Abstract
Glioblastomas (GBMs) are the most aggressive tumor type of the central nervous system, mainly due to their high invasiveness and innate resistance to radiotherapy and chemotherapy, with temozolomide (TMZ) being the current standard therapy. Recently, brachyury was described as a novel tumor suppressor gene in gliomas, and its loss was associated with increased gliomagenesis. Here, we aimed to explore the role of brachyury as a suppressor of glioma invasion, stem cell features, and resistance to TMZ. Using gene-edited glioma cells to overexpress brachyury, we found that brachyury-positive cells exhibit reduced invasive and migratory capabilities and stem cell features. Importantly, these brachyury-expressing cells have increased expression of differentiation markers, which corroborates the results from human glioma samples and in vivo tumors. Glioma cells treated with retinoic acid increased the differentiation status with concomitant increased expression of brachyury. We then selected TMZ-resistant (SNB-19) and TMZ-responsive (A172 and U373) cell lines to evaluate the role of brachyury in the response to TMZ treatment. We observed that both exogenous and endogenous brachyury activation, through overexpression and retinoic acid treatment, are associated with TMZ sensitization in glioma-resistant cell lines. In this study, we demonstrate that brachyury expression can impair aggressive glioma features associated with treatment resistance. Finally, we provide the first evidence that brachyury can be a potential therapeutic target in GBM patients who do not respond to conventional chemotherapeutic drugs.
Electronic supplementary material
The online version of this article (10.1007/s13311-020-00911-9) contains supplementary material, which is available to authorized users.
Key Words: Gliomas, brachyury, EMT, stem, temozolomide, therapy, TBXT
Introduction
Gliomas are the most common primary brain tumors and present high morbidity and mortality [1]. Glioblastoma (WHO grade IV; GBM) is the most aggressive and the most common form of glioma [2]. Temozolomide (TMZ) is the only chemotherapeutic drug that has been confirmed to significantly prolong the overall survival of GBM patients, yet the median survival after receiving standard treatment is only 12–15 months [2–4]. Innate resistance to current treatments, including TMZ, is a major cause of this dismal scenario, which indicates that overcoming TMZ resistance is critical to improving treatment outcomes. The existence of glioma stem cells (GSCs) has been suggested as one of the causes of resistance to chemo- and radiotherapy [5, 6]. Another mechanism of therapy resistance may be epithelial-to-mesenchymal transition (EMT), a process by which cells acquire a more invasive and infiltrative phenotype, which can exhibit crosstalk with GSC features [7]. The high infiltrative and invasive capacities of gliomas hamper complete tumor resection, leading to tumor recurrence and, ultimately, patient death [3].
The transcription factor brachyury (TBXT) has been described to be upregulated in several epithelial solid tumors [8–16]. Moreover, brachyury acts as a master regulator of tumor cell migration, invasion, and metastasis through EMT [8, 11, 12, 17]. It increases tumor stem cell properties [18–20] and promotes resistance to cytotoxicity-based therapy [19, 21, 22]. Consequently, an anti-brachyury vaccine was developed [23–26] and is being tested in a phase II clinical trial (www.clinicaltrials.gov, 2015 – NCT02383498) in patients with chordomas. However, we recently provided evidence that in gliomas, brachyury acts as a tumor suppressor, promoting cell death through autophagy [27]. Importantly, brachyury is an independent favorable prognostic biomarker for glioma patients [27]. Due to its suppressive behavior in gliomas and its role in invasion and resistance to therapy in other tumor types, in this work, we explored the potential role of brachyury in features associated with resistance to conventional TMZ treatment in gliomas.
Materials and Methods
Cell Lines and Brachyury Transfection
Three different GBM cell line models were used for functional studies: SNB-19 (DSMZ—German Collection of Microorganisms and Cell Cultures), A172 (ATCC-American Type Culture Collection, MD), and U373 (JF Costello Laboratory). Cell culture conditions were as described previously [27]. GBM cell lines were transfected with full-length human brachyury cloned into the pcDNA4/T0 vector (Invitrogen S.A., Barcelona, Spain), designated pcBrachy, or with pcDNA4/T0 empty vector (4/T0) as previously described [27].
Transcript Analysis by Semiquantitative PCR and qRT-PCR
Total RNA was isolated from tumor samples and cell lines using TRIzol Reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA). One microgram of RNA was reverse-transcribed using the Phusion RT-PCR Kit (Finnzymes, Thermo Fisher Scientific, Waltham, MA) as recommended by the manufacturer. For semiquantitative PCR, cDNA was amplified as previously described [8], and the products were separated on a 2% agarose gel. The qRT-PCR steps were performed using Sso Fast EvaGreen Supermix (Bio-Rad, Hercules, CA) and a CFX96 detection system (Bio-Rad). Optimized cycling conditions for qRT-PCR were as follows: enzyme activation for 30 s at 95 °C; 40 cycles of denaturation at 95 °C for 5 s and annealing/extension at 59 °C for 5 s. A melting curve was generated at 65 °C–95 °C (in 0.5 °C steps, each for 5 s/step). The expression levels were normalized to those of ACTB by ΔCT (relative expression) or by the ΔΔCT method (fold change). Further details about primer sequences are available in Table S1.
Western Blot
Cells were lysed in buffer containing 50 mM Tris pH 7.6–8, 150 mM NaCl, 5 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 10 mM Na4P2O7, 1% NP-40, and 1/7 protease cocktail inhibitors (Roche, Basel, Switzerland). Proteins were resolved on standard 12% SDS-PAGE gels and transferred onto nitrocellulose membranes. Immunodetection was achieved using antibodies against human β-actin (1:1000, sc-1616, Santa Cruz Biotechnology, Heidelberg, Germany), MGMT (1:500, 357000, Zymed, ThermoFisher Scientific, Waltham, MA), MLH1 (1:500, 554073, BD Pharmingen, BD Biosciences, San Jose, CA), MLH3 (1:500, sc-25313, Santa Cruz Biotechnology), MSH2 (1:500, NA27, Calbiochem, Merck Millipore, Darmstadt, Germany), MSH6 (1:500, 610919, BD Biosciences), PMS2 (1:500, BD Biosciences), PARP1/2 (1:1000, 9532S, Cell Signaling, Leiden, The Netherlands), BCL2 (1:200, 2870S, Cell Signaling), caspase-3 (1:1000, 9662P, Cell Signaling), and cleaved caspase-3 (1:1000, 9664P, Cell Signaling). Blots were visualized with peroxidase-conjugated secondary anti-rabbit (1:2000, sc-2004, Santa Cruz Biotechnology), anti-goat (1:2000, sc-2020, Santa Cruz Biotechnology), or anti-mouse (1:2000, sc-2031, Santa Cruz Biotechnology) antibody and chemiluminescence (SuperSignal West Femto Chemiluminescent Substrate; ThermoFisher Scientific).
Immunofluorescence Microscopy
Cells were plated on glass coverslips placed into 12-well plates and allowed to adhere overnight. Cells were fixed with 4% paraformaldehyde (PFA) followed by permeabilization with 0.1% Triton X-100 in 1× PBS. Cells were blocked in 10% FBS and labeled for 1 h at room temperature with different primary antibodies as follows: anti-Nestin (1:1000, MAB5326, Merck Millipore), MAP2 (1:100, M4403, Sigma-Aldrich, St. Lousis, MO), SOX2 (1:200, AB5603, Millipore), β-tubulin (1:500, sc-5274, Santa Cruz Biotechnology), or F-actin (1:100, Alexa Fluor® 568 Phalloidin, Life Technologies-Molecular Probes, Waltham, MA). Then, the cells were washed 3× for 5 min in 1× PBS and incubated again at room temperature for 1 h with a secondary antibody (anti-rabbit Alexa-488, anti-goat Alexa-568, anti-mouse Alexa-488, or Alexa-594 antibody, all diluted at 1:1000) obtained from Life Technologies-Molecular Probes. Coverslips were mounted on microscope slides with Vectashield with DAPI (Vector Laboratories, Burlingame, CA). Digital images were recorded with an Olympus DP30BW cooled CCD camera (Olympus BX61, Olympus Corporation, Hamburg, Germany).
Laminin Adhesion Assay
Culture 96-well plates were coated overnight at 4 °C with human laminin (10 μg/ml, Life Technologies) dissolved in 1× PBS. The wells were then washed with 0.1% bovine serum albumin (BSA) dissolved in DMEM and blocked for 1 h (37 °C) with 1% BSA in DMEM. Cells were seeded into extracellular matrix (ECM)-coated 96-well plates (2 × 105 cells/well). After 30, 60, and 90 min of incubation under standard culture conditions, the nonadherent cells were removed by washing with 1× PBS washing. Attached cells were fixed (4% PFA) and stained with crystal violet (5 mg/ml in 2% ethanol). The stained cells were washed, and crystal violet was eluted with 2% SDS. The amount of stain was analyzed by optical density in a microplate reader at 550 nm. Cell attachment was calculated by the difference in the percentage of adherent cells in comparison to controls (ECM-coated well without cells).
Wound Healing Migration Assay
The cells were seeded in 12-well plates and cultured to at least 95% confluence. Monolayer cells were washed with 1× PBS, scraped with a plastic 200 μl pipette tip, and then incubated with fresh DMEM. The “wounded” areas were imaged by phase-contrast microscopy at different time points. The relative migration distance was calculated as previously described [8, 28–30].
Matrigel Invasion Assay
Matrigel invasion assays were performed using 8 μm pore size BD BioCoat Matrigel Invasion Chambers (BD Biosciences). Briefly, 2.5 × 104 cells were seeded in the upper part of the chamber in DMEM that was serum and antibiotic-free, whereas the lower compartment contained fresh medium supplemented with 10% FBS and 10 ng/mL EGF (PeproTech, London, UK). After 22 h of incubation at 37 °C, the filter was washed with 1× PBS and fixed with 4% PFA. Then, residual cells were cleared with a cotton swab, the filter was washed with 1× PBS, and invasive cells attached to the lower filter surface were mounted with Vectashield with DAPI. Images were recorded on an Olympus BX61 microscope (Olympus Corporation, Tokyo, Japan), and invasive cells were counted using the ImageJ software (version 1.44), as previously described [8, 30].
Zymography Assay
To detect MMP enzymatic activity, transfected cells with and without brachyury expression were cultured for 24 h in serum-free DMEM that was subsequently used as a conditioned medium. Ten micrograms of protein from the conditioned medium of these cultures was loaded on 10% SDS-PAGE gels containing 1 mg/ml gelatin as substrate for MMP2 and MMP9. Zymograms were run in Tris/glycine SDS running buffer under nondenaturing conditions. After electrophoresis, gels were washed twice in 2% Triton X-100 to remove SDS. Zymograms were subsequently incubated for 16 h at 37 °C in the appropriate MMP substrate buffer (10 mM CaCl2 in 50 mM Tris-HCl, pH 7.5), as described [29, 30]. Proteolytic activity was visualized as the presence of clear bands against a blue background of Coomassie Blue-stained gelatin substrate.
Neurosphere and Self-Renewal Assay
A limiting dilution assay was performed to evaluate the capacity of glioma cells with and without brachyury expression to form neurospheres. Cells were trypsinized and plated in 12-well plates at different densities (100, 250, 500, and 1000 cells/well) in serum-free medium consisting of neurobasal medium (Invitrogen S.A.) supplemented with 1× B27 (Invitrogen S.A.), human recombinant bFGF, and EGF (20 ng/ml each; R&D Systems). The formed neurospheres were counted at 7, 9, and 11 days.
Only the U373 cell line formed spheres and was used for the following experiments. The best cell density for neurosphere formation was used (500 cells/ml), and cells were grown as described above. Neurospheres were imaged over time (0, 7, and 14 days) using an Olympus BX61 microscope (Olympus Corporation) and manually counted. For the self-renewal assay, spheres were collected and centrifuged at 900 rpm for 5 min. The supernatant was removed, and spheres were mechanically dissociated in 1 ml of fresh medium to obtain a single-cell suspension. Cells were counted and replated at a density of 1 × 103 cells/well to evaluate the capacity of self-renewal.
Pharmacologic Exposure
GBM cells, with or without brachyury expression, were exposed to 0.1 μM trans-retinoic acid (RA, Sigma) for 24 and 60 h to induce glioma differentiation.
The half-maximal inhibitory concentration (IC50) of TMZ in combination with or without RA was determined as follows: 4 × 103 cells were plated into 96-well plates in triplicate and allowed to adhere overnight. Cells were exposed to different concentrations of TMZ (10, 25, 50, 100, 200, 500, and 1000 μg/ml) for 72 h, with or without pretreatment with 0.1 μM RA for 24 h. The IC50 was measured by the MTS test (Promega, Madison, WI), and IC50 values were calculated by a nonlinear regression (curve fit) based on sigmoidal dose–response (variable slope) using Prism GraphPad-v5.0a (GraphPad Software, La Jolla, CA). The influence of RA alone on glioma cell viability and morphology was also evaluated.
To analyze the resistance mechanisms associated with TMZ, 2 × 104 cells were plated in a 6-well plate and treated with 600 μg/ml TMZ (the IC50 for pcBrachy cells) or vehicle (1% DMSO) for 72 h. Cells were then collected for protein extraction and analyzed by Western blot analysis.
In Silico Glioma Expression Analysis: Functional Annotation and Correlation Profiles
The expression profiles of genes associated with TBXT in GBM samples (n = 440) were extracted from The Cancer Genome Atlas (TCGA) using the cBioPortal for Cancer Genomics database (www.cbioportal.org/) [31, 32]. Genes with Spearman > 0.25 and q value < 0.01 were considered positively correlated with TBXT. Genes with Spearman < − 0.25 and q value < 0.001 were considered inversely correlated with TBXT. Then, TBXT positively correlated or inversely correlated gene profiles were subjected to KEGG pathway analysis and Gene Ontology functional annotation analysis using the DAVID 6.8 tool (https://david.ncifcrf.gov/home.jsp) [33, 34] against the Homo sapiens background reference. The pathways and biological pathways were considered significant when there was > 2-fold enrichment and p < 0.05. Correlation analysis between TBXT, epithelial (CDH1), mesenchymal (CDH2, SNAI1, SNAI2, VIM, FN1, JUP, NANOG), stemness (CD133, CD15, NES, MSI1), and differentiation (GFAP, OLIG2, TUBB3, MAP2, RBFOX3/NeuN) markers was performed using the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/). RNA sequencing data (represented as log2 (transcripts per kilobase million + 1)) of 681 glioma samples were analyzed (TCGA: GBM, n = 163; LGG, n = 518).
Statistical Analysis
For in vitro assays, simple comparisons between two different conditions were analyzed using Student’s t test and two-way ANOVA (Bonferroni posttest) for comparison of two conditions over time using Prism GraphPad-v5.0a. For the pharmacological analysis, we constructed a dose–response curve using GraphPad v5.0a and calculated the IC50 using a nonlinear regression curve. The level of significance in the statistical analyses is indicated as * for p < 0.05, as ** for p < 0.01, or as *** for p < 0.001.
Results
TBXT Gene Profiling in GBM Patients
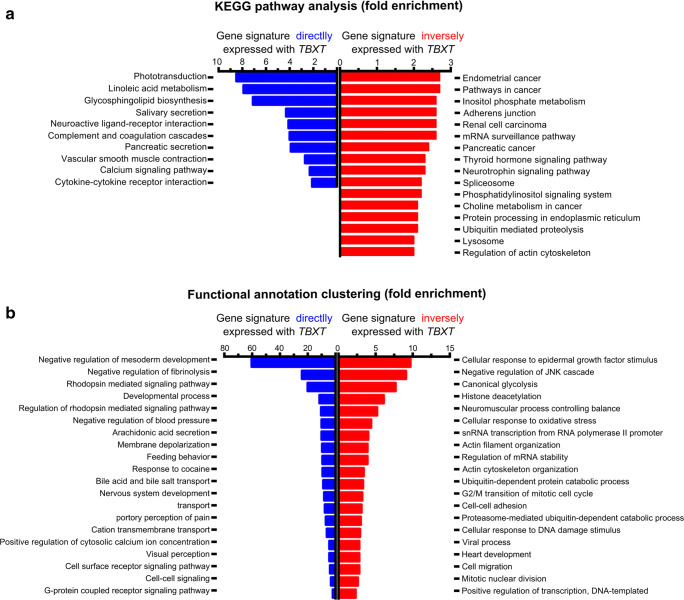
Recently, we reported that brachyury behaves as a suppressor gene in gliomas through the induction of cell death, indicating that brachyury can act in a tissue-dependent manner [27]. As such, we first explored the gene profile associated with the presence of TBXT (coexpressed) and the gene profile inversely associated with TBXT expression in GBM patients. Gene expression analysis was performed in 440 GBM patients from TCGA using the cBioPortal database. The gene signature coexpressed with TBXT (Spearman R > 0.25 and p value < 0.001) and the gene signature inversely expressed with TBXT (Spearman R < − 0.25 and p value < 0.001) were extracted and clustered based on the KEGG pathway and functional annotation pathway using the DAVID 6.8 tool (Fig. 1).
Fig. 1.
The brachyury (TBXT) gene expression profile is associated with nontumorigenic pathways and nervous system development. (a) KEGG pathway analysis of gene profiles coexpressed with TBXT (blue) or inversely expressed with TBXT (red). The positive TBXT gene profile (blue) is associated with nontumorigenic pathways, whereas the inverse TBXT gene profile (red) is associated with cancer pathways. (b) Classification of TBXT gene-associated profiles into functional processes using the DAVID classification system. The gene profile coexpressed with TBXT (blue) is associated with brain development and function. The gene profile inversely expressed with TBXT (red) is associated with tumorigenic functions. All the KEGG and functional clusters shown are with at least 2-fold enrichment and a p value < 0.05
We found that the gene signature coexpressed with TBXT is enriched in nontumoral and developmental processes, including several functions related to nervous system development (Fig. 1a, b, blue). In contrast, the gene signature associated with low levels of TBXT, which reflects the majority of GBM patients (low or no expression of TBXT), is specifically enriched in cancer pathways such as migration, actin cytoskeleton regulation, and response to DNA damage stimuli (Fig. 1a, b, red). The TBXT-associated gene profiles indicate that TBXT may be important in migration, invasion, and EMT-related processes, as well as in differentiation/stemness and response to TMZ (DNA damage stimuli) treatment.
Brachyury Inhibits Glioma Cell Migration and Invasion
Based on the in silico results, we further explored the role of brachyury expression in glioma cell migration, adhesion, migration, and EMT processes in vitro using three glioma cell models modulated to express brachyury (Fig. S1A). We observed that brachyury overexpression (pcBrachy) markedly decreased cell migration (Fig. 2a; Fig. S1B) and invasion (Fig. 2b; Fig. S1C) in the extracellular matrix when compared with 4/T0 control cells in all in vitro cell models. Accordingly, brachyury negatively influenced the time required to adhere to the laminin substrate (Fig. 2c; Fig. S1D). However, no effect was observed on MMP2 and MMP9 activity by zymography in vitro (Fig. S1E), or on MMP2, MMP9, MMP14, or MMP24 expression in vitro (Fig. S1F), or in glioma samples (Spearman R < 0.25) (Fig. S1G).
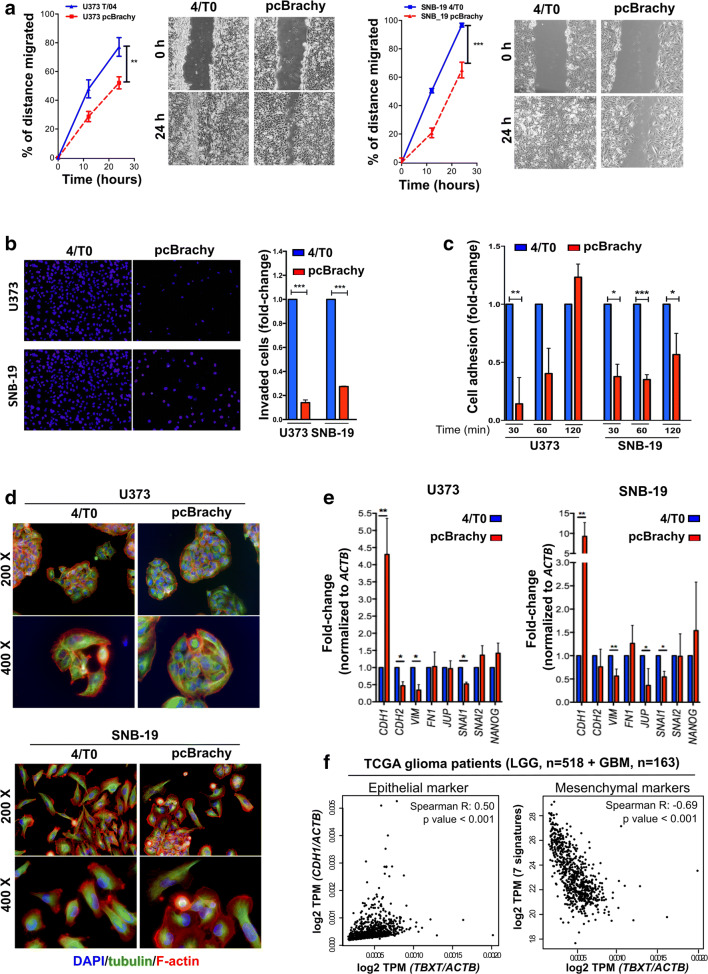
Fig. 2.
The presence of brachyury decreases glioma cell migration and invasiveness through a mesenchymal-epithelial transition phenotype. Brachyury (pcBrachy) inhibited glioma cell migration (a) and invasion (b) compared with that of the negative cell line control (4/T0). (c) Brachyury decreases the adhesion capacity of glioma cells to laminin. (d) Fluorescence microscopic staining of tubulin (green) and F-actin (red) and counterstaining with DAPI to visualize the nuclei (blue). Cells without brachyury (4/T0) present a more undifferentiated phenotype and redistribution of F-actin filaments to lamellipodium- and filopodia-like structures. Overexpression of brachyury (pcBrachy) changes the phenotype of glioma cells to a more cohesive-like phenotype. (e) Expression levels of epithelial markers (E-cadherin or CDH1) as well as mesenchymal markers (N-cadherin or CDH2, SNAIL, SLUG, VIM, FN1, NANOG, and JUP) were examined by qRT-PCR. The same genes were also analyzed in (f) human glioma samples by in silico analysis (TCGA, n = 681 gliomas) using the Gene Expression Profiling Interactive Analysis (GEPIA) program. In vitro data are presented as the means ± S.E.M. of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
We also observed that cells with brachyury overexpression had a more differentiated cellular state, which was characterized by loss of lamellipodium/filopodium projections on the cells (Fig. 2d). Indeed, brachyury promoted an increase in the epithelial marker E-cadherin (CDH1) and a decrease in several mesenchymal markers (N-cadherin or CDH2, SNAI1, SNAI2, JUP, VIM, FN1, NANOG) (Fig. 2e), compatible with a mesenchymal-epithelial transition (MET) phenotype. The same trends were observed in the TCGA glioma dataset (n = 681), in which TBXT was positively correlated with CDH1 (Spearman R = 0.5, p value < 0.001) and inversely correlated with the mesenchymal gene signature (7 genes: CDH2, SNAI1, SNAI2, JUP, VIM, FN1, NANOG) (Fig. 2f).
Brachyury Expression Attenuates the Glioma Stem Cell-like Phenotype
Due to the putative role of brachyury in brain differentiation processes (Fig. 1), we next performed an expression analysis for stem cell markers by qRT-PCR in our cell models with (pcBrachy) or without (4/T0) brachyury expression. We found that brachyury overexpression decreased the expression of the glioma stem cell markers L1CAM, MSI1 (Musashi), NES (Nestin), and SMEK2 (Fig. 3a). Similar results were observed at the protein level both in vitro and in vivo for Nestin and SOX2 staining (Fig. 3b). Importantly, in glioma samples (n = 681), TBXT was also inversely correlated with NES, MSI1, CD133, and CD15 stem cell markers (Fig. 3c), indicating that loss of brachyury is associated with more undifferentiated tumors, as described previously [27].
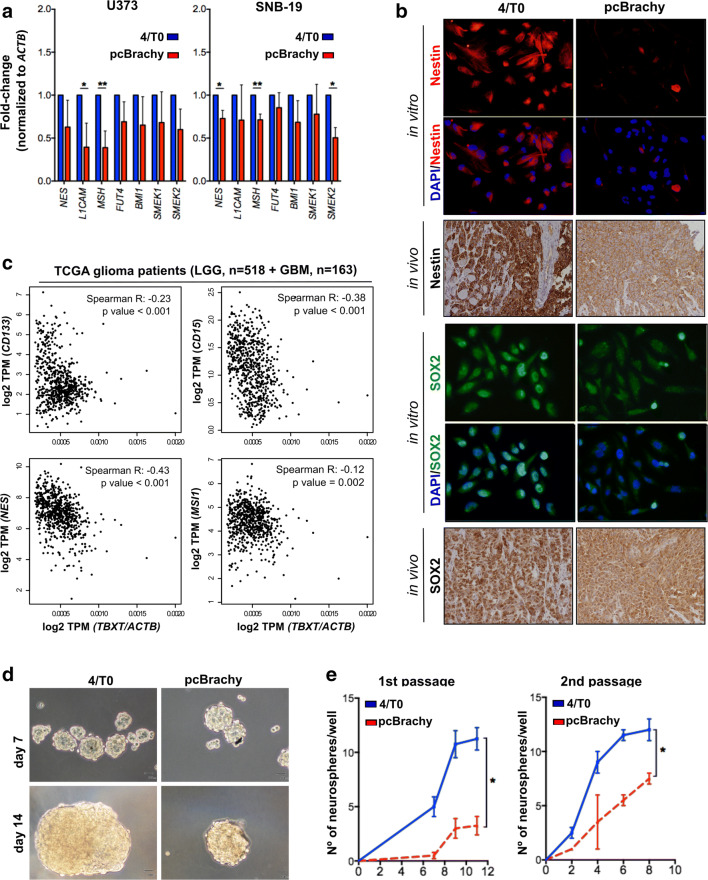
Fig. 3.
Brachyury is inversely correlated with glioma stem cell-like properties. Glioma cells with brachyury overexpression (pcBrachy) show lower expression of stem cell markers at both the mRNA (a) and protein levels (b). The same phenotype is observed in mouse tumors (b, in vivo) formed by glioma cells negative and positive for brachyury [27]. (c) Correlation analysis of stem cell markers (CD133, CD15, Nestin-NES, and Musashi-MSI1) in human glioma samples (TCGA, n = 681 gliomas) using the Gene Expression Profiling Interactive Analysis (GEPIA) program. (d) Representative images of the neurospheres formed by the U373 glioma cell line without (4/T0) or with brachyury (pcBrachy) expression. Magnification at × 20. (e) Quantification of the neurospheres formed by the self-renewal assay. In vitro data are presented as the means ± S.E.M. of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Then, to better understand whether brachyury has a functional role in stem cell-like properties, we performed a limiting dilution assay in U373, A172, and SNB-19 cells. However, A172 and SNB-19 cell lines did not form neurospheres, so we performed the limiting dilution assay only in U373 cells with (pcBrachy) or without (4/T0) brachyury expression (Fig. S2A). U373 4/T0 cells (brachyury negative) were able to form spheres with all cell densities tested in the timeframe of the assay, whereas pcBrachy cells showed a significant decrease in the number and size of spheres formed (Fig. 3d; Fig. S2A). In agreement with these findings, brachyury-positive cells (pcBrachy) exhibited a decreased self-renewal capacity compared with brachyury-negative cells (4/T0) (Fig. 3e). Curiously, when we assessed TBXT expression in the formed neurospheres, we found that pcBrachy neurospheres had lower expression levels compared with those developed under adherent conditions (Fig. S2B), suggesting a negative regulatory mechanism in stem cell-like glioma cells.
Brachyury-Positive Cells Present Increased Differentiation Markers
The association of brachyury with more differentiated tumors was observed in our in vitro models, in which brachyury-positive glioma cells showed an increased expression of the differentiation markers NeuN (Fig. 4a) and MAP2 (Fig. 4a, b). Using an in silico approach, we found that in glioma samples, TBXT was positively correlated with the differentiation markers GFAP, OLIG2, TUBB3, MAP2 (Fig. 4c), and NeuN (RBFOX3 gene) (Fig. S2C). The positive correlation between TBXT and all types of neuronal cells, including astrocytes, oligodendrocytes, and neurons, seems to reflect the heterogeneity of the TCGA dataset, which includes not only cancer cells but also other cells in the microenvironment. These data corroborate our previous findings that TBXT is associated with more differentiated or normal brain tissues [27]. Altogether, these results suggest that TBXT has a role in neuronal differentiation.
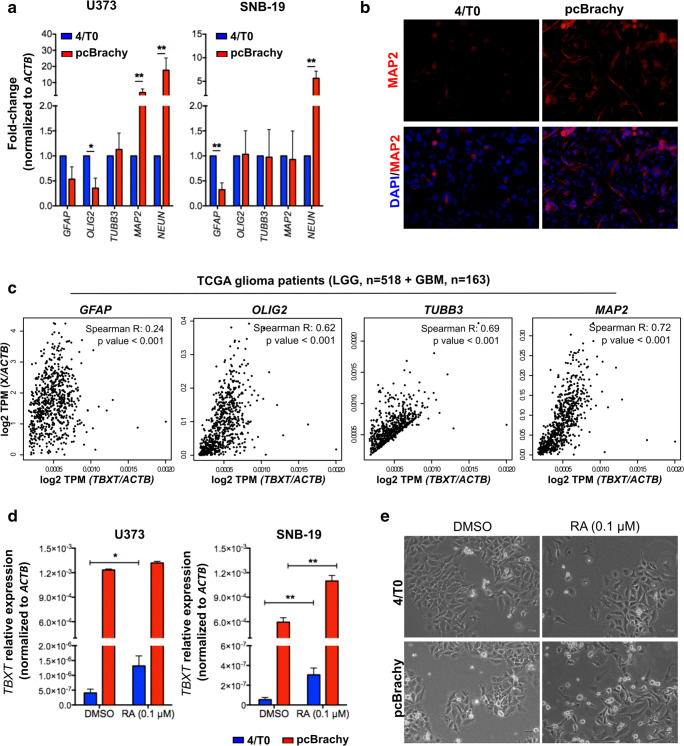
Fig. 4.
Brachyury is associated with an increased differentiation expression profile and is reactivated by retinoic acid treatment. (a) Brachyury overexpression (pcBrachy) in glioma cells is able to increase neural differentiation markers at the mRNA level, as assessed by qPCR. (b) Representative image of MAP2 staining in the SNB-19 cell lines with brachyury expression. (c) Correlation analysis of the brain differentiation markers (GFAP for astrocytes, OLIG2 for oligodendrocytes, TUBB3 and MAP2 for neurons) in human glioma samples (TCGA, n = 681 gliomas) using the Gene Expression Profiling Interactive Analysis (GEPIA) program. (d) Treatment with 0.1 μM retinoic acid (RA) induced endogenous TBXT expression in both control (4/T0) cell lines (after 24 h of RA exposure). (e) Representative images of glioma cells without (4/T0) or with brachyury (pcBrachy) expression treated with 0.1 μM RA after 60 h. pcBrachy cells present major morphological alterations (more differentiated cells) and more cells in suspension (indicative of cell death) compared with control cells (4/T0). In vitro data are presented as the means ± S.E.M. of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
To further assess whether brachyury could be modulated upon cell differentiation, glioma cells were treated with 0.1 μM retinoic acid (RA). We observed that RA treatment was able to induce endogenous TBXT expression after 24 h (Fig. 4b, d) until the end of the experiment (60 h; Fig. S2D), especially in negative cell lines (4/T0). Moreover, pcBrachy cells treated with RA had more evident morphological changes compared with 4/T0 cells, indicating that brachyury-expressing cells undergo faster differentiation, with a concomitant increase in cells in the medium (indicative of cell death) (Fig. 4e). Images of control cells at time 0 h are shown in Fig. 2e.
Brachyury Sensitizes the TMZ-Resistant SNB-19 Glioma Cell Line
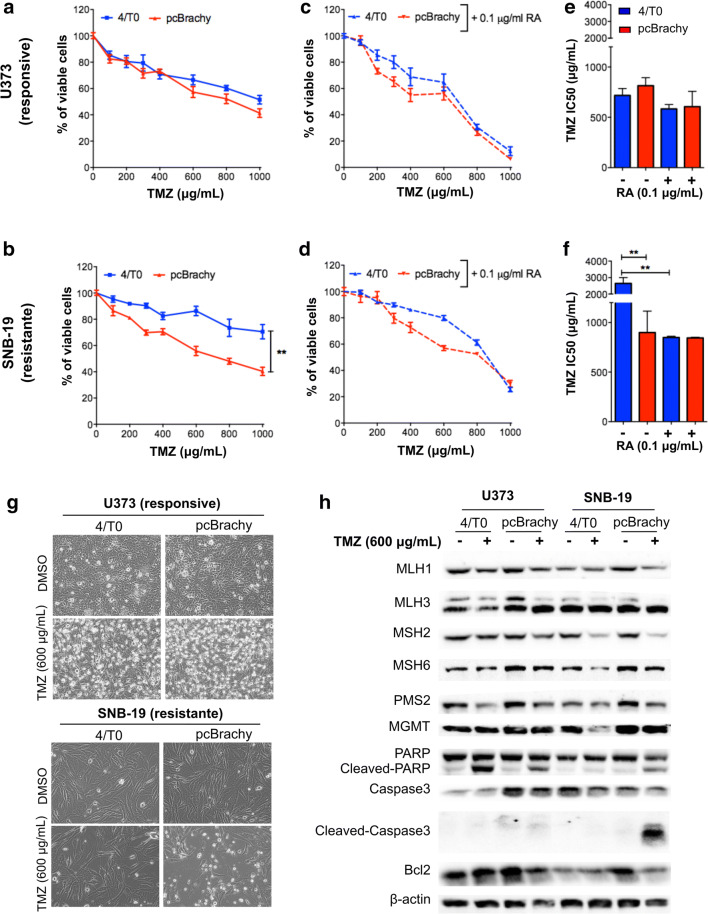
We further asked whether brachyury could influence the glioma response to TMZ-based chemotherapy. For that, we first exposed the wild-type cell models (U373, SNB-19, A172) to different concentrations of TMZ for 72 h, after which their viability was measured, and the IC50 was determined (Fig. S3A, B). We observed that the SNB-19 cell line was highly resistant to TMZ, whereas U373 and A172 were considered less resistant or responsive to TMZ, as previously described [35]. The same approach was used in glioma cells without (4/T0) and with brachyury (pcBrachy) expression. In less resistant glioma cells (U373 and A172), brachyury did not affect the TMZ response (Fig. 5a, e; Fig. S3C). However, in highly resistant cells (SNB-19), brachyury overexpression significantly increased TMZ-mediated cytotoxicity, resulting in a lower IC50 compared with that in brachyury-negative cells (4/T0) (Fig. 5b, f).
Fig. 5.
Brachyury sensitizes resistant glioma cells to temozolomide treatment. (a, b) Effect of temozolomide (TMZ) on cell viability in responsive (A—U373) and resistant (b)—SNB-19) glioma cells with brachyury expression. Brachyury overexpression promotes a higher sensitization to TMZ only in the resistant cells (SNB-19 pcBrachy). (c, d) Effect of TMZ on cell viability after 24 h of pretreatment with 0.1 μM retinoic acid (RA). An increased response to TMZ was observed in all cells treated with RA. (e, f) Brachyury has no effect on the sensitive cell line (e), and it decreases the IC50 in SNB-19-resistant TMZ cells (f). RA treatment decreases the IC50 in brachyury-negative SNB-19 (4/T0)-resistant TMZ cells. (g) Bright-light microscopy images of cells with (pcBrachy) or without (4/T0) brachyury expression treated (72 h) with the IC50 TMZ concentration for both pcBrachy cells (600 μg/mL). The control used was DMSO. (h) Western blot analysis of molecules involved in TMZ resistance. In vitro data are presented as the mean ± S.E.M. of three independent experiments in triplicate. **p < 0.01
We observed that RA exposure could induce endogenous TBXT expression (Fig. 4d), so we pretreated glioma cells with 0.1 μM RA for 24 h to first re-express endogenous brachyury in all cell line models before TMZ treatment. This allowed us to better understand whether endogenous brachyury is enough to sensitize resistant glioma cells to TMZ, as observed in the SNB-19 pcBrachy overexpressing cells. The concentration of RA used (0.1 μM) did not significantly affect glioma viability (Fig. S3D). The combination of RA and TMZ did not significantly affect the therapeutic response of responsive cell lines (Fig. 5c, e). However, this therapeutic combination resulted in a higher response to TMZ in the brachyury-negative resistant TMZ cell line (SNB-19 4/T0), with a similar IC50 found for the SNB-19 pcBrachy cells (Fig. 5d, f).
Aiming to understand the mechanisms by which brachyury may affect TMZ sensitization, we investigated some well-established mechanisms of TMZ resistance, including MGMT, base-excision repair (BER), and mismatch repair (MMR) proteins. To that end, cells were treated with 600 μg/ml TMZ (the IC50 determined for all pcBrachy cell models) for 72 h, and protein was extracted. We observed that both U373 4/T0 and pcBrachy cells showed reduced cell viability, as expected, whereas in the resistant SNB-19 cell line, only the cells with pcBrachy exhibited cell death at that concentration (Fig. 5g). We showed that brachyury expression did not influence MGMT either at the promoter methylation or protein level (Fig. S3E, F; Fig. 5h). In addition to the alterations expected by TMZ treatment, some MMR proteins (such as MSH6, PMS2) were decreased, and cell death proteins (cleaved PARP, caspase-3) were increased in the responsive cells (U373, A172) (Fig. 5h; Fig. S3F), and no other significant alterations were found. Regarding the role of brachyury in the resistant SNB-19 cell line, the significant change found in our analysis was the increased cleaved PARP and caspase-3 levels in SNB-19 pcBrachy cells, which was absent in the resistant SNB-19 4/T0 cells (Fig. 5h). This result indicates that brachyury can prime cells to respond to cell death stimuli.
Discussion
A novel role of brachyury as a tumor suppressor gene was recently reported in gliomas [27]. Importantly, brachyury was shown to be an independent favorable prognostic factor for glioma patients [27]. These results, together with the well-established oncogenic role described in other solid tumors [8–17, 19, 20, 36], indicate that brachyury may have a tissue-specific function.
In the present study, we explored the role of brachyury in glioma cell invasion, stemness, differentiation, and resistance to temozolomide (TMZ) therapy. As such, we investigated the glioma gene signatures associated with the presence or absence of brachyury (TBXT) expression in more than 440 GBM patients from a TCGA dataset. We found that the gene signature associated with TBXT expression was enriched in nontumoral pathways, including brain development functions. On the other hand, the gene signature inversely associated with TBXT was highly enriched for cancer-associated pathways such as migration, adhesion, actin cytoskeleton modulation, and response to DNA damage stimuli.
Previous studies in epithelial cancers have provided evidence that brachyury promotes cancer progression by inducing EMT and by increasing stem cell-like properties [8, 17, 19, 20] that ultimately result in increased resistance to conventional chemotherapy [19, 21, 22]. However, brachyury gene signatures in gliomas suggest that low levels or absence of brachyury is associated with more invasive tumors and potentially with resistance to the DNA damage chemotherapeutic agent TMZ. In contrast, high levels of brachyury are associated with more differentiated tumors. Indeed, we observed that brachyury-negative glioma cells present increased motility, adhesion, and invasion capabilities than brachyury-overexpressing cells. Our results show that brachyury in gliomas can induce MET with an increased expression of CDH1 and a decrease in several mesenchymal markers both in cell lines and clinical samples. Importantly, brachyury overexpression was significantly associated with decreased stem cell-like features and increased cell differentiation (both in vitro, in vivo, and in human samples). Interestingly, we found that when we forced glioma differentiation with retinoic acid (previously described to induce neuronal differentiation [37]), brachyury was re-expressed in brachyury-negative glioma cell lines, indicating a potential role in central nervous system development. These results are concordant with its expression in healthy brain tissues [18, 27].
Taking into consideration our results and in silico analysis, in which brachyury seems to be associated with DNA damage response stimuli, we hypothesized that brachyury could influence the response to TMZ therapy. We found that brachyury overexpression was able to sensitize a highly TMZ-resistant glioma cell line (SNB-19) but did not affect the less resistant or more responsive glioma cell lines (U373 and A172). Brachyury expression activates cell death (PARP and caspase-3 cleavage) after TMZ treatment in the highly TMZ-resistant cell line. Additionally, endogenous brachyury was induced by RA treatment in the brachyury-negative resistant cell line (4/T0). Indeed, a recent report showed that trans-retinoic acid is able to enhance TMZ-induced autophagy in the human U251 glioma cell line [38]. Clinically, the use of RA as an effective chemosensitizing agent has been controversial. Some clinical studies have indicated that RA in combination with TMZ did not significantly increase overall patient survival [39], whereas others have shown a therapeutic effect in individual patients with malignant gliomas [40, 41]. It is still unknown whether clinically administered RA is able to target all cancer cells or if it is sufficient to reactivate and maintain brachyury expression in an in vivo context and therefore has a clinical impact. Although the exact mechanism is still unknown, we hypothesize that brachyury is not a significant driver for therapy response but may only confer “competence” to resistant glioma cells to respond to the DNA damage promoted by the action of TMZ. Indeed, we have shown that brachyury in glioma cells increases the expression of several proapoptotic proteins (pRAD, p-P53, BAX, FADD) and decreases antiapoptotic proteins (cIAP, XIAP, HSP70), without observed cleavage of caspase-3 at basal conditions [27], as also observed here.
In conclusion, our study demonstrated that brachyury expression in gliomas could decrease cell motility, invasion, and stem cell-like properties with a concomitant increase in cell differentiation. Importantly, the data presented here allowed the identification of a potential therapeutic alternative for glioma patients who do not respond to TMZ chemotherapy through induction of brachyury expression with RA pretreatment. Although our data have the limitation of using only one cell line resistant to TMZ, we hypothesize that brachyury reactivation in patients resistant to conventional chemotherapy may have potential utility in the clinic. Future research is urgently needed to clinically determine the role of brachyury as a chemosensitizing agent in vivo.
Electronic Supplementary Material
(DOC 3275 kb)
(PDF 483 kb)
Acknowledgments:
This study was supported by the ICVS and Barretos Cancer Hospital internal research founds, by the Portuguese Fundação para a Ciência e a Tecnologia (FCT) projects PTDC/BEX-BID/5410/2014 and UID/BIM/04773/2013 CBMR to RP Andrade and by PTDC/SAU-TOX/114549/2009-FCOMP-01-0124-FEDER-016057, PTDC/SAU-ONC/115513/2009-FCOMP-01-0124-FEDER-015949, and PTDC/MED-ONC/31423/2017-POCI-01-0145-FEDER-031423 to RM Reis. The work was also supported by the Project ON.2 SR&TD Integrated Program (NORTE-07-0124-FEDER-000017) cofinanced by Programa Operacional Regional do Norte (ON.2—O Novo Norte), Quadro de Referência Estratégico Nacional (QREN), Fundo Europeu de Desenvolvimento Regional (FEDER). This study was partially developed under the scope of the Brazilian FAPESP grant (2012/19590-0) to RM Reis. F Pinto received a fellowship from FCT cofinanced by Fundo Social Europeu-FSE (SFRH/BD/81369/2011 and SFRH/BPD/115730/2016). AM Costa was funded by a postdoctoral fellow (PTDC/SAU-TOX/114549/2009).
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Aldape K, Brat DJ, et al. Announcing cIMPACT-NOW: the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy. Acta Neuropathol. 2017;133:1–3. doi: 10.1007/s00401-016-1646-x. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Wang R, Yu Y, et al. Glioblastoma Treatment Modalities besides Surgery. J Cancer. 2019;10:4793–4806. doi: 10.7150/jca.32475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas AA, Brennan CW, DeAngelis LM, et al. Emerging therapies for glioblastoma. JAMA Neurol. 2014;71:1437–1444. doi: 10.1001/jamaneurol.2014.1701. [DOI] [PubMed] [Google Scholar]
- 5.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 6.Prieto-Vila M, Takahashi RU, Usuba W, et al. Drug resistance driven by cancer stem cells and their niche. Int J Mol Sci 2017; 18. [DOI] [PMC free article] [PubMed]
- 7.Wang SS, Jiang J, Liang XH, et al. Links between cancer stem cells and epithelial-mesenchymal transition. Onco Targets Ther. 2015;8:2973–2980. doi: 10.2147/OTT.S91863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinto F, Pertega-Gomes N, Pereira MS, et al. T-box transcription factor brachyury is associated with prostate cancer progression and aggressiveness. Clin Cancer Res. 2014;20:4949–4961. doi: 10.1158/1078-0432.CCR-14-0421. [DOI] [PubMed] [Google Scholar]
- 9.Pinto F, Campanella NC, Abrahao-Machado LF, et al. The embryonic Brachyury transcription factor is a novel biomarker of GIST aggressiveness and poor survival. Gastric Cancer. 2016;19:651–659. doi: 10.1007/s10120-015-0505-0. [DOI] [PubMed] [Google Scholar]
- 10.Pinto F, Carcano FM, da Silva ECA, et al. Brachyury oncogene is a prognostic factor in high-risk testicular germ cell tumors. Andrology. 2018;6:597–604. doi: 10.1111/andr.12495. [DOI] [PubMed] [Google Scholar]
- 11.Fernando RI, Litzinger M, Trono P, et al. The T-box transcription factor brachyury promotes epithelial-mesenchymal transition in human tumor cells. J Clin Invest. 2010;120:533–544. doi: 10.1172/JCI38379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du R, Wu S, Lv X, et al. Overexpression of brachyury contributes to tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. J Exp Clin Cancer Res. 2014;33:105. doi: 10.1186/s13046-014-0105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang XR, Ng D, Alcorta DA, et al. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nat Genet. 2009;41:1176–1178. doi: 10.1038/ng.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palena C, Roselli M, Litzinger MT, et al. Overexpression of the EMT driver brachyury in breast carcinomas: association with poor prognosis. J Natl Cancer Inst 2014; 106. [DOI] [PMC free article] [PubMed]
- 15.Kilic N, Feldhaus S, Kilic E, et al. Brachyury expression predicts poor prognosis at early stages of colorectal cancer. Eur J Cancer. 2011;47:1080–1085. doi: 10.1016/j.ejca.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Haro A, Yano T, Kohno M, et al. Expression of brachyury gene is a significant prognostic factor for primary lung carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S509–516. doi: 10.1245/s10434-013-2914-9. [DOI] [PubMed] [Google Scholar]
- 17.Shimoda M, Sugiura T, Imajyo I, et al. The T-box transcription factor brachyury regulates epithelial-mesenchymal transition in association with cancer stem-like cells in adenoid cystic carcinoma cells. BMC Cancer. 2012;12:377. doi: 10.1186/1471-2407-12-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jezkova J, Williams JS, Pinto F, et al. Brachyury identifies a class of enteroendocrine cells in normal human intestinal crypts and colorectal cancer. Oncotarget. 2016;7:11478–11486. doi: 10.18632/oncotarget.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto F, Pertega-Gomes N, Vizcaino JR, et al. Brachyury as a potential modulator of androgen receptor activity and a key player in therapy resistance in prostate cancer. Oncotarget. 2016;7:28891–28902. doi: 10.18632/oncotarget.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar D, Shields B, Davies ML, et al. Brachyury confers cancer stem cell characteristics on colorectal cancer cells. Int J Cancer. 2012;130:328–337. doi: 10.1002/ijc.26029. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi Y, Sugiura T, Imajyo I, et al. Knockdown of the T-box transcription factor Brachyury increases sensitivity of adenoid cystic carcinoma cells to chemotherapy and radiation in vitro: implications for a new therapeutic principle. Int J Oncol. 2014;44:1107–1117. doi: 10.3892/ijo.2014.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang B, Cohen JR, Fernando RI, et al. The embryonic transcription factor brachyury blocks cell cycle progression and mediates tumor resistance to conventional antitumor therapies. Cell Death Dis. 2013;4:e682. doi: 10.1038/cddis.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heery CR, Singh BH, Rauckhorst M, et al. Phase I trial of a yeast-based therapeutic cancer vaccine (GI-6301) targeting the transcription factor brachyury. Cancer Immunol Res. 2015;3:1248–1256. doi: 10.1158/2326-6066.CIR-15-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heery CR, Palena C, McMahon S, et al. Phase I study of a poxviral TRICOM-based vaccine directed against the transcription factor brachyury. Clin Cancer Res. 2017;23:6833–6845. doi: 10.1158/1078-0432.CCR-17-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton DH, Litzinger MT, Jales A, et al. Immunological targeting of tumor cells undergoing an epithelial-mesenchymal transition via a recombinant brachyury-yeast vaccine. Oncotarget. 2013;4:1777–1790. doi: 10.18632/oncotarget.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatti-Mays ME, Redman JM, Donahue RN, et al. A phase I trial using a multitargeted recombinant adenovirus 5 (CEA/MUC1/brachyury)-based immunotherapy vaccine regimen in patients with advanced cancer. Oncologist 2019, 10.1634/theoncologist.2019-0608. [DOI] [PMC free article] [PubMed]
- 27.Pinto F, Costa AM, Santos GC, et al. The T-box transcription factor brachyury behaves as a tumor suppressor in gliomas. J Pathol 2020, 10.1002/path.5419. [DOI] [PubMed]
- 28.Martinho O, Pinto F, Granja S, et al. RKIP inhibition in cervical cancer is associated with higher tumor aggressive behavior and resistance to cisplatin therapy. PLoS One. 2013;8:e59104. doi: 10.1371/journal.pone.0059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira MS, Celeiro SP, Costa AM, et al. Loss of SPINT2 expression frequently occurs in glioma, leading to increased growth and invasion via MMP2. Cell Oncol (Dordr) 2020;43:107–121. doi: 10.1007/s13402-019-00475-7. [DOI] [PubMed] [Google Scholar]
- 30.Costa AM, Pinto F, Martinho O, et al. Silencing of the tumor suppressor gene WNK2 is associated with upregulation of MMP2 and JNK in gliomas. Oncotarget. 2015;6:1422–1434. doi: 10.18632/oncotarget.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed]
- 32.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1-13. [DOI] [PMC free article] [PubMed]
- 34.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44-57. [DOI] [PubMed]
- 35.Gaspar N, Marshall L, Perryman L, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010;70:9243–9252. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jezkova J, Williams JS, Jones-Hutchins F, et al. Brachyury regulates proliferation of cancer cells via a p27Kip1-dependent pathway. Oncotarget. 2014;5:3813–3822. doi: 10.18632/oncotarget.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janesick A, Wu SC, Blumberg B. Retinoic acid signaling and neuronal differentiation. Cell Mol Life Sci. 2015;72:1559–1576. doi: 10.1007/s00018-014-1815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L, Li H, Zhan Y. All-trans retinoic acid enhances temozolomide-induced autophagy in human glioma cells U251 via targeting Keap1/Nrf2/ARE signaling pathway. Oncol Lett. 2017;14:2709–2714. doi: 10.3892/ol.2017.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pitz MW, Lipson M, Hosseini B, et al. Extended adjuvant temozolomide with cis-retinoic acid for adult glioblastoma. Curr Oncol. 2012;19:308–314. doi: 10.3747/co.19.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaeckle KA, Hess KR, Yung WK, et al. Phase II evaluation of temozolomide and 13-cis-retinoic acid for the treatment of recurrent and progressive malignant glioma: a North American Brain Tumor Consortium study. J Clin Oncol. 2003;21:2305–2311. doi: 10.1200/JCO.2003.12.097. [DOI] [PubMed] [Google Scholar]
- 41.See SJ, Levin VA, Yung WK, et al. 13-cis-Retinoic acid in the treatment of recurrent glioblastoma multiforme. Neuro Oncol. 2004;6:253–258. doi: 10.1215/S1152851703000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 3275 kb)
(PDF 483 kb)