Abstract
Proteasomal activator 28 gamma (PA28γ), an essential constituent of the 20S proteasome, is frequently overexpressed in hepatocellular carcinoma. Hepatitis C virus (HCV) core protein is recently known to activate PA28γ expression in human hepatocytes via upregulation of p53 levels; however, its role in HCV tumorigenesis remains unknown. Here, we found that HCV core-activated PA28γ downregulates p16 levels via ubiquitin-independent proteasomal degradation. As a result, HCV core protein activated the Rb-E2F pathway to stimulate cell cycle progression from G1 to S phase, resulting in an increase in cell proliferation. The potential of HCV core protein to induce these effects was almost completely abolished by either PA28γ knockdown or p16 overexpression, confirming the role of the PA28γ-mediated p16 degradation in HCV tumorigenesis.
Keywords: Cell cycle, Hepatitis C virus, Core, PA28γ, Proteasome, p16
Cell cycle, Hepatitis C virus, Core, PA28γ, Proteasome, p16.
1. Introduction
Chronic infection with hepatitis C virus (HCV) is a major cause of human hepatic diseases, including hepatitis, cirrhosis and hepatocellular carcinoma (HCC) [1]. As a member of the Flaviviridae family, HCV contains a positive stranded RNA genome of 9.5 kb encoding a large polyprotein, which is proteolytically processed into 4 structural proteins and 6 nonstructural proteins [2]. Several HCV proteins including core, NS2, NS3, NS5A, and NS5B are implicated in HCV tumorigenesis [3, 4]. In particular, HCV core protein has been reported to alter diverse signaling pathways, transcriptional activation, and modulation of immune responses, apoptosis, and lipid metabolism [3, 4]. It has been also demonstrated that HCV core protein cooperates with ras oncogene in the transformation of rodent fibroblasts [5], stimulates cell growth and proliferation [6, 7, 8], promotes immortalization of primary human hepatocytes [9], and induces HCC in transgenic mice [10]. Despite the steadily accumulating evidence on the role of HCV core protein in HCC formation, its action mechanism is still a controversial topic.
Proteasomes are protein complexes that degrade unneeded or damaged proteins through proteolysis. Two distinct proteasomes differentially target proteins for degradation. The 26S proteasome responsible for the degradation of the majority of cellular proteins through an ubiquitin- and ATP-dependent pathway is formed by association of the 20S catalytic core composed of α and β subunits with the 19S regulator [11]. Alternatively, the 20S proteasome responsible for ubiquitin- and ATP-independent protein degradation is generated by a combination of a 20S catalytic core and a member of proteasomal activator 28 (PA28) family [11]. Unlike its family members, PA28α and PA28β, PA28γ is localized within the nucleus and has been implicated in tumorigenesis, as it affects cell proliferation and apoptosis via nuclear proteolysis of target proteins. For example, PA28γ lowers p53 levels by facilitating its interaction with mouse double minute 2 (MDM2), thereby inhibiting DNA damage-induced apoptosis [12]. PA28γ also promotes degradation of several negative cell cycle regulators, including p14, p16, and p21, to stimulate cell cycle progression from G1 to S phase [13]. Interestingly, PA28γ is frequently overexpressed in human cancers, including HCC [14]. In addition, PA28γ knockout impairs development of both hepatic steatosis and HCC in transgenic mice [15]. Despite increasing evidence supporting the oncogenic role of PA28γ in human cancers, its role and action mechanism in HCV tumorigenesis remain unknown.
Previous studies have shown that HCV core protein upregulates p53 levels via activation of the ataxia telangiectasia mutated (ATM)-checkpoint kinase 2 (CHK2) pathway and enhances its transcriptional activity [16, 17]. In addition, p53 activates PA28γ transcription through p53 response elements located within its promoter [18]. Furthermore, it has recently been reported that HCV core protein stimulates PA28γ expression in human hepatoma cells via activation of p53 [17]. In the present study, we further explored the biological significance of the p53-dependent PA28γ activation in HCV core-associated tumorigenesis, focusing on the PA28γ-mediated proteasomal degradation of p16 and its effects on the Rb-E2F pathway and cell proliferation.
2. Materials and methods
2.1. Plasmids
The HCV core expression plasmid, pCMV-3 × HA1-core, encodes the full-length HCV core (genotype 1b) downstream of three copies of the influenza virus hemagglutinin (HA) epitope [19]. The pCMV-3 × HA1-p16, encoding full-length HA-tagged p16, was described previously [20]. Scrambled (SC) shRNA, p53 shRNA, and PA28γ shRNA plasmids were purchased from Santa Cruz Biotechnology. Plasmid pCH110, encoding the Escherichia coli β-galactosidase (β-Gal) gene under the control of the SV40 promoter, and pCMV6 PSME3, encoding full-length human Myc-DDK-tagged PA28γ, were purchased from Pharmacia (Cat. No. 27-4508-01) and OriGene (Cat. No. SC321554), respectively. The E2F1-luc and pCMV p53-WT were gifts from Dr. Chang-Woo Lee (Sungkyunkwan University, Suwon, Korea). The pHA-Ub, encoding HA-tagged ubiquitin, was kindly provided by Y. Xiong (University of North Carolina at Chapel Hill, USA).
2.2. Cell lines and transfection
HepG2 (KCLB No. 58065) and Hep3B (KCLB No. 88064) obtained from the Korean Cell Line Bank were maintained in Dulbecco's modified Eagle's medium (WELGENE, Cat. No. LM 001–05) with 10% fetal bovine serum (Gibco, Cat. No. 16000-044) and antibiotics (penicillin and streptomycin; Gibco, Cat. No. 15140122). For transient expression, 4 × 105 cells per 60-mm dish were transfected with 2 μg of appropriate plasmid(s), using the TurboFect™ transfection reagent (Thermo Fisher Scientific, Cat. No. R0532) according to the manufacturer's instructions. Stable cell lines, HepG2-vector and HepG2-core, were established by transfection with pCMV-3 × HA1 and pCMV-3 × HA1-core, respectively, followed by selection with 750 μg ml−1 G418 (Gibco, Cat. No. 11811-031) for 4 weeks, as previously described [20]. The colonies formed on the dishes were amplified for the examination of HCV core expression by western blotting. The stable cell lines were maintained in the culture medium containing 250 μg ml−1 G418. Cells were treated with MG132 (Sigma, Cat. No. M7449) to inhibit cellular proteasomes for 4 h before harvesting.
2.3. Luciferase reporter assay
Approximately 2 × 105 cells per well in 6-well plate were transfected with 0.2 μg of E2F1-luc along with the indicated plasmids in Figures 3B, 3D, and S1B. To control for transfection efficiency, 0.1 μg of pCH110 was cotransfected as an internal control. At 48 h after transfection, a luciferase assay was performed using the Luciferase Reporter 1000 Assay System (Promega, Cat. No. E1910). The β-Gal activity was measured using a β-Gal activity (Thermo Fisher Scientific, Cat. No. 75705). The luciferase activity was normalized to the β-Gal activity measured in the corresponding cell extract.
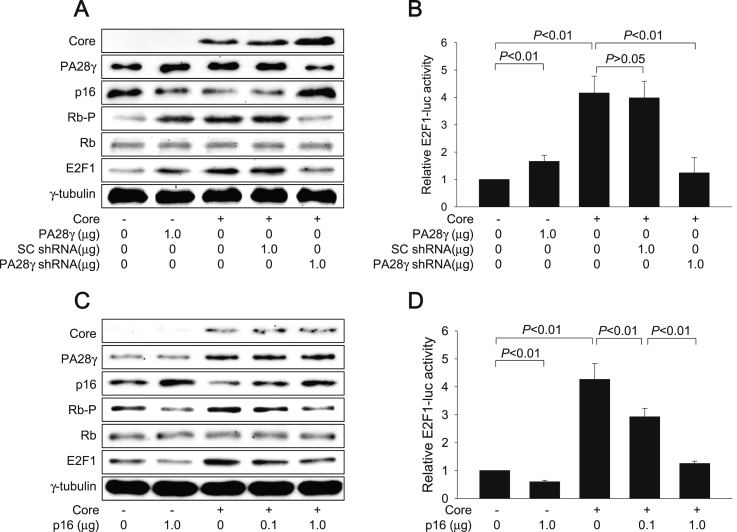
Figure 3.
HCV core protein activates the Rb-E2F pathway via PA28γ-mediated downregulation of p16 levels. (A, C) HepG2-vector and HepG2-core cells were transfected with the indicated amount of PA28γ expression plasmid, SC shRNA plasmid, PA28γ shRNA plasmid, and p16 expression plasmid along with an empty vector for 48 h, followed by western blotting. The uncropped versions of figures are presented in Fig. S3A. (B, D) HepG2-vector and HepG2-core cells were transfected with 0.2 μg of E2F1-luc along with the indicated amount of PA28γ expression plasmid, SC shRNA plasmid, PA28γ shRNA plasmid, and p16 expression plasmid, followed by luciferase assay. Results are shown as means ± standard deviation (SD) obtained from four independent experiments (n = 4).
2.4. Cell viability analysis
For the determination of cell viability, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed as previously described [21]. Briefly, cells grown in 96-well plates were treated with 10 μM MTT (USB, Cat. No. 19265) for 4 h at 37 °C. The formazan compounds derived from MTT by mitochondrial reductases present in the living cells were then dissolved in dimethyl sulfoxide, and quantified by measuring absorbance at 550 nm.
2.5. BrdU incorporation assay
For determination of DNA synthesis rate, the amount of BrdU incorporated into DNA was measured by a colorimetric immunoassay (Roche, Cat. No. 11647229001). Briefly, 1 × 104 cells per well in 96-well plates were incubated for 48 h under the indicated conditions and treated with 10 μM BrdU for an additional 24 h. Fixed cells were reacted with anti-BrdU-peroxidase for 2 h, and the color that developed after addition of trimethyl benzidine was measured at 490 nm and 405 nm.
2.6. Western blot analysis
Cells were lysed in buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, and 1% NP-40) supplemented with protease inhibitors (Roche, Cat. No. 04 693 132 001). Cell extracts were separated by SDS-PAGE and transferred onto a nitrocellulose membrane (Amersham, Cat. No. 10600004). Membranes were then incubated with antibodies against p16 (Abcam, Cat. No. ab51243); p53 (Cat. No. sc-126), E2F1 (Cat. No. sc-251), PA28γ (Cat. No. sc-136025) and ubiquitin (Cat. No. sc-9133) (Santa Cruz Biotechnology); HA (Cell Signaling, Cat. No. 9301S); phosphorylated Rb (Cell signaling, Cat. No. 9301S); Rb (Oncogene, Cat. No. OP77-100UG); HCV core protein (Thermo Fisher Scientific, Cat. No. MA1-080); and γ-tubulin (Sigma, Cat. No. T6557), and subsequently with an appropriate horseradish peroxidase-conjugated secondary antibody: anti-mouse or anti-rabbit IgG (H + L)-HRP (Bio-Rad, Cat. No. 1706516 and 170–6515, respectively). The ECL kit (Advansta, Cat. No. K-12045-D50) was used to visualize the protein bands with the ChemiDoc XRS imaging system (Bio-Rad).
2.7. Immunoprecipitation (IP)
An IP assay was performed using a Classic Magnetic IP/Co-IP kit (Pierce, Cat. No. 88804) according to the manufacturer's specifications. Briefly, 4 × 105 cells were transfected with the indicated plasmids along with pHA-Ub for 48 h under the indicated conditions. Whole cell lysates were incubated overnight at 4 °C with an anti-p16 antibody. Protein A/G magnetic beads (Pierce) were then added, and the lysates were incubated for an additional hour. Beads were collected using a magnetic stand (Pierce), and the eluted antigen/antibody complexes were subjected to western blotting using an anti-HA antibody.
2.8. Statistical analysis
The values indicate means ± standard deviations from at least three independent experiments. A two-tailed Student's t-test was used for all statistical analyses. A P value of <0.05 was considered statistically significant.
3. Results
3.1. HCV core protein downregulates p16 levels via p53-dependent upregulation of PA28γ levels
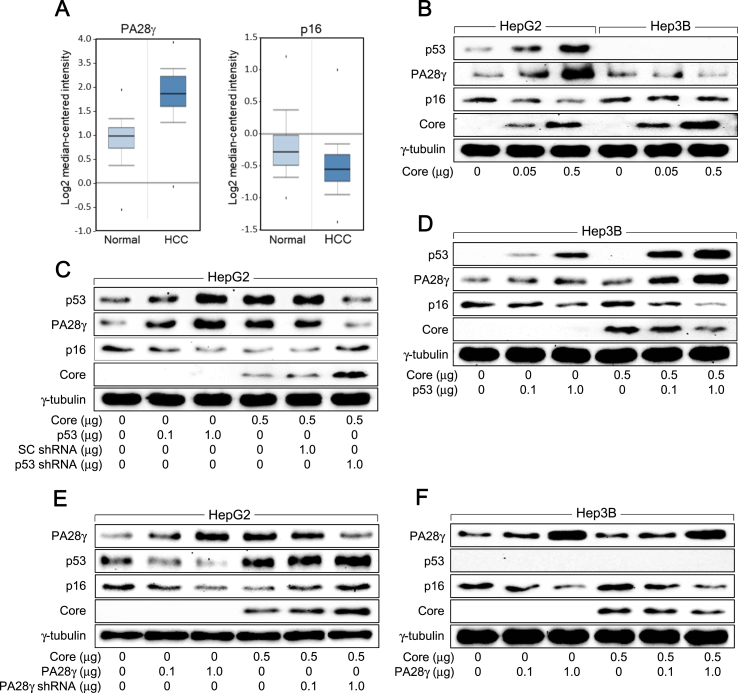
We analyzed the Roessler liver 2 statistics retrieved from Oncomine database to compare the mRNA levels of PA28γ and p16 between 220 normal liver and 225 HCC microarray datasets (Fig. S1A). Consistent to previous reports [14, 22, 23], PA28γ levels were found to be approximately 4-fold higher in HCC samples compared with normal liver samples, while p16 levels in HCC samples were less than 40% of the normal tissues (Figure 1A). Based on these data, we attempted to investigate the possible involvement of PA28γ in the regulation of p16 levels in HCV-associated HCC. Initially, it was examined whether HCV core protein differentially affects PA28γ levels in human hepatoma cells with or without p53 expression. Transient expression of HCV core protein dose-dependently upregulated p53 and PA28γ levels in HepG2 cells, while the effect on PA28γ was negligible in p53-negative Hep3B cells (Figure 1B). Additionally, overexpression of p53 in the absence of HCV core protein upregulated PA28γ levels in HepG2 cells, whereas knockdown of p53 in the presence of HCV core protein downregulated PA28γ levels in the same cells (Figure 1C). While overexpression of p53 in Hep3B cells also upregulated PA28γ levels in the presence and absence of HCV core protein, the effect was more dramatic in the presence of HCV core protein, definitely because HCV core protein upregulated ectopic p53 levels in these cells (Figure 1D). These results confirm our previous findings demonstrating that HCV core protein upregulates PA28γ levels via activation of p53 in human hepatoma cells [17].
Figure 1.
HCV core protein downregulates p16 levels via upregulation of p53 and PA28γ levels in human hepatoma cells. (A) Analysis of the expression levels of PA28γ and p16 in microarray datasets (Roessler liver 2) of normal liver (n = 220) and HCC (n = 225) retrieved from the Oncomine database. Unmodified datasheets are given in Fig. S1A. (B) HepG2 and Hep3B cells were transiently transfected with an increasing concentration of the HCV core expression plasmid for 48 h, followed by western blotting. The uncropped versions of figures are presented in Fig. S1B. (C, D) HepG2 and Hep3B cells were transfected with an empty vector or the HCV core expression plasmid along with the indicated amount of p53 expression plasmid, scrambled (SC) shRNA plasmid, and p53 shRNA plasmid, followed by western blotting. The uncropped versions of figures are presented in Fig. S1C and S1D. (E, F) HepG2 and Hep3B cells were transiently transfected with either an empty vector or the HCV core expression plasmid along with an increasing concentration of the PA28γ expression plasmid or PA28γ shRNA plasmid for 48 h, followed by western blotting. The uncropped versions of figures are presented in Fig. S1E and S1F.
Next, it was investigated whether HCV core protein also affects p16 levels in a p53-dependent manner. Consistent with a previous report [20], HCV core protein dose-dependently downregulated p16 levels in HepG2 cells, while the effect was marginal in Hep3B cells (Figure 1B). In addition, overexpression of p53 in the absence of HCV core protein downregulated p16 levels in HepG2 cells, whereas knockdown of p53 in the presence of HCV core protein upregulated p16 levels in HepG2 cells (Figure 1C). Moreover, ectopic expression of p53 downregulated p16 levels in Hep3B cells, an effect that was more dramatic in the presence of HCV core protein (Figure 1D), indicating that HCV core protein downregulates p16 levels via activation of p53 in human hepatoma cells.
It was attempted to provide direct evidence for the correlation between the upregulation of PA28γ levels and the downregulation of p16 levels, both of which were dependent on the activation of p53 by HCV core protein (Figure 1B to D). First, PA28γ levels were inversely proportional to those of p16 in both the presence and absence of HCV core protein in human hepatoma cells (Figure 1B to F). In addition, overexpression of PA28γ in the absence of HCV core protein downregulated p53 and p16 levels, whereas knockdown of PA28γ in the presence of HCV core protein upregulated p53 and p16 levels in HepG2 cells (Figure 1E); these results were consistent to previous reports demonstrating that PA28γ downregulates p53 [12] and p16 levels [13]. Moreover, ectopic expression of PA28γ equally downregulated p16 levels in the presence and absence of HCV core protein in Hep3B cells (Figure 1F), indicating that PA28γ can downregulate p16 levels irrespective of p53 and HCV core protein. Taken together, we conclude that HCV core protein downregulates p16 levels via p53-dependent upregulation of PA28γ levels in human hepatoma cells.
3.2. HCV core-activated PA28γ induces ubiquitin-independent proteasomal degradation of p16
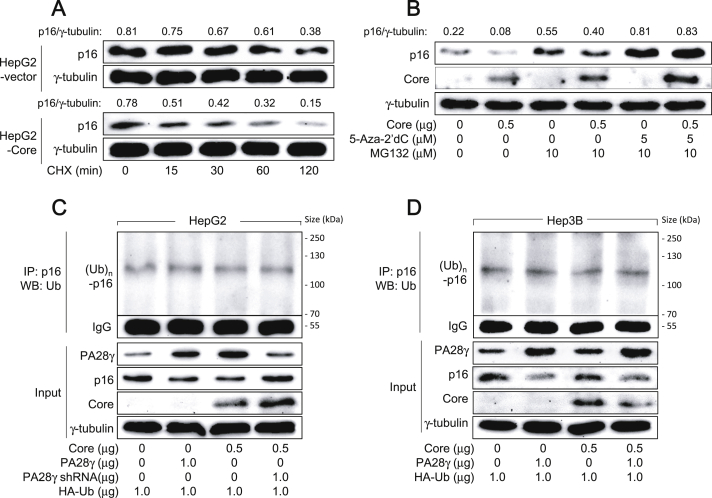
It was investigated whether upregulation of PA28γ levels by HCV core protein alters the protein stability of p16 by comparing its half-life in the presence or absence of HCV core protein. For this purpose, we treated HepG2 cells with or without HCV core protein expression with cycloheximide (CHX) to block further protein synthesis while measuring p16 and γ-tubulin levels in these cells. As shown in Figure 2A, the half-life of p16 in the presence of HCV core protein was shorter (t1/2 = 38.7 min) than that in the absence of HCV core protein (t1/2 = 128.4 min). In addition, treatment with a universal proteasomal inhibitor, MG132, upregulated p16 levels in both the presence and absence of HCV core protein to impair the potential of HCV core protein to downregulate p16 levels (Figure 2B), suggesting that HCV core protein downregulates p16 levels via proteasomal degradation. However, co-treatment with 5-Aza-2′dC, a potent DNA methyltransferase (DNMT) inhibitor, was required to completely abolish the potential of HCV core protein to downregulate p16 levels, indicating that HCV core protein also downregulates p16 levels via DNA methylation, as previously demonstrated [20].
Figure 2.
HCV core protein upregulates PA28γ levels to induce ubiquitin-independent proteasomal degradation of p16. (A) HepG2 cells stably transfected with either an empty vector or the HCV core expression plasmid were treated with 50 μg ml−1 of cycloheximide (CHX) for the indicated time, followed by western blotting. Each band was quantified using ImageJ image-analysis software (NIH, USA); the values indicate p16 levels relative to those of the loading control (γ-tubulin). The uncropped versions of figures are presented in Fig. S2A. (B) HepG2 cells were transfected with either an empty vector or the HCV core expression plasmid for 44 h and then either mock-treated or treated with 10 μM MG132 for an additional 4 h, followed by western blotting. The uncropped versions of figures are presented in Fig. S2B. (C, D) HepG2 and Hep3B cells were transfected with the indicated plasmids, along with pHA-Ub, for 48 h, followed by IP (using anti-p16 antibody)-coupled western blotting (using anti-HA antibody) to detect HA-ubiquitin-complexed p16. Levels of PA28γ, p16, HCV core protein, and γ-tubulin in the total cell lysates (input) were determined by western blotting. The uncropped versions of figures are presented in Fig. S2C and S2D.
Next, we investigated the possible mechanism by which HCV core protein induces the PA28γ-mediated proteasomal degradation of p16 in human hepatoma cells. Ectopic overexpression of PA28γ in the absence of HCV core protein in HepG2 cells (Figure 2C), and in the presence and absence of HCV core protein in Hep3B cells (Figure 2D) downregulated p16 levels without affecting its ubiquitination. In addition, HCV core protein-activated PA28γ also downregulated p16 levels without affecting its ubiquitination in HepG2 cells (Figure 2C); this effect was not observed in Hep3B cells, where HCV core protein was unable to upregulate PA28γ levels (Figure 2D). Moreover, PA28γ knockdown in the presence of HCV core protein upregulated p16 levels in HepG2 cells without affecting its ubiquitination (Figure 2C). The main p16 band corresponding to the polyubiquitinated form of the protein was undetectable in a preliminary experiment using a human IgG as an IP antibody (data not shown). In addition, a similar pattern of p16 ubiquitination has been reported in a previous study [24]. Taken together, it was possible to conclude that HCV core protein induces ubiquitin-independent proteasomal degradation of p16 via upregulation of PA28γ levels in human hepatoma cells.
3.3. HCV core protein activates the Rb-E2F pathway through PA28γ-mediated downregulation of p16 levels
It was investigated whether the PA28γ-mediated downregulation of p16 levels in the presence of HCV core protein results in activation of the Rb-E2F pathway, which is essential for cell cycle progression from G1 to S phase [25]. Consistent with a previous report [13], ectopic PA28γ expression in the absence of HCV core protein downregulated p16 levels and inactivated Rb via phosphorylation without affecting total Rb protein levels, resulting in an increase of both E2F1 protein levels and its transcriptional activity in HepG2 cells (Figure 3A and B). HCV core protein-activated PA28γ similarly inactivated Rb via phosphorylation, resulting in activation of E2F1 in HepG2 cells (Figure 3A and B). In addition, knockdown of PA28γ in the presence of HCV core protein upregulated p16 levels and decreased Rb phosphorylation without affecting total Rb protein levels, resulting in a decrease of both E2F1 protein levels and its transcriptional activity in HepG2 cells (Figure 3A and B).
Ectopic expression of p16 in the absence of HCV core protein decreased Rb phosphorylation and thereby downregulated E2F1 levels in HepG2 cells (Figure 3C), which is consistent with its role as a negative regulator of the cell cycle [26]. In addition, restoration of p16 levels in HCV core protein-expressing cells via p16 overexpression almost completely abolished the potential of HCV core protein to activate the Rb-E2F pathway (Figure 3C and D). Therefore, we conclude that HCV core protein activates the Rb-E2F pathway via PA28γ-mediated downregulation of p16 levels.
3.4. HCV core protein stimulates cell growth by downregulating p16 levels via PA28γ-mediated proteasomal degradation
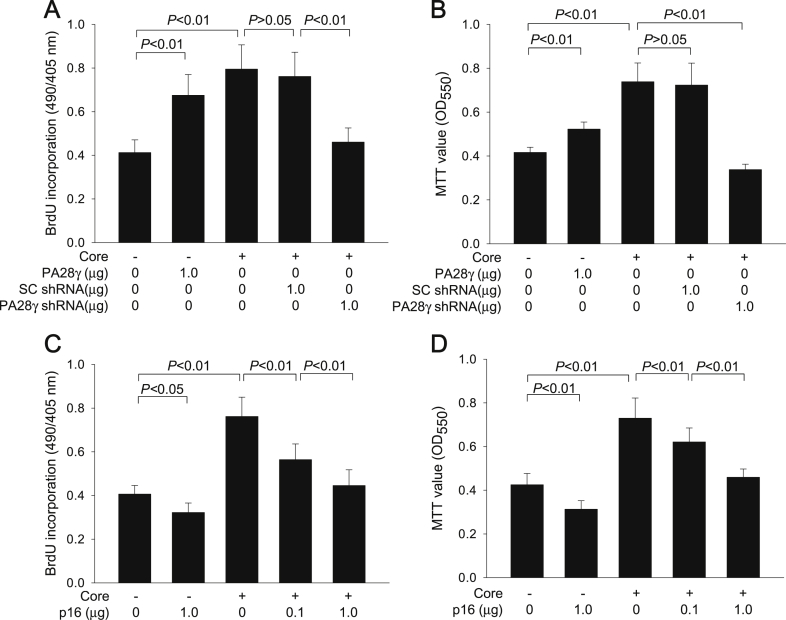
It was investigated whether the PA28γ-mediated downregulation of p16 level and subsequent activation of the Rb-E2F pathway in the presence of HCV core protein results in stimulation of cell growth. For this purpose, we first compared the proliferation rate of HepG2 cells with or without HCV core protein expression. According to data from the BrdU incorporation assay (Figure 4A), HCV core protein significantly increased DNA synthesis in HepG2 cells, which was consistent with its potential to activate the Rb-E2F pathway (Figure 3A and B). Accordingly, the growth rate of HCV core-expressing cells was 1.7-fold higher than that of control cells without HCV core expression, as demonstrated with MTT assay (Figure 4B).
Figure 4.
HCV core protein stimulates cell growth by downregulating p16 levels via PA28γ-mediated proteasomal degradation. HepG2-vector and HepG2-core protein cells seeded at 2 × 105 cells per well in 6-well plates were transfected with the indicated amount of PA28γ expression plasmid, SC shRNA plasmid, PA28γ shRNA plasmid, and p16 expression plasmid for 48 h. Cells were subjected to BrdU incorporation (A, C) and MTT assays (B, D) (n = 6).
In agreement with its potential to activate the Rb-E2F pathway (Figure 3A and B), ectopic expression of PA28γ in HepG2 cells without HCV core expression increased DNA synthesis rate and thereby stimulated cell growth (Figure 4A and B), which is consistent with a previous report [13]. Therefore, we investigated whether the PA28γ upregulation and subsequent activation of the Rb-E2F pathway is responsible at least in part for the HCV core-mediated stimulation of cell growth. Indeed, PA28γ knockdown significantly abolished the potential of HCV core protein to increase DNA synthesis and stimulate cell growth in HepG2 cells (Figure 4A to B), confirming that HCV core protein stimulates cell growth via upregulation of PA28γ levels.
It was further investigated whether PA28γ-mediated downregulation of p16 levels is responsible for the effect of HCV core protein on cell growth. Consistent with a previous report [27], ectopic expression of p16 in HepG2 cells without HCV core expression decreased DNA synthesis and thereby inhibited cell growth (Figure 4C and D), due to its potential to inactivate the Rb-E2F pathway (Figure 3C and D). In addition, ectopic expression of p16 in HCV core-expressing HepG2 cells (Figure 4C to D) significantly abolished the potential of HCV core protein to increase DNA synthesis and stimulate cell growth, as predicted from its effect on the Rb-E2F pathway in HCV core-expressing cells (Figure 3C and D). Therefore, we conclude that HCV core protein stimulates cell growth by downregulating p16 levels via PA28γ-mediated proteasomal degradation in human hepatoma cells.
4. Discussion
Some viral oncoproteins have evolved distinct strategies to inactivate p53, which is often critical for their roles in tumorigenesis [28]. The p53 protein was initially identified through its ability to interact with SV40 large T antigen [29, 30] and has since been shown to form a complex with several other viral proteins, including the adenovirus type 5 E1B 55K [31] and the E6 protein of oncogenic human papillomavirus types 16 and 18 [32]. HCV core protein also has been shown to affect p53 via several different mechanisms. First, it can inhibit or activate p53 via direct protein-protein interactions to execute its anti- or pro-apoptotic potential [16, 33, 34]. Second, HCV core protein indirectly counteracts p53-mediated growth suppression through activation of the MAPK and PI3K/Akt pathways [35]. Third, HCV core protein enhances the transcriptional activity of p53 via posttranslational modifications such as acetylation and phosphorylation [33]. Fourth, HCV core protein can upregulate p53 levels by inhibiting its ubiquitin-dependent proteasomal degradation. For example, HCV core protein inhibits p14 expression via DNA methylation and thereby inactivate the p14-MDM2 pathway, resulting in inhibition of the MDM2-mediated ubiquitination and proteasomal degradation of p53 [21]. Alternatively, HCV core protein induces oxidative stress via multiple mechanisms to activate the ATM-CHK2 pathway and upregulate p53 levels [17, 36], which is consistent with the present study. Further studies are required to clearly assess the contradictory roles of HCV core protein in the regulation of p53 levels, which might be affected by several factors, including the cell contexts, HCV core protein levels, model systems used, and other experimental conditions.
The biological significance of HCV core-mediated p53 activation in HCV pathogenesis is poorly understood. Considering the roles of p53 target proteins in the regulation of growth arrest, senescence, and apoptosis [37, 38, 39], it is little wonder that the HCV core-activated p53 negatively affects cell growth. Indeed, HCV core protein stimulates expression of p21 and Bax via activation of p53, resulting in cell cycle arrest and apoptotic cell death of the hepatocytes [16, 34, 40]. Interestingly, it has recently been demonstrated that HCV core protein transcriptionally activates the PA28γ gene through p53-response elements located within its promoter [17]. Unlike other p53 target proteins, PA28γ can act as a potent oncoprotein, promoting proteasomal degradation of negative regulators of cell growth, including p14, p16, p21, and p53 [12, 13]. Consistently, the present study showed that the p53-activated PA28γ downregulates p16 levels, resulting in activation of the Rb-E2F pathway and subsequent stimulation of cell cycle progression from G1 to S phase. The IP data shown in Figure 2C and D might be not enough to demonstrate that the HCV core-activated PA28γ induces ubiquitin-independent degradation of p16. However, several previous reports including reference [13] have already shown that PA28γ downregulates p16 via ubiquitin-independent proteasomal degradation. The present study focused on the downregulation of p16 levels by HCV core-activated PA28γ while simply showing the ubiquitination pattern of p16 in the presence or absence of HCV core and PA28γ.Inactivation of the p16 gene is one of the most common alterations in HCC [23]. In particular, DNA methylation appears to be the prominent cause for p16 inactivation in HCC [41, 42]. Higher frequencies of DNA methylation have been detected in the p16 promoter from HCCs with HCV infection compared to those without such infection [41, 42]. It has been further demonstrated that HCV core protein induces promoter hypermethylation of the p16 gene in human hepatocytes [20, 43]. Therefore, treatment with a universal DNMT inhibitor, 5-Aza-2′dC, effectively reactivates p16 expression in HCV core-expressing cells, resulting in inhibition of cell proliferation [20]. The present study provides another mechanism for the inactivation of p16 by HCV core protein, which is mediated by PA28γ of the cellular proteasome system. Therefore, it was possible to abolish the potential of HCV core protein to inhibit p16 expression by inhibiting both DNA methylation and proteasome with 5-Aza-2′dC and MG132, respectively (Figure 2B). Therefore, the HCV core-mediated p16 inactivation via modulation of cellular DNA methylation and proteasome systems may serve potential targets for the development of therapeutic schemes to treat HCC patients.
Declarations
Author contribution statement
Kyung Lib Jang: Conceived and designed the experiments; Wrote the paper.
Sungkyung Cha: Performed the experiments; Analyzed and interpreted the data.
Inbeom Park: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the National Research Foundation of Korea grant funded by the Korea government (MEST) (NRF-2019R1A2C2011478).
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Tornesello M.L., Buonaguro L., Izzo F., Buonaguro F.M. Molecular alterations in hepatocellular carcinoma associated with hepatitis B and hepatitis C infections. Oncotarget. 2016;7:25087–25102. doi: 10.18632/oncotarget.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki T., Aizaki H., Murakami K., Shoji I., Wakita T. Molecular biology of hepatitis C virus. J. Gastroenterol. 2007;42:411–423. doi: 10.1007/s00535-007-2030-3. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee A., Ray R.B., Ray R. Oncogenic potential of hepatitis C virus proteins. Viruses. 2010;2:2108–2133. doi: 10.3390/v2092108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vescovo T., Refolo G., Vitagliano G., Fimia G.M., Piacentini M. Molecular mechanisms of hepatitis C virus-induced hepatocellular carcinoma. Clin. Microbiol. Infect. 2016;22:853–861. doi: 10.1016/j.cmi.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Chang J., Yang S.H., Cho Y.G., Hwang S.B., Hahn Y.S., Sung Y.C. Hepatitis C virus core from two different genotypes has an oncogenic potential but is not sufficient for transforming primary rat embryo fibroblasts in cooperation with the H-ras oncogene. J. Virol. 1998;72:3060–3065. doi: 10.1128/jvi.72.4.3060-3065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki H., Hayashi J., Moriyama M., Arakawa Y., Hino O. Hepatitis C virus core protein interacts with 14-3-3 protein and activates the kinase Raf-1. J. Virol. 2000;74:1736–1741. doi: 10.1128/jvi.74.4.1736-1741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho J.W., Baek W.K., Suh S.I., Yang S.H., Chang J., Sung Y.C., Suh M.H. Hepatitis C virus core protein promotes cell proliferation through the upregulation of cyclin E expression levels. Liver. 2001;21:137–142. doi: 10.1034/j.1600-0676.2001.021002137.x. [DOI] [PubMed] [Google Scholar]
- 8.Erhardt A., Hassan M., Heintges T., Haussinger D. Hepatitis C virus core protein induces cell proliferation and activates ERK, JNK, and p38 MAP kinases together with the MAP kinase phosphatase MKP-1 in a HepG2 Tet-Off cell line. Virology. 2002;292:272–284. doi: 10.1006/viro.2001.1227. [DOI] [PubMed] [Google Scholar]
- 9.Ray R.B., Meyer K., Ray R. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology. 2000;271:197–204. doi: 10.1006/viro.2000.0295. [DOI] [PubMed] [Google Scholar]
- 10.Moriya K., Fujie H., Shintani Y., Yotsuyanagi H., Tsutsumi T., Ishibashi K., Matsuura Y., Kimura S., Miyamura T., Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 1998;4:1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 11.Tomko R.J., Jr., Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z., Zhang R. Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBO J. 2008;27:852–864. doi: 10.1038/emboj.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Barton L.F., Chi Y., Clurman B.E., Roberts J.M. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J., Cui L., Zeng Y., Wang G., Zhou P., Yang Y., Ji L., Zhao Y., Chen J., Wang Z., Shi T., Zhang P., Chen R., Li X. REGgamma is associated with multiple oncogenic pathways in human cancers. BMC Cancer. 2012;12:75. doi: 10.1186/1471-2407-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriishi K., Mochizuki R., Moriya K., Miyamoto H., Mori Y., Abe T., Murata S., Tanaka K., Miyamura T., Suzuki T., Koike K., Matsuura Y. Critical role of PA28gamma in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1661–1666. doi: 10.1073/pnas.0607312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu W., Lo S.Y., Chen M., Wu K., Fung Y.K., Ou J.H. Activation of p53 tumor suppressor by hepatitis C virus core protein. Virology. 1999;264:134–141. doi: 10.1006/viro.1999.9979. [DOI] [PubMed] [Google Scholar]
- 17.Kwak J., Tiwari I., Jang K.L. Hepatitis C virus core activates proteasomal activator 28gamma expression via upregulation of p53 levels to control virus propagation. J. Gen. Virol. 2017;98:56–67. doi: 10.1099/jgv.0.000655. [DOI] [PubMed] [Google Scholar]
- 18.Wan Z.X., Yuan D.M., Zhuo Y.M., Yi X., Zhou J., Xu Z.X., Zhou J.L. The proteasome activator PA28gamma, a negative regulator of p53, is transcriptionally up-regulated by p53. Int. J. Mol. Sci. 2014;15:2573–2584. doi: 10.3390/ijms15022573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arora P., Kim E.O., Jung J.K., Jang K.L. Hepatitis C virus core protein downregulates E-cadherin expression via activation of DNA methyltransferase 1 and 3b. Cancer Lett. 2008;261:244–252. doi: 10.1016/j.canlet.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 20.Park S.H., Lim J.S., Lim S.Y., Tiwari I., Jang K.L. Hepatitis C virus Core protein stimulates cell growth by down-regulating p16 expression via DNA methylation. Cancer Lett. 2011;310:61–68. doi: 10.1016/j.canlet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 21.Kwak J., Choi J.H., Jang K.L. Hepatitis C virus Core overcomes all-trans retinoic acid-induced apoptosis in human hepatoma cells by inhibiting p14 expression via DNA methylation. Oncotarget. 2017;8:85584–85598. doi: 10.18632/oncotarget.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liew C.T., Li H.M., Lo K.W., Leow C.K., Chan J.Y., Hin L.Y., Lau W.Y., Lai P.B., Lim B.K., Huang J., Leung W.T., Wu S., Lee J.C. High frequency of p16INK4A gene alterations in hepatocellular carcinoma. Oncogene. 1999;18:789–795. doi: 10.1038/sj.onc.1202359. [DOI] [PubMed] [Google Scholar]
- 23.Jin M., Piao Z., Kim N.G., Park C., Shin E.C., Park J.H., Jung H.J., Kim C.G., Kim H. p16 is a major inactivation target in hepatocellular carcinoma. Cancer. 2000;89:60–68. doi: 10.1002/1097-0142(20000701)89:1<60::aid-cncr9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Al-Khalaf H.H., Hendrayani S.F., Aboussekhra A. The atr protein kinase controls UV-dependent upregulation of p16INK4A through inhibition of Skp2-related polyubiquitination/degradation. Mol. Cancer. Res. 2011;9:311–319. doi: 10.1158/1541-7786.MCR-10-0506. [DOI] [PubMed] [Google Scholar]
- 25.Sun A., Bagella L., Tutton S., Romano G., Giordano A. From G0 to S phase: a view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J. Cell. Biochem. 2007;102:1400–1404. doi: 10.1002/jcb.21609. [DOI] [PubMed] [Google Scholar]
- 26.Serrano M., Hannon G.J., Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 27.Huang J.Z., Xia S.S., Ye Q.F., Jiang H.Y., Chen Z.H. Effects of p16 gene on biological behaviours in hepatocellular carcinoma cells. World J. Gastroenterol. 2003;9:84–88. doi: 10.3748/wjg.v9.i1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tornesello M.L., Annunziata C., Tornesello A.L., Buonaguro L., Buonaguro F.M. Human oncoviruses and p53 tumor suppressor pathway deregulation at the origin of human cancers. Cancers. 2018;10 doi: 10.3390/cancers10070213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane D.P., Crawford L.V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 30.Linzer D.I., Levine A.J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 31.Sarnow P., Ho Y.S., Williams J., Levine A.J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 32.Werness B.A., Levine A.J., Howley P.M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 33.Kao C.F., Chen S.Y., Chen J.Y., Wu Lee Y.H. Modulation of p53 transcription regulatory activity and post-translational modification by hepatitis C virus core protein. Oncogene. 2004;23:2472–2483. doi: 10.1038/sj.onc.1207368. [DOI] [PubMed] [Google Scholar]
- 34.Otsuka M., Kato N., Lan K., Yoshida H., Kato J., Goto T., Shiratori Y., Omata M. Hepatitis C virus core protein enhances p53 function through augmentation of DNA binding affinity and transcriptional ability. J. Biol. Chem. 2000;275:34122–34130. doi: 10.1074/jbc.M000578200. [DOI] [PubMed] [Google Scholar]
- 35.Jahan S., Khaliq S., Siddiqi M.H., Ijaz B., Ahmad W., Ashfaq U.A., Hassan S. Anti-apoptotic effect of HCV core gene of genotype 3a in Huh-7 cell line. Virol. J. 2011;8:522. doi: 10.1186/1743-422X-8-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov A.V., Smirnova O.A., Petrushanko I.Y., Ivanova O.N., Karpenko I.L., Alekseeva E., Sominskaya I., Makarov A.A., Bartosch B., Kochetkov S.N., Isaguliants M.G. HCV core protein uses multiple mechanisms to induce oxidative stress in human hepatoma Huh7 cells. Viruses. 2015;7:2745–2770. doi: 10.3390/v7062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mihara M., Erster S., Zaika A., Petrenko O., Chittenden T., Pancoska P., Moll U.M. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 38.Vousden K.H., Lu X. Live or let die: the cell's response to p53. Nat. Rev. Canc. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 39.Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 40.Shen S., Li C., Dai M., Yan X. Induction of Huh7 cell apoptosis by HCV core proteins via CK1alphap53Bid signaling pathway. Mol. Med. Rep. 2018;17:7559–7566. doi: 10.3892/mmr.2018.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneto H., Sasaki S., Yamamoto H., Itoh F., Toyota M., Suzuki H., Ozeki I., Iwata N., Ohmura T., Satoh T., Karino Y., Satoh T., Toyota J., Satoh M., Endo T., Omata M., Imai K. Detection of hypermethylation of the p16(INK4A) gene promoter in chronic hepatitis and cirrhosis associated with hepatitis B or C virus. Gut. 2001;48:372–377. doi: 10.1136/gut.48.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X., Hui A.M., Sun L., Hasegawa K., Torzilli G., Minagawa M., Takayama T., Makuuchi M. p16INK4A hypermethylation is associated with hepatitis virus infection, age, and gender in hepatocellular carcinoma. Clin. Canc. Res. 2004;10:7484–7489. doi: 10.1158/1078-0432.CCR-04-1715. [DOI] [PubMed] [Google Scholar]
- 43.Lim J.S., Park S.H., Jang K.L. Hepatitis C virus Core protein overcomes stress-induced premature senescence by down-regulating p16 expression via DNA methylation. Cancer Lett. 2012;321:154–161. doi: 10.1016/j.canlet.2012.01.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.