity). The nucleotide sequence was submitted to GenBank as P. thermoglucosidasius strain Pharon1 under accession number MG965879. Phylogenetic analysis with the alignment of the obtained gene sequence is illustrated in Fig. 1.
Fig. 1.
Neighbor joining phylogenetic tree of 16S rRNA genes. The numbers at the nodes are bootstrap values recovered from 100 trees, the bar indicates 0.2% nucleotide substitution
Selection of the best production media
Medium containing 1% soluble starch, 0.2% yeast extract, 0.5% peptone, 0.05% MgSO4, 0.05% NaCl, and 0.015% CaCl2 (pH 7.0) [7] exhibited the highest amylase activity by P. thermoglucogenesis Pharaon1. Moreover, The DNS assay was carried out at different temperatures indicated that 70°C was the optimum temperature for the assay.
Full factorial design
Table 2 presents the full factorial design matrix for the tested variables, given in uncoded values, plus the experimental and predicted data of amylase activity obtained by P. thermoglucogenesis Pharaon1. As shown, the highest enzyme activity was recorded in trial (12) when the bacterial strain was cultured at pH 5 and incubated aerobically at 50°C for 96 h.
Table 2.
The full factorial design matrix showing the observed amylase activity of P. thermoglucosidasius Pharon1 exerted by the design trails
| Run | Coded parameters | Mean amylase activity (U/mL/min) | ||||
|---|---|---|---|---|---|---|
| A | B | C | D | Observed | Expected | |
| 1 | 8 | 50 | 96 | Aerobic | 0.0 | 0.3 |
| 2 | 5 | 50 | 96 | Anaerobic | 18.2 | 18.5 |
| 3 | 8 | 50 | 24 | Anaerobic | 0.0 | 0.3 |
| 4 | 5 | 75 | 96 | Anaerobic | 6.1 | 5.8 |
| 5 | 8 | 50 | 24 | Aerobic | 0.0 | − 0.3 |
| 6 | 5 | 75 | 24 | Anaerobic | 2.1 | 2.4 |
| 7 | 8 | 75 | 96 | Anaerobic | 0.0 | 0.3 |
| 8 | 5 | 50 | 24 | Anaerobic | 5.0 | 4.7 |
| 9 | 8 | 75 | 96 | Aerobic | 0.0 | − 0.3 |
| 10 | 5 | 75 | 96 | Aerobic | 17.4 | 17.7 |
| 11 | 5 | 50 | 24 | Aerobic | 32.0 | 32.3 |
| 12 | 8 | 75 | 24 | Aerobic | 0.0 | 0.3 |
| 13 | 8 | 75 | 24 | Anaerobic | 0.0 | − 0.3 |
| 14 | 5 | 50 | 96 | Aerobic | 59.4 | 59.1 |
| 15 | 8 | 50 | 96 | Anaerobic | 0.0 | − 0.3 |
| 16 | 5 | 75 | 24 | Aerobic | 3.3 | 3.0 |
P. thermoglucosidasius Pharon1 was inoculated in M4 medium [7] containing 1% soluble starch. Runs were triplicated, and the mean amylase activity was calculated
The ANOVA results in Table (S1) show that all of the tested factors of growth medium pH (A), incubation temperature (B), incubation period (C), and aeration (D) significantly (P<0.05) affect the amylase activity. Additionally, the 2-way and 3-way interacted terms of A*B, A*D, A*C, B*D, and A*B*D showed significant effects on the enzyme activity, while the interacted terms of B*C*D, B*C, A*B*C, C*D, and A*C*D showed non-significant effects on the same response under the same conditions.
Results also indicated that the regression model for this experimental design was highly significant (P<0.05). The coefficient of determination (R2) value was 0.9997 indicating a high correlation between the experimental and the predicted data at a confidence level of 95%.
The obtained results were fitted in a linear polynomial equation as the following:
Response surface optimization
Factors of growth medium pH, temperature, and incubation period were represented in a CCD to determine their exact optimum levels for maximum amylase activity, while the aeration factor was categorical, and thus, it was kept at its optimum level, aerobic conditions as obtained from the tested full factorial model.
The design matrix of CCD is shown in Table 3. The results of amylase activity obtained in the CCD trials were evaluated through analysis of variance ANOVA results showed the significant effect (P<0.05) of each tested variable, A, B, and C in addition to all the squared variables, A2, B2, and C2 on the amylase activity by P. thermoglucosidasius. It was also noticed that all of 2-way interacted variables, A*B, A*C, and B*C showed a non-significant effect on amylase production. This model has a high F value and low P value (P < 0.05) that representing a good predictor for the responses (Table S2).
Table 3.
Central composite design (CCD) matrix by the three tested variables besides the observed and expected amylase activity exerted by the model trials
| Run | Coded parameters | Mean amylase activity (U/mL/min) | |||
|---|---|---|---|---|---|
| A | B | C | Observed | Expected | |
| 1 | 8.0 | 50.0 | 96.0 | 0 | 12.9 |
| 2 | 6.5 | 62.5 | 60.0 | 65 | 63.3 |
| 3 | 6.5 | 41.4 | 60.0 | 24.4 | 30.8 |
| 4 | 6.5 | 62.5 | 60.0 | 65.5 | 63.3 |
| 5 | 3.9 | 62.5 | 60.0 | 0 | 19.1 |
| 6 | 6.5 | 62.5 | 60.0 | 62.3 | 63.3 |
| 7 | 5.0 | 50.0 | 96.0 | 80.7 | 71.2 |
| 8 | 5.0 | 75.0 | 24.0 | 3.9 | − 12.5 |
| 9 | 5.0 | 50.0 | 24.0 | 32.2 | 20.9 |
| 10 | 8.0 | 75.0 | 24.0 | 0 | 6.1 |
| 11 | 5.0 | 75.0 | 96.0 | 18.2 | 20.6 |
| 12 | 9.0 | 62.5 | 60.0 | 0 | − 14.2 |
| 13 | 8.0 | 50.0 | 24.0 | 0 | − 5.9 |
| 14 | 6.5 | 62.5 | 120.5 | 68.3 | 58.5 |
| 15 | 6.5 | 62.5 | 60.0 | 62.2 | 63.3 |
| 16 | 8.0 | 75.0 | 96.0 | 0 | 7.8 |
| 17 | 6.5 | 62.5 | 0.0 | 0 | 14.7 |
| 18 | 6.5 | 62.5 | 60.0 | 62.9 | 63.3 |
| 19 | 6.5 | 83.5 | 60.0 | 0 | − 1.6 |
| 20 | 6.5 | 62.5 | 60.0 | 62.9 | 63.3 |
P. thermoglucosidasius Pharon1 was inoculated in M4 medium [7] containing 1% soluble starch under aerobic condition. Runs were triplicated, and the mean amylase activity was calculated
The coefficient of determination (R2) was 0.9081 reflecting 0.1% of the total differences in the responses could not be explained by this model. The quadratic model for predicting the optimal point was expressed according to a second-order polynomial equation as following:
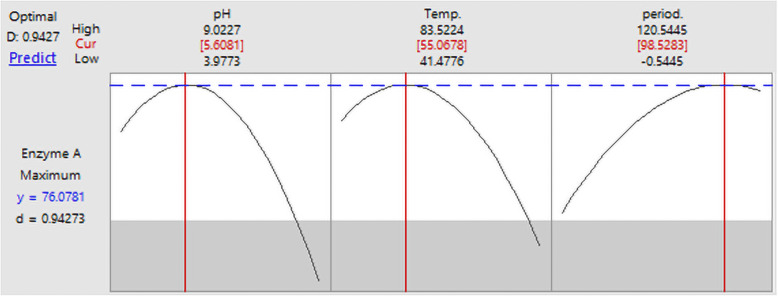
The maximum amylase activity achieved was 76.08 U/mL/min at pH 5.6, incubation temperature of 55°C for 98.5 h with a desirability of 94.27% as demonstrated by the response optimizer in Fig. 2. The 2D contour and 3D response surface plots showing the optimum level for each tested variable were represented in Fig. 3.
Fig. 2.
Predicted solution for the maximum amylase activity by P. thermoglucosidasius using the response optimizer of Minitab Software v. 18 defining the optimum levels of the selected three variables, growth medium pH, temperature, and incubation period. (Cur) is the curvature value (optimum), y is the amylase activity predicted, and D is the desirability value
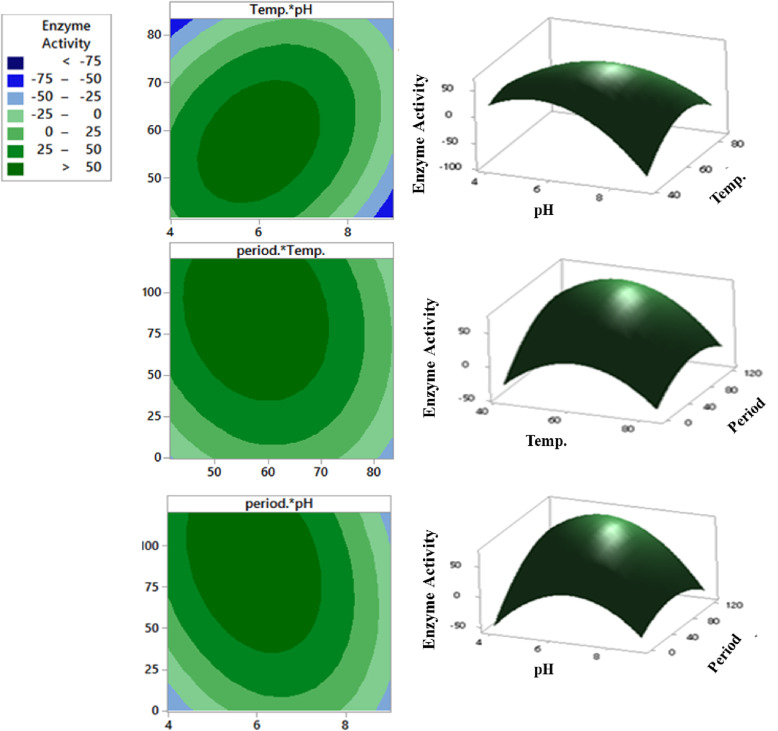
Fig. 3.
2D contour (on the left) and 3D surface (on the right) plots show the interaction effect and the optimum levels of the three significant factors on amylase activity
Thermal stability
The thermo-stability of the crude enzyme was determined. It was found that the enzyme activity at room temperature was relatively low (100 %); but the highest activity (286 ± 4.65 %) was recorded after heating the enzyme at 70°C for 30 min, and the crude showed thermal stability as it was active even after exposure to 100°C for 120 min and lost about 13% from its activity (Fig. 4).
Fig. 4.
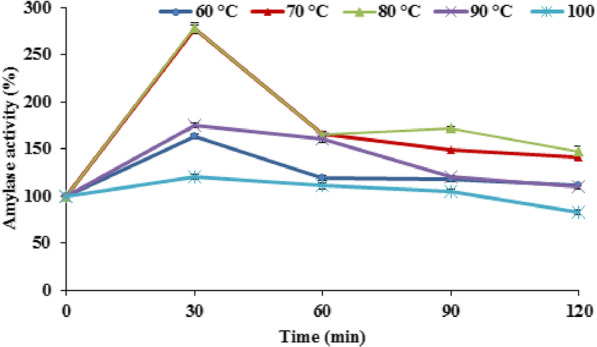
Thermo-stability of the produced amylase enzyme at different time intervals. Error bars represent SE (n = 3)
Discussion
Thermophilic microorganisms with an optimum growth temperature of 50°C or above are sources of thermostable enzymes such as amylases, cellulases, chitinases, pectinases, xylanases, proteases, lipases, and DNA polymerases.
In this study, a thermophilic isolate was recovered from the hot springs region, Pharaoh’s Bath, Sinai, Egypt. Previous work by Saeed et al. [23] reported the isolation of thermophilic bacteria from the same site. Mohammad et al. [24] illustrated the isolation of closely related thermophiles from hot springs and deep sea in Jordon. Thermophiles exhibit maximum growth at 75°C [25]; therefore, the isolated bacteria recovered from this study are considered an extreme thermophile.
In this study, the isolate was screened for the production of amylase, cellulase, and caseinase production. The isolate showed its ability to produce the three enzymes which have significant biotechnological applications.
The selected isolate was identified by 16S rRNA gene sequencing as P. thermoglucosidasius strain Pharon1 (MG965879). P. thermoglucosidasius is a facultative anaerobic thermophilic bacterium which is frequently isolated from high-temperature environments including hot springs and compost [26]. The genus Geobacillus and its members have become potential for applications in biotechnology and bioremediation [27, 28] with a continually increasing industrial interest for their thermostable gene products [29, 30]. Thermophilic microorganisms with an optimum growth temperature of 50°C or above are sources of thermostable enzymes such as amylases, cellulases, chitinases, pectinases, xylanases, proteases, lipases, and DNA polymerases. These enzymes are appropriate for performing biotechnological processes at elevated temperatures [31].
Four different starch-containing media were inoculated with P. thermoglucosidasius strain Pharon1. After incubation for 24 h, the highest amylase activity was recorded using M4 medium as measured using the DNS assay method. The M4 medium was selected for the optimization of amylase production.
To optimize the physical conditions of the enzyme production, statistical approaches namely full factorial and central composite design were applied. Statistical optimization allows quick screening of large experimental domain and explains accurately the role of each tested variable and the combined effects of tested factors. The full factorial approach confirmed the significance of the four tested physical variables (pH, temperature, incubation period, and aeration) on the production of amylase enzyme. The obtained significant and numeric variables (pH, temperature, and incubation period) were used to design the central composite model which is an accurate tool in determining the optimum level for each factor with respect to the effect of the other factors [12, 32].
The maximum enzyme activity (80.6 U/mL/min) was accomplished when the bacterial strain was grown at a temperature at 54.1°C. This agreed with Arfah et al. [33] and Tiwari et al. [34] who reported that the optimum temperature of amylase production was at 55°C by Bacillus sp. RSII-1b and Bacillus tequilensis RG-01, respectively. However, other studies reported the production of amylases at a wider range of temperatures as mentioned and reported by Bekler et al. who obtained the maximum level of amylase from Bacillus paralicheniformis at 60°C [13].
Commercially, the interest has been focused on thermophilic amylase capable of functioning at a low pH range of 4.5–5.5 [35]. A similar result was obtained by Antrim et al. [36] who reported a thermostable α-amylase with activity at pH 5.5. Uguru et al. [37] isolated a strain of Thermoactinomyces thalpophilus which produced an extracellular amylase with optimum pH of 5.0. The thermostable amylases have extensive applications in a number of industrial processes [38].
The incubation of the crude enzyme preparation at 70°C for 30 min before adding the substrate resulted in 2.86 fold increase in its amylase activity compared to the enzyme preparation kept at room temperature, which means that the enzyme required thermal activation energy to exert its full activity. Thermostable amylase produced from P. thermoglucosidasius strain Pharon1 continued to be active over a temperature range. The thermostable enzyme produced in this study lost only 13% of its activity after 120 min of incubation at 100°C. Previous studies on Bacillus subtilis showed that 67% of the original activities were lost at 90°C [39].
It was also reported that α-amylases from B. subtilis KIBGE and B. subtilis, 65 showed optimal activities at 50°C and 60°C, respectively, and 28% reduction in enzyme activity was observed at 70°C, whereas incubation at 80°C resulted in complete inactivated of the enzyme [40].
Thermostable amylolytic enzymes are of great importance in modern biotechnological applications. They are used to produce many valuable products in different industries such as glucose, dextrose syrup, crystalline dextrose, maltose, and maltodextrins [41–43].
In conclusion, the enzyme produced by the novel local isolate, P. thermoglucosidasius strain Pharon1 (MG965879) showed strong degradative ability under extreme environmental conditions mark it as a potential candidate for biotechnological applications. Further characterization of the produced enzymes needs to be carried on.
Conclusions
The novel bacterial strain P. thermoglucosidasius strain Pharon1 can be used in amylase production because it could be considered a cost-effective source for the enzyme. Where it requires a relatively low incubation temperature (55oC) to produce the thermophilic amylase at acidic pH of 5.6 within about 98 h of incubation; also, the enzyme produced showed thermostability after exposure to 100°C for 120 min.
Supplementary Information
Additional file 1: Table S1. Statistical analysis for amylase activity in the full factorial design. Table S2. Statistical analysis in the central composite design.
Acknowledgements
The authors are greatly thankful to the Department of Microbiology, Faculty of Science, Ain Shams University, Egypt, and to Ms. Amira Gamal, Assistant lecturer at Egyptian Atomic Energy Authority for her collaboration in Statistical factorial designs.
Abbreviations
- DNS
Dinitrosalicylic acid
- CCD
Central composite design
- RSM
Response surface methodology
- R2
Coefficient of determination
Authors’ contributions
AMS and HAES designed and carried out the entire experimental work, analyzed the data, and wrote the manuscript. EHS supervised the entire work and reviewed the final manuscript. The authors have read and approved the final manuscript.
Funding
The entire work was carried out in Ain shams University with the available resource; some analyses were done with self-funding.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that there are no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mageswari A, Subramanian P, Chandrasekaran S, Sivashanmugam K, Babu S, Gothandam KM. Optimization and immobilization of amylase obtained from halotolerant bacteria isolated from solar salterns. J Genet Eng Biotechnol. 2012;10:201–208. doi: 10.1016/j.jgeb.2012.09.001. [DOI] [Google Scholar]
- 2.Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B. Microbial α-amylases: a biotechnological perspective. Process Biochem. 2003;38:1599–1616. doi: 10.1016/S0032-9592(03)00053-0. [DOI] [Google Scholar]
- 3.Ashraf H, Iqbal J, Qadeer MA. Production of alpha amylase by Bacillus licheniformis using an economical medium. Bioresour Technol. 2003;87:57–61. doi: 10.1016/S0960-8524(02)00198-0. [DOI] [PubMed] [Google Scholar]
- 4.Apar DK, Özbek B. α-Amylase inactivation by temperature during starch hydrolysis. Process Biochem. 2004;39:1137–1144. doi: 10.1016/S0032-9592(03)00224-3. [DOI] [Google Scholar]
- 5.Saxena RK, Dutt K, Agarwal L, Nayyar P. A highly thermostable and alkaline amylase from a Bacillus sp. PN5. Bioresour Technol. 2007;98:260–265. doi: 10.1016/j.biortech.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Vaikundamoorthy R, Rajendran R, Selvaraju A, Moorthy K, Perumal S. Development of thermostable amylase enzyme from Bacillus cereus for potential antibiofilm activity. Bioorg Chem. 2018;77:494–506. doi: 10.1016/j.bioorg.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Fooladi J, Sajjadian A. Screening the thermophilic and hyperthermophilic bacterial population of three Iranian hot-springs to detect the thermostable α-amylase producing strain. Iran J Microbiol. 2010;2:46–50. [PMC free article] [PubMed] [Google Scholar]
- 8.Mantiri FR, Rumende RRH, Sudewi S. Identification of α-amylase gene by PCR and activity of thermostable α-amylase from thermophilic Anoxybacillus thermarum isolated from Remboken hot spring in Minahasa, Indonesia. IOP Conf Series Earth Environ Sci. 2019;217:012045. doi: 10.1088/1755-1315/217/1/012045. [DOI] [Google Scholar]
- 9.Fentahun M, Kumari PV. Isolation and screening of amylase producing thermophilic spore forming Bacilli from starch rich soil and characterization of their amylase activity. Afr J Microbiol Res. 2017;11:851–859. [Google Scholar]
- 10.Indriati G, Megahati RRP. Isolation of thermophilic bacteria and optimizing the medium growth conditions. Int J Curr Microbiol Appl Sci. 2018;7:1457–1464. doi: 10.20546/ijcmas.2018.701.177. [DOI] [Google Scholar]
- 11.Ojha SK, Singh PK, Mishra S, Pattnaik R, Dixit S, Verma SK. Response surface methodology based optimization and scale-up production of amylase from a novel bacterial strain, Bacillus aryabhattai KIIT BE-1. Biotechnol Rep. 2020;27:e00506. doi: 10.1016/j.btre.2020.e00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gangadharan D, Sivaramakrishnan S, Nampoothiri KM, Sukumaran RK, Pandey A. Response surface methodology for the optimization of alpha amylase production by Bacillus amyloliquefaciens. Bioresour Technol. 2008;99:4597–4602. doi: 10.1016/j.biortech.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 13.Bekler FM, Yalaz S, Guven RG, Guven K. Optimization of the thermostable alkaline and Ca-dependent α−amylase production from Bacillus paralicheniformis by statistical modeling. J Serb Chem Soc. 2019;84:1093–1104. doi: 10.2298/JSC190227021B. [DOI] [Google Scholar]
- 14.Garg D, Kaur DM. Extraction, purification and characterization of enzyme amylase from Bacillus amyloliquefaciens. Int J Adv Eng Sci. 2013;3:158–159. [Google Scholar]
- 15.Azadian F, Badoei-dalfard A, Namaki-Shoushtari A, Karami Z, Hassanshahian M. Production and characterization of an acido-thermophilic, organic solvent stable cellulase from Bacillus sonorensis HSC7 by conversion of lignocellulosic wastes. J Genet Eng Biotechnol. 2017;15:187–196. doi: 10.1016/j.jgeb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazanas N. Proteolytic activity of microorganisms isolated from freshwater fish. Appl Environ Microbiol. 1968;16:128–132. doi: 10.1128/AM.16.1.128-132.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Thomas L, Madden AA, Schäffer ZZ, Miller W, Lipman DJ. Gapped BLAST and PSI BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurumurthy DM, Neelagund SE. Geobacillus sp. Iso 5, a novel amylase-producing thermophile from thermal springs in Konkan region of Southern India. J Earth Sci. 2010;21:319–322. doi: 10.1007/s12583-010-0248-0. [DOI] [Google Scholar]
- 19.Rao JUM, Satyanarayana T. Biophysical and biochemical characterization of a hyperthermostable and Ca2+ independent α-amylase of an extreme thermophile Geobacillus thermoleovorans. Appl Biochem Biotechnol. 2008;150:205–219. doi: 10.1007/s12010-008-8171-x. [DOI] [PubMed] [Google Scholar]
- 20.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 21.Ballschmiter M, Fütterer O, Liebl W. Identification and characterization of a novel intracellular alkaline α-amylase from the hyperthermophilic bacterium Thermotoga maritima MSB8. Appl Environ Microbiol. 2006;72:2206–2211. doi: 10.1128/AEM.72.3.2206-2211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uzyol KS, Akbulut BS, Denizci AA, Kazan D. Thermostable alpha-amylase from moderately halophilic Halomonas sp. AAD21. Turk J Biol. 2012;36:327–338. [Google Scholar]
- 23.Saeed AM, Sayed HAE, El-Shatoury EH. Optimizing the reduction of molybdate by two novel thermophilic bacilli isolated from Sinai, Egypt. Curr Microbiol. 2020;77:786–794. doi: 10.1007/s00284-020-01874-y. [DOI] [PubMed] [Google Scholar]
- 24.Mohammad BT, Al Daghistani HI, Jaouani A, Abdel-Latif S, Kennes C. Isolation and characterization of thermophilic bacteria from jordanian hot springs: Bacillus licheniformis and Thermomonas hydrothermalis isolates as potential producers of thermostable enzymes. Int J Microbiol. 2017;2017:1–12. doi: 10.1155/2017/6943952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta GN, Srivastava S, Khare SK, Prakash V. Extremophiles: an overview of microorganism from extreme environment. Int J Agric Environ Biotechnol. 2014;7:371–380. doi: 10.5958/2230-732X.2014.00258.7. [DOI] [Google Scholar]
- 26.Mohr T, Aliyu H, Küchlin R, Polliack S, Zwick M, Neumann A, Cowan D, De Maayer P. CO-dependent hydrogen production by the facultative anaerobe Parageobacillus thermoglucosidasius. Microb Cell Factories. 2018;17:108–119. doi: 10.1186/s12934-018-0954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houde A, Kademi A, Leblanc D. Lipases and their industrial applications. Appl Biochem Biotechnol. 2004;118:155–170. doi: 10.1385/ABAB:118:1-3:155. [DOI] [PubMed] [Google Scholar]
- 28.Meintanis C, Chalkou KI, Kormas KA, Karagouni AD. Biodegradation of crude oil by thermophilic bacteria isolated from a volcano island. Biodegradation. 2006;17:3–9. doi: 10.1007/s10532-005-6495-6. [DOI] [PubMed] [Google Scholar]
- 29.Lama L, Calandrelli V, Gambacorta A, Nicolaus B. Purification and characterization of thermostable xylanase and β-xylosidase by the thermophilic bacterium Bacillus thermantarcticus. Res Microbiol. 2004;155:283–289. doi: 10.1016/j.resmic.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Schallmey M, Singh A, Ward OP. Developments in the use of Bacillus species for industrial production. Can J Microbiol. 2004;50:1–17. doi: 10.1139/w03-076. [DOI] [PubMed] [Google Scholar]
- 31.Singh G, Bhalla A, Kaur P, Capalash N, Sharma P. Laccase from prokaryotes: a new source for an old enzyme. Rev Environ Sci Biotechnol. 2011;10:309–326. doi: 10.1007/s11157-011-9257-4. [DOI] [Google Scholar]
- 32.Anderson-Cook CM, Borror CM, Montgomery DC. Response surface design evaluation and comparison. J Stat Plann Inference. 2009;139:629–641. doi: 10.1016/j.jspi.2008.04.004. [DOI] [Google Scholar]
- 33.Arfah RA, Ahmad A, Djide MN, Anis M, Zakir M. Production optimization and characterization of amylase enzyme isolated from termofil bacteria Bacillus sp RSAII-1b from Lejja hot spring South Sulawesi. Am J Biomed Life Sci. 2015;3:115–119. doi: 10.11648/j.ajbls.20150306.13. [DOI] [Google Scholar]
- 34.Tiwari S, Shukla N, Mishra P, Gaur R. Enhanced production and characterization of a solvent stable amylase from solvent tolerant Bacillus tequilensis RG-01: thermostable and surfactant resistant. Sci World J. 2014;2014:1–11. doi: 10.1155/2014/972763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkelmann G. Microbial degradation of natural products. Weinheim: VCH, Germany; 1992. pp. 85–191. [Google Scholar]
- 36.Antrim RL, Solheim BA, Solheim L, Auterinen AL, Cunefare J, Karppelin S. A new Bacillus licheniformis alpha-amylase capable of low pH liquefaction. Starch-Stärke. 1991;43:355–360. doi: 10.1002/star.19910430907. [DOI] [Google Scholar]
- 37.Uguru GC, Akinyanju JA, Sani A. The use of sorghum for thermostable amylase production from Thermoactinomyces thalpophilus. Lett Appl Microbiol. 1997;25:13–16. doi: 10.1046/j.1472-765X.1997.00158.x. [DOI] [Google Scholar]
- 38.Kumar RSS, Singh SA, Rao AA. Thermal stability of α-amylase from malted jowar (Sorghum bicolor) J Agric Food Chem. 2005;53:6883–6888. doi: 10.1021/jf0501701. [DOI] [PubMed] [Google Scholar]
- 39.Shafaat S, Akram M, Rehman A. Isolation and characterization of a thermostable-amylase from Bacillus subtilis. Afr J Microbiol Res. 2011;5:3334–3338. doi: 10.5897/AJMR11.666. [DOI] [Google Scholar]
- 40.Bano S, Qader SAU, Aman A, Syed MN, Azhar A. Purification and characterization of novel α-amylase from Bacillus subtilis KIBGE HAS. AAPS Pharm Sci Tech. 2011;12:255–261. doi: 10.1208/s12249-011-9586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamford TLM, Stamford NP, Coelho LCBB, Araujo JM. Production and characterization of a thermostable α-amylase from Nocardiopsis sp. endophyte of yam bean. Bioresour Technol. 2001;76:137–141. doi: 10.1016/S0960-8524(00)00089-4. [DOI] [PubMed] [Google Scholar]
- 42.Gomes I, Gomes J, Steiner W. Highly thermostable amylase and pullulanase of the extreme thermophilic eubacterium Rhodothermus marinus: production and partial characterization. Bioresour Technol. 2003;90:207–214. doi: 10.1016/S0960-8524(03)00110-X. [DOI] [PubMed] [Google Scholar]
- 43.Asgher M, Asad MJ, Rahman SU, Legge RL. A thermostable α-amylase from a moderately thermophilic Bacillus subtilis strain for starch processing. J Food Eng. 2007;79:950–955. doi: 10.1016/j.jfoodeng.2005.12.053. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Statistical analysis for amylase activity in the full factorial design. Table S2. Statistical analysis in the central composite design.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.