Abstract
Disease outcomes are heterogeneous in Parkinson’s disease and may be predicted by gene variants. This study investigated if the BDNF rs6265 single nucleotide polymorphism (SNP) is associated with differential outcomes with specific pharmacotherapy treatment strategies in the “NIH Exploratory Trials in PD Long-term Study 1” (NET-PD LS-1, n = 540). DNA samples were genotyped for the rs6265 SNP and others (rs11030094, rs10501087, rs1491850, rs908867, and rs1157659). The primary measures were the Unified Parkinson’s Disease Rating Scale (UPDRS) and its motor component (UPDRS-III). Groups were divided by genotype and treatment regimen (levodopa monotherapy vs levodopa with other medications vs no levodopa). T allele carriers were associated with worse UPDRS outcomes compared to C/C subjects when treated with levodopa monotherapy (+ 6 points, p = 0.02) and to T allele carriers treated with no levodopa treatment strategies (UPDRS: + 8 points, p = 0.01; UPDRS-III: + 6 points, p = 0.01). Similar effects of worse outcomes associated with levodopa monotherapy were observed in the BDNF rs11030094, rs10501087, and rs1491850 SNPs. This study suggests the levodopa monotherapy strategy is associated with worse disease outcomes in BDNF rs6265 T carriers. Pending prospective validation, BDNF variants may be precision medicine factors to consider for symptomatic treatment decisions for early-stage PD patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13311-020-00965-9.
Keywords: Parkinson disease, Brain-derived neurotrophic factor, Levodopa, Subthalamic nucleus deep brain stimulation, rs6265
Introduction
Parkinson’s disease (PD) therapies result in varying degrees of success for any given patient. Indeed, the heterogeneity in clinical outcomes is notable, including the responses to the mainstay therapy levodopa [1]. Heterogeneity in phenomenology and therapeutic outcomes may arise from genetic variants [2, 3]. Single nucleotide polymorphisms (SNPs), particularly those associated with increased PD incidence, have received the most study. One SNP that is not associated with increased risk of PD [4] but is associated with a milder early clinical phenotype in the unmedicated state is the rs6265 SNP in the gene BDNF [5, 6]. In addition, the rs6265 SNP (T allele) is associated with earlier development of levodopa-induced dyskinesias [7, 8], though not in every population [9]. The rs6265 SNP (C>T), a Val66Met substitution in the prodomain of brain-derived neurotrophic factor (BDNF), results in decreased activity-dependent release of BDNF [10]. T allele carriers (C/T or T/T), approximately one-third of the general population [11], have reduced plasticity and volume of the motor cortex and the caudate nucleus [12–14]. BDNF signaling may also contribute to the efficacy of subthalamic nucleus (STN) DBS [15, 16]. Therefore, we hypothesized the BDNF rs6265 T variant may alter antiparkinsonian treatment outcomes.
Retrospective examination of completed, rigorously conducted clinical trials provides an opportunity to test whether the rs6265 T variant alters patient outcomes. The US National Institutes of Health (NIH) Exploratory Trials in PD Long-term Study 1 (NET-PD LS-1) enrolled early-stage PD subjects, all of whom were receiving dopaminergic treatment at entry, with no restriction on the type of dopaminergic pharmacotherapy [17]. Subjects could receive levodopa monotherapy, levodopa in combination with other dopaminergic medications, or no levodopa (e.g., dopamine agonists). All NET-PD LS-1 subjects were followed systematically for an average of 3.5 years, and a subset consented for genomic DNA banking for future analyses, offering a well-characterized dataset for BDNF SNP genotyping and analysis. In addition, NET-PD LS-1 treating neurologists were allowed to adjust baseline antiparkinsonian medications as they saw fit (i.e., not stipulated by study protocol), allowing for separate analyses for the most common, symptomatic drug therapies employed in early-stage PD.
Methods
We retrospectively analyzed available DNA samples and clinical data from the NET-PD LS-1 trial. All clinical and genetic data from NET-PD LS-1 are accessible through the US National Institute of Neurological Disorders and Stroke (NINDS). Analyses were determined by the Michigan State University Biomedical and Health Institutional Review Board not to meet the definition of human subject research, as defined by the U.S. Department of Health and Human Services regulations for the protection of human research subjects.
NET-PD LS-1 Subjects and Trial Design
The NET-PD LS-1 trial design and results have been published previously [17]. Briefly, LS-1 was a multicenter, randomized efficacy trial that determined creatine was not disease-modifying. Subjects were less than 5 years since diagnosis of PD and had taken levodopa or a dopamine agonist for at least 90 days but not longer than 2 years. A subset of subjects consented for genomic DNA collection and storage for examination separate from the planned LS-1 analysis. Outcome measures included the UPDRS, its first three parts (UPDRS-I (mentation, behavior, and mood), UPDRS-II (activities of daily living), and UPDRS-III (motor)), and the Parkinson’s Disease Questionnaire (PDQ)-39, a patient-reported, quality-of-life measure.
SNP Genotyping
DNA samples from NET-PD LS-1 subjects were obtained from the Coriell Institute for Medical Research biorepository. Samples were genotyped for the BDNF rs6265 SNP using the 5′ exonuclease allelic discrimination TaqMan assay. We also examined five other BDNF SNPs that are not in the coding sequence nor known to alter BDNF protein structure but are associated with modest hippocampal or whole-brain atrophy—viz., rs908867, rs11030094, rs10501087, rs1157659, and rs1491850 (all SNPs examined are in Supplementary Figure 1) [18]. Genotyping was performed with a minimum 20 ng of genomic DNA in a 25-μl reaction volume in duplicate using TaqMan Genotyping Mastermix (Applied Biosystems, Waltham, MA; 4371353). Reactions were run using a Real-Time PCR instrument (Viia 7, Applied Biosystems). Data were analyzed for genotype determination calls made by TaqMan Genotyper software (Applied Biosystems).
NET-PD LS-1 Analyses
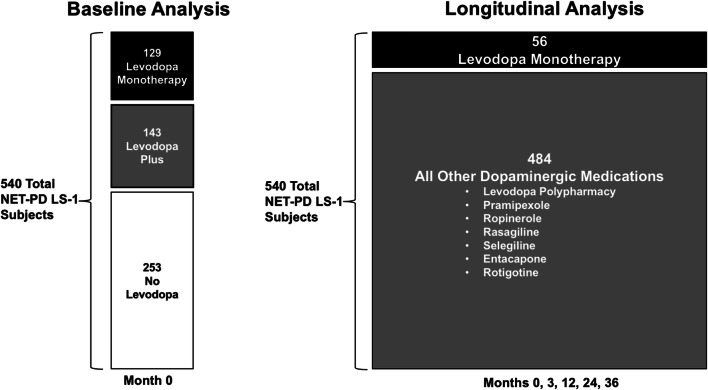
Two analyses were conducted using the LS-1 dataset (Fig. 1): the baseline analysis for initial hypothesis testing and the longitudinal analysis used for testing the hypothesis over time. The 540 subjects for whom samples were available comprise the population for both analyses. The baseline analysis consists of subject data from the first time point of the NET-PD LS-1 trial (month 0) and included three main treatment strategies: 129 subjects treated with levodopa monotherapy (“LD”), 143 subjects treated with levodopa-inclusive polypharmacy (including dopamine agonists, “LD Plus”), and 253 subjects treated with dopamine agonists along with other medications (i.e., no levodopa, “No LD”). The longitudinal analysis consisted of the same treatment strategies but with a differing distribution: 56 subjects remained on LD for 36 months, and 484 subjects were treated with either LD Plus or No LD during that same period. Full assessments in the longitudinal cohort were conducted at 0, 3, 12, 24, and 36 months with interim visits occurring at 6 and 18 months.
Fig. 1.
Baseline and longitudinal analyses of NET-PD LS-1. A schematic showing the number of subjects by group for the baseline and longitudinal analyses. Dopaminergic medications prescribed other than levodopa are listed
NET-PD LS-1 Statistics
Subject data were categorized by BDNF SNP and analyzed using SAS (v9.4; Cary, NC). Data were categorized by each genotype (major/major, major/minor, and minor/minor) for analysis and then pooled into “risk allele” groups based on either previous association with detriment (rs6265) or calculated means from UPDRS measurements. Subjects within the creatine and placebo treatment arms were pooled due to no significant differences in the primary endpoint reported at the conclusion of the NET-PD LS-1 study [17].
Both the baseline and longitudinal analyses are used to examine the effects of the BDNF SNPs according to two perspectives. First, genotypes are compared within-treatment-strategy groups (like levodopa monotherapy) as a way to examine genotype-related variance. Second, treatment strategy groups are compared within-genotype as a “precision medicine” approach. The impact of the BDNF SNPs is discussed with each perspective separately, but it should be emphasized that only one statistical model was used to test the data, including correcting for multiple comparisons for the hypothesis-driven analysis of the rs6265 SNP.
The baseline within-treatment-strategy and within-genotype approaches included analysis of covariance (ANCOVA) models with baseline value of the UPDRS and UPDRS-III scores in the ON state as outcomes, a genotype (variant carrier or not)-by-treatment strategy (LD, LD Plus, or No LD) interaction as the factor of interest, controlling for study site, treatment arm, age, gender, race/ethnicity, duration of PD, LEDD, genotype, and medication. The longitudinal within-medication and within-genotype approaches included repeated measures ANCOVA models with UPDRS and UPDRS-III scores in the ON state, UPDRS-I and UPDRS-II scores and the PDQ-39 score as outcomes, a genotype-by-time interaction for the within-treatment-strategy approach, and a treatment-by-time interaction for the within-genotype approach controlling for study site, treatment arm, age, gender, race/ethnicity, duration of PD, LEDD, time, and genotype (for the within-treatment-strategy approach) or treatment (for the within-genotype approach). Models assessing the effect of the non-rs6265 SNPs were adjusted further for the rs6265 genotype. The covariance matrices for the within-subject observations were modeled using an unstructured pattern. Least square means and standard errors (SE) are reported for all models. The least square means are adjusted estimates, so the sum of the individual UPDRS-I-III estimates may not equal the adjusted total UPDRS estimate.
Of importance, the rs6265 genotype was considered the primary SNP of interest and multiple pairwise differences in least square means for within-treatment and within-genotype comparisons in the baseline cohort, and further for within-time comparisons for the longitudinal analysis, were accounted for by adjusting the corresponding p values with a Bonferroni correction for multiple comparisons. Results for the other SNPs were considered exploratory and not corrected for multiple comparisons. Statistical significance was set at p < 0.05 with the intention to interpret all analyses as requiring validation by a randomized, prospective cohort.
Results
Baseline Analysis
For all six BDNF SNPs, demographics were examined for the baseline analysis (Supplementary Tables 1-6). A difference in minority race for the rs1157659 SNP was revealed. Differences in disease duration since time of diagnosis are reported with small numerical differences for the rs908867 and rs1157659 SNPs. With medical regimens combined, UPDRS, part III, and PDQ-39 were not different between rs6265 genotypes at baseline (data not shown).
BDNF rs6265 T Allele Is Associated with Worse Scores on LD and Better Outcomes with No LD
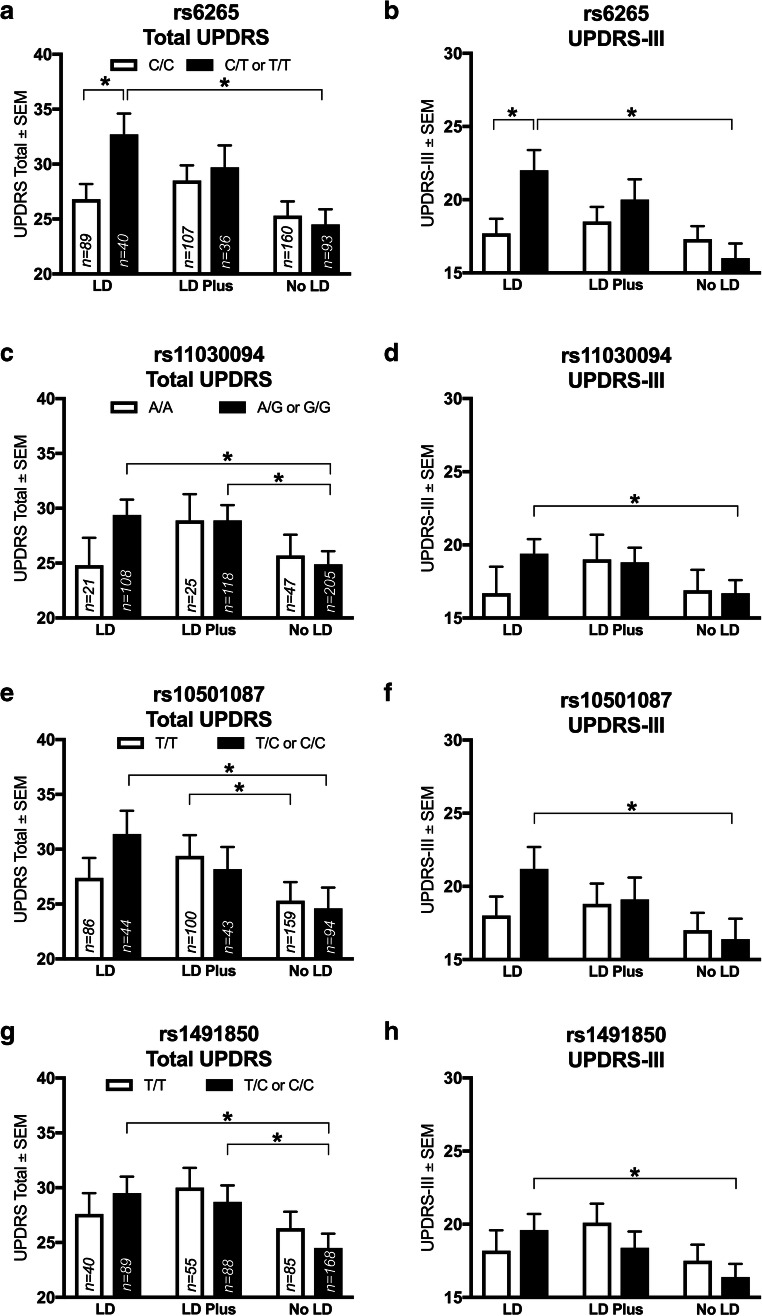
Subjects prescribed levodopa monotherapy (“LD”) had different UPDRS scores depending on rs6265 genotype. The UPDRS score for LD subjects carrying the T allele (C/T or T/T) was ≈ 6 points worse than C/C subjects (32.7 (1.9) vs 26.8 (1.4), respectively; Fig. 2a, p = 0.02). This difference is preserved when comparing UPDRS-III scores for C/C subjects (17.7 (1.0)) to T allele carriers (22.0 (1.4); Fig. 2b, p = 0.03). In contrast, subjects prescribed a combination of levodopa-inclusive polypharmacy (“LD Plus”) or a regimen not including levodopa (“No LD”) did not have different UPDRS scores between genotypes (Fig. 2a, b).
Fig. 2.
BDNF rs6265 SNP and others in the NET-PD LS-1 baseline analysis are associated with differences in outcomes with specific treatment strategies. In subjects on levodopa monotherapy (“LD”), the BDNF rs6265 T allele (C/T or T/T) is associated with worse UPDRS total (a) and part III (b) scores (p = 0.02 and p = 0.03, respectively). Within rs6265 T allele carriers, LD was associated with a worse UPDRS than a no levodopa regimen (“No LD”, A, p = 0.01); similarly, No LD was superior to LD by UPDRS-III in T allele carriers (b, p = 0.01). The rs11030094 G allele also showed a within-genotype difference, where LD is worse than No LD by UPDRS (c, p = 0.002) and UPDRS-III (d, p = 0.01); further, LD plus was worse than No LD by UPDRS (c, p = 0.01). The rs10501087 C/T or C/C subjects show a worse score in association with LD by both UPDRS (e, p = 0.01) and UPDRS-III (F, p = 0.001). In T/T subjects, LD Plus was worse than No LD by UDPRS (e, p = 0.01). For the rs1491850 SNP, LD was worse than No LD within T/C or C/C subjects by UPDRS (g, p = 0.002) and UPDRS-III (h, p = 0.01); further, LD Plus was worse than No LD (g, p = 0.01). Values represent the mean ± SEM. * represents a significant comparison. Darkened bars correspond to “risk” allele carriers; clear bars correspond to no risk allele. Group n values are listed within corresponding bars
From a within-genotype perspective, the three treatment strategy groups (LD, LD Plus, and No LD) were compared within rs6265 genotype. For T allele carriers (C/T or T/T), LD was associated with worse UPDRS scores than No LD (32.7 (1.9) vs 24.5 (1.4), respectively; Fig. 2a, p = 0.01). This difference remains when examining UPDRS-III scores with a better score in No LD (16.0 (1.0)) compared to LD (22.0 (1.4); Fig. 2b, p = 0.01). All differences for the above rs6265 analysis were present with a Bonferroni correction.
BDNF rs11030094 G Allele Is Associated with Better Scores with No LD over LD
LD and LD Plus in rs11030094 G allele carriers (A/G or G/G) were associated with worse total UPDRS scores than those with No LD (29.4 (1.4) and 28.9 (1.4) vs 24.9 (1.2), respectively; Fig. 2c, LD p = 0.002, LD Plus p = 0.01). UPDRS-III scores also were different when comparing G allele carriers treated with LD (16.7 (0.9)) to No LD (19.4 (1.0); Fig. 2d, p = 0.01). For LD subjects, G allele carriers’ UPDRS and UPDRS-III average scores were not higher than for A/A subjects (Fig. 2c, d).
BDNF rs10501087 C Allele Is Associated with Better Scores with No LD over LD
C allele carriers for the rs10501087 SNP (T/C or C/C) treated with LD were associated with worse UPDRS scores than No LD subjects (31.4 (2.1) vs 24.6 (1.9), respectively; Fig. 2e, p = 0.01). UPDRS scores were different in T/T subjects between LD Plus (29.4 (1.9)) and No LD treatment strategies (25.3 (1.7); Fig. 2e, p = 0.01). UPDRS-III scores also were worse for C allele carriers with LD (16.4 (1.4)) compared to No LD (21.2 (1.5); Fig. 2f, p = 0.001). Within LD, C allele carriers’ UPDRS scores were not different compared to T/T subjects (Fig. 2e).
BDNF rs1491850 C Allele Is Associated with Better UPDRS-III Scores with No LD Versus LD
Similar to SNPs evaluated above, rs1491850 C allele carriers (T/C or C/C) prescribed LD or LD Plus had worse average UPDRS scores compared to No LD subjects (29.5 (1.5) and 28.7 (1.5) vs 24.5 (1.3), respectively; Fig. 2g; LD p = 0.002, LD Plus p = 0.01). This difference between LD and No LD in C allele carriers also was present in UPDRS-III scores (19.6 (1.1) and 16.4 (0.9), respectively; Fig. 2h, p = 0.01). No difference between treatment strategies was observed in T/T subjects. In contrast to the rs6265 SNP, LD subjects did not have different average UPDRS or UPDRS-III scores dependent on rs1491850 genotype (Fig. 2g, h).
BDNF rs908867 and rs1157659 SNPs Not Associated with Pharmacogenetic Effects
LD subjects did not have different average UPDRS or UPDRS-III scores dependent on genotype (Supplementary Figure 2a,b). Furthermore, within-genotype comparisons did not reveal differences between strategies as assessed by UPDRS or UPDRS-III scores (Supplementary Figure 2c,d).
Longitudinal Analysis: Genotype Associations Within Treatment Strategy
BDNF rs6265 T Allele Is Not Associated with Significantly Worse Scores When on LD over 3 years
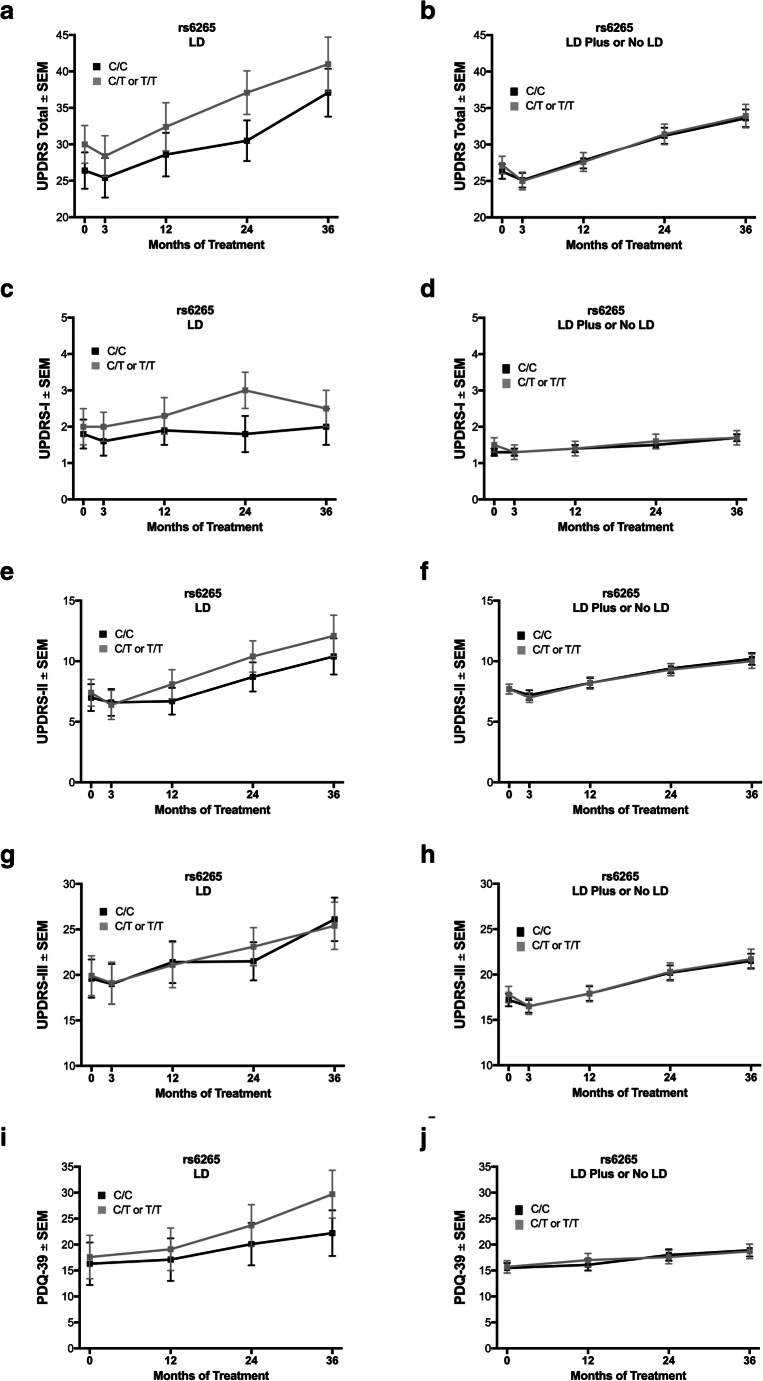
To examine if baseline findings between rs6265 genotypes for those on LD remained consistent over time, a longitudinal analysis (Fig. 1) was conducted over the first 3 years of the study from time of enrollment. LD subjects over this extended interval were examined for an effect of rs6265 on disease outcomes over time. T allele carriers (C/T or T/T) were not associated with a worse UPDRS score with LD, compared to C/C subjects (Fig. 3a; p = 0.2, including Bonferroni correction for multiple comparisons), nor were differences in UPDRS subparts detected (Fig. 3c, e, g). The T allele had no effect on UPDRS or UPDRS-I-III in subjects with LD Plus or No LD strategies over the 3-year course of treatment of the longitudinal analysis (Fig. 3b, d, f, h).
Fig. 3.
The BDNF rs6265 SNP in the longitudinal analysis. With levodopa monotherapy, carrying the rs6265 T allele (C/T or T/T) was not associated with statistically significant differences between genotypes by UPDRS, its parts I–III, or PDQ-39 (a, c, e, g, i, respectively). For subjects treated with strategies other than levodopa monotherapy, no differences between rs6265 genotypes were observed by UPDRS, its parts I–III, or PDQ-39 (b, d, f, h, j, respectively). Values represent the mean ± SEM. Gray boxes correspond to T allele carriers; black boxes correspond to C/C subjects
Lastly, the PDQ-39 was used as a patient-reported, quality-of-life measure to determine if findings from the UPDRS in the baseline cohort were replicated by a subject-reported metric over time. The PDQ-39 average score for T allele carriers was numerically higher over longitudinal treatment than for C/C subjects (+ 7.4 points at 36 months, Fig. 3i). In addition, the PDQ-39 was similar between genotypes for LD Plus and No LD subjects treated over 3 years (Fig. 3j).
BDNF rs11030094 G Allele Is Associated with Worse UPDRS Scores with LD over 3 years
LD subjects were examined for an association of the rs11030094 G allele and disease outcomes over 3 years. At 24 months, G allele carriers (A/G or G/G) treated with LD were associated with worse UPDRS scores (34.7 (2.4)) compared to A/A subjects (23.8 (4.5); Fig. 4a, p = 0.02). UPDRS-III scores between G allele carriers and A/A subjects were similar (Fig. 4g). The UPDRS-I, but not UPDRS-II, scores were on average 1.5 points worse in G allele carriers (2.2 (0.3)) vs A/A subjects (0.7 (0.6); Fig. 4c, p = 0.02). In addition, G allele carriers treated with LD were associated with worse baseline PDQ-39 scores (16.9 (1.8)) compared to A/A subjects (7.9 (4.0); Fig. 4i, p = 0.04).
Fig. 4.
The BDNF rs11030094 SNP in the longitudinal analysis. Possession of the rs11030094 G allele (A/G or G/G) is associated with worse performance on UPDRS when treated with levodopa monotherapy, becoming significant at 24 months (a, p = 0.02), and this difference is also significant at 24 months in UPDRS-I (c, p = 0.02) but not UPDRS-II (e) or UPDRS-III (g). A difference is observed in PDQ-39 scores at baseline between genotypes (i, p = 0.04). For subjects treated with strategies other than levodopa monotherapy, A/A subjects have lower UPDRS score at 36 months compared to G allele carriers (b, p < 0.05); further, a difference was observed in UPDRS-II at 24 months (f, p = 0.04). No differences between genotypes are observed by UPDRS-I, UPDRS-III, or PDQ-39 (d, h, j, respectively). Values represent the mean ± SEM. * represents a significant comparison. Black boxes correspond to G allele carriers; gray boxes correspond to A/A subjects
Examining subjects on the LD Plus or No LD treatment strategies revealed a small difference between A/A subjects and G allele carriers by UPDRS at 36 months (30.4 (2.0) vs 34.3 (1.2), respectively; Fig. 4b; p < 0.05). UPDRS-I, UPDRS-III, and PDQ-39 scores were not associated with differences between genotypes (Fig. 4d, h j). However, at 24 months, A/A subjects had worse UPDRS-II scores (9.8 (0.8)) compared to G allele carriers (7.3 (1.9); Fig. 4f, p = 0.04).
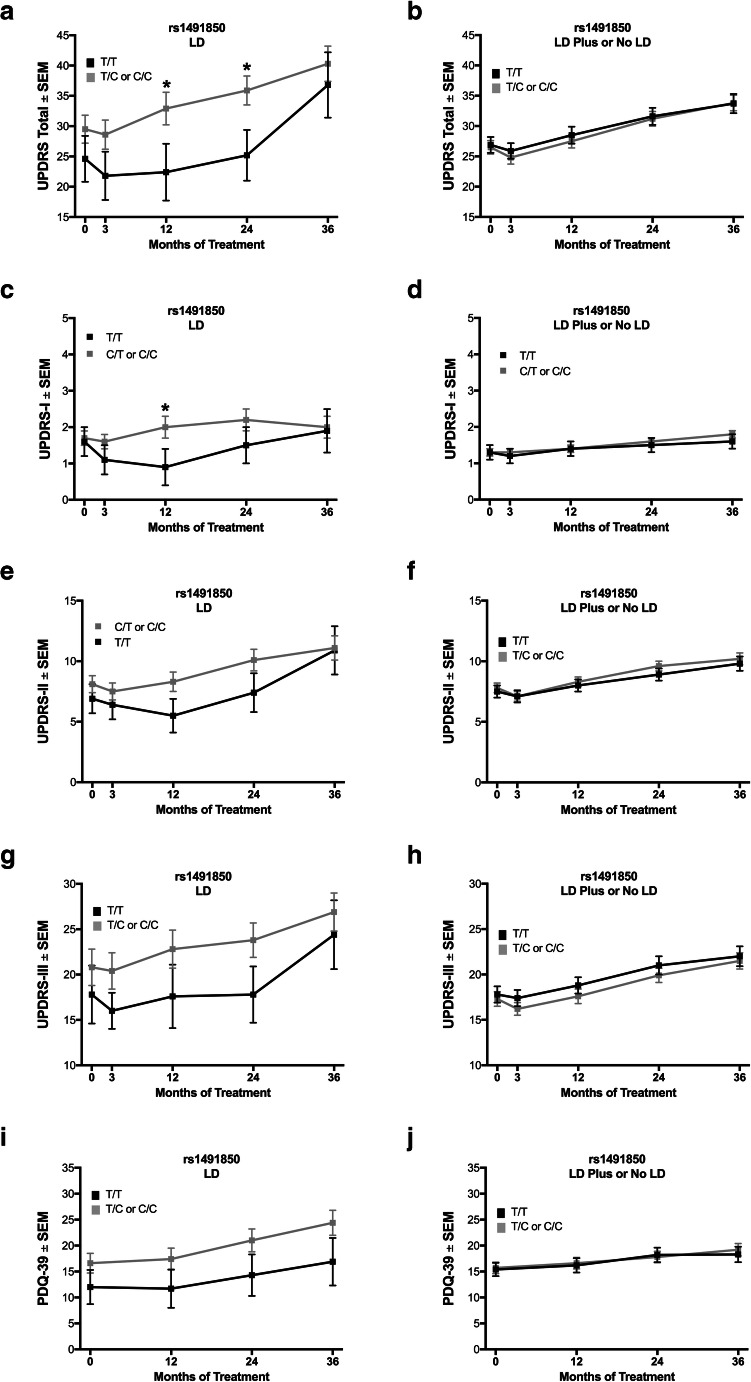
BDNF rs1491850 C Allele Is Associated with Worse Scores with LD over 3 years
LD subjects who possessed the rs1491850 C allele (T/C or C/C) were associated with worse UPDRS scores than T/T subjects (at 12 months, 32.9 (2.7) vs 22.4 (4.7), respectively, p = 0.04; at 24 months, 35.9 (2.4) vs 25.2 (4.2), respectively, p = 0.02; Fig. 5a). In examining UPDRS-I-III for LD subjects, UPDRS-I was higher at 12 months for C allele carriers (2.0 (0.3) vs T/T subjects (0.9 (0.5); Fig. 5c, p = 0.04). The PDQ-39 was not different between C allele carriers and T/T subjects in the LD group (Fig. 5i). In contrast to the LD subjects, no differences between rs1491850 genotypes were observed in UPDRS, its subparts, or PDQ-39 when examining No LD or LD Plus subjects (Fig. 5b, d, f, h, j).
Fig. 5.
The BDNF rs1491850 SNP in the longitudinal analysis. Carrying the rs1491850 C allele (T/C or C/C) is associated with worse performance on UPDRS when treated with levodopa monotherapy, becoming significant at 12 and 24 months (a, p = 0.04 and p = 0.02, respectively), and this difference is also significant at 12 months in UPDRS-I (c, p = 0.04) but not UPDRS-II (e), UPDRS-III (g), or PDQ-39 (i). For subjects treated with strategies other than levodopa monotherapy, no differences between genotypes are observed by UPDRS, its parts I–III, or PDQ-39 (b, d, f, h, j, respectively). Values represent the mean ± SEM. * represents a significant comparison. Gray boxes correspond to C allele carriers; black boxes correspond to T/T subjects
BDNF rs10501087, rs908867, and rs1157659 SNPs Are Not Associated with Differences in Outcomes with LD over 3 years
No differences between rs10501087 genotypes were detected by UPDRS or UPDRS-III for LD or LD Plus subjects (Supplementary Figure 3). Similarly, no differences between genotypes for either the rs908867 or rs1157659 SNPs were observed by UPDRS or UPDRS-III for either LD or LD Plus subjects (Supplementary Figures 4, 5).
Discussion
Using a hypothesis-driven approach, two potential conclusions can be made about disease outcomes for PD subjects and the BDNF rs6265 SNP. First, the rs6265 SNP may contribute to the heterogeneous outcomes of the treatment strategies commonly employed in early-stage PD, particularly for levodopa monotherapy. In LS-1 subjects treated with levodopa monotherapy at study entry, carrying the rs6265 T allele is associated with a clinically meaningful [19] six-point higher UPDRS score on average, and a four-point difference in UPDRS-III on average, compared to C/C subjects. Second, rs6265 T allele carriers have a better disease outcome with a treatment strategy without levodopa compared to levodopa monotherapy. Both of these conclusions are supported by findings in the baseline analysis and were consistent with the results from the longitudinal analysis. In addition, when accounting for the effects of rs6265, other BDNF SNPs with previously described neurological effects in non-PD contexts were associated with parallel findings, either on the differences in outcomes with levodopa monotherapy or on better outcomes with a treatment strategy without levodopa.
No association exists between the rs6265 T allele and PD risk [4]. In late-stage PD, BDNF genotype does not affect the limited remaining response to medication [20]. However, the rs6265 T allele is associated with earlier development of levodopa-induced dyskinesia (LID) [7], suggesting an earlier failure of therapeutic benefit. Of importance, the higher UPDRS score that was observed in medically treated rs6265 T allele carriers cannot be attributed to LID, as UPDRS-IV scores were low overall with no differences observed. This indicates the suboptimal efficacy of T allele carriers on medical therapy precedes the earlier emergence of LID. In addition, the very low UPDRS-IV scores suggest that rs6265 T allele carriers are no different in their ON time percentages. Differences in disease progression rates between genotypes may contribute to these results, though this is unlikely for several reasons. First, inclusion criteria selected a relatively narrow group of subjects with respect to disease duration. Second, disease duration was a covariate in the statistical model. Third, the primary findings are present robustly at baseline when differences in disease progression would result in relatively small differences in disease stage. Lastly, annualized rates of change in UPDRS (total or its parts) are the same between the three rs6265 genotypes when compared separately in early-stage, unmedicated PD subjects in both the DATATOP and PPMI cohorts [6]. A final explanation for the present results is T allele carriers may require higher doses for optimal antiparkinsonian effect, whereas dosing policies and practice may favor C/C patients; however, this is partly mitigated by including LEDD as a covariate. Even if dosing policies tend to favor different genotypes in a way that is not addressed statistically, these data still implicate BDNF in the neurobiology of dopamine transmission.
Carrying the rs6265 T allele results in reduced activity-dependent release of BDNF [10], and in association with our observations of worse disease outcomes with LD, it is possible that adequate BDNF release is necessary for an optimal effect of levodopa. In a heterozygous BDNF knockout mouse, reduction of BDNF results in decreased endogenous dopamine release and dopamine transporter function that is reversed partly with acute BDNF administration [21]. Further, mice carrying the rs6265 SNP exhibit increased glutamatergic transmission and reduced synaptic plasticity in the dorsolateral striatum [22]. Carriers of the rs6265 T allele may have a compromised ability to release dopamine in response to oral levodopa. Further, the symptomatic benefits of exercise may be due to BDNF expression [23, 24]. Conversely, carrying the rs6265 T allele may confer an advantage to the brain’s ability to compensate to dopamine depletion in the absence of oral replacement [6]. The possibility for the rs6265 SNP to account for some of the variance in a sample of PD patients prescribed levodopa might allow the use of SNP status in trial design, potentially reducing the number needed to enroll and particularly if the primary outcome is based on UPDRS.
With approximately one-third of the population carrying at least one of the rs6265 T alleles [11], the present study may have implications for a large proportion of PD patients. Precision medicine attempts to differentiate patients in order to guide clinical decisions. This study is the first to suggest that in early-stage PD patients, those carrying the rs6265 T allele have better outcomes when not treated with levodopa monotherapy. Current guidelines suggest that initial treatment strategies can include either levodopa or a dopamine agonist [25, 26]. Some advocate for initiating symptomatic therapy with levodopa but using a dose-limiting strategy through the addition of a dopamine agonist [27]. This study suggests that one-third of patients may benefit from treatment strategies that avoid levodopa monotherapy, for example, either starting with or adding dopamine agonists early in treatment. With this in mind, rs6265 SNP genotyping may be the first precision medicine approach for any neurodegenerative disease that influences a treatment recommendation, if these results are replicated in a prospective clinical trial.
Other intronic SNPs in the gene BDNF were examined. After controlling for rs6265 genotype, the rs11030094, rs10501087, and rs1491850 SNPs’ minor alleles (G, C, and C, respectively) were associated with better outcomes from treatment strategies without levodopa. The neurobiology of these SNPs has received less attention than the rs6265 SNP; therefore, their impact on BDNF function requires additional study before the mechanisms by which these SNPs interact with treatments may be hypothesized. Given the outcomes of the minor alleles for rs11030094, rs10501087, and rs1491850, the effect on BDNF function may be similar to the effect of the rs6265 T allele. Regardless of their mechanisms, these additional alleles may be associated with clinically important effects.
This study was conducted retrospectively with a specific a priori hypothesis regarding the BDNF rs6265 SNP. As such, subjects were not stratified by genotype as part of the trials’ designs. The treating neurologists were not restricted to specific protocols for treatment strategies, so subjects were not randomized nor forced to adhere to particular regimens. While this design may better reflect community practice, it limited the power of the longitudinal analysis and added the confound of selection bias for transitioning minor allele carriers away from levodopa monotherapy over time. However, genotypes were not known to any site investigators; therefore, treatment decisions were made blind to the primary variable of interest to this study. As such, the baseline analysis is likely to be less susceptible to this selection bias than the longitudinal analysis, likely explaining the loss of significant differences between rs6265 genotypes over time. The NET-PD LS-1 trial was conducted across many sites by experienced investigators, so the outcome measures are of high-quality and are generalizable to the PD patient population. This study’s findings were first generated from a cross-sectional, baseline analysis that was extended over 3 years in a separate longitudinal analysis. Lastly, as this study was exploratory and at times used less stringent statistical criteria, some of the findings may be spurious; however, the NET-PD LS-1 rs6265 SNP analyses used a Bonferroni correction for multiple comparisons. To strengthen further the evidence for this study’s conclusions, similar analyses in a prospective trial are the suggested next step.
In summary, the BDNF rs6265 T allele is associated with worse disease outcomes in those treated with levodopa monotherapy in early-stage PD patients. Individuals with the rs6265 T allele (approximately one-third of PD patients) may have improved early therapeutic outcomes when treated with a regimen that includes a dopamine agonist, with or without levodopa. If this observation is replicated, accounting for BDNF SNPs in clinical practice and in planning clinical trials may be warranted. Further investigation of the interaction of the BDNF SNPs and antiparkinsonian treatment strategies also may yield new targets for drug development.
Supplementary Information
BDNF gene schematic showing the relative locations of the examined six SNPs. The gene BDNF is shown in the forward and antisense (AS) directions with the relative locations of the six examined SNPs marked. (PNG 132 kb)
BDNF rs908867 and rs1157659 SNPs in baseline analysis are not associated with differences in outcomes from specific treatment strategies. Analyzing UPDRS total and part III for the rs908867 SNP showed no differences in the baseline cohort (A and B, respectively). No differences were detected by UPDRS total or part III for the rs1157659 SNP as well (C and D, respectively). Values represent the mean ± SEM. (PNG 298 kb)
The BDNF rs10501087 SNP in the longitudinal analysis. Subjects carrying the rs10501087 C allele do not exhibit differences from non-risk allele carriers when treated with levodopa monotherapy by UPDRS or UPDRS-III (A and C, respectively). Further, no differences between genotypes were observed in subjects not on levodopa monotherapy by UPDRS or UPDRS-III (B and D, respectively). Values represent the mean ± SEM. (PNG 335 kb)
The BDNF rs908867 SNP in the longitudinal analysis. Subjects carrying the rs908867 T allele do not exhibit differences from non-risk allele carriers when treated with levodopa monotherapy by UPDRS or UPDRS-III (A and C, respectively). Further, no differences between genotypes were observed in subjects not on levodopa monotherapy by UPDRS or UPDRS-III (B and D, respectively). Values represent the mean ± SEM. (PNG 339 kb)
The BDNF rs1157659 SNP in the longitudinal analysis. Subjects carrying the rs1157659 G allele do not exhibit differences from A/A subjects when treated with levodopa monotherapy by UPDRS or UPDRS-III (A and C, respectively). Further, no differences between genotypes were observed in subjects not on levodopa monotherapy by UPDRS or UPDRS-III (B and D, respectively). Values represent the mean ± SEM. (PNG 341 kb)
(DOCX 23 kb)
(PDF 1225 kb)
Acknowledgments
This research was supported by the grants from the United States National Institute of Neurological Disorders and Stroke (NINDS, #NS095656) and the Saint Mary’s Foundation (Grand Rapids, MI, USA).
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Authors’ Roles
This research project was conceived by DLF, JLG, and CES; organized by DLF and CES; and executed by DLF, AC-S, JWL, and CES. Statistical analyses were designed and executed by PA and JJE and were reviewed and critiqued by DLF, PA, KK, JJE, and CES. The manuscript was first written by DLF and CES, and it was reviewed and critiqued by all authors.
Data Availability
All NET-PD LS-1 data used in this study are accessible through the United States National Institute of Neurological Disorders and Stroke (NINDS) and the NET-PD Executive Committee.
Compliance with Ethical Standards
Disclosures
Mallory L. Hacker receives research funding from the NIH and the American Parkinson Disease Association.
John L. Goudreau receives research funding from the NIH and the Michael J. Fox Foundation, and he also receives income for reviewing for the NIH.
Karl Kieburtz has received research grant support from the NIH, the Michael J Fox Foundation, has consulted with Clintrex, Novartis, Roche/Genentech, and has stock ownership in Clintrex.
Jordan J. Elm receives research funding from NIH and the Michael J. Fox Foundation.
Jack W. Lipton receives research funding from the NIH and the Michael J. Fox Foundation, and he also receives income for reviewing for the NIH.
P. David Charles receives income from Allergan, Alliance for Patient Access, Ipsen, Revance, and Medtronic for consulting services. Vanderbilt University Medical Center receives income from grants or contracts with Abbott, Abbvie, Allergan, Boston Scientific, Dart, Intec, Ipsen, Lundbeck, Medtronic, Merz, and USWorldmeds for research or educational programs led by David Charles.
Caryl E. Sortwell receives research funding from the NIH, Michael J. Fox Foundation, the Saint Mary’s Foundation, and the U.S. Department of Defense. She also receives income for reviewing for the NIH, the Michael J. Fox Foundation, and the Weston Brain Institute.
The other authors declare that they have no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hauser RA, et al. Levodopa response in early Parkinson’s disease. Mov Disord. 2009;24(16):2328–2336. doi: 10.1002/mds.22759. [DOI] [PubMed] [Google Scholar]
- 2.Espay AJ, et al. Biomarker-driven phenotyping in Parkinson’s disease: A translational missing link in disease-modifying clinical trials. Mov Disord. 2017;32(3):319–324. doi: 10.1002/mds.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurzawski M, Bialecka M, Drozdzik M. Pharmacogenetic considerations in the treatment of Parkinson’s disease. Neurodegener Dis Manag. 2015;5(1):27–35. doi: 10.2217/nmt.14.38. [DOI] [PubMed] [Google Scholar]
- 4.Zintzaras, E. and G.M. Hadjigeorgiou, The role of G196A polymorphism in the brain-derived neurotrophic factor gene in the cause of Parkinson’s disease: a meta-analysis. J Hum Genet, 2005. 50(11): p. 560-566. [DOI] [PubMed]
- 5.Karamohamed S, et al. BDNF genetic variants are associated with onset age of familial Parkinson disease: GenePD Study. Neurology. 2005;65(11):1823–1825. doi: 10.1212/01.wnl.0000187075.81589.fd. [DOI] [PubMed] [Google Scholar]
- 6.Fischer DL, et al. Bdnf variant is associated with milder motor symptom severity in early-stage Parkinson’s disease. Parkinsonism Relat Disord. 2018;53:70–75. doi: 10.1016/j.parkreldis.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Foltynie T, et al. BDNF val66met influences time to onset of levodopa induced dyskinesia in Parkinson’s disease. J Neurol Neurosurg Psychiatry. 2009;80(2):141–144. doi: 10.1136/jnnp.2008.154294. [DOI] [PubMed] [Google Scholar]
- 8.Kusters CDJ, et al. Dopamine receptors and BDNF-haplotypes predict dyskinesia in Parkinson’s disease. Parkinsonism Relat Disord. 2018;47:39–44. doi: 10.1016/j.parkreldis.2017.11.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan N, et al. Sequence variants in SLC6A3, DRD2, and BDNF genes and time to levodopa-induced dyskinesias in Parkinson’s disease. J Mol Neurosci. 2014;53(2):183–188. doi: 10.1007/s12031-014-0276-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen ZY, et al. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24(18):4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan MF, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–269. doi: 10.1016/S0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 12.Kleim JA, et al. BDNF val66met polymorphism is associated with modified experience-dependent plasticity in human motor cortex. Nat Neurosci. 2006;9(6):735–737. doi: 10.1038/nn1699. [DOI] [PubMed] [Google Scholar]
- 13.McHughen SA, et al. BDNF val66met polymorphism influences motor system function in the human brain. Cereb Cortex. 2010;20(5):1254–1262. doi: 10.1093/cercor/bhp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pezawas L, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24(45):10099–11102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer DL, et al. Subthalamic Nucleus Deep Brain Stimulation Employs trkB Signaling for Neuroprotection and Functional Restoration. J Neurosci. 2017;37(28):6786–6796. doi: 10.1523/JNEUROSCI.2060-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, D.L. and C.E. Sortwell, BDNF provides many routes toward STN DBS-mediated disease modification. Mov Disord, 2019. 34(1): p. 22-34. [DOI] [PMC free article] [PubMed]
- 17.Writing Group for the NINDS Exploratory Trials in Parkinson Disease Investigators et al. Effect of creatine monohydrate on clinical progression in patients with Parkinson disease: a randomized clinical trial. JAMA. 2015;313(6):584–593. doi: 10.1001/jama.2015.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honea RA, et al. Characterizing the role of brain derived neurotrophic factor genetic variation in Alzheimer’s disease neurodegeneration. PLoS One. 2013;8(9):e76001. doi: 10.1371/journal.pone.0076001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath K, et al. Minimal clinically important difference on the Motor Examination part of MDS-UPDRS. Parkinsonism Relat Disord. 2015;21(12):1421–1426. doi: 10.1016/j.parkreldis.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Svetel M, et al. No association between brain-derived neurotrophic factor G196A polymorphism and clinical features of Parkinson’s disease. Eur Neurol. 2013;70(5-6):257–262. doi: 10.1159/000352033. [DOI] [PubMed] [Google Scholar]
- 21.Bosse KE, et al. Aberrant striatal dopamine transmitter dynamics in brain-derived neurotrophic factor-deficient mice. J Neurochem. 2012;120(3):385–395. doi: 10.1111/j.1471-4159.2011.07531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing D, Lee FS, Ninan I. The BDNF Val66Met polymorphism enhances glutamatergic transmission but diminishes activity-dependent synaptic plasticity in the dorsolateral striatum. Neuropharmacology. 2017;112(Pt A):84–93. doi: 10.1016/j.neuropharm.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackay CP, Kuys SS, Brauer SG. The Effect of Aerobic Exercise on Brain-Derived Neurotrophic Factor in People with Neurological Disorders: A Systematic Review and Meta-Analysis. Neural Plast. 2017;2017:4716197. doi: 10.1155/2017/4716197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis T, Rochester L. Mobilizing Parkinson’s Disease: The Future of Exercise. J Parkinsons Dis. 2018;8(s1):S95–S100. doi: 10.3233/JPD-181489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyasaki JM, et al. Practice parameter: initiation of treatment for Parkinson’s disease: an evidence-based review: report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology. 2002;58(1):11–17. doi: 10.1212/WNL.58.1.11. [DOI] [PubMed] [Google Scholar]
- 26.Fox, S.H., et al., International Parkinson and movement disorder society evidence-based medicine review: Update on treatments for the motor symptoms of Parkinson’s disease. Mov Disord, 2018. 33(8): p. 1248-1266. [DOI] [PubMed]
- 27.Okun, M.S., Management of Parkinson Disease in 2017: Personalized Approaches for Patient-Specific Needs. JAMA, 2017. 318(9): p. 791-792. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BDNF gene schematic showing the relative locations of the examined six SNPs. The gene BDNF is shown in the forward and antisense (AS) directions with the relative locations of the six examined SNPs marked. (PNG 132 kb)
BDNF rs908867 and rs1157659 SNPs in baseline analysis are not associated with differences in outcomes from specific treatment strategies. Analyzing UPDRS total and part III for the rs908867 SNP showed no differences in the baseline cohort (A and B, respectively). No differences were detected by UPDRS total or part III for the rs1157659 SNP as well (C and D, respectively). Values represent the mean ± SEM. (PNG 298 kb)
The BDNF rs10501087 SNP in the longitudinal analysis. Subjects carrying the rs10501087 C allele do not exhibit differences from non-risk allele carriers when treated with levodopa monotherapy by UPDRS or UPDRS-III (A and C, respectively). Further, no differences between genotypes were observed in subjects not on levodopa monotherapy by UPDRS or UPDRS-III (B and D, respectively). Values represent the mean ± SEM. (PNG 335 kb)
The BDNF rs908867 SNP in the longitudinal analysis. Subjects carrying the rs908867 T allele do not exhibit differences from non-risk allele carriers when treated with levodopa monotherapy by UPDRS or UPDRS-III (A and C, respectively). Further, no differences between genotypes were observed in subjects not on levodopa monotherapy by UPDRS or UPDRS-III (B and D, respectively). Values represent the mean ± SEM. (PNG 339 kb)
The BDNF rs1157659 SNP in the longitudinal analysis. Subjects carrying the rs1157659 G allele do not exhibit differences from A/A subjects when treated with levodopa monotherapy by UPDRS or UPDRS-III (A and C, respectively). Further, no differences between genotypes were observed in subjects not on levodopa monotherapy by UPDRS or UPDRS-III (B and D, respectively). Values represent the mean ± SEM. (PNG 341 kb)
(DOCX 23 kb)
(PDF 1225 kb)
Data Availability Statement
All NET-PD LS-1 data used in this study are accessible through the United States National Institute of Neurological Disorders and Stroke (NINDS) and the NET-PD Executive Committee.